ABSTRACT
Lung cancer is one of the most common cancers in the world, which accounts for about 27% of all cancer deaths. However, the mechanisms underlying the pathogenesis of lung cancer cells remain largely elusive. In this study, we examined the role of the Forkhead box protein P1 (FOXP1) in lung cancer development. Our Oncomine analysis shows that FOXP1 is downregulated in lung adenocarcinoma compared with normal lung tissue. Knockdown of FOXP1 promotes the growth and invasion of PC9 and A549 cells by regulating genes of chemokine signaling molecules, including CCR1, ADCY5, GNG7, VAV3, and PLCB1. Simultaneous knockdown of CCR1 and FOXP1 attenuated FOXP1 knockdown-induced increase of lung cancer cell growth. Finally, knockdown of FOXP1 in PC9 cells promotes the tumorigenesis via CCR1 signaling in xenograft mouse model. Taken together, our data suggest that FOXP1 plays important roles in preventing lung adenocarcinoma development via suppressing chemokine signaling pathways.
Keywords: lung cancer, FOXP1, proliferation, chemokine signaling
Introduction
Lung cancer is the leading cause of cancer-related mortality in the world. In 2014, more than 150,000 deaths were reported in the United States.1 There are two major subtypes of lung cancer, non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Approximately 85% of lung cancers are NSCLC, which is composed of adenocarcinoma (40%), squamous-cell carcinoma (25–30%), and large-cell carcinoma (10–15%).2 Some frequently mutated genes, such as TP53, Kras, EGFR PTEN, FGFR, KEAP1 and ALK, have been investigated previously,3–7 benefiting the treatment of subsets of NSCLC patient. However, the prognosis of NSCLC patients is still poor due to the complexity of the mechanisms underlying the tumorigenesis of lung cancer. Better understanding of the mechanisms will help to improve the therapeutic outcomes.
The Forkhead box protein P1 (FOXP1) belongs to a family of winged-helix transcription factors that are involved in a broad range of functions, such as cell cycle progression, proliferation, differentiation, and apoptosis.8 FOXP1 was originally cloned from mouse B cell leukemia cell line and subsequently been found to be expressed in many different types of tissues in human and in a variety of cancers,9,10 where FOXP1 can serve as a tumor suppressor or oncogene in different cancers.11–14 Shu et al. showed that FOXP1 are highly expressed in the developing airway epithelium and is crucial for the lung development.15 The expression of FOXP1 has been shown to be downregulated in lung adenocarcinoma and high expression of FOXP1 is associated with improved survival in NSCLC patients.13 However, the role of FOXP1 in lung cancer cells remains elusive.
In this study, we demonstrated that the expression of FOXP1 was decreased in the human lung adenocarcinoma compared with normal lung tissue. Moreover, knockdown of FOXP1 promoted the growth of lung cancer cells PC9 and A549 via inhibiting apoptosis. The migration and invasion of lung cancer cells were enhanced after the FOXP1 knockdown. Furthermore, RNA-sequence analysis revealed that chemokine signaling genes, including CCR1, ADCY5, GNG7, VAV3, and PLCB1, were upregulated in PC9 and A549 cells transfected with FOXP1 siRNA. Concomitant knockdown of CCR1 with FOXP1 attenuated FOXP1 knockdown-induced increase of lung cancer cell proliferation. Finally, knockdown of FOXP1 in PC9 cells promotes the tumorigenesis in xenograft mouse model. Taken together, our data suggest that FOXP1 serves as a tumor suppressor of lung adenocarcinoma via downregulating chemokine signaling pathways.
Results
FOXP1 is downregulated in lung adenocarcinoma
To examine the role of FOXP1 in lung adenocarcinoma, we compared the expression of FOXP1 in normal lung tissue and lung adenocarcinoma obtained from four studies in Oncomine microarray database.16–19 As shown in Figure 1, the expression of FOXP1was dramatically downregulated in lung adenocarcinoma compared with normal lung tissue from different datasets, indicating that FOXP1 plays important roles in lung adenocarcinoma. Furthermore, the expression of FOXP1 was further reduced in grade 3 than grade 2 of lung adenocarcinoma (Figure 1d), indicating that the expression of FOXP1correlates with the severity of lung adenocarcinoma.
Figure 1.
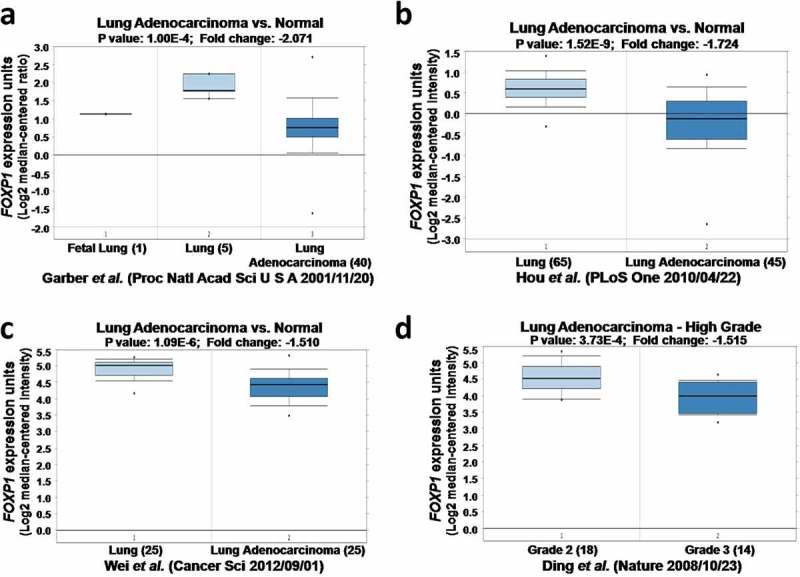
The expression of FOXP1 in normal lung tissues and lung cancer by Oncomine analysis. FOXP1 expression is decreased in lung cancer compared with normal lung. a-d figures were from different Oncomine databases.
Knockdown of FOXP1 promotes growth and invasion of lung cancer cells
Cancer cell proliferation is an important step in cancer development. First of all, we examined the effect of FOXP1 on lung cancer cell growth by using the siRNA specific for FOXP1. As shown in Figure 2, after the FOXP1 siRNA transfection, the FOXP1 expression was significantly reduced in both PC9 and A549 cells compared with that in control siRNA-transfected cells as evidenced by the western blot analysis, indicating the successful knock-down of FOXP1 in these cells. Furthermore, MTT assay demonstrated that the cell growth of PC9 and A549 cells was significantly enhanced beginning at 24 hrs after the knock-down of FOXP1, suggesting that FOXP1 exhibits inhibitory effect on lung cancer cell growth (Figure 2b & 2e). Furthermore, flow cytometer analysis demonstrated that the apoptosis of lung cancer cells was significantly decreased after the knockdown of FOXP1 (Figure 2c & 2f). Next, we examined the effect of FOXP1 on invasion of lung cancer cells by employing the Boyden chamber invasion assay. As shown in Figure 2g & h, after FOXP1 siRNA treatment, the number of cells penetrated through the Matrigel was dramatically increased in both PC9 and A549 cells transfected with FOXP1 siRNA. These results indicate that knockdown of FOXP1 promotes the growth and invasion of lung cancer cells.
Figure 2.
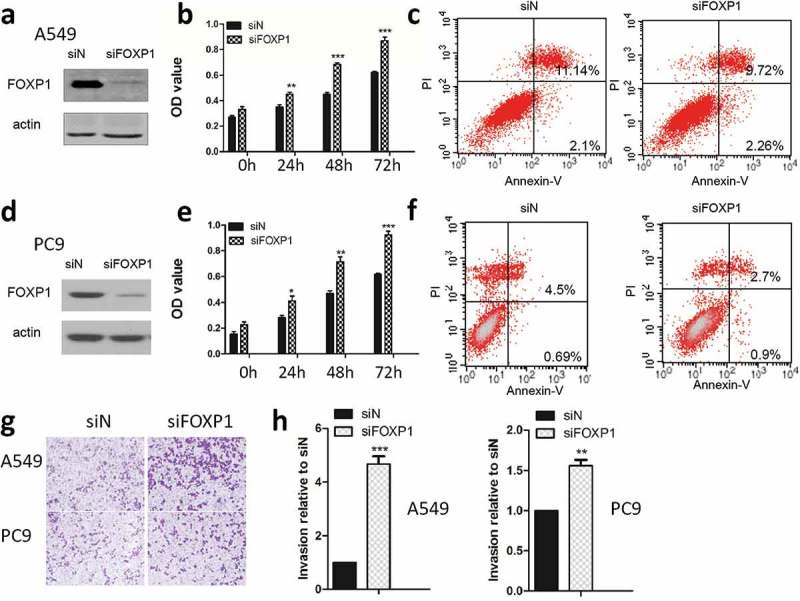
Knockdown of FOXP1 promotes the growth and invasion of lung cancer cells.
Western blot shows the protein expression of FOXP1 in A549 (a) and PC9 (d) cells 48 hrs after siFOXP1 treatment. Bar graphs show OD values of A549 (b) and PC9 (e) cells at 24h, 48h, and 72h after the knockdown of FOXP1. Flow cytometry analysis shows the apoptosis of A549 (c) and PC9 (f) cells 48 hrs after the knockdown of FOXP1. Representative pictures (g) and summarized data (h) show that the invasion of A549 and PC9 cells 48 hrs after the knockdown of FOXP1.* p < 0.05, ** p < 0.01, *** p < 0.001.
Chemokine signaling pathway is involved in FOXP1-mediated tumor suppressing effects
To reveal the mechanism underlying FOXP1-mediated suppression of lung cancer cells, we performed RNA-Seq analysis in PC9 cells transfected with control or FOXP1 siRNA. As shown in the heatmap, the gene expression profiles of PC9 cells transfected with FOXP1 siRNA clustered together and were distinguishable from that of control siRNA-treated cells (Figure 3a). The GO enrichment analysis demonstrated that the differentially expressed genes were categorized into 17 functional categories (Figure 3b). Of particular interest is the chemokine signaling pathway, which is known to play important roles in non-small cell lung cancer progression.20,21 Genes related to chemokine signaling pathway are dramatically altered by the knock down of FOXP1, such as CCR1, ADCY5, GNG7, VAV3, and PLCB1. The changes of these genes were further validated by quantitative RT-PCR experiments in both A549 and PC9 cells (Figure 3c & 3d). These results indicate that multiple genes downstream of FOXP1 might be involved in FOXP1-mediated suppressing of lung tumor cell growth and invasion.
Figure 3.
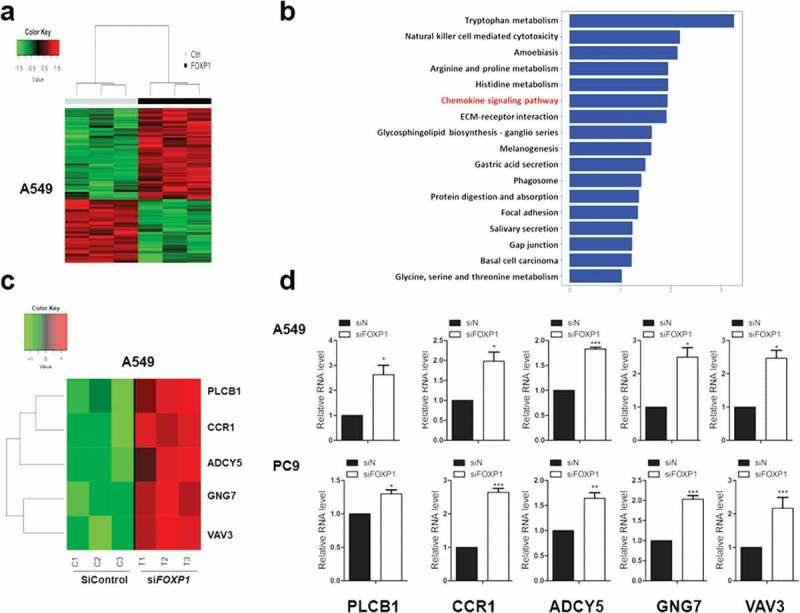
Chemokine signaling pathways are involved in FOXP1-mediated effect.
(a) Hierarchical clustering analysis reveals the alterations of transcripts in A549 cells treated with control or FOXP1 siRNA. (b) KEGG analysis of those significantly changed genes shows their enrichment in chemokine signaling pathway. (c and d) The heatmap of representative genes (c) and the RT-PCR validations (d), showing the alterations of CCR1, ADCY5, GNG7, VAV3, and PLCB1 after the knockdown of FOXP1. * p < 0.05, *** p < 0.001.
CCR1 is required for FOXP1 sirna-induced lung cancer cell growth and invasion
Chemokine (C-C motif) receptor 1 (CCR1) has been shown to promote cancer cell proliferation and invasion of cancer cells, including lung cancer cell lines.22 Of important note, CCR1 also was identified to be regulated by FOXP1 in colon cancer cell lines.23 As such, we attempted to determine the role of CCR1 in FOXP1 siRNA-induced growth and invasion of lung cancer cells, we transfected A549 and PC9 cells with CCR1 siRNA in addition to FOXP1 siRNA transfection and measured the cancer cell growth and invasion. As shown in Figure 4, knockdown of CCR1 reduced the growth and invasion of lung cancer cells. Furthermore, co-transfection of siRNAs targeting CCR1 and FOXP1 reversed the increase of A549 and PC9 cells growth induced by FOXP1 siRNA. Similarly, the enhanced invasion of A549 and PC9 cells induced by FOXP1 siRNA was also dramatically attenuated by the co-transfection of CCR1 siRNA. In line with the critical role of CCR1, overexpression of CCR1 dramatically decreased the expression of ADCY5, GNG7, and VAV3. However, the expression of PLCB1 was not affected by CCR1 overexpression (Figure 4e). To further explore the mechanism underlying the regulation of CCR1 by FOXP1, we performed luciferase assay. As shown in Figure 4f and g, the CCR1 promoter activity was significantly enhanced by the knockdown of FOXP1. These results indicate that CCR1-mediated downstream signaling pathway plays important roles in FOXP1-mediated tumor suppression.
Figure 4.
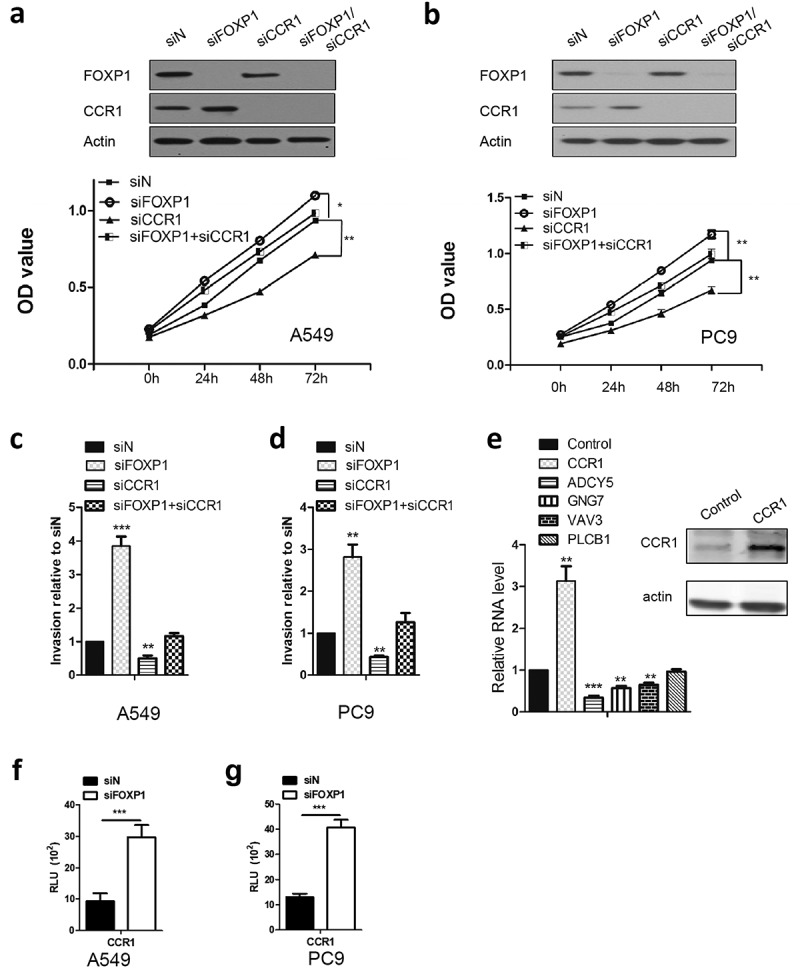
CCR1 is required for FOXP1-mediated effect.
(a and b) OD values of A549 (a) and PC9 (b) cells at 24h, 48h, and 72h after siFOXP1, siCCR1, or siFOXP1+ siCCR1 treatment. Upper panels are Western blot images showing the FOXP1 and CCR1 expression 48 hrs after the treatment of different siRNAs. Actin serves as loading control. (c and d) Bar graphs show the relative invasion of A549 (c) and PC9 (d) cells 48 hrs after siFOXP1, siCCR1, or siFOXP1+ siCCR1 treatment. (e) RT-PCR shows the expression levels of ADCY5, GNG7 and VAV3 but not PLCB1 48 hrs after the overexpression of CCR1. Inset is the gel picture of Western blot examining the expression of CCR1 in overexpressed cells. (f and g) Bar graphs show the luciferase activity in A549 (f) and PC9 (g) cells after 36 hrs of FOXP1 siRNA treatment. * p < 0.05, ** p < 0.01, *** p < 0.001.
Knockdown of FOXP1 increased the tumor growth
To examine the role of FOXP1 in lung cancer development in vivo, we utilized xenograft mouse model by injecting PC9 cells transfected control or FOXP1 and CCR1 shRNAs into both flanks of nude mice. As shown in Figure 5, 28 days after the injection, the size and weight of tumors induced by FOXP1 shRNA-transfected PC9 cells were significantly larger than that induced by control shRNA-transfected PC9 cells. Furthermore, PC9 cells co-transfected with FOXP1 and CCR1 shRNAs failed to increase the size and weight of tumors compared with control shRNA-transfected PC9 cells. These results further confirm that FOXP1 suppresses the lung cancer development via CCR1-mediated signaling pathway.
Figure 5.
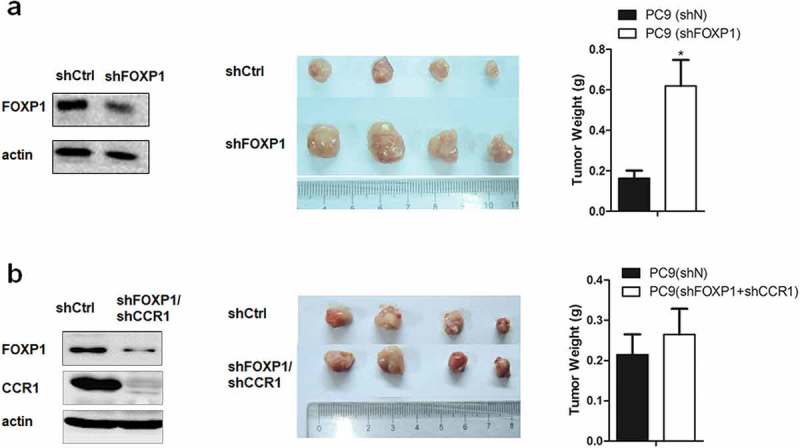
Downregulation of FOXP1 promotes the tumorigenesis via CCR1 in xenograft mouse model.
(a) Left panel shows the representative Western Blot image of PC9 cells treated with shFOXP1. Actin serves as the loading control. Middle panel shows the representative picture of tumors induced by PC9 cells treated with shCtrl or shFOXP1. Bar charts in the right panel show the weight of tumors generated by PC9 cells treated with shCtrl or shFOXP1. (b) Left panel shows the representative Western Blot image of PC9 cells treated with shFOXP1+ shCCR1. Actin serves as the loading control. Middle panel shows the picture of tumors induced by PC9 cells treated with shCtrl or shFOXP1+ shCCR1. Bar charts in the right panel show the weight of tumors generated by PC9 cells treated with shCtrl or shFOXP1+ shCCR1. * p < 0.05.
Discussion
FOXP1 is a member of the widely expressed FOXP subfamily of “forkhead” transcription factors that have a variety of functions in cellular proliferation, differentiation and neoplastic transformation,24 indicating that FOXP1 plays important roles in cancer pathogenesis. In supporting the role of FXOP1 in lung adenocarcinoma, we demonstrate that the expression of FOXP1was decreased in lung adenocarcinoma tissue using Oncomine analysis. Like other cancers, lung cancer is characterized by high growth rate and strong invasion of cancer cells. Downregulation of FOXP1 in lung cancer cells by siRNA transfection dramatically increased the growth and invasion of lung cancer cells, PC9 and A549. Furthermore, the apoptosis of lung cancer cells was decreased when the expression of FOXP1 was downregulated. In line with these in vitro data, we further demonstrated that downregulation of FOXP1 enhanced the tumorigenesis of PC9 cells in cells an in vivo xenograft mouse model. These results indicate that FOXP1 may play a protective role in lung cancer development via promoting apoptosis and inhibiting growth of cancer cells.
Chemokines have originally been recognized as critical mediators regulating the recruitment of immune cells.25 Several lines of evidence demonstrated that cancer cells express multiple chemokines and chemokine receptors, which regulate the behaviors of cancer cells upon activation by chemokines.26 Chemokine signaling pathways play important roles in cancer progression including tumor growth, senescence, epithelial mesenchymal transition, and immune evasion.27 On the other hand, diverse chemokines regulate the function of T-cell, macrophage, and dendritic cells, which regulate the function of cancer cells in the microenvironment.28 In the present study, we demonstrated that downregulation of FOXP1 altered the expression of many genes, which are significantly enriched in chemokine signaling pathways, including CCR1, ADCY5, GNG7, VAV3, and PLCB1, indicating that chemokine signaling pathways are involved in FOXP1-mediated effects. CCR1 has been shown to promote the tumor cell invasion and metastasis of colorectal cancer and liver cancer.29,30 Consistently, we demonstrated that knockdown of FOXP1 resulted in the increased expression of CCR1, which promotes the growth and invasion of lung cancer cells. Simultaneous knockdown of CCR1 attenuated the pro-tumoral effect of FOXP1 siRNA in lung cancer cells. These results indicate that downregulation of FOXP1 promotes the tumorigenesis via the downstream chemokine signaling CCR1. In addition, we cannot rule out the possibility of that CCR1 may also act its function independent of FOXP1 in regulation of cancer development.
GNG7 is a highly specific promoter methylated gene associated with head and neck cancer. It has been shown that the expression of GNG7 was reduced in esophageal cancer and pancreatic cancer, indicating that GNG7 functions as a tumor suppressor.31,32 However, we demonstrated that the expression of GNG7 was increased in lung cancer cells after the knockdown of FOXP1, suggesting that unlike in other cancers, GNG7 may exert oncogenic effect in lung cancer. VAV3 is a guanine nucleotide exchange factor for Rho GTPases.33 VAV3 has been shown to promote breast cancer, prostate cancer, and skin tumor.34–36 Similarly, we showed here that VAV3 was increased after the knockdown of FOXP1 and promoted the growth and invasion of lung cancer cells. Together with ADCY5, GNG7 and VAV3 are downstream targets of CCR1 as overexpression of CCR1 resulted in the decreased expression of ADCY5, GNG7, and VAV3. However, PLCB1 was not affected by the overexpression of CCR1, suggesting that PLCB1 might be involved in a distinct pathway following the FOXP1 downregulation. Together, these results revealed previously unrecognized roles of CCR1, ADCY5, GNG7, VAV3, and PLCB1 in FOXP1-mediated antitumor effect in lung cancer.
In summary, our study demonstrates that FOXP1 is downregulated in lung cancer. Knockdown of FOXP1 in lung cancer cells promotes the growth and invasion activities. Genes related to chemokine signaling pathways CCR1, ADCY5, GNG7, VAV3, and PLCB1 are the downstream events of FOXP1-mediated effects. These results suggest that FOXP1 many play a protective role in lung adenocarcinoma development and can be targeted for the development of novel diagnostic and therapeutic strategies for lung adenocarcinoma.
Materials and methods
Oncomine analysis
Oncomine, a cancer microarray database and web-based data mining platform, was used to compare mRNA level of FOXP1 in lung cancer vs. normal patient datasets. The relative expression levels of genes were plotted using Graphpad Prism software.
Cell culture and sirna transfection
PC9 and A549 cells were cultured in RPMI and MEM, both of which were supplemented with 10% fetal bovine serum (FBS), 2 mmol/l glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. Cells less than 30 passages were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37°C. For transfection of siRNA, Lipofectamine® RNAiMAX Reagent (Invitrogen) was mixed with the siRNA construct according to the manufacturer’s instructions. All the siRNAs used in this study were purchased from Sigma. The MISSION® siRNA Universal Negative Control (siN) obtained from Sigma was used as control siRNA. In detail, siFOXP1 (NM_001012505 > SASI_Hs01_00228672) and siCCR1 (NM_001295 > SASI_Hs01_00119718) were used here.
Luciferase assay
The promoter sequence of CCR1 (Sup. Table 1) was cloned into pGL4.10 reporter plasmid (Promega, USA) by PCR using forward primer (5′-3′ CGGCTAGCGAAGGAAGAACACCAGAGACC) and reverse primer (5′-3′ ATGATATCGGTATTTGTCATTGGCCTGG) with the digestion of the NheI/EcoRV sites. Firefly luciferase and Renilla luciferase were quantified with the Dual-Luciferase Reporter Assay system and the Stop & Glo Reagent kit according to the manufacturer’s instruction (Promega, USA) after 36 hours of control siRNA or FOXP1 siRNA transfection.
MTT assay
PC9 and A549 cells were seeded in 96-well plates with 5000 cells/well. The cells were then treated with control or target siRNAs for 48 hr. MTT was added to the cells which were then cultivated for another 4 hr. Following the removal of the supernatant, DMSO (100 μl/well) was added to the cells which were agitated for 15 min. The absorbance was measured at 570 nm by an ELISA reader. Each assay was repeated three times.
Detection of apoptosis
Activation of apoptosis signaling in A549 and PC9 cells was assessed by Annexin V/PI double staining assay using FITC Annexin V Apoptosis Detection kits (Beyotime Institute of Biotechnology, Haimen, China). After treatments, cells were trypsinized, washed, and resuspended in Annexin V/PI binding buffer subjected to staining according to the manufacturer’s instructions. The stained cells were analyzed by flow cytometry (BD Biosciences).
Western blot
Whole-cell lysates from PC9 and A549 cells were prepared and 20 µg of protein was separated using 10% SDS-PAGE gel. After blocking with Tris-buffered saline/0.05%Tween 20 containing 5% skim milk or 3% bovine serum albumin (Sigma, St. Louis, MO), blots were incubated at 4 °C overnight with the primary antibodies against human FOXP1 protein (Abcam, ab16645), CCR1 (Abcam, ab13240), and actin (Abcam, ab8227), respectively. A horseradish peroxidase conjugated IgG was used as secondary antibody according to manufacturer’s instruction.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
The total RNA from PC9 or A549 cells was prepared using the Trizol reagent (Invitrogen, Carlsbad, CA). 1 µg of total RNA was treated with DNase I (Invitrogen) and the cDNA was synthesized in vitro from the mRNA template using SuperScript® III First-Strand Synthesis Kit (Invitrogen). cDNA was amplified by PCR using specific primer pairs. Taqman probes were taken to examine the gene expression, including CCR1 (Hs00928897_s1, ADCY5 (Hs00766287_m1), GNG7 (Hs00192999_m1), VAV3 (Hs00916818_m1), PLCB1 (Hs01001930_m1). Actin served as the internal control. The relative gene expression was calculated using the equation: 2−ΔΔCt.
Invasion assay
Lung cancer cell invasion was measured by using the Matrigel-coated transwell culture chambers as described previously.37 Briefly, PC9 or A549 cells transfected with control or FOXP1 siRNA were maintained in serum-free-F12 medium in the upper chamber. The lower chamber was filled with 10% FBS-containing culture medium. Cells were incubated for 18 h at 37°C in a humidified atmosphere with 95% air and 5% O2. The invaded cells penetrated through the Matrigel in the lower chamber were fixed and counted under a light microscope.
RNA sequencing and differentially expressed genes (DEGs) analysis
Three control and three FOXP1 knockdown A549 cell lines were subjected to RNA sequencing. Total RNA was extracted 48 h after the FOXP1 siRNA or control siRNA transfected. With the RNA sequencing data prepared, the expression abundance (FPKM) value of each gene was estimated by running cufflinks,38 and the differential expressed genes were assessed by cuffdiff. Only those genes with |fold change|> 2 and adjusted p value < 0.01 were recognized as statistically differentially expressed between two groups. The adjusted p value was obtained through applying Benjamini and Hochberg’s (BH) false discovery rate correction on the original p value, and fold change threshold was selected based on our purpose of focusing on significantly differentially expressed genes.
Hierarchical clustering and GO analysis
We performed hierarchical clustering to classify analyzed samples based on gene expression profiles.39 Hierarchical clustering was carried out using differentially expressed genes to observe the global gene expression patterns. We utilized R packages – GO.db to detect Gene Ontology categories with significant enrichment in DEGs comparing to which across all measured genes. The significantly enriched biological processes were identified as p value less than threshold value 0.01.
Xenograft mouse model
Following the methods described in the reference to implant PC9 cells subcutaneously into both flanks of nude mice at 2 × 106 cells in 100 μl per spot, the tumors were collected after the implantation about 28 days.40 Control cells or those with either shFOXP1 or sFOXP1/shCCR1 were injected into opposite flanks. All animal experiments were approved by the Animal Care and Use Committee. ShRNA-FOXP1 (Sigma, SHCLNV-NM_032682> TRCN0000015664) and ShRNA-CCR1 (Sigma, SHCLNV-NM_009912> TRCN0000027778) were introduced to cell using Lentiviral system
Statistical analysis
Data are presented as mean ± s.e.m. of three independent experiments. Significance of means between two groups is determined by student’s t-test. Difference in cell growth between control and PRAME siRNA-treated groups was evaluated by repeated measures analysis of variance (ANOVA). A P-value of < 0.05 was considered significantly different.
Funding Statement
This work is supported by Project of Shanghai Municipal Commission of Health and Family Planning (No. 20134084).
Acknowledgments
Not applicable.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin 2014; 64(1):9–29. doi: 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 2.Read WL, Page NC, Tierney RM, Piccirillo JF, Govindan R. The epidemiology of bronchioloalveolar carcinoma over the past two decades: analysis of the SEER database. Lung Cancer 2004; 45(2):137–142. doi: 10.1016/j.lungcan.2004.01.019 [DOI] [PubMed] [Google Scholar]
- 3.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001; 15(24):3243–3248. doi: 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Cho SN, Akkanti B, Jin N, Mao J, Long W, Chen T, Zhang Y, Tang X, Wistub II, et al. ErbB2 Pathway activation upon Smad4 loss promotes lung tumor growth and metastasis. Cell Rep 2015; 10(9):1599–1613. doi: 10.1016/j.celrep.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511(7511):543–550. doi: 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489(7417):519–525. doi: 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Cho SN, Wu SP, Jin N, Moghaddam SJ, Gilbert JL, Wistuba I, DeMayo FJ. Mig-6 deficiency cooperates with oncogenic Kras to promote mouse lung tumorigenesis. Lung Cancer 2017; 112:47–56. doi: 10.1016/j.lungcan.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013; 13(7):482–495. doi: 10.1038/nrc3539 [DOI] [PubMed] [Google Scholar]
- 9.Streubel B, Vinatzer U, Lamprecht A, Raderer M, Chott A. T(3;4)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005; 19(4):652–658. doi: 10.1038/sj.leu.2403644 [DOI] [PubMed] [Google Scholar]
- 10.Banham AH, Connors JM, Brown PJ, Cordell JL, Ott G, Sreenivasan G, Farinha P, Horsman DE, Gascoyne RD. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res 2005; 11(3):1065–1072. [PubMed] [Google Scholar]
- 11.Qiu DP, Han F, Zhuang H, Li QJ, Zhang XZ. Overexpression of FoxP1 is a novel biomarker of malignant human pancreatic cancer. Int J Clin Exp Med 2016; 9(6):9054–9063. [Google Scholar]
- 12.Flori M, Schmid CA, Sumrall ET, Tzankov A, Law CW, Robinson MD, Muller A. The hematopoietic oncoprotein FOXP1 promotes tumor cell survival in diffuse large B-cell lymphoma by repressing S1PR2 signaling. Blood 2016; 127(11):1438–1448. doi: 10.1182/blood-2015-08662635 [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Zhang XS, Zhu HJ, Wang XD, Ni SS, Huang JF. High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am J Clin Pathol 2012; 138(2):230–235. doi: 10.1309/Ajcpdhqfnyjz01yg [DOI] [PubMed] [Google Scholar]
- 14.Choi EJ, Seo EJ, Kim DK, Lee SI, Kwon YW, Jang IH, Kim KH, Suh DS, Kim JH. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget 2016; 7(3):3496–3509. doi: 10.18632/oncotarget.6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu WG, Lu MM, Zhang YZ, Tucker PW, Zhou DY, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 2007; 134(10):1991–2000. doi: 10.1242/dev.02846 [DOI] [PubMed] [Google Scholar]
- 16.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001; 98(24):13784–13789. doi: 10.1073/pnas.241500798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou J, Aerts J, Den Hamer B, van Ijcken W, Den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010; 5(4):e10312. doi: 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei TYW, Juan CC, Hisa JY, Su LJ, Lee YCG, Chou HY, Chen JMM, Wu YC, Chiu SC, Hsu CP, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci 2012; 103(9):1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455(7216):1069–1075. doi: 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas-Fuentes S, Salgado-Aguayo A, Pertuz Belloso S, Gorocica Rosete P, Alvarado-Vasquez N, Aquino-Jarquin G. Role of chemokines in non-small cell lung cancer: angiogenesis and inflammation. J Cancer 2015; 6(10):938–952. doi: 10.7150/jca.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arenberg D. Chemokines in the biology of lung cancer. J Thorac Oncol 2006; 1(4):287–288. doi: 10.1016/S1556-0864(15)31582-3 [DOI] [PubMed] [Google Scholar]
- 22.Wang CL, Sun BS, Tang Y, Zhuang HQ, Cao WZ. CCR1 knockdown suppresses human non-small cell lung cancer cell invasion. J Cancer Res Clin Oncol 2009; 135(5):695–701. doi: 10.1007/s00432-008-0505-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Smedt L, Palmans S, Govaere O, Moisse M, Boeckx B, De Hertogh G, Prenen H, Van Cutsem E, Tejpar S, Tousseyn T, et al. Expression of FOXP1 and Colorectal Cancer Prognosis. Lab Med 2015; 46(4):299–311. doi: 10.1309/LM7IHV2NJI1PHMXC [DOI] [PubMed] [Google Scholar]
- 24.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol 2002; 250(1):1–23. doi: 10.1006/dbio.2002.0780 [DOI] [PubMed] [Google Scholar]
- 25.Zabel BA, Zuniga L, Ohyama T, Allen SJ, Cichy J, Handel TM, Butcher EC. Chemoattractants, extracellular proteases, and the integrated host defense response. Exp Hematol 2006; 34(8):1021–10321. doi: 10.1016/j.exphem.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 26.Hembruff SL, Cheng N. Chemokine signaling in cancer: implications on the tumor microenvironment and therapeutic targeting. Cancer Therapy 2009; 7:254–267. [PMC free article] [PubMed] [Google Scholar]
- 27.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget 2013; 4(12):2171–2185. doi: 10.18632/oncotarget.1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013; 14(10):1014–1022. doi: 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamura T, Fujishita T, Loetscher P, Revesz L, Hashida H, Kizaka-Kondoh S, Aoki M, Taketo MM. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc Natl Acad Sci U S A 2010; 107(29):13063–13068. doi: 10.1073/pnas.1002372107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodero MP, Auvynet C, Poupel L, Combadiere B, Combadiere C. Control of both myeloid cell infiltration and angiogenesis by CCR1 promotes liver cancer metastasis development in mice. Neoplasia 2013; 15(6):641–648. doi: 10.1593/neo.121866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta M, Mimori K, Fukuyoshi Y, Kita Y, Motoyama K, Yamashita K, Ishii H, Inoue H, Mori M. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. Br J Cancer 2008; 98(2):410–417. doi: 10.1038/sj.bjc.6604124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata K, Mori M, Tanaka S, Kitano S, Akiyoshi T. Identification and cloning of human G-protein gamma 7, down-regulated in pancreatic cancer. Biochem Biophys Res Commun 1998; 246(1):205–209. doi: 10.1006/bbrc.1998.8581 [DOI] [PubMed] [Google Scholar]
- 33.Movilla N, Bustelo XR. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol 1999; 19(11):7870–7885. doi: 10.1128/MCB.19.11.7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F, Paramio JM, Bustelo XR. The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal 2012; 5(244):ra71. doi: 10.1126/scisignal.2002962 [DOI] [PubMed] [Google Scholar]
- 35.Menacho-Marquez M, Garcia-Escudero R, Ojeda V, Abad A, Delgado P, Costa C, Ruiz S, Alarcon B, Paramio JM, Bustelo XR. The Rho exchange factors Vav2 and Vav3 favor skin tumor initiation and promotion by engaging extracellular signaling loops. PLoS Biol 2013; 11(7):e1001615. doi: 10.1371/journal.pbio.1001615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marques RB, Dits NF, Erkens-Schulze S, van Weerden WM, Jenster G. Bypass mechanisms of the androgen receptor pathway in therapy-resistant prostate cancer cell models. PLoS One 2010; 5(10):e13500. doi: 10.1371/journal.pone.0013500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu KC, Huang AC, Wu PP, Lin HY, Chueh FS, Yang JS, Lu CC, Chiang JH, Meng M, Chung JG. Gallic acid suppresses the migration and invasion of PC-3 human prostate cancer cells via inhibition of matrix metalloproteinase-2 and −9 signaling pathways. Oncol Rep 2011; 26(1):177–184. doi: 10.3892/or.2011.1264 [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28(5):511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet 1999; 22(3):281–285. doi: 10.1038/10343 [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Yu G, Zhao Y, Zhao D, Wang Y, Wang L, Liu J, Li L, Zeng Y, Dang Y, et al. REGgamma modulates p53 activity by regulating its cellular localization. J Cell Sci 2010; 123(Pt 23):4076–4084. doi: 10.1242/jcs.067405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


