ABSTRACT
The response of cold shock proteins to exercise and environmental temperature in human skeletal muscle is not known. The purpose of this study was to determine the early mRNA response of human stress proteins to endurance exercise and environmental temperatures. Seven recreationally trained males cycled for 1 hour at 60% VO2peak in 7°C, 20°C, and 33°C with biopsies taken pre- and 3 hours post-exercise. Gene expression for heat shock and cold shock proteins were analyzed using qRT-PCR on muscle biopsy samples from the vastus lateralis. RBM3 mRNA was reduced 1.43 ± 0.10 fold (p = 0.006) while there was a trend for CIRP to decrease1.27 ± 0.14 fold (p = 0.059) from pre- to 3 h post-exercise. CIRP and RBM3 mRNA were not different between temperatures (p = 0.273 and p = 0.686, respectively). HSP70 mRNA was 2.27 ± 0.23 fold higher 3 h post-exercise when compared to pre-exercise (p = 0.002) but was not significantly different between temperatures (p = 0.103). HSP27, HSP90, and HSF1 mRNA did not change from pre- to post-exercise (p = 0.052, p = 0.324, p = 0.795) and were not different between temperatures (p = 0.247, p = 0.134, p = 0.808). These data indicate that exposure to mild heat and cold during aerobic exercise have limited effect on the skeletal muscle mRNA expression of heat shock and cold shock proteins. However, skeletal muscle mRNA of cold shock proteins decrease, while HSP70 mRNA increases in response to a low to moderate intensity aerobic exercise bout.
KEYWORDS: Human, gene expression, physical activity, endurance, CIRP, RBM3
Introduction
Stress proteins work to maintain homeostasis in response to a variety of external and internal stresses. They act as chaperones assisting with proper protein folding, assembly, and transport [1–3]. Heat shock proteins have been widely studied in animals and humans and are found in most tissues including skeletal muscle [4]. However, the two known cold shock proteins, cold-inducible RNA binding protein (CIRP) and RNA binding motif protein 3 (RBM3), have received relatively little attention in the literature on their response to stress in humans. Furthermore, there is little background information on the exact function of these genes. Therefore, the significance of this novel proof-of-concept pilot study is to document the mRNA response of cold shock proteins to exercise in different environmental temperatures in humans.
CIRP mRNA expression increases in mouse and human cells when placed directly in cold temperatures ranging from 25°C through 32°C as compared to 37°C [5–7]. These are common environmental temperatures, but they are significantly lower than the survivable limits of core body temperature in humans. Several studies measured the response of cold shock proteins in skeletal muscle tissue to hypothermia and atrophy. CIRP and RBM3 mRNA increase in the soleus muscle following hind-limb suspension in young and old rats with no impact of age [8]. This response indicates that there could be a defensive effect of the cold shock proteins in order to protect skeletal muscle during disuse. RBM3 protein abundance in C2C12 myoblasts of mice increase following 6 hours at 32°C [9]. Additionally, RBM3 mRNA expression increases in muscle of male hibernating black bears that were contained in environmental temperatures ranging from −10°C to −35°C when compared to non-hibernating bears [10]. Bears have a very different physiology that helps protect their muscle mass when hibernating, and cold shock proteins may play a key role. The role of cold shock proteins in humans is not well understood but appears to play a role in maintenance of muscle mass during disuse and cold exposure. More descriptive and mechanistic research in physiologically relevant environmental conditions and the impact of exercise is needed.
The response of heat shock proteins 70, 27, and 90 (HSP70, HSP27, HSP90) in skeletal muscle tissue to exercise or temperature in humans has been studied extensively. Heat shock proteins increase when exposed to temperatures up to 44°C [11,12], whereas exposure to cold shock produces a decrease in heat shock protein content [13]. There are different responses when comparing trained and untrained individuals, as well as when comparing the intensity and type of aerobic exercise performed by trained individuals. Exercise that causes damage to skeletal muscle tissue, specifically eccentric contractions, triggers an increase in HSP27 and HSP70 mRNA [14–17]. Similarly, HSP70 mRNA increases in untrained individuals after moderate aerobic exercise [18–20], but only after high intensity aerobic training in trained individuals [21]. The response of heat shock factor-1 (HSF1), the primary activator of heat shock proteins, similarly depends on the severity and type of stress the subject is exposed to [22]. A slight increase in HSF1 in response to exercise in humans may be detected even in the absence of an effect on the other heat shock proteins. It is not clear if cold shock proteins have a similar response to exercise and temperature as heat shock proteins or if these stress proteins respond to the stress in a temperature dependent manner as their names suggest.
While the transcriptional response of heat shock proteins has been well studied, the response of the cold shock proteins under physiologic conditions is not well known. Thus, the purpose of this study is to determine the early human skeletal muscle transcriptional response of heat shock and cold shock proteins to exercise stress in 20°C, 7°C, and 33°C environmental temperatures.
Methods
Design
All protocols were approved by the Institutional Review Board. A total of seven males aged 21–29 who engaged in at least 30 minutes of moderate physical activity three days a week participated in this study. The subjects had no prior medical history and were free from medical or health conditions that would prevent them from completing the full research protocol. Participants arrived at the laboratory after an overnight fast and refrained from unaccustomed exercise 48 hours prior to coming into the laboratory. Participants recorded their exercise activities 48 hours and their dietary intake 24 hours prior to the initial trial so they could repeat these activities for each trial. The 3 experimental trials consisted of cycling at 60% of the participant’s VO2peak for 1 hour in an environmentally controlled chamber (Darwin Chambers Company, St. Louis, MO) set to 20°C, 7°C, or 33°C environment with 60% humidity. The participants were required to consume 500 mL of room temperature water during each exercise trial. Muscle biopsies were taken pre- and 3 hours post-exercise during each experimental trial for the analysis of mRNA of selected heat and cold shock proteins. While little is known of the time-course of activation of transcription of cold shock protein mRNA, the 3 hour timepoint was chosen based on the response of many early response genes associated with exercise and temperature under similar conditions [23].
The initial visit consisted of measuring the subject’s height, weight, and aerobic capacity (VO2peak). To measure their VO2peak, the subjects performed a graded exercise test on an electronically braked Velotron Cycle ergometer (RacerMate Inc., Seattle, WA). The first stage began at 95 watts (W) and increased by 35 W every 3 minutes until volitional fatigue. The maximum power output was calculated by taking the highest completed stage in watts, adding the proportion of time spent in the last stage multiplied by the 35 watts per stage increment. A flow and gas calibrated metabolic cart (Parvomedics Inc., Salt Lake City, Utah) was used to continuously measure expired gases.
Core temperature, skin temperature, and heart rate
Core temperature was measured during exercise using an ingestible capsule thermistor which transmitted to an EQo2 LifeMonitor sensor module (Hildalgo, Cambridge, UK). The thermistor capsule was ingested 1 hour before exercise with 125 ml of water and a fiber bar (Fiber One, General Mills, Minneapolis, MN) to facilitate movement of the capsule out of the stomach. The LifeMonitor module also measured heart rate and skin temperature using an infared themister. The data were obtained every 15 s. and averaged for the entire 60 minute exercise bout during each of the trials.
Muscle biopsies
Muscle biopsies were obtained from the vastus lateralis muscle using a 5 mm Bergstrom percutaneous muscle biopsy needle with the aid of suction [24]. After excess blood and connective tissue was removed, the muscle samples were immersed in RNAlater solution (Qiagen, Valencia, CA). Samples were stored at 4°C overnight then at −80°C for later analysis. Subsequent biopsies during a given trial were performed 2 cm proximal to the previous biopsy on the same leg. The order of the leg biopsied was randomized and alternated for each following trial.
mRNA Analysis
Muscle samples were analyzed using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) to quantify the expression of mRNA. Approximately 15–25 mg of skeletal muscle was homogenized in 500 µl of Trizol reagent (Invitrogen, Carlsbad, CA) in 2 ml Red Rino tubes using the Bullet Blender Storm 24 at speed setting of 4 for 2 minutes (Next Advance, Troy, NY). Samples were then incubated at room temperature for 5 minutes before 100 µl of chloroform per 500 µl of Trizol was added and then shaken by hand. Samples were incubated for 3 minutes at room temperature, centrifuged at 12,000 g for 15 minutes at 4°C, then the aqueous solution was transferred to a fresh tube with 1 µl of glycogen to aid in the visualization of the pellet. Then, 250 µl of isopropyl alcohol was added and incubated overnight at −20°C. The next morning samples were centrifuged for 12,000 g for 10 minutes at 4°C and the RNA was washed by removing the supernatant and adding 800 µl of 75% ethanol. Samples were vortexed then centrifuged at 7,500 g for 5 minutes at 4°C. The supernatant was removed and the pellet was dried for approximately 5 minutes. Next, the mRNA was dissolved in 30 µl RNase-free water. RNA was quantified using a nano-spectrophotometer (nano-drop ND-1000, Wilmington, DE). First-strand cDNA synthesis was achieved using a Superscript-first-strand system kit for RT-PCR (Invitrogen, Carlsbad, CA). Each sample within a given subject was adjusted to contain the same RNA concentration (189.51 ng) using RNase free water. Each 20 µL RT-PCR reaction volume contained 1µl of primers (2.5 nmol/L), 10 µL of SYBR Green Supermix (Bio-Rad, CA), 2.5 µL of sample cDNA, and 6.5 µl of nuclease free water. Samples were then analyzed using the Agilent Technologies Stratagene Mx3005p real time PCR detection system (Agilent Technologies Inc, Santa Clara, CA) with a fast 2-step protocol (1 cycle at 98°C for 30 seconds followed by 60 cycles at 98°C for 15 seconds then 1 cycle at 60°C for 30 seconds).
Quantification of mRNA for genes of interest were performed for pre-exercise and 3 hours post-exercise muscle samples using the 2−ΔΔCT [25]. The most stable reference genes were analyzed using NormFinder software (MOMA, Denmark) [26] and the geometric mean of the combination of the stable reference genes were used. The reference genes used were glyceraldehyde 3-phosphate dehydrogenase (GAPDH), beta2-microglobulin (B2M), and beta-actin (ACTB). The genes of interest were CIRP, RBM3, HSP70, HSP90, HSP27, and HSF1. The primers for these genes were designed and obtained from Integrated DNA Technologies (Coralville, IA). See Table 1 for primer sequences. The genes of interest were measured and normalized to the reference genes at the pre-exercise condition and following 3 hours of recovery. Thus, when using this method all pre-values are normalized to 1.0 fold change (mathematically represents no change) and the 3 h post values indicate the relative fold change from pre. For graphical purposes the redundant pre-values for each trial are not reported and instead the x-intercept is adjusted to ±1.0 fold change to indicate no change from pre.
Table 1.
Primer sequences used for RT-qPCR.
| Gene | Primer 1 | Primer 2 |
|---|---|---|
| CIRP | GAC CTG CCC GAC TCA GT | CAC AAC CAC CAC TTC AGA GAT |
| RBM3 | CAG TTG CCA TGA GAG CCA T | CAC CTC TAG AGT AGC TGC GA |
| HSP70 | GTC CAC TAC CTT TTT CGA GAG T | CTG GAA ACG GAA CAC TGG AT |
| HSP90 | GTC TCT GCA TTC CCT GTC AC | GGT CTT GGG TCT GGG TTT C |
| HSP27 | TCA AAC GGG TCA TTG CCA T | CTG GCT GAC TCT GCT CT |
| HSF1 | GAA GCA GGA GTG CAT GGA C | AGA TCA GGA ACT GAA TGA GCT TG |
| GAPDH | ACA TCG CTC AGA CAC CAT G | TGT AGT TGA GGT CAA TGA AGG G |
| B2M | GGA CTG GTC TTT CTA TCT CTT GT | ACC TCC ATG ATG CTG CTT AC |
| ACTB | GTC CCC CAA CTT GAG ATG TAT G | AAG TCA GTG TAC AGG TAA GCC |
Statistical analysis
Differences in gene expression were analyzed with a repeated measures two-way ANOVA (time x trial). When significance was indicated with the initial ANOVA, a Fisher’s protected least significant difference post-hoc test was used to evaluate where significant differences occurred. The average heart rate, core temperature, and skin temperature data were analyzed with a repeated measures one-way ANOVA. A probability of type I error of less than 5% was considered significant (p < 0.05) for all analysis. All data was analyzed using the Statistical Package for Social Sciences software (SPSS 23.0, Chicago, IL). All data are reported as mean ± SE.
Results
Subjects
Descriptive data of the seven recreationally active males who participated in this study are shown in Table 2. The physiological parameters during exercise in 20°C, 7°C, and 33°C trials are shown in Table 3. Heart rate was higher in the 33°C than in the 20°C and 7°C trials (p < 0.001). VO2 was higher in 33°C than 20°C and 7°C trials (p = 0.002 and p = 0.001, respectively). Core temperature was not different between trials (p = 0.190), while skin temperature was higher in the 33°C trial than the 20°C trial (p = 0.033) and the 7°C trial (p = 0.011). Furthermore, skin temperature was higher in the 33°C trial than the 7°C trial (p = 0.009). The physiological parameters during exercise in 20°C, 7°C, and 33°C trials are shown in Table 3.
Table 2.
Descriptive data (n = 7).
| Age (y) | 24 ± 1.2 |
|---|---|
| Height (cm) | 178 ± 1.7 |
| Weight (kg) | 76.8 ± 1.9 |
| VO2peak (L · min−1) | 4.5 ± 0.2 |
| Power at VO2peak (W) | 290.4 ± 7.8 |
Data are mean ± SE
Table 3.
Physiological parameters during exercise in 20°C, 7°C, and 33°C environments.
| 20°C | 7°C | 33°C | |
|---|---|---|---|
| Heart Rate (bpm) | 155 ± 3 | 153 ± 3 | 171 ± 3 *† |
| VO2 (L · min−1) | 2.9 ± 0.1 | 2.7 ± 0.1 | 3.1 ± 0.1 |
| Power (W) | 174 ± 5 | 174 ± 5 | 174 ± 5 |
| Core Temperature (°C) | 37.8 ± 0.2 | 37.9 ± 0.2 | 38.1 ± 0.2 |
| Skin Temperature (°C) | 36.7 ± 0.7 | 35.3 ± 10.8* | 37.9 ± 0.2*† |
Data are mean ± SE; * p < 0.05 from 20°C, † p < 0.05 from 7°C
Gene expression
There was a trend that did not reach statistical significance for CIRP mRNA to be 1.27 ± 0.14 fold lower at 3 hours post-exercise compared to pre-exercise (p = 0.059, main effect of exercise). RBM3 mRNA was 1.43 ± 0.10 fold lower post-exercise compared to pre-exercise (p = 0.006, main effect of exercise). There were no differences between 7°C, 20°C, and 33°C conditions for CIRP mRNA (p = 0.273, main effect of temperature) and RBM3 mRNA (p = 0.686, main effect of temperature) (Figure 1).
Figure 1.
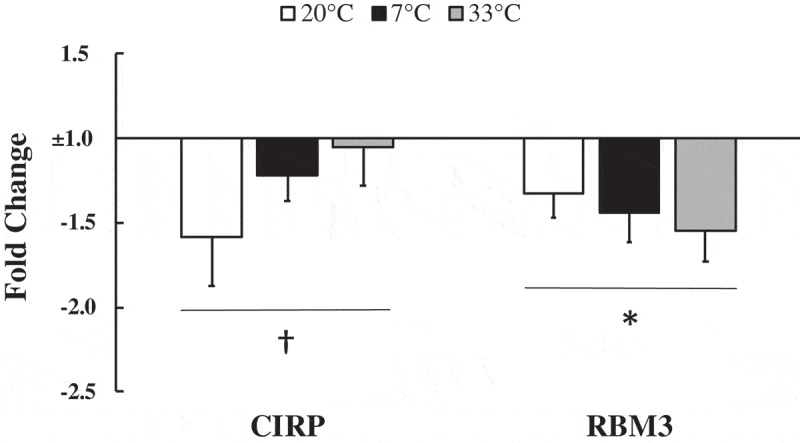
Cold shock protein response to exercise.
Cold-inducible RNA binding protein (CIRP) and RNA binding motif protein-3 (RBM3) mRNA at 3 hours post-exercise relative to pre-exercise values normalized to 1 fold (no change) in 20°C, 7°C, and 33°C environmental conditions. Data are expressed as mean ± SE. * p < 0.05, † p = 0.059.
HSP70 mRNA increased 2.27 ± 0.23 fold at 3 hours after exercise compared to pre-exercise (p = 0.002), and approached a trend but did not reach statistical significance for 33°C to be higher than 20°C and 7°C when comparing temperatures (ANOVA main effect; p = 0.103). HSP27 mRNA demonstrated a non-statistically significant 1.27 ± 0.13 fold decrease at 3 hours post-exercise compared to pre-exercise (p = 0.052) with no differences between the different temperature trials (ANOVA main effect; p = 0.247). HSP90 mRNA and HSF1 mRNA were not different at 3 hours post-exercise relative to pre-exercise (p = 0.324 and p = 0.795, respectively) and were not different between temperatures (ANOVA main effect; p = 0.134 and p = 0.808, respectively) (Figure 2).
Figure 2.
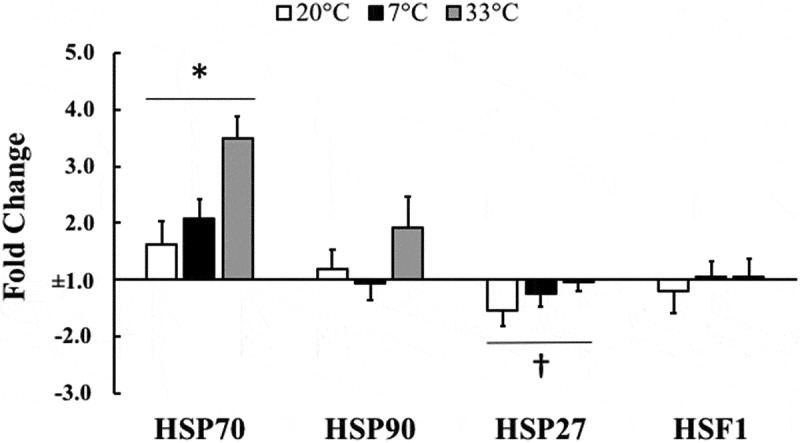
Heat shock protein response to exercise.
Heat shock protein 70 (HSP70), heat shock protein 90 (HSP90), heat shock protein 27 (HSP27), and heat shock factor-1 (HSF1) mRNA at 3 hours post-exercise relative to pre-exercise values normalized to 1 fold (no change) in 20°C, 7°C, and 33°C environmental conditions. Data are expressed as mean ± SE. * p < 0.05, † p = 0.052.
Discussion
Not only is this the first study to measure the response of cold shock proteins mRNA in human skeletal muscle tissue, it is the first study describing the mRNA response of cold shock proteins to exercise and temperature in a human physiologically relevant environment. The novel findings of this study were that there was a trend for a decrease in CIRP mRNA and a significant decrease in RBM3 mRNA in response to exercise. Additionally, the temperature in which exercise took place did not alter the response of either CIRP or RBM3. The relatively small alterations in the mRNA of heat shock proteins in this study was not surprising based on previous literature and our current protocol. None-the-less, the heat shock mRNA data allows an anchor point in which to put the cold shock mRNA data in context. This initial proof-of-concept pilot study indicates that the mRNA for cold shock proteins appear to decline in response to aerobic exercise performed in physiologically relevant conditions.
CIRP and RBM3 predominately respond to hypothermia in mammalian cells. Specifically, RBM3 increases in response to hypothermia which leads to a decrease in apoptosis [27] which could prevent a decrease in skeletal muscle mass [9]. The typical range of temperatures tested in vitro are 32°C to 37°C, though few in vivo mammalian cells other than the testes and skin reach temperatures as low as 32°C without other intervention [28]. In the current study, there was a decrease in RBM3 mRNA following 3 hours of recovery post-exercise when compared to pre-exercise, but there was no difference when comparing the response at 7°C and 33°C to 20°C. CIRP also appeared to follow this same pattern although it did not reach statistical significance for a decrease with exercise. The human body responds to hypothermia in several ways to maintain core body temperature, one of which is through skeletal muscle contractions in the form of shivering. Humans would not survive with a core body temperature as low as 32°C and thus shivering would increase temperature thereby minimizing the response from cold shock proteins at rest. In this same manner, exercise increases heat production and may limit any cold related response. Indeed, our data indicate an increase in core temperature during exercise with only very minimal differences between environments, which is supported by previous literature [23,29]. This core temperature response could explain the limited effect of environmental temperatures observed in the current study on CIRP and RBM3 mRNA. Specifically, core temperature is increased even in the cold and thus no cold stress is imposed on the muscle during exercise. It was beyond the scope of the current study to include no exercise control trials for each environment. However, in absence of the exercise and presence of a temperature alteration, these genes may respond differently. Additionally, there may be a differential response if exercise or recovery occurred in more extreme environments that could potentially lower core body temperature warranting a cold shock protein response. While the different temperatures did not alter core temperature, skin temperature was different between each of the trials. It is possible that thermos-reception from the skin may ultimately lead to a differential mRNA response and that skeletal muscle mRNA may be altered. Indeed, we have previously observed an altered mRNA response to exercise in different environmental temperatures when core temperature was unaltered [23]. However, it appears that exercise may prevent the need for a protective response by CIRP and RBM3 mRNA in physiologically relevant environmental conditions whether due to heat produced by working skeletal muscle or other exercise induced mechanisms.
Our findings for the response of the heat shock related proteins mRNA was not surprising given the nature of the subjects and protocol that was utilized. We did not observe a change in HSF1, HSP90, or HSP27 mRNA in the current study in response to exercise or temperature. However, HSP70 mRNA did increase with exercise regardless of trial. Furthermore, both HSP70 and HSP90 mRNA approached a statistical trend (p = 0.105 and p = 0.134, respectively) to be further elevated in the hot environmental condition. These data provide insight into the differential response of cold shock proteins than heat shock proteins with regard to the exercise response in a human model. The response of heat shock protein mRNA to exercise appears to be related to the fitness of the participant as well as the intensity and mode of the exercise bout [14–16,19,20]. The current study incorporated aerobically fit subjects (mean VO2peak = 58.9 ml • kg−1 • min−1) working at a moderate intensity of 60% VO2peak. A greater increase in heat shock protein response is expected when untrained subjects perform moderate or vigorous aerobic exercise [14–16] and even more so when the exercise consists of muscle damaging eccentric muscular contractions [19,20]. Indeed there was an increase in HSP70 mRNA in response to exercise in the current study which supports previous results using a similar exercise protocol [30]. Additionally, our observation of no change in HSP27 mRNA is in agreement with other literature that typically reports no change following exercise [14–16,31]. Therefore, the moderate 2–4 fold increase in HSP70 mRNA and no change in HSF1, HSP90, and HSP27 mRNA in this study following exercise was expected considering the use of fit subjects and an exercise protocol that did not include high intensity aerobic or muscle damaging exercise. If less fit subjects engaged in higher intensity muscle damaging exercise, then we may have expected a greater increase in these heat shock protein mRNA’s. Although we did not observe a statistically significant response to temperature, it may be possible that HSP70 and HSP90 mRNA do increase to a greater extent in the heat compared to other temperature environments. Both HSP70 and HSP90 mRNA approached a statistical trend (p = 0.103 and p = 0.134, respectively) to have a greater increase in the heat and very well may have reached significance should the sample size have been larger. Indeed, heat shock proteins increase when exposed to heat [11,13,22,32,33] which supports the trend of a difference between environmental temperatures found in this study. The human body responds differently to decreased temperatures when compared to cell cultures, however this demonstrates a difference in the response of heat shock proteins to different environmental temperatures.
Conclusion
The novelty of this study is the measurement of the early response of cold shock protein mRNA in human skeletal muscle tissue to exercise and environmental temperature. The current data indicates that low to moderate intensity exercise, regardless of the environmental temperature, decreases the transcription of RBM3 mRNA and trends toward decreasing CIRP mRNA in skeletal muscle tissue at 3 hours after exercise. Additionally, given the nature of this protocol, the current data supports previous work on the heat shock protein response and clearly differentiates the response of heat shock and cold shock proteins to exercise. This initial proof-of-concept study provides an initial look at the cold shock protein mRNA response and clearly indicates the need for continued investigations which can further detail the mechanisms and outcomes of cold shock proteins.
Funding Statement
This work was supported by the National Institute for General Medical Science (NIGMS P20GM103427), Nebraska IDeA Networks of Biomedical Research Excellence (INBRE) and the University of Nebraska at Omaha University Committee on Research and Creative Activity graduate student grant.
Abbreviations
- CIRP
cold-inducible RNA-binding protein
- HSF1
heat shock factor-1
- HSP70
heat shock protein 70
- HSP90
heat shock protein 90
- HSP27
heat shock protein 27
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- RBM3
RNA-binding motif protein 3
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Fujita J. Cold shock response in mammalian cells. J Mol Microbiol Biotechnol. 1999;1(2):243–255. [PubMed] [Google Scholar]
- [2].Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92(5):2177–2186. [DOI] [PubMed] [Google Scholar]
- [3].Welch WJ. Mammalian stress response: cell physiology, structure/function of stress proteins, and implications for medicine and disease. Physiol Rev. 1992;72(4):1063–1081. [DOI] [PubMed] [Google Scholar]
- [4].Koh TJ. Do small heat shock proteins protect skeletal muscle from injury? Exerc Sport Sci Rev. 2002;30(3):117–121. [DOI] [PubMed] [Google Scholar]
- [5].Danno S, Nishiyama H, Higashitsuji H, et al. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236(3):804–807. [DOI] [PubMed] [Google Scholar]
- [6].Nishiyama H, Itoh K, Kaneko Y, et al. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137(4):899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Saito K, Fukuda N, Matsumoto T, et al. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010;1358:20–29. [DOI] [PubMed] [Google Scholar]
- [8].Dupont-Versteegden EE, Nagarajan R, Beggs ML, et al. Identification of cold-shock protein RBM3 as a possible regulator of skeletal muscle size through expression profiling. Am J Physiol Regul Integr Comp Physiol. 2008;295(4):1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ferry AL, Vanderklish PW, Dupont-Versteegden EE. Enhanced survival of skeletal muscle myoblasts in response to overexpression of cold shock protein RBM3. Am J Physiol Cell Physiol. 2011;301(2):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fedorov VB, Goropashnaya AV, Toien O, et al. Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus). Physiol Genomics. 2009;37(2):108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Landry J, Chrétien P, Lambert H, et al. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J Cell Biol. 1989;109(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Welch WJ, Suhan JP. Cellular and biochemical events in mammalian cells during and after recovery from physiological stress. J Cell Biol. 1986;103(5):2035–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martínez-Paz P, Morales M, Martín R, et al. Characterization of the small heat shock protein Hsp27 gene in Chironomus riparius (Diptera) and its expression profile in response to temperature changes and xenobiotic exposures. Cell Stress Chaperones. 2014;19(4):529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mikkelsen UR, Paulsen G, Schjerling P, et al. The heat shock protein response following eccentric exercise in human skeletal muscle is unaffected by local NSAID infusion. Eur J Appl Physiol. 2013;113(7):1883–1893. [DOI] [PubMed] [Google Scholar]
- [15].Paulsen G, Vissing K, Kalhovde JM, et al. Maximal eccentric exercise induces a rapid accumulation of small heat shock proteins on myofibrils and a delayed HSP70 response in humans. Am J Physiol Regul Integr Comp Physiol. 2007;293(2):844. [DOI] [PubMed] [Google Scholar]
- [16].Thompson HS, Scordilis SP, Clarkson PM, et al. A single bout of eccentric exercise increases HSP27 and HSC/HSP70 in human skeletal muscle. Acta Physiol Scand. 2001;171(2):187–193. [DOI] [PubMed] [Google Scholar]
- [17].Thompson HS, Maynard EB, Morales ER, et al. Exercise‐induced HSP27, HSP70 and MAPK responses in human skeletal muscle. Acta Physiol Scand. 2003;178(1):61–72. [DOI] [PubMed] [Google Scholar]
- [18].Febbraio MA, Koukoulas I. HSP72 gene expression progressively increases in human skeletal muscle during prolonged, exhaustive exercise. J Appl Physiol . 2000;89(3):1055–1060. [DOI] [PubMed] [Google Scholar]
- [19].Khassaf M, Child RB, McArdle A, et al. Time course of responses of human skeletal muscle to oxidative stress induced by nondamaging exercise. J Appl Physiol . 2001;90(3):1031–1035. [DOI] [PubMed] [Google Scholar]
- [20].Morton JP, MacLaren DP, Cable NT, et al. Time course and differential responses of the major heat shock protein families in human skeletal muscle following acute nondamaging treadmill exercise. J Appl Physiol . 2006;101(1):176–182. [DOI] [PubMed] [Google Scholar]
- [21].Vogt M, Puntschart A, Geiser J, et al. Molecular adaptations in human skeletal muscle to endurance training under simulated hypoxic conditions. J Appl Physiol. 2001;91(1):173–182. [DOI] [PubMed] [Google Scholar]
- [22].Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13(3):1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shute RJ, Heesch MW, Zak RB, et al. Effects of exercise in a cold environment on transcriptional control of PGC-1α. Am J Physiol Regul Integr Comp Physiol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bergstrom J. Muscle electrolytes in man determined by neutron activation analysis on needle biopsy specimens. Scand J Clin Lab Invest (England). 1962;14(Suppl):68. [Google Scholar]
- [25].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [26].Exposito-Rodriguez M, Borges AA, Borges-Perez A, et al. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chip S, Zelmer A, Ogunshola OO, et al. The RNA-binding protein RBM3 is involved in hypothermia induced neuroprotection. Neurobiol Dis. 2011;43(2):388–396. [DOI] [PubMed] [Google Scholar]
- [28].Lleonart ME. A new generation of proto-oncogenes: cold-inducible RNA binding proteins. Biochim Biophys Acta (BBA)- Rev Cancer. 2010;1805(1):43–52. [DOI] [PubMed] [Google Scholar]
- [29].Bubak MP, Heesch MW, Shute RJ, et al. Irisin and fibronectin type III domain-containing 5 responses to exercise in different environmental conditions. Int J Exercise Sci. 2017;10(5):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Watkins AM, Cheek DJ, Harvey AE, et al. Heat shock protein (HSP-72) levels in skeletal muscle following work in heat. Aviat Space Environ Med. 2007;78(9):901–905. [PubMed] [Google Scholar]
- [31].Gjøvaag TF, Dahl HA. Effect of training and detraining on the expression of heat shock proteins in m. triceps brachii of untrained males and females. Eur J Appl Physiol. 2006;98(3):310–322. [DOI] [PubMed] [Google Scholar]
- [32].Gibson OR, Dennis A, Parfitt T, et al. Extracellular Hsp72 concentration relates to a minimum endogenous criteria during acute exercise-heat exposure. Cell Stress Chaperones. 2014;19(3):389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McClung JP, Hasday JD, He JR, et al. Exercise-heat acclimation in humans alters baseline levels and ex vivo heat inducibility of HSP72 and HSP90 in peripheral blood mononuclear cells. Am J Physiol Regul Integr Comp Physiol. 2008;294(1):185. [DOI] [PubMed] [Google Scholar]


