Abstract
Objective:
We employed a high-dimensional covariate adjustment method in microbiome analysis to better control for behavioral and clinical confounders, and in doing so examine the effects of HIV on the rectal microbiome.
Design:
Three hundred eighty-three men who have sex with men were grouped into four HIV viremia categories: HIV negative (n = 200), HIV+ undetectable (HIV RNA <20 copies/mL; n = 66), HIV+ suppressed (RNA 20–200 copies/mL; n = 72) and HIV+ viremic (RNA >200 copies/mL; n = 45).
Methods:
We performed 16S rRNA gene sequencing on rectal swab samples and used inverse probability of treatment-weighted marginal structural models to examine differences in microbial composition by HIV viremia category.
Results:
HIV viremia explained a significant amount of variability in microbial composition in both unadjusted and covariate-adjusted analyses (R2 = .011, p = .02). Alterations in bacterial taxa were more apparent with increasing viremia. Relative to the HIV negative group, HIV+ undetectable participants showed depletions in Brachyspira, Campylobacter, and Parasutterella while suppressed participants demonstrated depletions in Barnesiella, Brachyspira and Helicobacter. The microbial signature of viremic men was most distinct, showing enrichment in inflammatory genera Peptoniphilus, Porphyromonas, and Prevotella and depletion of Bacteroides, Brachyspira, and Faecalibacterium, among others.
Conclusions:
Our study shows that, after accounting for the influence of multiple confounding factors, HIV is associated with dysbiosis in the gastrointestinal microbiome in a dose-dependent manner. This analytic approach may allow for better identification of true microbial associations by limiting the effects of confounding, and thus improve comparability across future studies.
Keywords: HIV, Microbiome, Dysbiosis, Men who have sex with men, IPTW, Propensity scores, Causal inference
Introduction
The trillions of bacteria, viruses, and fungi inhabiting the human gastrointestinal (GI) tract have a profound impact on our health and the development of disease. Disruption in the homeostasis of the these microbes, a state of “dysbiosis,” has been associated with a broad range of illnesses, including localized GI conditions, neurocognitive disorders, cancer, autoimmune disorders, and cardiovascular disease [1]. There is tremendous variability in the diversity and composition of the microbiome, even between healthy individuals [2], and the effects of different exposures, behaviors, and personal characteristics on the composition and function of the microbiome are incompletely understood.
Chronic inflammation is a hallmark of HIV infection and continues despite suppressive antiretroviral therapy (ART). The GI tract is a primary site for HIV replication resulting in significant loss of CD4+ T-cells vital to a healthy mucosal immune system. Depletion of regulatory immune cells and pathways leads to decreased epithelial barrier function allowing translocation of microbes and microbial products which contributes to the chronic inflammatory response [3, 4]. HIV replication may also result in a state of dysbiosis [5–7], which has been correlated with increases in markers of disease progression, microbial translocation, and immune activation [5, 7–9]. It has been hypothesized that HIV-associated immune dysfunction induces this dysbiosis, and dysbiosis causes further dysfunction [5], thereby driving persistent systemic inflammation in HIV-infected individuals [10].
Many studies have found that overall microbial diversity is reduced in HIV-infected individuals [11–13], and HIV has been associated with a shift from commensals such as Bacteroides to pro-inflammatory taxa such as Prevotella [11, 14, 15]. However, results have been inconsistent, with some studies showing the opposite or no effects of HIV on microbial diversity [16–18] and little effect on composition [19, 20]. Certainly, differences in sample collection, sequencing, post-processing, and analytic methodology may be responsible for much of the heterogeneity in results [21]. While some research has focused on the role of sampling variability and type-I error on irreproducibility of findings [22, 23], comparatively little attention has been paid to sources of systematic error such as incomparable study cohorts or confounders such as sexual behavior, substance use, diet, race/ethnicity, and age. Such confounders are highly prevalent in observational human studies of the microbiome and may have a larger effect than HIV itself [19]. Due to these limitations, the specific effects of HIV on the microbiome warrant further investigation.
Increased relative abundance of pro-inflammatory bacteria has been correlated with increased viremia, whereas the opposite has been found for potentially beneficial bacteria [9, 13, 14]. Given these findings and the effects of viral replication on mucosal immunity, it stands to reason that the level of viremia may be a significant determinant of HIV-associated dysbiosis. However, the effect of viremia has not been thoroughly explored. Numerous studies comparing cohorts of HIV-infected individuals that are either on ART or ART-naïve have shown that ART does not result in full “reconstitution” of the microbiome, even if the virus is suppressed [13, 24, 25]. Additional studies focused on elite controllers showed that the microbial composition among individuals with controlled viremia is more similar to HIV-uninfected than viremic individuals [26, 27]. However, there are likely to be important biological differences between elite controllers and other HIV-infected individuals that may limit the generalizability of these findings. In order to accurately characterize the effects of HIV on the microbiome, a more detailed examination of the effects of viremia is needed.
To this end, we compared intestinal microbial composition between HIV-uninfected, HIV-infected with undetectable viremia (HIV RNA <20 copies/ml), HIV-infected with suppressed viremia (HIV RNA ≥20–200 copies/ml), and HIV-infected viremic individuals (HIV RNA >200 copies/ml). We utilized data from a cohort comprised entirely of men who have sex with men (MSM) and employed inverse probability of treatment weighting (IPTW) to control for a robust set of clinical and behavioral confounders. We hypothesized that alterations to the microbiome would be present in all HIV-infected subgroups as compared to HIV-uninfected controls, and the severity of dysbiosis would increase with increasing viremia.
Methods
Study Population
Specimens for this study were obtained from an ongoing prospective cohort (The mSTUDY, NIDA U01 DA036267). The mSTUDY was approved by a UCLA Institutional Review Board (IRB) and all subjects provided written informed consent at study entry. Participants are recruited from community clinics in Los Angeles and complete biannual assessments including a comprehensive physical examination and medical history, urine toxicology panel, clinical laboratory tests including plasma HIV RNA, specimen collection, and detailed behavioral questionnaire. Data presented in this manuscript were collected from baseline study visits completed between August 2014 and July 2017. Additional details on sample selection and HIV RNA quantification are provided in the supplemental content.
Specimen collection and DNA preparation
The majority (76%) of rectal swabs (FLOQSwabs, Copan Diagnostics, Murrieta, CA) were collected via anoscopy under direct mucosal visualization and without preparatory enema at approximately 8 cm from the anal verge. Due to an mSTUDY protocol change, others (24%) were participant self-collected at approximately 4–5 cm from the anal verge. Collection method was taken into account in the analysis (see Tables 1 and S1). Swabs were immediately frozen neat at −80°C until processing in bulk. For DNA processing the samples were transferred to Lysing Matrix E tubes (MP Biomedicals, Burlingame, CA) containing RLT lysis buffer (Qiagen, Hilden, Germany) and bead-beated on a TissueLyser (Qiagen). DNA was then extracted using the AllPrep DNA/RNA/Protein kit (Qiagen) per manufacturer’s protocol.
Table 1.
Participant characteristics, N = 383 men who have sex with men in Los Angeles, CA
| HIV-Neg ative mean (sd)/ n (%) |
Undetectable (HIV RNA < 20 copies/mL) |
Suppressed (HIV RNA ≥20 - 200 copies/mL) |
Viremic (HIV RNA > 2 00 copies/mL) |
SMDa
(unwei ghted) |
SMD (weig hted) |
|
|---|---|---|---|---|---|---|
| n | 200 | 66 | 72 | 45 | ||
| Age | 28.91 (6.43) | 33.41 (6.55) | 34.10 (6.36) | 33.18 (6.86) | .41 | .17 |
| Employment | .54 | .25 | ||||
| Student | 26 (13.0) | 5 (7.6) | 3 (4.2) | 1 (2.2) | ||
| Unemployed | 61 (30.5) | 23 (34.8) | 50 (69.4) | 27 (60.0) | ||
| Full/part time | 113 (56.5) | 38 (57.6) | 19 (26.4) | 17 (37.8) | ||
| Race/Ethnicity | .48 | .37 | ||||
| Black Non-Hispanic | 82 (41.0) | 17 (25.8) | 25 (34.7) | 26 (57.8) | ||
| Hispanic | 98 (49.0) | 37 (56.1) | 35 (48.6) | 19 (42.2) | ||
| Other Non-Hispanic | 20 (10.0) | 12 (18.2) | 12 (16.7) | 0 | ||
| Country of origin | .12 | .08 | ||||
| United States | 171 (85.5) | 53 (80.3) | 55 (76.4) | 36 (80.0) | ||
| Other | 29 (14.5) | 13 (19.7) | 17 (23.6) | 9 (20.0) | ||
| Homeless in past 6 months | 65 (32.5) | 20 (30.3) | 21 (29.2) | 22 (48.9) | .21 | .18 |
| RAI in past 7 days | 88 (44.0) | 32 (48.5) | 30 (41.7) | 19 (42.2) | .08 | .07 |
| Number of RAI acts in past month | 2.06 (4.19) | 2.48 (4.68) | 2.33 (5.45) | 4.42 (8.03) | .19 | .07 |
| Methamphetamine use in past 6 months | .54 | .20 | ||||
| Daily/Weekly | 21 (10.5) | 14 (21.2) | 18 (25.0) | 21 (46.7) | ||
| Monthly/less | 32 (16.0) | 21 (31.8) | 19 (26.4) | 9 (20.0) | ||
| Never | 147 (73.5) | 31 (47.0) | 35 (48.6) | 15 (33.3) | ||
| Marijuana use | .30 | .14 | ||||
| Daily/Weekly | 71 (35.5) | 19 (28.8) | 21 (29.2) | 19 (42.2) | ||
| Monthly/less | 57 (28.5) | 10 (15.2) | 16 (22.2) | 10 (22.2) | ||
| Never | 72 (36.0) | 37 (56.1) | 35 (48.6) | 16 (35.6) | ||
| Cocaine use | .16 | .09 | ||||
| At least once | 53 (26.5) | 11 (16.7) | 21 (29.2) | 13 (28.9) | ||
| Never | 147 (73.5) | 55 (83.3) | 51 (70.8) | 32 (71.1) | ||
| Tobacco smoking | .26 | .15 | ||||
| >1 pack/day | 10 (5.0) | 3 (4.5) | 5 (6.9) | 2 (4.4) | ||
| <1 pack/day | 68 (4.0) | 24 (36.4) | 31 (43.1) | 25 (55.6) | ||
| Nonsmoker | 122 (61.0) | 39 (59.1) | 36 (50.0) | 18 (40.0) | ||
| Binge drinking in past 6 monthsb | .40 | .15 | ||||
| Weekly | 24 (12.0) | 15 (22.7) | 8 (11.1) | 8 (17.8) | ||
| Monthly/less | 111 (55.5) | 26 (39.4) | 20 (27.8) | 16 (35.6) | ||
| Never | 65 (32.5) | 25 (37.9) | 44 (61.1) | 21 (44.7) | ||
| Obesec | 64 (32.0) | 16 (24.2) | 13 (18.1) | 10 (22.2) | .17 | .15 |
| Antibiotic use | 11 (5.5) | 5 (7.6) | 7 (9.7) | 8 (17.8) | .21 | .08 |
| Sample collection method | .21 | .08 | ||||
| Anoscopy | 152 (77.0) | 47 (71.2) | 53 (73.6) | 39 (86.7) | ||
| Self-collected | 46 (23.0) | 19 (28.8) | 19 (26.4) | 6 (13.3) | ||
| Years since HIV diagnosis d | N/A | 7 (6) | 7 (5) | 8 (6) | N/A | N/A |
| HIV RNA copies/mL (median, IQR) | N/A | N/A | 20 (30) | 15,730 (48,680) | N/A | N/A |
| CD4 cells/mm3 | N/A | 708.95 (279.6) | 645.21 (262.9) | 470.02 (280.1) | N/A | N/A |
| ART regimen | N/A | N/A | ||||
| INSTI + NRTI | 0 | 30 (45.5) | 30 (41.7) | 8 (17.8) | ||
| NNRTI + NRTI | 0 | 21 (31.8) | 20 (27.8) | 7 (15.6) | ||
| NRTI + PI | 10 (15.2) | 11 (15.3) | 9 (20) | |||
| Other | 0 | 5 (7.6) | 8 (10.2) | 4 (8.8) | ||
| Missing/Not reported/NA | 166 (83) | 0 | 3 (4.2) | 17 (37.8) | ||
| PrEP usere | 37 (19) | N/A | N/A | N/A | N/A | N/A |
SMD = Standardized mean difference; RAI = Receptive anal intercourse; ART = Antiretroviral therapy; INSTI = Integrase strand transfer inhibitor; NRTI = Nucleoside reverse transcriptase inhibitor; NNRTI = Non-nucleoside reverse transcriptase inhibitor; PI = Protease inhibitor
SMD is a measure of imbalance across groups; higher SMDs indicate greater imbalance. Average SMD before weighting = .29, after weighting = .15.
Binge drinking defined as 6 or more drinks on one occasion.
Obese defined as BMI > 30 or BMI > 25 and waist circumference > 40 inches.
Years since HIV diagnosis, HIV RNA, CD4 cell count and ART regimen were not included in the inverse probability of treatment weight model (as they are generally not relevant to HIV negative participants), all other variables in the table were included.
HIV negative men taking tenofovir disoproxil fumarate/emtricitabine for pre-exposure prophylaxis (PrEP).
16S rRNA gene sequencing and data processing
Microbiome profiling was performed by sequencing of the V4 region of the 16S rRNA gene as previously described [28, 29]. Briefly, the V4 region was amplified in triplicate reactions using Golay-barcoded primers 515F/806R. Negative controls from the DNA extraction and PCR steps, as well as independent aliquots of a bacterial mock community [30] were processed alongside the samples to identify contaminant sequences and ensure data reproducibility. PCR products were then pooled and sequenced on the Illumina MiSeq platform using 2×150bp v2 chemistry. The sequences were demultiplexed with Golay error correction using QIIME v1.9.1 [31], and Divisive Amplicon Denoising Algorithm (DADA2) version 1.8 was used for error correction, exact sequence inference, read merging, and chimera removal [32]. Following contaminant removal (see supplemental content), the amplicon sequence variant (ASV) table comprised 19,955,039 total merged read pairs (mean per sample = 52,375; range 10,906 to 124,889). Taxonomic assignment was performed using RDP trainset 16 [33]. Rarefaction was performed at a depth of 10,906 reads for alpha diversity analyses. For all other analyses, estimates of relative library sizes (“size factors”) were obtained by calculating geometric means of pairwise read count ratios [34].
Behavioral and clinical covariates
Demographic and behavioral covariates included in the analyses were age, race/ethnicity, employment status, country of origin, a dichotomous variable for homelessness in past month, number of receptive anal intercourse (RAI) acts in past month, a dichotomous variable for RAI within the past seven days, frequency of methamphetamine, marijuana, and cocaine use in the past 6 months, tobacco smoking, and binge drinking. All demographic and behavioral data were self-reported by participants using a computer-aided self-interview (CASI); measures are described in the supplemental content. Dichotomous variables for obesity (defined as BMI > 30 or waist circumference > 40 inches), and antibiotic use in the past month were also included in the analyses; these data were collected by clinical staff.
Statistical analyses
The primary analyses were unadjusted and inverse probability of treatment-weighted comparisons of microbiome diversity and composition between HIV-, HIV+ undetectable, HIV+ suppressed, and HIV+ viremic participants. The R package ‘phyloseq’ was used to calculate alpha diversity statistics, distance matrices, and for ordination. Differences in alpha diversity between groups were examined with Kruskal-Wallis tests followed by comparisons of median values using quantile regression (R package ‘quantreg’). Permutational Multivariate ANOVA (PERMANOVA) was used to test for overall differences in microbial composition between HIV groups (R package ‘vegan’). Zero-inflated negative binomial (ZINB) models were fit in order to test for differential abundance in bacterial genera between groups with multinomial least absolute shrinkage and selection operator (LASSO) models employed as a confirmatory analysis (R packages ‘pscl’ and ‘glmnet’). ZINB and LASSO model selection and analytic procedures are described in the supplemental content.
IPTW [35] is a method of confounder control where the study sample is re-weighted in order to create a “pseudo-population” in which treatment/exposure, here referring to the four HIV viremia groups, is independent of confounding variables (see supplemental content). We used IPTW to control for all variables described in the Behavioral and Clinical Covariates section. Weights were estimated using generalized boosted models (R package ‘twang’), and balance between groups was assessed by computing standardized mean differences for each covariate in the weighted sample (R package ‘tableone’). Table 1 and Table S1 provide information on covariate balance before and after weighting. Robust variance estimates for inference tests in weighted ZINB analyses were obtained via the sandwich estimator (R package ‘sandwich’). Additional detail about the IPTW estimation and modeling procedures is provided in the supplemental content. Missing covariate data were imputed using the Chained Equations method [36] (R package ‘mice’); the proportion of missing data for each covariate is shown in Table S1. In order to account for multiple testing, alpha diversity and ZINB p values were corrected with the Benjamini-Hochberg false discovery rate (FDR) method [37]; FDR adjusted p values are labelled as q values. We utilized a threshold of two-sided p or q < 0.1 for significance testing; accordingly, we also display false coverage rate (FCR)-adjusted 90% confidence intervals [38] where relevant. All statistical analyses were completed using R v.3.4.3
Results
Sample characteristics
N = 383 participants were included in this study; 200 were HIV-, 66 were HIV+ undetectable (HIV RNA <20 copies/ml), 72 were HIV+ suppressed (HIV RNA ≥20–200 copies/ml), and 45 were HIV+ viremic (HIV RNA >200 copies/ml). All participants were MSM with an average age 31 (standard deviation = 7). Most were Hispanic (49%) or non-Hispanic Black (39%). Table 1 provides further detail on participant characteristics. Generally, HIV-infected participants, especially those with higher levels of viremia, were more likely to be older, unemployed, recently homeless, and to report methamphetamine use and frequent binge drinking than their HIV-uninfected peers. Among the HIV-infected participants, the mean number of years since diagnosis was 7.5 (sd = 5.7), mean log10 viral load was 2.0 (sd = 1.2), and mean CD4 cell count was 625 cells/mm3 (sd = 287). No participants were ART-naïve and ninety percent of participants reported current ART.
Effects of HIV viremia on overall microbial composition
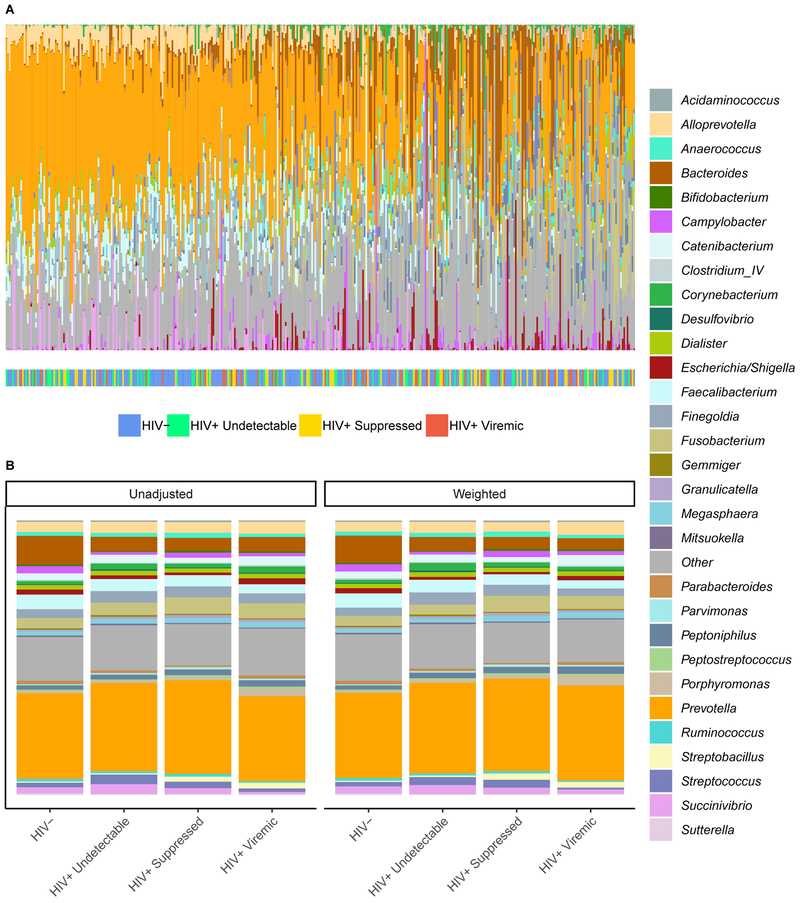
The relative composition of each individual’s microbiome is displayed in Figures 1A and S1, and average composition within each HIV viremia category in Figure 1B. Prevotella is the most highly represented bacterial genus among most participants, with increasing relative amounts of Bacteroides, Bifidobacteria, and Fusobacteria in those towards the right side of the axis (Figure 1A). Higher levels of Alloprevotella and Porphyromonas are apparent in the HIV+ viremic group, and lower levels of Bacteroides are apparent in all HIV+ groups relative to HIV- controls (Figure 1B).
Figure 1.
Rectal microbial composition of study participants, N = 383. (A) Columns represent the relative composition of each subject’s microbiome at the genus level. HIV status of the subjects is indicated by a colored line below their microbial composition. Subjects are ordered by the first principal coordinate of a Bray-Curtis pairwise distance matrix. Genera representing less than 1% of the composition on average across samples were combined into “Other.” (B) Average microbial composition within each HIV viremia category. Unadjusted and inverse probability of treatment weighted compositions are shown. Bacterial genera representing less than 1% of the overall relative composition or present in less than 20% of the samples were grouped into “Other.”
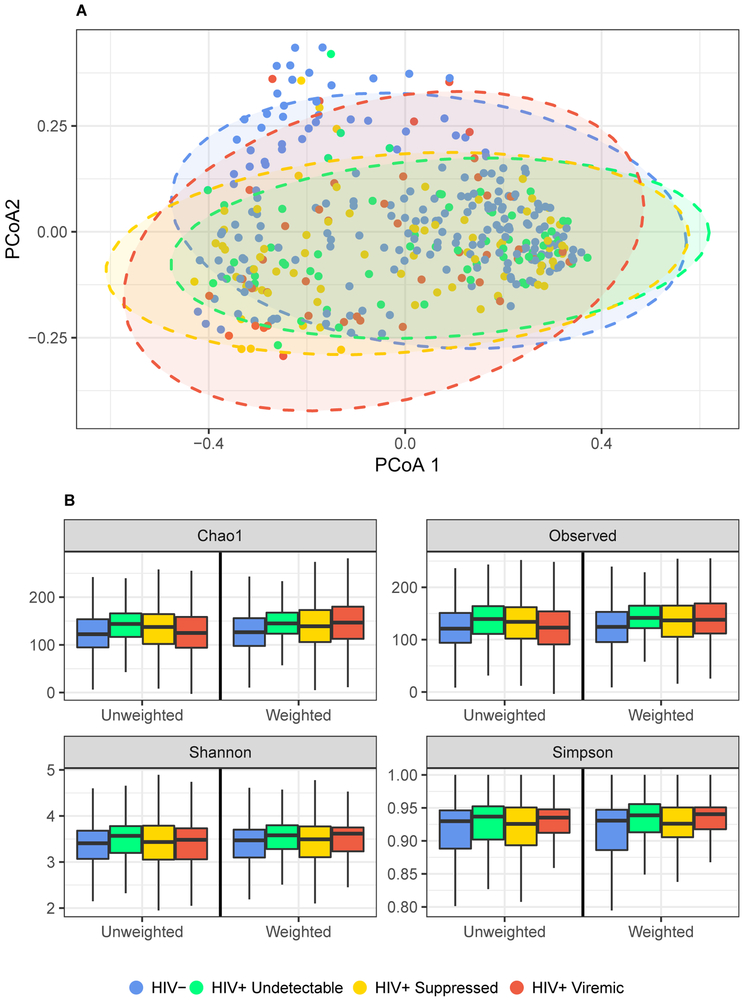
To quantitatively examine the influence of HIV viremia on differences in microbial composition between-subjects we used PERMANOVA with Bray-Curtis distance. HIV viremia explained a significant amount of variability in microbial composition in both unadjusted (R2 = .014, p = .001) and covariate-adjusted analyses (R2 = .011, p = .017) (Table S2). Figure 2A displays ordination of the samples by principal coordinates analysis (Bray-Curtis distance), where HIV- and HIV+ viremic groups are distinct while HIV+ undetectable and HIV+ suppressed are more similar.
Figure 2.
Associations between HIV viremia and overall microbial composition. (A) Ordination of the Bray-Curtis distance between samples using principal coordinates analysis. PCoA = Principal coordinate axis. Ellipses are 95% confidence regions for each group assuming points follow a multivariate t distribution. (B) Boxplots of richness metrics. Boxes represent the lower, median, and upper quartile of the data and whiskers are 1.5*interquartile range.
Comparisons of alpha diversity suggest a tendency for HIV+ individuals to have higher diversity in metrics that do not account for evenness (observed count and Chao1 statistic) (Figure 2B). Kruskal-Wallis analyses revealed significant differences in observed and Chao1 richness by HIV group (Table S3). Quantile regression was further used to investigate these differences and revealed higher median observed and Chao1 values for HIV+ suppressed versus HIV- individuals (q = .022 in IPTW-adjusted analyses). No other significant differences were found in any group. Shannon and Simpson indices did not vary greatly between groups.
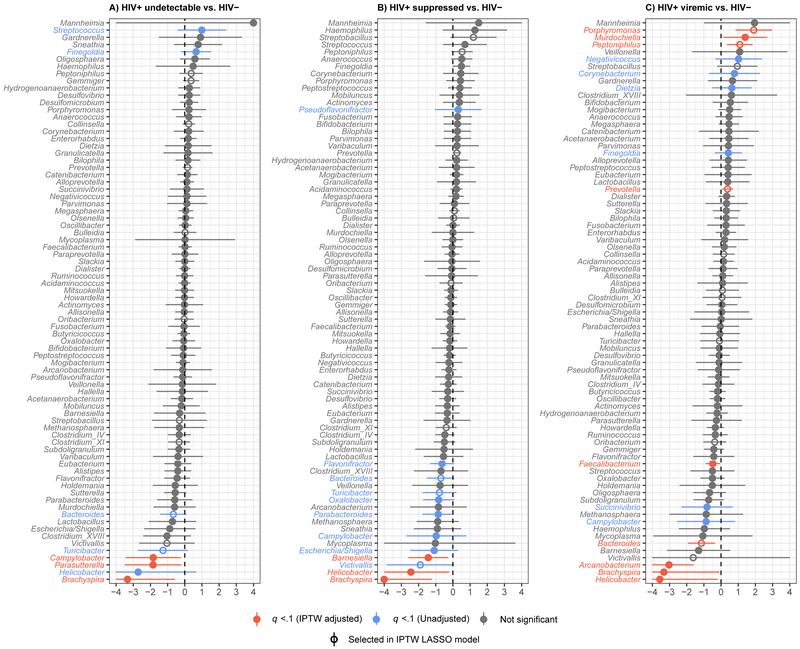
Differences in specific bacterial taxa associated with HIV viremia
Zero-inflated binomial (ZINB) models were utilized to identify bacterial genera that were differentially abundant among the HIV viremia groups. HIV+ undetectable showed significant enrichment in Finegoldia and Streptococcus and depletion in Bacteroides, Brachyspira, Campylobacter, Helicobacter, Parasutterella, and Turicibacter when compared to HIV-uninfected. After IPTW adjustment for behavioral and clinical confounders, depletions in Campylobacter, Parasutterella and Brachyspira remained significant (Figure 3A; Table S4). HIV+ suppressed participants (HIV RNA ≤200 copies/ml) had increased Pseudoflavonifractor and decreased Bacteroides, Barnsiella, Brachyspira, Campylobacter, Escherichia/Shigella, Flaonifractor, Helicobacter, Oxalobacter, Parabacteroides, Turicibacter, and Victivallis relative to HIV-negative subjects. Following IPTW adjustment, depletions in Barnesiella, Helicobacter, and Brachyspira remained significant (Figure 3B; Table S4).
Figure 3.
Comparisons of individual bacterial genera between HIV viremia categories. Forest plots of results of zero-inflated negative binomial models comparing genus-level bacterial counts between HIV-negative and (A) HIV+ undetectable (HIV RNA <20 copies/ml), (B) HIV+ suppressed (HIV RNA >20 and ≤200 copies/ml) and (C) HIV+ viremic (HIV RNA >200 copies/ml) participants. Inverse probability of treatment-weighted effect sizes and 90% false coverage rate-adjusted confidence intervals (truncated at −4, 4) are plotted, with statistical significance (q < 0.1) indicated in color. Effect sizes are log ratios of normalized genera counts.
HIV+ viremic men (HIV RNA >200 copies/ml) had the most distinct microbial signature relative to HIV-negative, showing significant enrichment of Corynebacterium, Dietzia, Finegolda, Murdochiella, Negativicoccus, Peptoniphilus, Porphyromonas, and Prevotella as well as depletion of Arcanobacterium, Brachyspira, Bacteroides, Campylobacter, Faecalibacterium, Helicobacter and Succinivibrio. With IPTW adjustment, enrichment of Murdochiella, Peptoniphilus, Porphyromonas, and Prevotella and depletion of Arcanobacterium, Bacteroides, Brachyspira, Faecalibacterium, and Helicobacter were significant (Figure 3C; Table S4). For some bacteria, including Faecalibacterium, Peptoniphilus, Porphyromonas, Prevotella, and Streptococcus, effect size (i.e., degree of enrichment or depletion) increased with increasing viremia (Figure S2).
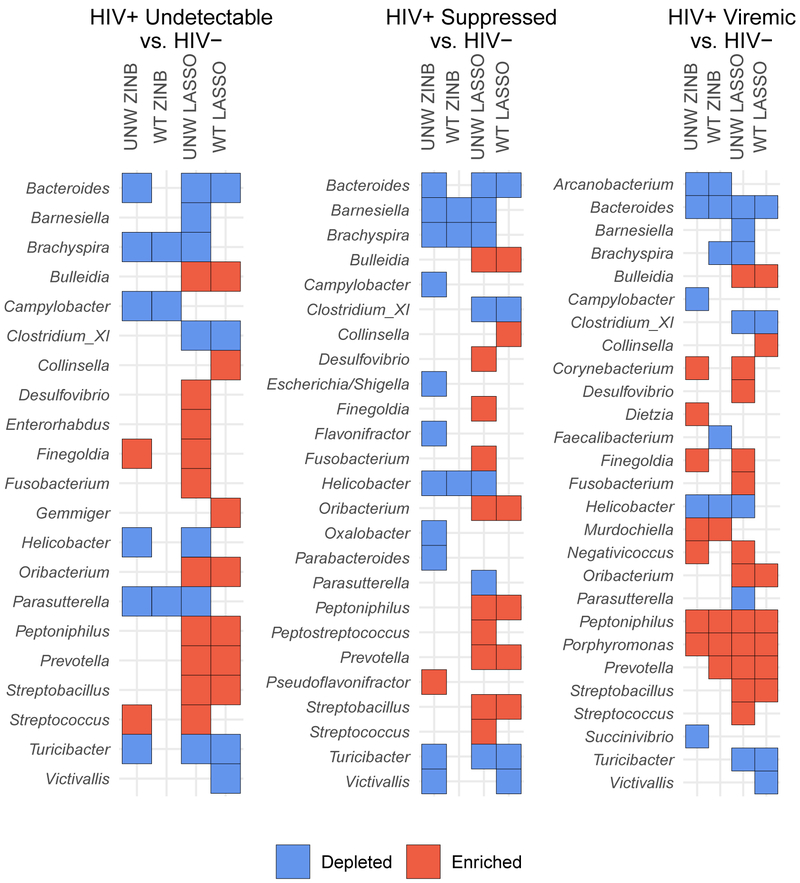
An IPTW-adjusted multinomial least absolute shrinkage and selection operator (LASSO) model was also used as an additional method of feature selection to compare with ZINB findings. Among HIV+ participants with undetectable or suppressed viremia, no genera were significant in both the adjusted ZINB and LASSO models. However, among the HIV+ viremic group, differences in Bacteroides, Peptoniphilus, Porphyromonas, and Prevotella were consistent across analytic strategies (Figure 4).
Figure 4.
Summary of zero-inflated negative binomial (ZINB) and least absolute shrinkage and selection operator (LASSO) model results. Enriched taxa are those with positive effect sizes (relative to HIV-), depleted are those with negative effect sizes. Genera with no effect in either analysis are not shown. UNW = unadjusted, WT = IPTW adjusted.
It was also of interest to determine whether HIV+ participants with low levels of viremia (HIV RNA < 200 copies/mL) had distinct microbial signatures from those who were HIV+ but undetectable (HIV RNA <20 copies/mL). One genus, Sneathea, was significantly different between these groups in adjusted ZINB analyses (q < .1). The LASSO model identified depletions in Gemmiger in HIV+ suppressed as compared to undetectable participants (Figure S3).
Discussion
In this study examining the effects of HIV on the rectal microbiome in a cohort of 383 young, mostly minority MSM, we found important differences in microbial composition between HIV-uninfected and HIV-infected men which varied depending on level of viremia. HIV viremia category accounted for about 1% of the variability in microbiome composition, an effect size that is consistent with previous studies [19]. As hypothesized, microbiome perturbations were most evident among HIV+ viremic men, and least evident in HIV+ men with undetectable viremia. Importantly, we utilized IPTW to account for multiple confounding factors in our analyses, which decreased the likelihood that the results we report are attributable to clinical or behavioral covariates affecting the microbiome such as sexual behavior, substance use, or obesity.
High diversity is generally associated with a healthy rectal microbiome [2], and reduced richness and diversity has been reported in studies comparing HIV-infected and uninfected persons [11, 12, 25, 39–41]. Still other studies report no differences in diversity associated with HIV-infection [16, 17], while others have suggested that differences in diversity may be related to sampling location [14], or HIV-treatment status [24, 40]. We found few significant differences in diversity metrics in our study, and findings did not follow a clear dose-response pattern with level of viremia. As we were able to adjust for multiple confounders in our analyses, we can be reasonably confident that previously reported determinants of diversity such as sexual behavior [19] and substance use [42] had limited influence on our findings. Our results suggest that once these confounding factors are taken into account, bacterial diversity and richness may not be substantially impacted by HIV infection itself.
One of the more consistent findings across studies of HIV and the microbiome has been enrichment in Prevotella and depletion in Bacteroides among both untreated and treated HIV-infected individuals [9, 11, 14]. Prevotella species are considered pro-inflammatory [43, 44], while Bacteroides species have been shown to induce regulatory T-cell differentiation and IL-10 production [45, 46]. Previous work has suggested that observed alterations to the Prevotella/Bacteroides ratio may have been due to sexual behavior rather than HIV [19]; however, others have shown decreased Bacteroides among HIV-infected MSM who were matched with MSM controls [9]. Our study examined exclusively MSM and controlled for recency and frequency of receptive anal intercourse in our analyses, therefore, our study provides additional evidence that HIV may directly alter the Prevotella/Bacteroides ratio independent of sexual behavior. Although we found decreased Bacteroides in all HIV-infected individuals, the effect was similar between undetectable and suppressed participants and only statistically significant after adjustment for confounding in the viremic group. In addition, we found increasing relative amounts of Prevotella with increasing levels of viremia, which were only significant in the viremic group. Our findings are consistent with previous research showing that Prevotella may normalize with ART [13] whereas depletions in Bacteroides persist even with therapy [24].
Of the 78 genera tested, ZINB and LASSO models identified Porphyromonas as the genus with the largest difference between HIV+ viremic and HIV- individuals. Porphyromonas is a well-known modifier of inflammatory cytokines [47]. In fact, Porphyromonas gingivalis has been identified as a potential cause of systemic inflammation and metabolic disorders associated with periodontal disease [48] and implicated in inflammatory processes leading to the development of atherosclerosis [49]. Furthermore, administration of P. gingivalis to mice was shown to induce GI dysbiosis and contribute to intestinal permeability [50]. The association between increasing levels of HIV viremia and Porphyromonas may therefore represent an important mechanism behind HIV-associated chronic inflammation deserving of further study.
Of particular interest in our study is the examination of low level viremia individuals who are not undetectable (HIV RNA ≥20–200 copies/ml). It is notable that this group, while distinct from HIV-uninfected, was very similar to the undetectable viremia (HIV RNA <20 copies/ml) group in ordination analysis. Only a single genus, Sneatha, was statistically significantly different between these two groups when directly compared. Clinically, persistent low level viremia may increase risk of subsequent virologic failure [51], but a recent large study showed no difference in progression to AIDS or incidence of non-AIDS events in persons with low level viremia compared to undetectable [52]. Our analysis suggests that microbial composition is similar between those with low level and undetectable viremia, but remains distinguishable from HIV-uninfected individuals. While those with low level viremia can still have microbial translocation and inflammatory biomarkers [53], the overall decreased dysbiosis in low level viremia may correspond to reduced chronic inflammation which lessens clinical progression.
Our results should be interpreted with consideration of the following limitations. First, we did not have diet information on our cohort. We included race/ethnicity, country of origin, employment status, obesity, and homelessness (all of which may impact dietary intake) in our covariate adjustment set to mitigate this limitation to the best of our ability. Second, the IPTW procedure will only achieve perfect balance between exposure groups in nonparametric settings with large sample sizes relative to the number of relevant confounders, and there is potential for residual confounding even by variables we controlled for in our study. However, we note that many of the most significant confounders (e.g. age, antibiotic use, sexual behavior, alcohol use, obesity) were well-balanced after weighting. The ability of 16S sequencing to provide species-level resolution is limited, thus we conducted analyses at the genus level. We acknowledge that differentially abundant genera do not necessarily indicate differences in functionally important species. Finally, we did not have time since ART initiation for our cohort, and thus cannot determine if participants were viremic because they recently started treatment or because treatment was failing.
Our study also has numerous strengths. Primarily, we utilized data from a large cohort of regionally, socioeconomically, and behaviorally similar individuals, increasing the internal validity of our findings. We employed a novel technique, IPTW, to incorporate a large amount of clinical and behavioral data into our analyses. With IPTW, analyses are “marginal structural models” instead of conditional on covariates, as in multiple regression. Modeling microbiome data marginally offers several advantages including the ability to control for many confounding factors without inducing overfitting bias [54] or losing efficiency due to overstratification [55]. Addressing sources of systematic error using IPTW may improve reproducibility in future studies of HIV and the microbiome. We also stratified our HIV-infected participants by level of viremia, allowing us to examine differences between HIV-uninfected and HIV+ undetectable, suppressed, and viremic individuals. This stratification leads to better understanding of the relationship between active viral replication and dysbiosis, namely, that dysbiosis increases with increasing viremia. Finally, we were able to replicate our major findings using two distinct analytic strategies. Genera identified as differentially abundant in both analyses may be more likely to be true discoveries.
This study contributes to a growing body of literature describing the effects of HIV on microbial dysbiosis. We show that, even when taking into account the influence of multiple confounding factors, HIV is associated with intestinal dysbiosis in a dose-dependent manner. Although great strides have been made in the management of chronic HIV infection, the life expectancy among HIV-infected individuals remains reduced relative to their HIV-negative peers [56]. This reduction has largely been attributed to increased rates of inflammation-related comorbidities observed among people living with HIV [57], and the microbiome likely plays a key role in modulating interactions between HIV and the immune system. Therefore, understanding the ways in which HIV and the microbiome interact may be a crucial step towards developing intervention strategies to reduce the burden of HIV-associated morbidity and mortality.
Supplementary Material
Acknowledgements
Sources of Funding: This work was supported by the National Institute on Drug Abuse (1R36 DA046310 and 2U01 DA036267) and the UCLA Center for HIV Identification, Prevention, and Treatment Services (CHIPTS; National Institute of Mental Health P30 MH58107). Additional support provided by the UCLA AIDS Institute and UCLA CFAR Microbiome and Mucosal Immunology Core (P30 AI028697). J.A.F. was supported in part by National Institute of Allergy and Infectious Diseases (NIAID) (K08 AI124979). Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
Supplemental Content
Supplemental Digital Content 1.docx
References
- 1.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016; 375(24):2369–2379. [DOI] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortum. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol 2012; 30:149–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev 2013; 26(1):2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. Aids 2016; 30(18):2737–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gootenberg DB, Paer JM, Luevano JM, Kwon DS. HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation. Curr Opin Infect Dis 2017; 30(1):31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 2016; 11(2):182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d’Ettorre G, Ceccarelli G, Giustini N, Serafino S, Calantone N, De Girolamo G, et al. Probiotics Reduce Inflammation in Antiretroviral Treated, HIV-Infected Individuals: Results of the “Probio-HIV” Clinical Trial. PLoS One 2015; 10(9):e0137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5(193):193ra191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med 2011; 62:141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10(2):e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. Aids 2014; 28(5):753–760. [DOI] [PubMed] [Google Scholar]
- 13.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. Aids 2015; 29(18):2409–2418. [DOI] [PubMed] [Google Scholar]
- 14.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7(4):983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrus ML, Madrid N, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8(4):760–772. [DOI] [PubMed] [Google Scholar]
- 16.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volpe GE, Ward H, Mwamburi M, Dinh D, Bhalchandra S, Wanke C, et al. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs 2014; 75(2):347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14(3):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadella M, Nowak P, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine 2016; 5:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoel H, Hove-Skovsgaard M, Hov JR, Gaardbo JC, Holm K, Kummen M, et al. Impact of HIV and Type 2 diabetes on Gut Microbiota Diversity, Tryptophan Catabolism and Endothelial Dysfunction. Sci Rep 2018; 8(1):6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollock J, Glendinning L, Wisedchanwet T, Watson M. The Madness of Microbiome: Attempting To Find Consensus “Best Practice” for 16S Microbiome Studies. Appl Environ Microbiol 2018; 84(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsen J, Brejnrod A, Mortensen M, Rasmussen MA, Stokholm J, Al-Soud WA, et al. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome 2016; 4(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawinkel S, Mattiello F, Bijnens L, Thas O. A broken promise: microbiome differential abundance methods do not control the false discovery rate. Brief Bioinform 2017. [DOI] [PubMed] [Google Scholar]
- 24.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014; 5(4):562–570. [DOI] [PubMed] [Google Scholar]
- 25.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manner IW, Baekken M, Kvale D, Oektedalen O, Pedersen M, Nielsen SD, et al. Markers of microbial translocation predict hypertension in HIV-infected individuals. HIV Med 2013; 14(6):354–361. [DOI] [PubMed] [Google Scholar]
- 27.Vesterbacka J, Rivera J, Noyan K, Parera M, Neogi U, Calle M, et al. Richer gut microbiota with distinct metabolic profile in HIV infected Elite Controllers. Sci Rep 2017; 7(1):6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med 2016; 8(349):349ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pannaraj PS, Ly M, Cerini C, Saavedra M, Aldrovandi GM, Saboory AA, et al. Shared and Distinct Features of Human Milk and Infant Stool Viromes. Front Microbiol 2018; 9:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender JM, Li F, Adisetiyo H, Lee D, Zabih S, Hung L, et al. Quantification of variation and the impact of biomass in targeted 16S rRNA gene sequencing studies. Microbiome 2018; 6(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Reeve J, Zhang L, Huang S, Wang X, Chen J. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ 2018; 6:e4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000; 11(5):550–560. [DOI] [PubMed] [Google Scholar]
- 36.Van Buuren S, Brand JPL, Groothuis-Oudshoorn CGM, Rubin DB. Fully conditional specification in multivariate imputation. Journal of Statistical Computation and Simulation 2006; 76(12):1049–1064. [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300. [Google Scholar]
- 38.Benjamini Y, Yekutieli D. False Discovery Rate–Adjusted Multiple Confidence Intervals for Selected Parameters. Journal of the American Statistical Association 2005; 100(469):71–81. [Google Scholar]
- 39.Sun Y, Ma Y, Lin P, Tang YW, Yang L, Shen Y, et al. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Infect 2016; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe 2016; 19(3):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubourg G, Lagier JC, Hue S, Surenaud M, Bachar D, Robert C, et al. Gut microbiota associated with HIV infection is significantly enriched in bacteria tolerant to oxygen. BMJ Open Gastroenterol 2016; 3(1):e000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Xie Z, Wang H, Shen Z, Guo Y, Gao Y, et al. Bacterial Diversity of Intestinal Microbiota in Patients with Substance Use Disorders Revealed by 16S rRNA Gene Deep Sequencing. Sci Rep 2017; 7(1):3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol 2016; 9(1):24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci (Landmark Ed) 2010; 15:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008; 453(7195):620–625. [DOI] [PubMed] [Google Scholar]
- 47.Wang PL, Ohura K. Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts-CD14 and Toll-like receptors. Crit Rev Oral Biol Med 2002; 13(2):132–142. [DOI] [PubMed] [Google Scholar]
- 48.Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep 2014; 4:4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi J, Chung SW, Kim SJ, Kim SJ. Establishment of Porphyromonas gingivalis-specific T-cell lines from atherosclerosis patients. Oral Microbiol Immunol 2001; 16(5):316–318. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, et al. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS One 2015; 10(7):e0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laprise C, de Pokomandy A, Baril JG, Dufresne S, Trottier H. Virologic failure following persistent low-level viremia in a cohort of HIV-positive patients: results from 12 years of observation. Clin Infect Dis 2013; 57(10):1489–1496. [DOI] [PubMed] [Google Scholar]
- 52.Bernal E, Gomez JM, Jarrin I, Cano A, Munoz A, Alcaraz A, et al. Low level viremia is associated with clinical progression in HIV-infected patients receiving antiretroviral treatment. J Acquir Immune Defic Syndr 2018. [DOI] [PubMed] [Google Scholar]
- 53.Reus S, Portilla J, Sanchez-Paya J, Giner L, Frances R, Such J, et al. Low-level HIV viremia is associated with microbial translocation and inflammation. J Acquir Immune Defic Syndr 2013; 62(2):129–134. [DOI] [PubMed] [Google Scholar]
- 54.Babyak MA. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med 2004; 66(3):411–421. [DOI] [PubMed] [Google Scholar]
- 55.De Stavola BL, Cox DR. On the consequences of overstratification. Biometrika 2008; 95(4):992–996. [Google Scholar]
- 56.Harrison KM, Song R, Zhang X. Life expectancy after HIV diagnosis based on national HIV surveillance data from 25 states, United States. J Acquir Immune Defic Syndr 2010; 53(1):124–130. [DOI] [PubMed] [Google Scholar]
- 57.Guaraldi G, Orlando G, Zona S, Menozzi M, Carli F, Garlassi E, et al. Premature age-related comorbidities among HIV-infected persons compared with the general population. Clin Infect Dis 2011; 53(11):1120–1126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.