ABSTRACT
Approximately 75 MAP kinase kinase kinases (MAPKKKs) have been identified in the rice genome. However, only a few of them have been functionally characterized. In this paper, we report the function of a rice MAPKKK, OsMAPKKK63. OsMAPKKK63 was found to be induced by several abiotic stresses, including high salinity, chilling and drought. Our data indicate that OsMAPKKK63 possesses in vitro kinase activity and that it interacts with rice MAP kinase kinase OsMKK1 and OsMKK6. The two rice MKKs are known mediator of the salt stress response, implying that OsMAPKKK63 may be involved in the high salinity response. Our analysis of an OsMAPKKK63 knockout mutant indeed demonstrated that it is necessary for normal response to high salt. On the other hand, OsMAPKKK63 OX lines exhibited viviparous phenotype in both rice and Arabidopsis. The result suggests that OsMAPKKK63 may also be involved in seed dormancy control.
KEYWORDS: Rice, MAP kinase kinase kinase (MAPKKK), abiotic stress, vivipary, abscisic acid (ABA)
Results and discussion
Mitogen-activated protein (MAP) kinase cascades are signaling modules ubiquitous among eukaryotes.1,2 The signaling module consists of three kinases: MAP kinase kinase kinase (MAPKKK), MAP kinase kinase (MAPKK), and MAP kinase (MAPK). Within the signaling module, the three kinases function sequentially. MAPKKK, which is activated by various internal and external stimuli, phosphorylates MAPKK to activate it. Activated MAPKK then phosphorylates MAPK, which, in turn, phosphorylates downstream targets, including transcription factors, enzymes, and other cellular proteins. The phosphorylation relay system plays important roles in development, hormone signaling, abiotic stress response and pathogen defense in plants.
In rice, approximately 75 MAPKKKs, 8 MAPKKs, and 17 MAPKs have been identified.1,3-5 Many studies have shown that MAPKKs and MAPKs are involved in diverse cellular processes, such as biotic and abiotic stress responses, plant development, and secondary metabolism. Compared with MAPKKs and MAPKs, relatively few MAPKKKs of rice in particular and plant, in general, have been characterized. DSM1 (Drought Hypersensitive Mutant1), a Raf-like MAPKKK, regulates drought response by controlling reactive oxygen species scavenging.6 ILA1 (Increased Leaf Angle1), which is a Group C Raf-like MAPKKK, is involved in the formation of leaf laminal joint.7 OsACDR1 (Accelerated Cell Death and Resistance1) and OsEDR1 (Enhanced Disease Resistance1) are rice MAPKKKs involved in disease resistance.8,9 Thus, only a few rice MAPKKKs have been functionally characterized, and the functions of most rice MAPKKKs remain to be determined.
In this paper, we report the in vitro and in vivo function of a rice MAPKKK, OsMAPKKK63 (LOC_Os01g50370.1). We first examined the stress induction pattern of OsMAPKKK63 gene expression by carrying out Real-Time RT-PCR. The result (Figure 1a) indicated that OsMAPKKK63 expression was induced by cold and drought treatments, 14-fold and 12-fold, respectively. It was also induced by high salt (NaCl) to some degree (ca. threefold), but not by high osmolarity (mannitol). We also determined the effects of stress hormones on OsMAPKKK63 expression. Figure 1b shows that its expression was not affected by ABA but slightly decreased by ethylene or methyl jasmonate treatment. Thus, OsMAPKKK63 expression was not significantly affected by the hormone treatments. We then investigated its subcellular localization by a transient expression assay. OsMAPKKK63-YFP fusion construct was prepared and, after introducing into tobacco (Nicotiana benthamiana) leaves by Agrobacterium-mediated infiltration, its localization was determined by epifluorescence microscopy. Figure 1c shows that it is localized both in the nucleus and cytosol, similar to the vector (YFP) control.
Figure 1.
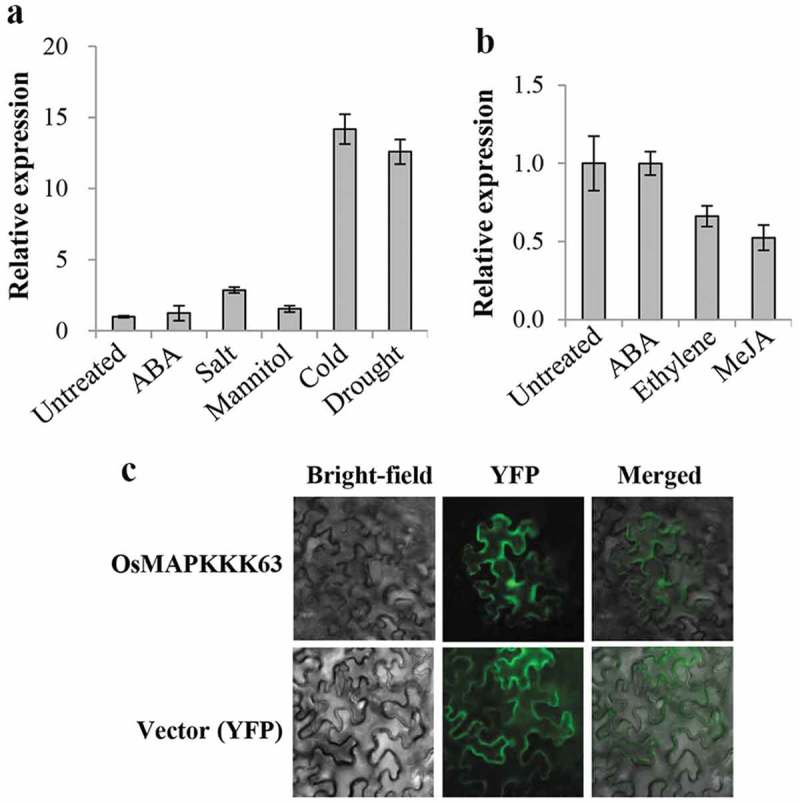
Expression pattern and subcellular localization of OsMAPKKK63. (a) Stress induction pattern of OsMAPKKK63 expression. RNA was isolated from 4-week-old seedlings treated with ABA (100 μM), NaCl (200 mM) (Salt), mannitol (500 mM), chilling (8 hr at 9°C) (Cold), and water-deficit (10 hr on the bench) (Drought). Real-Time RT-PCR was carried out in duplicates. Bars indicate standard errors. (b) Hormone-induced expression of OsMAPKKK63. Real-Time RT-PCR was performed as in (a) using RNA isolated from 3-week-old seedlings treated with 100 μM each of ABA, 1-aminocyclopropane-1-carboxylic acid (Ethylene), and methyl jasmonate (MeJA). Bars indicate standard errors. (c) Subcellular localization of OsMAPKKK63 was examined by epifluorescence using Tobacco (N. benthmiana) leaves infiltrated with Agrobacterium containing an OsMAPKKK63-YFP fusion construct. Images obtained with bright-field (Bright-field), YFP filter (YFP), or merging (Merged) are shown.
To address the function of OsMAPKKK63, we first investigated its in vitro kinase activity. Recombinant proteins containing the full-length OsMAPKKK63 (amino acids 1–484) or its kinase domain (amino acids 1–257) were prepared as fusion proteins to the maltose binding protein (MBP) tag, and kinase assays were performed using myelin basic protein (MyBP) as a substrate. Figure 2a shows that full-length OsMAPKKK63 could phosphorylate the substrate (lane 1), and the kinase domain (amino acids 1–257) of OsMAPKKK63 also could phosphorylate MyBP (lane 2). In the assay, the kinase domain, which can be considered a constitutively active form, displayed stronger activity than the full-length OsMAPKKK63. MAPKKKs are known to be serine/threonine kinases. To confirm its kinase activity, we carried out an inhibitor study employing a broad-spectrum kinase inhibitor, staurosporine.10 In the assay (Figure 2b), 32P incorporation by OsMAPKKK63 was reduced significantly by staurosporine in a concentration-dependent manner (lanes 2–4), and its activity was abolished almost completely at micromolar concentrations of the inhibitor. Together, our findings indicated that OsMAPKKK63 possesses in vitro kinase activity.
Figure 2.
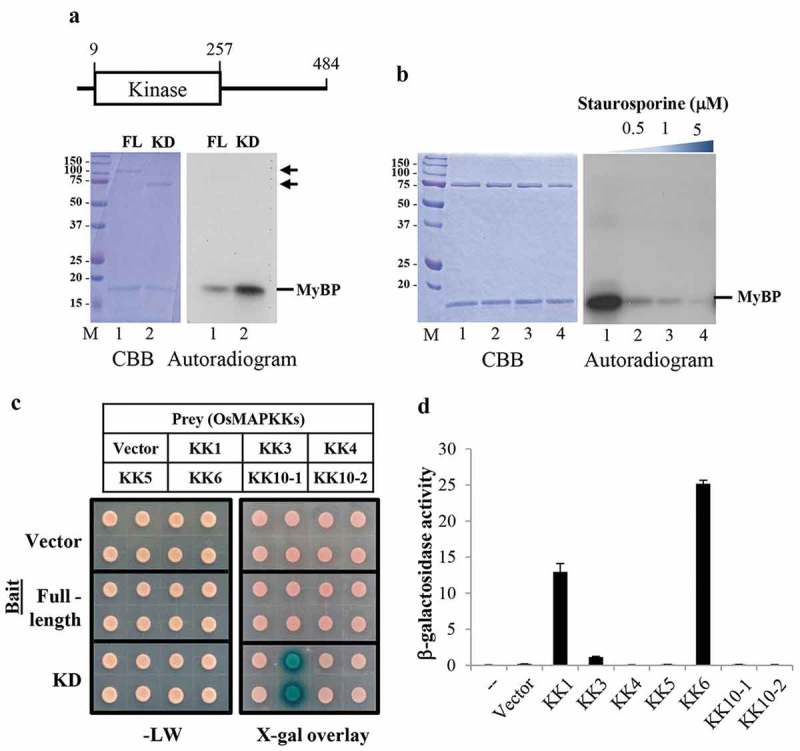
Kinase activity of OsMAPKKK63 and its interaction with OsMKKs. (a) Top, schematic diagram of OsMAPKKK63 domain structure. The numbers indicate amino acid position. Bottom, Kinase assay gel pictures. 0.5 μg of full-length (FL) or kinase domain (KD) recombinant OsMAPKKK63 were used in the assay. Coumassie brilliant blue-stained gel (CBB) and autoradiogram showing 32P incorporation are shown. M, size markers. MyBP, myelin basic protein. FL, full-length. KD, kinase domain. (b) Inhibition of OsMAPKKK63 kinase activity by staurosporine was examined. Kinase assay was performed as in (A), except that staurosporine was added as indicated. (c) Yeast two-hybrid assay to determine interactions between OsMAPKKK63 and OsMKKs. Full-length or kinase domain (KD) of OsMAPKKK63 was employed as bait and OsMKKs as prey, as indicated. Bottom left panel shows yeast colonies grown on CM-Leu-Trp medium, without reporter selection. Bottom right panel shows the result of X-gal overlay assay of yeast grown on CM-Leu-Trp medium. (d) β-galactosidase reporter activity was determined by the liquid assay using o-nitrophenyl-β-D-galactopyranoside as a substrate. The numbers indicate Miller units.
We next asked whether OsMAPKKK63 could interact with rice MAPKKs. Because OsMAPKKK63 is a MAPKKK, it would be expected to interact with MAPKKs, if it is functional. We carried out two-hybrid assays to examine the interactions between OsMAPKKK63 and OsMAPKKs. As shown in Figure 2c, full-length OsMAPKKK63 did not interact with any of the seven OsMAPKKs we tested. However, the kinase domain of OsMAPKKK63 interacted strongly with OsMKK1 and OsMKK6 (Figure 2c and d). Thus, our result suggested that the constitutive active form (i.e. kinase domain) of OsMAPKKK63 might interact with OsMAPKK1 and OsMAPKK6. OsMAPKK1 is a known regulator of the salt stress response, and its expression is induced by high salt.11 OsMAPKK6, on the other hand, is involved in salt and chilling stress tolerance.12,13 These observations, together with its stress induction pattern (Figure 1a), imply that OsMAPKKK63 function might be associated with high salt or other stress response.
To investigate the in planta function of OsMAPKKK63, we acquired its knockout (KO) mutant (PFG-4A-03730) from the T-DNA insertion database.14,15 In the mutant, T-DNA is inserted into the 5’ untranslated region (−87 from the initiation codon) of OsMAPKKK63. After confirming the T-DNA insertion at the annotated position and the abolishment of OsMAPKKK63 expression (Figure 3a), we investigated the abiotic stress tolerance of the mutant plants. Among the several stresses, we examined, most clear-cut result was observed in the high salt test. Figure 3b and C show that shoot growth of the mutant seedlings was less sensitive (i.e. more tolerant) to high salt inhibition than wild type plants. For instance, shoot growth rates of wild type seedlings at 150 mM and 175 mM NaCl decreased to 36% and 22%, respectively, of the control rate. At the same condition, growth rates of the mutant seedlings were 49% and 45%, respectively, of the control rate. The higher growth rates of the mutant plants indicates that OsMAPKKK63 is required for normal response to high salinity stress and that its KO line is partially insensitive to salt (i.e. salt-tolerant). Thus, our result demonstrated that OsMAPKKK63 is a regulator of high salt response. It is noteworthy that the OsMAPKKK63 KO mutant was salt-tolerant. The finding suggests that OsMAPKKK63 can be utilized for the generation of salt-tolerant rice variety.
Figure 3.
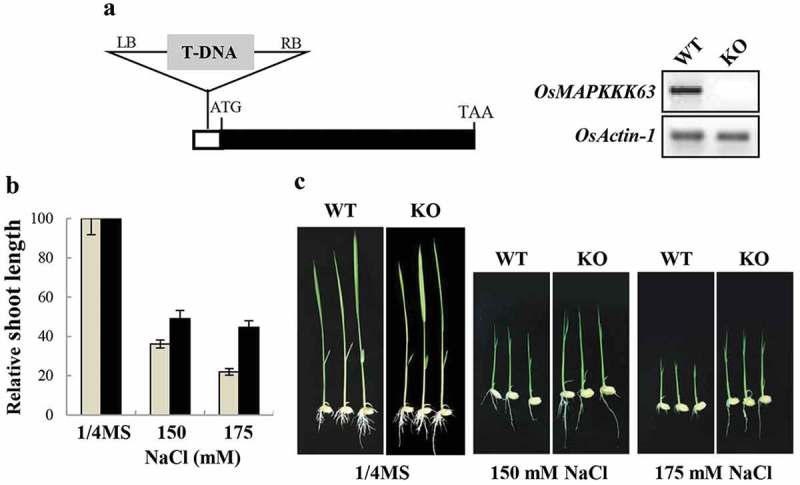
Salt tolerance of OsMAPKKK63 KO line. (a) Left, Schematic diagram of OsMAPKKK63 gene structure. T-DNA is inserted in the 5’-untranslated region (−87 from the initiation codon). Right, expression of OsMAPKKK63 determined by semiquantitative RT-PCR. Rice actin-1 gene was used as a reference gene. (b) The effect of high salt (NaCl) on shoot growth. Bars indicate standard errors. (c) Photographs showing representative seedlings. Plants were grown for 7 days before taking pictures. WT, wild type. KO, knockout.
We attempted to generate OsMAPKKK63 overexpression (OX) lines to gain further information about its in planta function. However, for unknown reason, it was difficult to obtain its OX lines. We managed to generate one OX line, and, in the course of our experiment to analyze its phenotypes, we found that the OX line was viviparous. We do not know whether the vivipary was the main reason for the difficulty we encountered in the generation of OsMAPKKK63 OX lines. However, as shown in Figure 4a, precocious germination of seeds was observed under high humidity condition. For example, seeds of freshly harvested panicle have a tendency to sprout more easily than wild type seeds (Figure 4b). Moreover, precocious germination of seeds was observed also under field condition (Figure 4c): ca. 70% of the transgenic seeds germinated in the panicle during maturation, whereas the vivipary was not observed with wild type plants. To validate the OsMAPKKK63 OX phenotype, we acquired an activation-tagged mutant line (PFG_3D-02878), in which an enhancer-containing T-DNA was inserted in the 5 flanking region of OsMAPKKK63 (Figure 4a). OsMAPKKK63 expression level was lower in the mutant compared with that of the OX line (Figure 4a). Nonetheless, the viviparous phenotype was observed also with the activation-tagged mutant (Figure 4b). Thus, both the OX line and activation-tagged line of OsMAPKKK63 exhibited vivipary.
Figure 4.
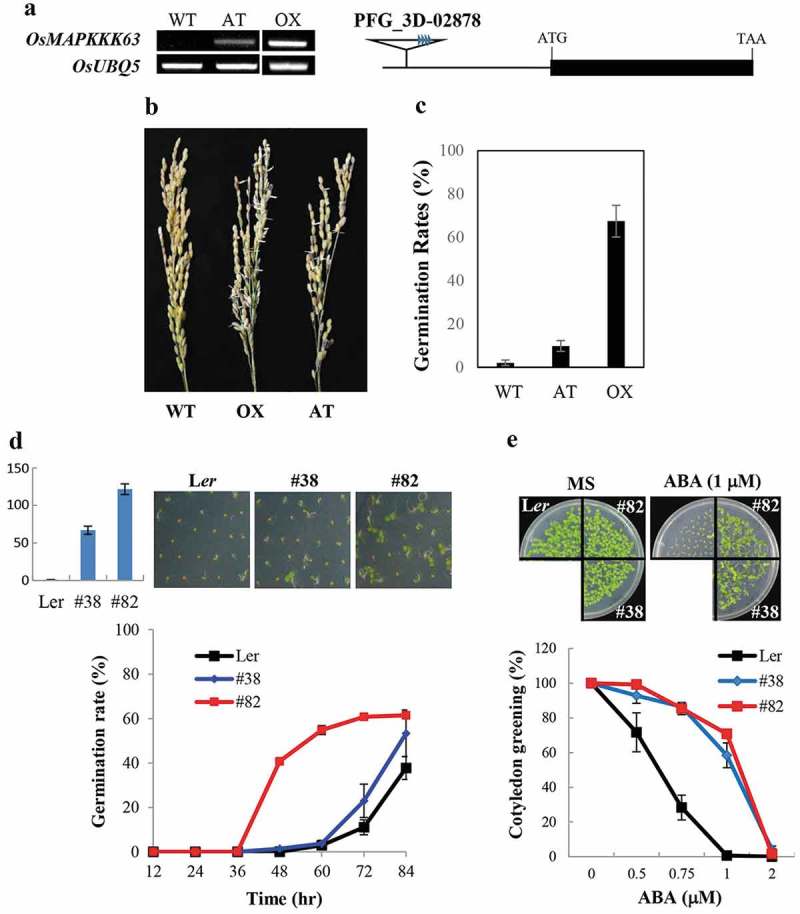
Seed dormancy of OsMAPKKK63 OX lines. (a) Left, OsMAPKKK63 expression levels in the transgenic rice plants determined by semiquantitative RT-PCR. AT, activation-tagged line. OX, OX line. Rice ubiquitin5 gene was used as a reference gene. Right, a schematic diagram of rice activation-tagged line (PFG_3D-02878). (b) Rice panicles harvested from the field-grown plants and kept in high humidity (99%) condition for two days. Please note that OX and AT line seeds sprouted. (c) Vivipary of OX and AT lines. Preharvest sprouting (i.e. germination of seeds in the live panicle of field-grown plants) is presented as germination rates. (d) Seed dormancy of Arabidopsis OX lines. Top left, Expression levels of OsMAPKKK63 in the transgenic lines determined by Real-Time RT-PCR. Top right, Photographs showing representative germinating seeds. Bottom, Germination rates of freshly harvested seeds. Seeds were plated without stratification and germination (radicle emergence) rates were scores at indicated time points. The numbers indicate line numbers, and small bars indicate standard errors. (e) ABA sensitivity of Arabidopsis OX lines during cotyledon greening/expansion stage. Top panels show seedlings grown for four days in MS medium or MS medium containing 1 μM ABA. Bottom, cotyledon greening efficiency. Seeds were germinate and grown for four days before counting seedling with green cotyledons. Bars indicate standard errors.
To further confirm the role of OsMAPKKK63 in seed dormancy control, we employed a heterologous approach, i.e. we generated transgenic Arabidopsis plants overexpressing OsMAPKKK63 (Figure 4d). We then determined the seed dormancy of the transgenic plants by scoring the germination rates of freshly harvested seeds without stratification. Figure 4d shows that germination rates of the transgenic Arabidopsis seeds, especially those of #82 expressing OsMAPKKK63 at a higher level, were much higher than the wild type rates (e.g. 55% vs 4% at the 60 hr time point). The result indicated that seed dormancy was decreased in the OsMAPKKK63 OX lines, which was consistent with the viviparous phenotype observed with rice OX lines.
Phenotypes described above, namely salt-insensitivity and reduced seed dormancy are associated with reduced ABA sensitivity.16 We, therefore, hypothesized that ABA sensitivity might have been altered in the OsMAPKKK63 OX lines. To test the hypothesis, we examined ABA sensitivity of the transgenic Arabidopsis plants during several different developmental stages, such as germination, postgermination shoot development (cotyledon greening/expansion), and primary root elongation. The result showed that ABA sensitivity of the transgenic plants was significantly reduced during cotyledon greening stage (Figure 4e). For example, cotyledon greening/expansion of wild type seedlings was almost completely suppressed at 1 μM ABA. By contrast, the transgenic seedlings retained high cotyledon greening efficiency, i.e. 58% (#38) and 71% (#82), respectively, under the same condition. Thus, OsMAPKKK63 OX lines were ABA-insensitive during the shoot establishment stage.
In summary, our data indicate that OsMAPKKK63 expression is induced by several abiotic stresses. It possesses in vitro kinase activity, and its constitutively active form interacts with OsMKK1 and OxMAPKK6 in yeast. In planta, OsPMAPKKK63 is required normal salt stress response, and our data suggest that it probably plays an important role in the control of seed dormancy.
Materials and methods
Plant material and growth
Japonica rice (Oryza sativa L. cv Dongjin) and Arabidopsis (ecotype Landsberg erecta, Ler) were used in this study. Plants were grown under long day condition (14-hr-light/10-hr-dark cycle for rice, 16-hr-light/8-hr-dark cycle for Arabidopsis) at 28°C (rice) or 21°C (Arabidopsis). For normal axenic growth, ¼ X and ½ X Murashige and Skoog (MS) media were used for rice and Arabidopsis, respectively.
Entire coding region of OsMAPKKK63 was cloned into the OX vector pGA161117 for the generation of rice OX lines, and transformation was performed as described previously.17,18 For Arabidopsis transformation, the coding region of OsMAPKKK63 was cloned into the GUS-less pBI121 vector,19 and transformation was carried out according to Bechtold and Pelletier.20
RNA isolation and expression analysis
Rice RNA was isolated according to Chomczynski and Mackey,21 and Arabidopsis RNA was isolated employing the Qiagen RNeasy plant mini kit. For RT-PCR (coupled reverse transcription-polymerase chain reaction), RNA was treated with DNase I to remove possible contaminating DNA. cDNA was synthesized using the Superscript III (Invitrogen), according to the supplier’s manual. For semi-quantitative RT-PCR, cDNA amplification was performed within a linear range of amplification. For Real-Time RT-PCR, amplification of the first strand cDNA was carried out in a Bio-Rad CFX96 Real-time PCR Systems (Bio-Rad).
Preparation of recombinant proteins and kinase assay
Recombinant proteins were prepared as described previously,22 employing the pMAL system (NEB) with a modification. Kinase assay was performed according to Lee et al.22 0.5 μg of proteins were used in the assay, and the incorporation of 32P was examined by autoradiography of dried gels.
Yeast two-hybrid assay
Two-hybrid assay was performed as described previously.23 Bait constructs containing full-length or kinase domain of OsMAPKKK63 was prepared by cloning amplified coding regions into pPC62LexA bait (DNA-binding domain) vector. OsMKK coding regions were amplified and cloned into pYESTrp2 activation domain vector to be used as prey. Yeast transformants were grown in CM (complete minimal)-Leu-Trp medium, and the β-galactosidase reporter activity was assayed according to the X-gal overly protocol24 or standard Miller liquid assay protocol.25
Note: Primer sequences used in this study are available upon request.
Funding Statement
This work was supported in part by the Rural Development Administration, Republic of Korea [Next-Generation BioGreen 21 Program (grant PJ00819804 to S.Y.K.)].
Acknowledgments
The authors are grateful to the Kumho Life Science Laboratory of Chonnam National University for providing equipment and plant growth facilities.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Martin G.. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez MC, Petersen M, Mundy J.. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 3.Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010;17:139–153. doi: 10.1093/dnares/dsq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyna NS, Yang Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact. 2006;19:530–540. doi: 10.1094/MPMI-19-0530. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Ma H, Hong H, Yao W, Xie W, Xiao J, Wang S. Transcriptome-based analysis of mitogen-activated protein kinase cascades in the rice response to Xanthomonas oryzae infection. Rice. 2015;8:4. doi: 10.1186/s12284-014-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ning J, Li X, Hicks LM, Xiong L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010;152:876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning J, Zhang B, Wang N, Zhou Y, Xiong L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell. 2011;23:4334–4347. doi: 10.1105/tpc.111.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JA, Agrawal GK, Rakwal R, Han KS, Kim KN, Yun CH, Jwa NS. Molecular cloning and mRNA expression analysis of a novel rice (Oryzasativa L.) MAPK kinase kinase, OsEDR1, an ortholog of Arabidopsis AtEDR1, reveal its role in defense/stress signalling pathways and development. Biochem Biophys Res Commun. 2003;300:868–876. [DOI] [PubMed] [Google Scholar]
- 9.Kim JA, Cho K, Singh R, Jung YH, Jeong SH, Kim SH, Tamogami S. Rice OsACDR1 (Oryza sativa accelerated cell death and resistance 1) is a potential positive regulator of fungal disease resistance. Mol Cells. 2009;28:431–439. doi: 10.1007/s10059-009-0161-5. [DOI] [PubMed] [Google Scholar]
- 10.Ruegg UT, Burgess GM. Staurosporine, K-252 and UCN-01: potent but nonspecific inhibitors of protein kinases. Trends Pharmacol Sci. 1989;10:218–220. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Jing W, Zhang W. The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 2014;227:181–189. doi: 10.1016/j.plantsci.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Kumar K, Sinha AK. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice (N Y). 2013;6:25. doi: 10.1186/1939-8433-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie GS, Kato F, Imai R. Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J. 2012;443:95–102. doi: 10.1042/BJ20111792. [DOI] [PubMed] [Google Scholar]
- 14.Jeon J-S, Lee S, Jung K-H, Jun S-H, Jeong D-H, Lee J, Nam J. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. [DOI] [PubMed] [Google Scholar]
- 15.Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Kim J. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 16.Finkelstein R. Abscisic Acid synthesis and response. Arabidopsis Book. 2013;11:e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, Jeon J-S, Jung K-H, An G. Binary vectors for efficient transformation of rice. J Plant Biol. 1999;42:310. doi: 10.1007/BF03030346. [DOI] [Google Scholar]
- 18.Jeon JS, Chung YY, Lee S, Yi GH, Oh BG, An G. Isolation and characterization of an anther-specific gene, RA8, from rice (Oryza sativa L.). Plant Mol Biol. 1999;39:35–44. [DOI] [PubMed] [Google Scholar]
- 19.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;20:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Mackey K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- 22.Lee SJ, Lee MH, Kim JI, Kim SY. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 2015;56:84–97. doi: 10.1093/pcp/pcu148. [DOI] [PubMed] [Google Scholar]
- 23.Lee SJ, Cho DI, Kang JY, Kim SY. An ARIA-interacting AP2 domain protein is a novel component of ABA signaling. Mol Cells. 2009;27:409–416. doi: 10.1007/s10059-009-0058-3. [DOI] [PubMed] [Google Scholar]
- 24.Duttweiler HM. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 1996;12:340–341. [DOI] [PubMed] [Google Scholar]
- 25.Guthrie C, Fink GR, eds. Guide to yeast genetics and molecular biology, Methods in Enzymology Vol. 194 San Diego: Academic Press; 1991. [PubMed] [Google Scholar]


