Abstract
Protein interactions with nucleic acids are important for the synthesis, regulation, and stability of macromolecules. While a number of assays are available for interrogating these interactions, Differential Radial Capillary Action of Ligand Assay (DRaCALA) has been developed as an easy and flexible platform that allows for the study of individual interactions when carrying out high-throughput screening for novel binding proteins and small molecule inhibitors. In this article, we describe the principle of DRaCALA and methods that utilize DRaCALA to determine the affinity and specificity of individual protein-nucleic acid interactions as well as uses for screening for binding proteins and chemical inhibitors.
Keywords: Protein-nucleic acid interactions, binding assay, radiolabeled ligand, dissociation constant (Kd), off-rate (koff), high-throughput screening
INTRODUCTION
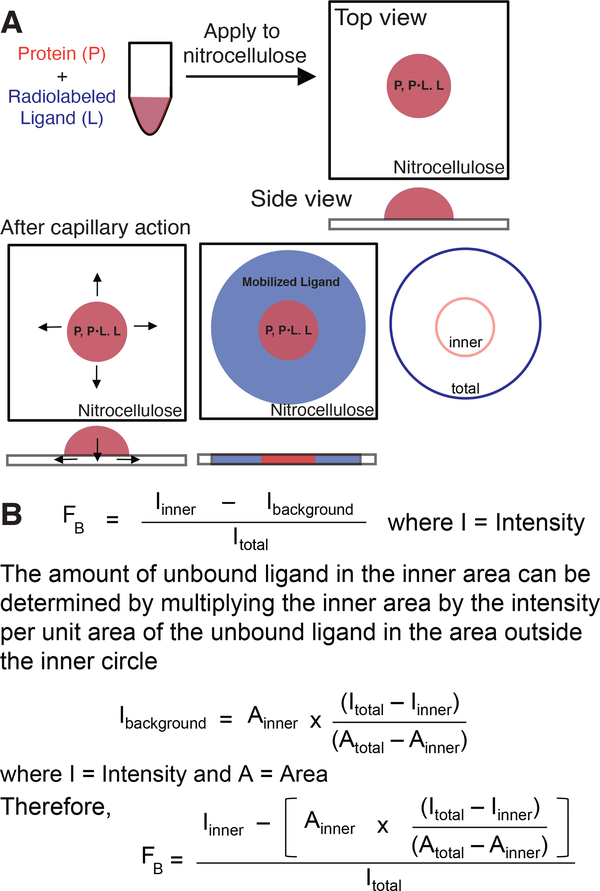
Differential Radial Capillary Action of Ligand Assay (DRaCALA) is the application of mixtures of protein and radiolabeled nucleic acids onto a nitrocellulose membrane. Nitrocellulose membranes are able to retain proteins based on hydrophobic interactions while free nucleic acids can pass through nitrocellulose unimpeded. The principle of DRaCALA is based on the differential mobility of unbound nucleic acids and nucleic acids in complex with proteins when applied to a dry nitrocellulose membrane (Roelofs, Wang, Sintim, & Lee, 2011). While DRaCALA is similar to filter binding assays (see discussion of other assays in Background Information), there are two key differences. First, the sample is applied to dry nitrocellulose, the liquid is absorbed into the membrane, and then moves radially from the site of application by capillary action (Figure 1A). Second, there is no wash step. Two important implications arise from these differences in sample processing. First, the absence of a wash means that the total amount of radiolabeled nucleic acid in each sample is known. Second, a radiolabeled nucleic acid ligand can physically be present in two different zones, i.e. the site of application (inner circle) and the zone of mobilization (total circle) (Figure 1). Since the application zone represents the immobilized protein-nucleic acid complex and the mobilization zone represents unbound nucleic acid, the distribution of the labeled nucleic acid in these two zones can be used to calculate the fraction bound (FB) at a particular concentration of protein and nucleic acid (See Figure 1). A simplistic interpretation of the equation is that the inner intensity represents the bound nucleic acid (Iinner) divided by the total intensity (Itotal). This approximation requires a correction factor since the inner intensity represents the summation of the protein-nucleic acid complex and the free nucleic acid that is distributed through the entire zone of mobilization. This background amount of unbound nucleic acid in the inner circle can be determined in two steps. First, determine the intensity per unit area by calculating the intensity in the total circle that excludes the zone of immobilization (Itotal – Iinner) and dividing by the area of the total circle (Atotal – Ainner) (Figure 1). Second, multiply the intensity per unit area by the area of the inner circle. Once this correction factor is subtracted from the intensity at the site of application, the fraction bound can be determined fairly accurately.
Figure 1.
Principle of DRaCALA.
(A) Schematic representation of DRaCALA assay on application of protein-ligand mixture (in red) onto nitrocellulose with consecutive ligand mobilization (in blue) by capillary action. Protein (P), ligand (L), and protein–ligand complex (P•L) distribution during the assay is shown. (B) Equations used to analyze DRaCALA data for fraction of the ligand bound (FB) for purified proteins. Taken with permission from Roelofs et al. (2011).
The ease of use of DRaCALA, which simply consists of applying liquid mixtures on a dry nitrocellulose membrane, means this assay is flexible for a number of applications. One interesting application of DRaCALA is the ability to detect specific protein-nucleic acid interactions in whole cell lysates. This allows an additional application for screening for protein-nucleic acid interactions. Here we will describe the following applications of DRaCALA: 1. Identifying proteins that bind nucleic acids, 2. Determining binding constants, 3. Characterizing binding specificity, 4. Determining the off-rate, 5. Screening for small molecule inhibitors, and 6. Screening for novel protein receptors of nucleic acids. The protocols as presented are sequential to allow the investigator to obtain additional information regarding the protein-ligand interaction in a stepwise manner.
STRATEGIC PLANNING
The series of protocols outlined below describe how protein-ligand interactions can be detected and characterized using DRaCALA. Selection of an appropriate ligand is important the success of the assay. Since DRaCALA relies on the differential migration of free ligand spotted on a nitrocellulose membrane, an appropriate ligand for this assay is one that migrates unimpeded when applied onto nitrocellulose in the absence of proteins. This is an absolute requirement since a ligand that fails to migrate cannot yield useful information in DRaCALA. Previously, we have shown that radiolabeled nucleic acids are mobile on nitrocellulose. Suitable ligands include single stranded oligo DNA and RNA, annealed DNA oligonucleotides, DNA PCR products, and plasmid DNA linearized by restriction enzymes (Donaldson, Roelofs, Luo, Sintim, & Lee, 2011). In addition, a number of signaling molecules two nucleotides or less, including mononucleotides, cyclic mononucleotides, cyclic dinucleotides, linear dinucleotides, and ppGpp, have been successfully used in DRaCALA (Corrigan, Bellows, Wood, & Gründling, 2016; Corrigan et al., 2013; Donaldson et al., 2011; Fang et al., 2014; Orr et al., 2015; Roelofs et al., 2015; Roelofs et al., 2011; W. Ross, Vrentas, Sanchez-Vazquez, Gaal, & Gourse, 2013; Zhang, Zbornikova, Rejman, & Gerdes, 2018). All of these ligands were radiolabeled with 32P or 14C since 3H cannot be detected. For signaling nucleotides less than two nucleotides in length, the radiolabel is introduced enzymatically using the appropriate synthase enzymes. For DNA or RNA longer than two nucleotides, a 32P label can be easily introduced on the 5’-OH using polynucleotide kinase (PNK) and 32P γ-ATP. Fluorescently labeled nucleic acids are not mobile on nitrocellulose membrane based on our experience (see supplemental figures in (Donaldson et al., 2011), so they are not appropriate ligands for DRaCALA. The suitability of any ligand for DRaCALA can be tested empirically by spotting the ligand in the reaction buffer on dry nitrocellulose. If the ligand is uniformly distributed across the mobilization zone, then the ligand is suitable for DRaCALA. If the ligand is concentrated at the zone of application in the absence of protein, then it should not be used for DRaCALA. Once an appropriate ligand is identified, the amount of radiolabeled ligand used should be at least 10-fold less than the dissociation constant (Kd) by experimental determination using the protocols described in this article. The recommended amount of radiolabeled ligand used should yield a signal intensity greater 3,000 photo-stimulated luminescence (PSL) units in a one-hour exposure to a phosphorimager screen. A concentration of the radiolabeled ligand less than 100 PSL per hour is insufficient for detection in a typical overnight exposure. Once these parameters for the radiolabeled ligand have been determined, the ligand can be used in the following protocols to assess protein-nucleic acid interactions.
BASIC PROTOCOL 1
Determination of protein binding by DRaCALA
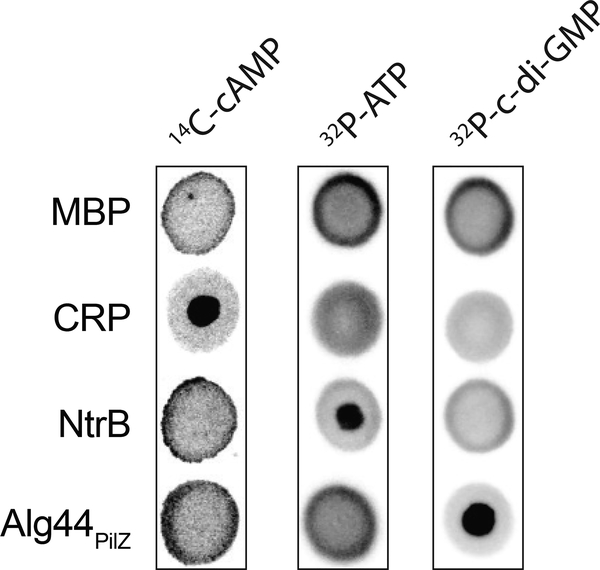
An important first step in designing a DRaCALA assay for detecting nucleic acid (ligand) binding to a protein is to ensure that ligand is mobilizable on nitrocellulose in the absence of protein. An additional control for binding specificity is to use a negative control protein, such as maltose binding protein (MBP), that does not sequester nucleic acids (Roelofs et al., 2011). The typical volume of each spot that is applied to nitrocellulose is 2–10 μL. Since there is flexibility in the volume of individual assay spots, the total volume of the binding mixture needs to be determined for each experiment. The suggested total volume of the binding assay should be 5–10x the volume of individual assay spots. This allows for conducting the assay at least in triplicate to ensure that the mixture is homogeneous. For Basic Protocol 1, the volumes stated in the protocol are for 2 μL spots. Basic Protocol 1 is required before conducting the experiments described in Basic Protocols 2–6. Example results are shown in Figure 2 for Escherichia coli cAMP receptor protein (CRP) binding to cyclic AMP (cAMP), E. coli NtrB kinase binding ATP, and Pseudomonas aeruginosa Alg44 binding cyclic-di-GMP (c-di-GMP) (Roelofs et al., 2011).
Consider carefully the buffer used for purified protein and the radiolableled ligand that is being tested. If the protein is enzymatically active towards the ligand, a failure to detect an interaction may be due to degradation of the ligand and subsequent release of the products. For some enzymes, removal of divalent cations or the use of calcium ions may prevent or reduce activity.
Figure 2.
DRaCALA allows detection of protein-nucleic acid interaction.
Representative DRaCALA images of interactions between indicated purified proteins (20 μM) with 14C-cAMP, 32P-ATP, or 32P –c-di-GMP. Taken with permission from Roelofs et al. (2011).
Materials
Radiolabeled nucleic acid (>3,000 PSL per hour exposure to phosphorimager screen)
Purified control and investigational proteins (concentration exceeding 20 μM is a good starting point)
5–10 X binding buffer (For our examples, the 10x binding buffer consists of 100 mM Tris pH 8.0, 1 M NaCl, 50 mM MgCl2. The specific buffer choice will depend on the protein under investigation.)
Nuclease-free double deionized water (ddH2O)
Appropriate nuclease free disposable plasticware for the number of samples (Microcentrifuge tubes, PCR strip tubes, PCR plates, or 96-well round bottom plates)
Pipette and pipette tips or pin tools (V&P Scientific) (For pin tools, a container with wash buffer (0.01% Tween in ddH2O) is required for cleaning between sample applications)
Nitrocellulose membrane with 0.45 μm pores (We have successfully used nitrocellulose membranes from several vendors.)
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
- Prepare 10 μL of 2x concentrated radiolabel ligand mix for each protein sample to be tested. Mix radiolabeled ligand, 10x binding buffer and ddH2O. Transfer 10 μl aliquots of the 2x radiolabel ligand mix to a nuclease free disposable plasticware tube or assay plate. An example for this mixture is as follows:
1 μl 10x binding buffer 8 μl ddH2O 1 μl Radiolabeled nucleic acid The amount of radioactive molecule needed will vary depending on the radioisotope and its specific activity. The concentration of radiolabeled nucleic acid should be experimentally determined by serial dilution to obtain the concentration such that the signal is >3,000 PSL in a one-hour exposure to phosphorimager screen. - Cut nitrocellulose membrane to the appropriate length.For 2 μL spots, the area of the spot is less than 1 cm × 1 cm. For triplicates, triple the area of the nitrocellulose. The nitrocellulose membrane is typically sandwiched between two layers of wax paper. Placing the nitrocellulose above both layers of wax paper ensures that the liquid does not bleed through the nitrocellulose.
- Aliquot 10 μL of most concentrated purified protein into each ligand mix from Step 1.Include reactions for positive and negative control. Good negative control for nucleic acids is MBP which does not bind nucleic acids.
- Mix by either pipetting at least 5 times with a pipette set to 10 μL volume or, if using a 96-well plate, place on a plate shaker at setting 5 for 30 sec.From our experience, the DRaCALA assay can show differences in intensity of multiple spots from the same assay mixture. These differences in intensity are often associated with the binding mixture not as homogeneous as expected since the samples were mixed.
- Pipette 2 μl of the protein-ligand mixture from Step 4 onto dry nitrocellulose placed on top of the wax paper backing.If using pipette tips, use new tips for every reaction. If using a pin tool, wash pin tool with wash buffer (0.01% Tween 20) three times by submerging into wash buffer and wicking dry on clean paper towel. After cleaning, place pin tool in sample for 2 seconds, remove from samples and press down vertically on nitrocellulose without dragging pins sideways. Hold on nitrocellulose for at least 10 seconds for the entire mixture to move into the membrane by capillary action.
Repeat the spotting twice more with each protein sample to obtain three replicates.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap spotted nitrocellulose membranes with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to the phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization, respectively.This process is easiest with appropriate positive controls. See Alg44 sample for 32P-c-di-GMP binding (Figure 2).
- Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).For the Fujifilm Multi Gauge software package, DRaCALA data can be collected as follows: open the image file to be analyzed, navigate to the “Measure” tab, and select the “Plate” menu. Within this menu, the user may customize the diameter, orientation, and spacing of an array of circles to align with the inner and outer DRaCALA spots. Once aligned, the user can navigate to the “Analysis” tab, which will generate a text file with the PSL and area (in square mm) of the circles.For the GE Amersham Typhoon, DRaCALA data can be collected as follows: scan the phsophorimager screen using the Phosphor Imaging setting. Using the ImageQuant TL software, open the image file to be analyzed, navigate to the “Analysis” tab, select “Areas” under “Shape Definition”, and choose the circle drawing tool. Individual circles may be drawn to align with DRaCALA spots. To generate a data table, select the “Area Window” tab. The “Volume” data is analogous to the signal intensity. Note that the area of the circles generated by the ImageQuant TL software is in pixels.
Calculate the fraction bound for each spot using the formula given in Fig 1B.
Protein binding to nucleic acid can be determined by analyzing the triplicate data between buffer only negative control and ligand treated with protein using an unpaired two-tailed t test.
BASIC PROTOCOL 2
Determination of binding affinity by DRaCALA
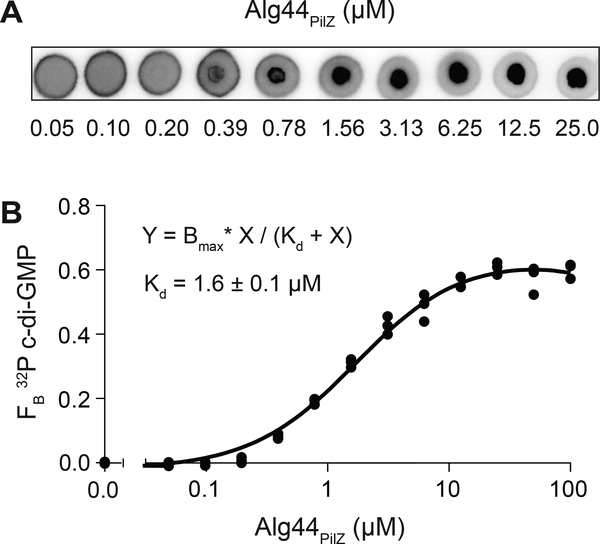
A key property of macromolecular interactions is the affinity of the molecules, or how tightly the molecules are associated. The dissociation constant (Kd) is the concentration of the protein required for 50% of the ligand to be bound. Kd is inversely related to affinity; lower Kd values indicate higher affinity and vice versa. If a protein from Basic Protocol 1 shows binding interaction, DRaCALA can be used to quantify the Kd of protein-ligand interaction by serially diluting the protein from stock concentrations (which should be above the Kd) to concentrations below the Kd. Each concentration of protein can be assessed for binding to radiolabeled nucleic acid by DRaCALA. An example outcome is Alg44 binding to c-di-GMP (Figure 3) (Roelofs et al., 2011).
If the stock concentration of the protein does not bind, then either the protein is not binding the nucleic acid ligand or other assays should be used. All Kd measurements obtained by DRaCALA should be considered apparent Kds since they are subject to the specific buffer and experimental conditions. While not absolute, the data provides a suitable basis of comparison to Kds obtained under similar conditions. For example, comparing the wild-type protein with a site-directed substitution of a specific residue within the protein can reveal the importance of the residue for the binding interaction.
Figure 3.
Determination of dissociation constant (Kd) by DRaCALA.
(A) Representative DRaCALA images of 32P-c-di-GMP interaction with indicated concentration of His-MBP-Alg44PilZ (μM). (B) FB calculated from (A) were plotted as a function of His-MBP-Alg44PilZ concentration. The best-fit line was determined by nonlinear regression using the indicated equation. Adapted with permission from Roelofs et al. (2011).
Materials
Radiolabeled nucleic acid (>3,000 PSL per hour exposure to phosphorimager screen)
Purified investigational protein
5–10 X binding buffer (For our examples, the 10x binding buffer consists of 100 mM Tris pH 8.0, 1 M NaCl, 50 mM MgCl2. The specific buffer choice will depend on the protein under investigation.)
Nuclease-free double deionized water (ddH2O)
Appropriate nuclease free disposable plasticware for the number of samples (Microcentrifuge tubes, PCR strip tubes, PCR plates, or 96-well round bottom plates)
Pipette and pipette tips or pin tools (V&P Scientific) (For pin tools, a container with 0.01% Tween in ddH2O is required for cleaning between sample applications)
Nitrocellulose membrane with 0.45 μm pores
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
Prepare 10 μL of 2x concentrated radiolabel ligand mix for each protein sample to be tested. Mix radiolabeled ligand, 10x binding buffer, and ddH2O. Transfer 10 μl aliquots of the 2x radiolabel ligand mix to an appropriate nuclease free disposable tube or assay plate.
- Cut nitrocellulose membrane to the appropriate length.For 2 μL spots, the area of the spot is less than 1 cm × 1 cm. For triplicates, triple the area of the nitrocellulose.
- Dilute proteins by serial 2-fold dilutions, typically 7–11 times.Include a buffer only negative control.
Transfer 10 μL of each protein dilution into each ligand mixture from step 1.
Mix by either pipetting at least 5 times with a pipette set to 10 μL volume or, if using a 96-well plate, place on a plate shaker at setting 5 for 30 sec.
- Pipette 2 μl onto dry nitrocellulose placed on top of the wax paper backing.If using a pin tool, wash pin tool with 0.01% Tween 20 three times by submerging into wash buffer and wicking dry on clean paper towel. Place pin tool in sample and press down vertically on nitrocellulose without dragging tips sideways. Hold for at least 10 seconds for the entire mixture to move into the membrane by capillary action.
Repeat the spotting twice more with each protein sample to obtain three replicates.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap nitrocellulose membrane with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager.
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization respectively.This process is easiest with appropriate positive (undiluted protein) control.
Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).
Calculate the fraction bound for each spot using the formula given in Fig 1B.
- Kds can be determined using nonlinear regression curve fit analysis (GraphPad Prism software).Other parameters can be added to the analysis including the number of specific binding sites and the maximum binding (Bmax). Another consideration arises from proteins that interact with themselves to form dimers and higher multimers. This can lead to cooperative binding to multimeric proteins with multiple binding sites. Another consideration is that the subunits of multimeric proteins have affinity for each other and these interactions contribute to the binding to the ligand.
BASIC PROTOCOL 3
Determination of specificity of binding by DRaCALA
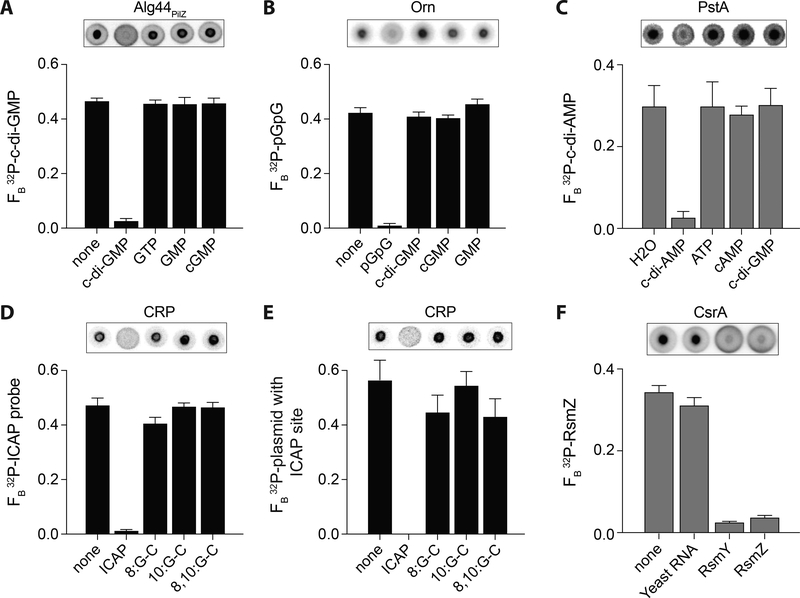
For proteins that can bind one or more ligands, binding interactions can occur at the same site or multiple sites in the protein. For example, transcription factors can bind to multiple operator sequences on the genome using the same DNA binding domain. In a second scenario, enzymes can bind the substrate, but other molecules can serve as competitive inhibitors. A third example is an enzyme with a catalytic site that binds its substrate and an allosteric site that can regulate enzymatic activity. In this latter example, binding of the two ligands occurs at two different sites. One famous example of this is the E. coli bacterial transcription factor CAP/CRP (catabolite activating protein/cAMP receptor protein) that binds cAMP to activate binding to DNA. To distinguish between the possibilities that the two ligands bind one binding pocket or two separate binding pockets, one can assess the ability of one ligand to compete for the same binding pocket with another ligand. To facilitate these assays, the ligand known to bind the receptor is radiolabeled and structurally similar, unlabeled ligands are added as competitors. DRaCALA can be used to rapidly assess the specificity of protein-nucleic acid interactions as shown in Figure 4. Examples of analyzing specific protein-ligand interactions using competition experiments include: Alg44 binding c-di-GMP (Roelofs et al., 2011), Oligoribonuclease binding pGpG (Orr et al., 2015), PstA binding to cyclic-di-AMP (c-di-AMP) (Corrigan et al., 2013), CAP binding annealed oligonucleotides that form the idealize CAP (ICAP) binding site, CAP binding to ICAP on a plasmid (Donaldson et al., 2011), and CsrA binding small RNAs RsmY and RsmZ (Patel, Gebbie, & Lee, 2014). The choice of competitors is important for interpreting the results from these types of experiments. The use of increasingly structurally related competitor molecules allows for more specific conclusions from competition experiments. Initial competition experiments utilize high concentrations of the competitors, providing them in excess to assess a relevant range. Additional experiments can be performed using decreasing concentrations, thereby enabling determination of the substrate preference of the ligand-binding pocket within the protein.
Figure 4.
DRaCALA detection of specific protein-nucleotide, protein-DNA, and protein-RNA interactions.
DRaCALA allows detection of specific protein-nucleotide, protein-DNA, and protein-RNA interactions in purified proteins (black bars) and overexpressed Escherichia coli T7Iq whole cell lysates (gray bars). Representative images of DRaCALA spots are shown above the graph. Competitors are indicated on the x-axes. (A) Binding of 32P-c-di-GMP by 2.5 μM His-MBP-Alg44PilZ in the presence of 500 μM of indicated unlabeled competitors. Adapted with permission from Roelofs et al. (2011). (B) Binding of 32P-pGpG by His-MBP-Orn (P. aeruginosa) in the presence of 250 μM of indicated unlabeled competitors. Adapted with permission from Orr et al. (2015). (C) Binding of 32P-c-di-AMP by E. coli T7Iq lysates overexpressing His-PstA (Staphylococcus aureus) in the presence of 400 μM of indicated unlabeled competitors. Adapted with permission from Corrigan et al. (2013). (D, E) Binding of 100 nM CRP with 200 μM cAMP to (D) 32P-labeled annealed oligonucleotide containing the ICAP sequence or (E) 32P-labeled plasmid with 1x ICAP site in the presence of the indicated unlabeled ICAP consensus probes or G-C substitutions at indicated positions at 1000-fold excess. Adapted with permission from Donaldson et al. (2011). (F) Binding of 32P-labeled RsmZ small RNA by E. coli T7Iq lysates (diluted 1:8) overexpressing His-CsrA (V. cholerae) in the presence of 10 μg/mL total Yeast RNA (Saccharomyces cerevisiae) or 400 nM unlabeled small RNAs RsmY and RsmZ (P. aeruginosa) competitors. Taken with permission from Patel et al. (2015).
Materials
Radiolabeled nucleic acid (>3,000 PSL per hour exposure to phosphorimager screen)
Purified investigational protein
5–10 X binding buffer (For our examples, the 10x binding buffer consists of 100 mM Tris pH 8.0, 1 M NaCl, 50 mM MgCl2. The specific buffer choice will depend on the protein under investigation.)
Unlabeled competitor (for final concentration >20x protein concentration)
Nuclease-free double deionized water (ddH2O)
Appropriate nuclease free disposable plasticware for the number of samples (Microcentrifuge tubes, PCR strip tubes, PCR plates, or 96-well round bottom plates)
Pipette and pipette tips or pin tools (V&P Scientific) (For pin tools, a container with 0.01% Tween in ddH2O is required for cleaning between sample applications)
Nitrocellulose membrane with 0.45 μm pores
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
Prepare 10 μL of 2x concentration of competitor mix containing the unlabeled nucleic acid (final concentration is greater than 20x of protein concentration) and radiolabel ligand mix for each protein sample to be tested. Mix unlabeled competitor, radiolabeled ligand, 10x binding buffer and ddH2O. Place each competitor mix into a nuclease free disposable tube or assay plate.
- Cut nitrocellulose membrane to the appropriate lengthFor 2 μL spots, the area of the spot is less than 1 cm × 1 cm. For triplicates, triple the area of the nitrocellulose.
Transfer 10 μL of purified protein at a final concentration two times higher than Kd (determined using Basic Protocol 2) into each ligand mix from step 1.
Mix by either pipetting at least 5 times with a pipette set to 10 μL volume or, if using a 96-well plate, place on a plate shaker at setting 5 for 30 sec.
- Pipette 2 μl onto dry nitrocellulose placed on top of the wax paper backing.If using a pin tool rather than a pipette, wash pin tool with 0.01% Tween 20 three times by submerging into wash buffer and wicking dry on clean paper towel. After cleaning, place pin tool in sample and press down vertically on nitrocellulose without dragging pins sideways. Hold on nitrocellulose for at least 10 seconds for the entire mixture to move into the membrane by capillary action.
Repeat the spotting twice more to obtain three replicates.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap nitrocellulose membrane with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization respectively.This process is easiest with appropriate positive (no competitor) control.
Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).
Calculate the fraction bound for each spot using formula given in Fig 1B.
Specificity of competition can be determined by analyzing the triplicate data between no competitor negative control and samples treated with unlabeled competitors using an unpaired two-tailed t test.
BASIC PROTOCOL 4
Determination of dissociation rate (koff) by DRaCALA
An important parameter for appreciating the duration of protein-nucleic acid interactions in the cell is to understand the rate of complex formation and dissociation. Most complexes with affinities in the micromolar or sub-micromolar range have off rates with sufficient time intervals that allow analysis by DRaCALA. Since koff and Kd is related to the on rate (kon) by the equation
| Kd = koff / kon |
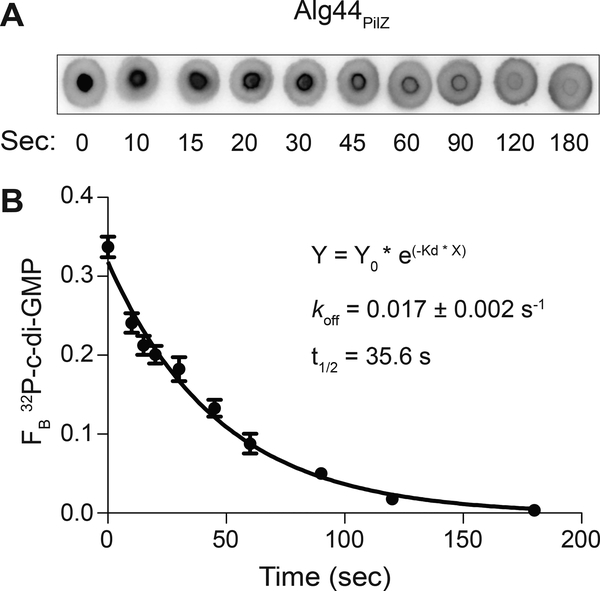
DRaCALA can be used to estimate the kon as well. To measure koff, the protein is first bound to the radiolabeled ligand. Subsequently, a specific unlabeled competitor is added in vast excess (>100x of protein concentration) and the reaction mixture is assayed by DRaCALA at specific time intervals. In the case of Alg44 interaction with c-di-GMP, the time course is complete by 180 seconds (Figure 5) (Roelofs et al., 2011). The specific timing for each interaction under study will need to be empirically determined.
Figure 5.
Determination of dissociation rate (koff) by DRaCALA.
(A) Representative DRaCALA images of protein-ligand spots taken at various time points after initiation of competition. The competition reaction was initiated by addition of 1 mM of unlabeled c-di-GMP to a mixture of 32P-c-di-GMP and His-MBP-Alg44PilZ. (B) FB of 32P-c-di-GMP is plotted against time after the addition of unlabeled competitor. koff is calculated by nonlinear regression analysis fit to a one phase exponential decay. Adapted with permission from Roelofs et al. (2011).
Materials
Radiolabeled nucleic acid (>3,000 PSL per hour exposure to phosphorimager screen)
Purified investigational protein
5–10 X binding buffer (For our examples, the 10x binding buffer consists of 100 mM Tris pH 8.0, 1 M NaCl, 50 mM MgCl2. The specific buffer choice will depend on the protein under investigation.)
Nuclease-free double deionized water (ddH2O)
Unlabeled specific competitor (at a sufficient concentration for a final concentration ≥100x protein concentration in the assay)
Timer
Nuclease free disposable microcentrifuge tubes
Pipette and pipette tips
Nitrocellulose membrane with 0.45 μm pores
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
Prepare 18 μL of 2x concentrated radiolabeled ligand mix. Mix radiolabeled ligand, 10x binding buffer and ddH2O. Place 18 μl of the 2x radiolabel ligand mix to a microcentrifuge tube.
- Cut nitrocellulose membrane to the appropriate length.For 2 μL spots, the area of the spot is less than 1 cm × 1 cm. For triplicate, triple the area of the nitrocellulose.
Transfer 20 μL of purified protein at a final concentration two times higher than Kd (determined by Basic Protocol 2) into sample to allow protein and radiolabeled ligand to bind.
Add 2 μL of the unlabeled specific competitor at a sufficient concentration so that the final concentration is >100 fold higher than the protein concentration. Mix immediately 5x using a pipette set to 20 μL.
Start timer in order to track the time elapse upon ligand addition.
- Remove 2 μl of the mixture and spot on nitrocellulose at regular time intervals.Time intervals depend on the affinity of the interaction. The higher the Kd, indicative of weaker interactions, requires shorter sampling times. For Kd ~1 μM, sampling at 15, 30, 60, 120, 300 seconds is a recommended starting point. Adjust accordingly.
Repeat preparation of the sample and time course to obtain triplicate data.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap nitrocellulose membrane with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager.
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization respectively.This process is easiest with appropriate positive (time 0) controls
Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).
Calculate the fraction bound for each spot using formula indicated in Fig 1B.
The koff can be determined using nonlinear regression curve fit with one phase exponential decay (GraphPad Prism software).
BASIC PROTOCOL 5
Screening for small molecule inhibitors by DRaCALA
DRaCALA is easily scalable for high-throughput applications such as screening for small molecule inhibitors of protein-nucleic acid interactions. Interactions that have been characterized using Protocol 1–3 are suitable for screening. To perform the screen, several pieces of equipment are required including a dedicated liquid dispenser (for radioactivity) and an appropriately formatted pin tool. The assay is performed by addition of the protein to the chemical library followed by the subsequent addition of the radiolabeled ligand (Lieberman, Orr, Wang, & Lee, 2014). A reduction in the FB is indicative of lead compounds in the chemical library that prevent binding of the protein to the nucleic acid. An example of a screen for small molecule inhibitors of Pseudomonas aeruginosa Alg44 binding to 32P-c-di-GMP is depicted in Figure 6 (Zhou et al., 2017). The use of positive and negative controls in every plate provides a measure of assay quality and reproducibility. Since most compounds are inactive, identification of compounds that reduce the FB more than 2 or 3 standard deviation from the average of the plate are considered as lead compounds for follow-up studies. When using 384-well plates, the corresponding pin tool produces 0.5 μL spots. While this reduced volume is sufficient for detection, the small areas can lead to spotting errors. These errors can be easily detected by visual inspection of the DRaCALA images.
Before conducting the screen on compound library, the Z-factor should be determined for a specific concentration of protein and unlabeled known competitor. By assaying a 384 well plate with interspaced positive and negative controls such that there are two columns of protein with no competitor with two columns of protein with the known specific competitor. Using the data collected, one can determine the mean and standard deviation of the positive and negative controls to calculate the Z-factor. The conditions are considered appropriate for high-throughput screening if the Z-factor is > 0.5.
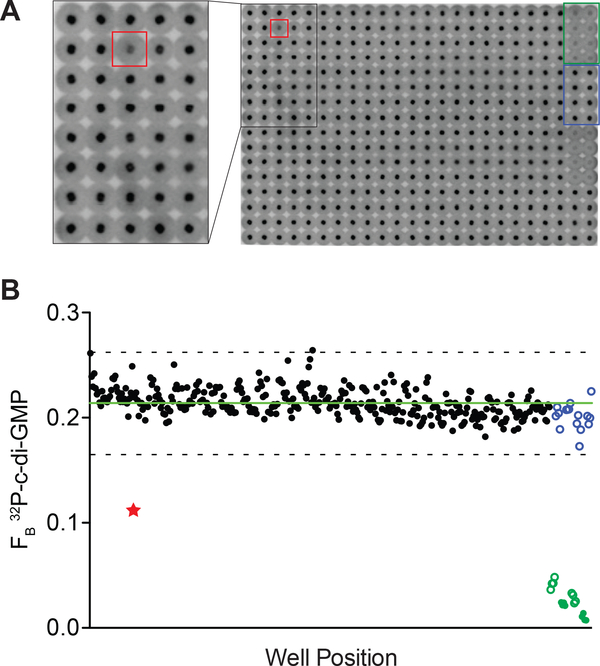
Figure 6.
Identification of a chemical inhibitor of Alg44PilZ binding to c-di-GMP.
(A) Representative image of DRaCALA spots from one screening plate. Spot boxed in red corresponds to a well that contained a compound that inhibited His-MBP-Alg44PilZ binding to 32P-c-di-GMP. Spots boxed in green and blue correspond to wells containing 5 μM unlabeled c-di-GMP and DMSO, respectively. Inset shows a zoomed image of the indicated area. (B) FB is plotted in order of position for the entire 384-well plate. Red star corresponds to the red-boxed well in (A). Blue circles correspond to wells containing DMSO solvent only. Green open and closed circles correspond to wells containing 5 μM and 10 μM unlabeled c-di-GMP, respectively. Green line represents the average FB of the wells containing test compounds. Dashed line represents the 3 standard deviation cutoff for classification of hit compounds. The compound identified from screening this plate was AC1LFHI9 (HI9), a derivative of thiol-benzo-triazolo-quinazolinone. Adapted with permission from Zhou et al. (2017).
Materials
Radiolabeled ligand of interest
Purified protein of interest
Appropriate buffer
Unlabeled known competitor (for final concentration >20x protein concentration)
Chemical library in 384-well plate format resuspended in appropriate buffer
Nuclease-free double deionized water (ddH2O)
Multiwell liquid dispenser
384-well Pin tool
0.01% Tween in ddH2O (for cleaning pin tool)
Nitrocellulose Membrane (0.45 μm pores)
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
- Ensure the compounds in the chemical library are at the bottoms of their respective wells by centrifuging for 1 – 2 minutes at 3,220 g. This will ensure the protein-ligand mixture comes into contact with the same amount of compound in each well.This protocol is based on a 20 μL assay volume in each well in 384-well plates. Adjust proportionally for different starting volumes. Ideally, 16–32 wells are left blank for positive and negative controls (see step 2). It is recommended that the assay be carried out in duplicate or triplicate.
- Add 20 μL of the control assays to each plate of the chemical library. For a negative competition control, use either buffer or DMSO instead of competitor. For a positive competition control, use an unlabeled version of the ligand of interest at a concentration that is sufficient for competition.Ideally, add 8–16 wells of each control per plate if sufficient number of empty wells are available.
- Prepare protein by diluting in appropriate buffer to a concentration that is 4x above Kd as determined by Basic Protocol 2.For each 384-well plate, prepare 5 mL of the diluted protein mixture. For experiments with less than 5 plates, preparation of an additional 5 mL is required for priming the dispenser.
- Prepare radiolabeled ligand mixture by diluting the ligand such that an overnight exposure results in >3,000 PSL.For each 384-well plate, prepare 5 mL of the diluted radiolabeled ligand. For experiments with less than 5 plates, preparation of an additional 5 mL is required for priming the dispenser.
- Using an automated dispenser such as the BioTek MultiFlo, add 10 μL diluted protein to each well and shake using a plate shaker at setting 5 for 2–5 minutes.It is important to use an automated dispenser for large-scale DRaCALA applications to minimize any potential timing effects caused by dispensing reagents into 384-wells.
Clean the automated dispenser with ddH2O.
Using an automated dispenser such as the BioTek MultiFlo, add 10 μL diluted radiolabeled ligand mixture to each well and shake using a plate shaker at setting 5 for 2 minutes.
Clean the automated dispenser with ddH2O.
Clean the 384-well pin tool by washing in a 0.01% detergent solution like Tween.
- Cut nitrocellulose membrane to the appropriate dimensions.For standard 384-well plates, the area of the nitrocellulose needed is 8 cm × 12 cm. It is recommended that the assay be performed in duplicate, so 16 cm × 12 cm of nitrocellulose is required per plate.
Align the pin tool with wells and carefully insert pins into the plate.
Stamp the pin tool onto the nitrocellulose in duplicate. Re-wash the pin tool when changing the plates to be stamped.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap nitrocellulose membrane with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager.
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization respectively.This process is easiest with appropriate positive and negative controls.
Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).
Calculate the FB for each spot using formula given in Fig 1B.
- To identify compounds that resulted in inhibition of ligand binding, determine the average and standard deviation of the FB for the library for all of the test compounds in the plate. A minimal threshold for a competitor is set 2 standard deviations below the average FB for the library. For more stringency, use 3 standard deviations below the average. The positive competitor control (unlabeled vs. radiolabeled ligand of interest) should be used as a reference.Keep in mind that spotting errors can result in false positive with an artificially high FB. These can be removed by visual identification.
Any hits should be independently re-tested by DRaCALAs. Validation can be performed on compounds that are obtained from an independent source.
BASIC PROTOCOL 6
Screening for novel binding proteins by DRaCALA
DRaCALA can be adapted for screening for proteins that can interact with a specific radiolabeled nucleic acid. Successful DRaCALA screens of this kind have utilized an arrayed library of lysates expressing individual genes. For genomes in which all open reading frames (ORFs) have been cloned, a screen of the ORF library (ORFeome) allows for systematic identification of proteins encoded within a genome that interact with a particular nucleic acid. This approach has been successfully applied to identify c-di-GMP binding proteins from E. coli ORFeome (Fang et al., 2014), c-di-AMP binding proteins from the Staphylococcus aureus ORFeome (Corrigan et al., 2013), c-di-GMP-binding proteins (Roelofs et al., 2015) and pGpG-binding proteins (Orr et al., 2015) from the Vibrio cholerae ORFeome. The approach is not limited to entire ORFeomes. Smaller sets of proteins or protein fragments can be screened for binding (Roelofs et al., 2015). In addition, genes subjected to site-directed or random mutagenesis can be expressed individually and screened for altered binding activity (Roelofs et al., 2015).
Prior to using this protocol, the cell lysates, each containing one overexpressed protein, need to be arrayed in a 96-well format. The particular example presented in this protocol utilizes E. coli cells and the T7 polymerase system to overexpress proteins (Orr & Lee, 2017). A screen conducted to identify protein binding partners of pAp from the V. cholerae ORFeome is depicted in Figure 7. The protocol describes the steps to perform a DRaCALA-based screen on the prepared ORFeome lysates.
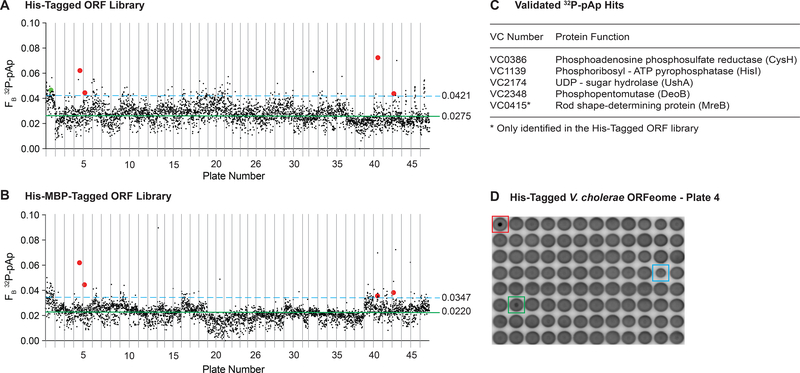
Figure 7.
A high-throughput DRaCALA screen of the V. cholerae ORF library identifies potential binding proteins for pAp.
3,866 His-tagged (A) or His-MBP-tagged (B) ORFs were assessed for binding to 2.5 nM 32P-pAp in the presence of 500 μM cold ATP competitor. The ORFs are arranged by ascending plate number from the BEI V. cholerae Gateway Clone Set. Plates 6 & 42, 13 & 25, and 31 & 36 are combined. The x-axes are modified to represent the absence of a complete plate 25. The average fraction bound for each library (solid green line) and 2 standard deviation above the mean (dashed blue line) are indicated. Red dots represent hits identified in both libraries and are validated. Green dot represents a hit only in the His-tag library and is validated. Black dots represent either wells below the 2 SD fraction bound cutoff, or those which were not validated. Validated hits appear in color. (C) A list of the validated 32P-pAp binding proteins with annotated functions. (D) A representative plate from the V. cholerae 32P-pAp screen. Red box indicates the purified NrnA control. Green box indicates a validated hit (VC2348). Blue box indicates a spotting error, which can cause a high fraction bound.
Materials
Radiolabeled ligand of interest
Library of cell lysates expressing different proteins in 96-well round bottom plates
Lysate of a positive control protein
Appropriate buffer
Nuclease-free double deionized water (ddH2O)
Multiwell liquid dispenser
96-well Pin tool
0.01% Tween in ddH2O (for cleaning pin tool)
Nitrocellulose Membrane (0.45 μm pores)
Plastic cling wrap
Phosphorimager screen or film with appropriate cassette
Phosphorimager or densitometer with quantification software (Fujifilm Multi Gauge software v3.0)
- Centrifuge the cell lysate library plates for 1 – 2 minutes at 3,220 g to ensure the lysate is at the bottom of the well.This protocol is based on a total 40 μL assay volume in each well in a 96-well plate. Adjust proportionally for different starting volumes.
- Add one negative and positive control to each plate. For a negative competition control, use 20 μL of lysate containing the vector control. For a positive competition control, use 20 μL of a lysate expressing a known positive binding protein.If there are no empty wells in the plates, use a separate plate for controls.
- Prepare radiolabeled ligand mixture by diluting the ligand such that an overnight exposure results in >3,000 PSL.For each 96-well plate, prepare 3 mL of the diluted radiolabeled ligand. For experiments with less than 5 plates, preparation of an additional 3 mL is required for priming the dispenser.
Using an automated dispenser such as the BioTek MultiFlo, add 20 μL diluted radiolabeled ligand mixture to each well and shake on a plate shaker at setting 5 for 1 minute.
Clean the automated dispenser with ddH2O.
Clean the 96-well pin tool by washing in a 0.01% detergent solution like Tween.
- Cut nitrocellulose membrane to the appropriate dimensions.For standard 96-well plates, the area of the nitrocellulose needed is 8 cm × 12 cm. It is recommended that the assay be performed in duplicate, so 16 cm × 12 cm of nitrocellulose is required per plate.
Align the pin tool with wells and carefully insert pins into the plate.
Stamp the pin tool onto the nitrocellulose in duplicate. Re-wash the pin tool when changing plates to be stamped.
Allow spots to dry for 5–10 minutes depending on temperature and humidity.
- Wrap the nitrocellulose membrane with plastic wrap.Ensure that wrapped membrane has no wrinkles which can cause distortions when exposing to phosphorimager screen.
Expose the phosphorimager screen for the appropriate time such that the intensity is greater than 3,000 PSL.
Scan using phosphorimager.
- Using the phosphorimager software, measure areas of inner and outer circles of the spots which correspond to the site of the application and the zone of mobilization respectively.This process is easiest with appropriate positive and negative controls.
Obtain numerical values for the areas and intensities of the zone of application (inner circle) and zone of migration (outer circle).
Calculate the fraction bound for each spot using the formula indicated in Fig 1B.
- To identify positive binding proteins, determine the average and standard deviation of the FB for the library for all of the test lysates in the plate. A minimal threshold for a binding protein is set 2 standard deviations above the average FB for the library. For more stringency, use 3 standard deviations above the average. The positive control (known binding protein vs. buffer control) should be used as a reference.Keep in mind that spotting errors can result in false positive with an artificially high FB. These can be removed by visual identification.
Any hits should be independently re-tested by DRaCALAs. Validation should be performed on ORFs that are identified as positive by purifying the protein and performing Basic Protocol 1–4.
REAGENTS AND SOLUTIONS
Unique reagents and solutions are required for specific proteins and nucleic acid combinations. The default buffer solution has been listed with each protocol. Proteins and lysates are stored at −80 ˚C. Radiolabeled and unlabeled nucleic acids are stored at −20 ˚C. All other solutions can be kept at room temperature.
COMMENTARY
Background Information
Protein interactions with nucleic acids are central to living processes and are required for the synthesis of macromolecules, including DNA, RNA, and proteins. Protein interactions with DNA are mediated by both sequence-specific interactions and interactions with the negative charge of the phospho-deoxyribose backbone. Protein interactions with RNA depend on the RNA primary sequence, secondary structures, and tertiary folds. In addition to interactions with macromolecular nucleic acids, proteins often bind to short nucleotides (typically less than three bases). Enzymes often utilize mononucleotide triphosphates to energize reactions. Besides mononucleotide triphosphates, proteins can be allosterically regulated by a number of secondary signaling nucleotide derivatives such as cyclic AMP (cAMP) (Emmer, deCrombrugghe, Pastan, & Perlman, 1970; Zubay, Schwartz, & Beckwith, 1970), cyclic GMP (cGMP) (Cobbs, Barkdoll, & Pugh, 1985; Erneux et al., 1981; Haynes & Yau, 1985; Kuo & Greengard, 1970; Nakamura & Gold, 1987), ppGpp (Cashel, 1969; Paul et al., 2004) (W. Ross et al., 2013), cyclic-di-GMP (c-di-GMP) (P. Ross et al., 1987), and many others. These secondary signaling nucleotides regulate diverse biological functions from bacteria to multicellular eukaryotes. Thus, protein interaction with nucleic acids is an important area of research. The ability to qualitatively and quantitatively assess these interactions is important for understanding fundamental biological processes.
A number of experimental approaches have been developed to observe and measure protein-nucleic acid interactions in vitro (Connors, 1987) and in vivo. The in vitro methods are described briefly below in order to compare and contrast them with DRaCALA. The methods differ by the choice of ligand: unmodified unlabeled ligand, unmodified radiolabeled ligand, fluorescently modified ligand, and affinity tagged ligand. There are benefits and drawbacks to measuring protein-ligand interactions with each type of ligand. Unmodified unlabeled ligands are in their biologically relevant forms and their interactions with proteins presumably best represent biological interactions in cells. However, these direct interactions require dedicated equipment that detects changes in heat (isothermal titration calorimetry) or association to surfaces (surface plasmon resonance). Unmodified radiolabeled ligands have the same benefit as unlabeled ligands in that they retain their natural chemical properties as their unlabeled counterparts. Radiolabeled ligands provide convenience for detection, but because of radioactivity, they require special handling and safety monitoring. Fluorescently modified ligands are also easy to detect; however, covalently modified ligands, particularly the low molecular weight second messenger nucleotides, can lead to altered binding properties since the fluorophore is often similar in molecular weight to the ligand. Due to space limitation, we provide only a brief summary of the most common biochemical methods used, including isothermal titration calorimetry (ITC) (Sturtevant, 1974), surface plasmon resonance (SPR) (Alfthan, 1998), filter binding (Nirenberg & Leder, 1964), equilibrium dialysis, electromobility shift assay (EMSA) (Fried & Crothers, 1981) and fluorescence anisotropy (Brand & Johnson, 2008; Lakowicz, 2006). The use of affinity tagged ligands to pull out binding proteins for subsequent MS/MS identification has been discussed elsewhere (Corrigan et al., 2013; Nesper, Reinders, Glatter, Schmidt, & Jenal, 2012; Sureka et al., 2014).
Unlabeled, unmodified ligands can be detected by biophysical changes that occur when the proteins interact with nucleic acids. One such readout is the change in heat upon physical interaction between proteins and nucleic acids, which can be detected by ITC. Serial addition of a known concentration of ligand to a known concentration of protein allows for calculation of binding affinity, stoichiometry of binding, and enthalpy of binding. While powerful, ITC is sensitive to subtle differences in the buffers used for proteins and nucleic acids (Duff, Grubbs, & Howell, 2011; Wiseman, Williston, Brandts, & Lin, 1989). Furthermore, ITC typically requires a relatively large amount of protein and ligand for assaying their interactions. Another technique based on physical changes is Surface Plasmon Resonance (SPR). SPR measures plasmons generated when incident light hits a surface coated with a metal film. Since this property is sensitive to local changes in the metal film, SPR can detect differences between unbound proteins and protein bound to ligands (in this case, nucleic acids). SPR can measure binding affinity and kinetics (Frostell, Vinterback, & Sjobom, 2013). The signal is typically proportional to the molecular weight of the attached protein and the ligand. For larger nucleic acids, SPR can generate a sufficient signal to noise ratio. However, for smaller nucleotides, the SPR signal can become vanishingly small. To bypass this problem, the nucleic acid can be fixed on the surface; however, this typically requires modified nucleic acids. Both ITC and SPR require specialized instruments and are difficult to scale for high-throughput uses.
The use of labeled ligands permits many additional modes of detection. Nucleic acids can be labeled using two main methodologies. One method is to incorporate radioactive isotopes directly into the nucleic acid of interest (see below for considerations for generation of radiolabeled ligands). Radiolabeled molecules typically have indistinguishable properties from their biological counterparts, but their use is complicated by safety concerns. A second method is to covalently modify nucleic acids with fluorophores or chromophores. These reagents can be easily obtained from any of the commercial sources that synthesize oligonucleotides. For DNA methods, a 5’ end labeled primer can be used in a polymerase chain reaction (PCR) to amplify any sequence, thereby generating a labeled probe. For RNA, the entire molecule would have to be synthesized with the label.
Radiolabeled molecules can be detected using a number of assays including filter binding, equilibrium dialysis, EMSA (electromobility shift assay) and DRaCALA. Each of the assays have particular strengths depending on the specific application. Filter binding takes advantage of the ability of nitrocellulose membrane to bind proteins, but not nucleic acids. When a mixture of protein and nucleic acid is passed through the membrane, only the protein will be retained; however, if the protein binds to the labeled nucleic acid, the radiolabeled nucleic acid will be also retained on the membrane. To prevent non-specific retention of the labeled nucleic acid, the nitrocellulose membrane is typically washed. Once dried, the amount of radioactivity on the filter can be measured by scintillation counting or exposure to phosphorimager screens or films. While filter binding is scalable (a variety of dot blot apparatuses are available), there are a few issues that complicate this assay. First, collection of all of the radiolabeled ligand in the bound and unbound fraction is difficult. Second, the wash step introduces time in which weakly bound ligands can dissociate. Third, there are technical issues with vacuuming devices that arise from air bubbles trapped in the device.
Equilibrium dialysis measures the movement of a labeled ligand between two chambers: one containing the binding protein and one with buffer only. The device utilizes a semipermeable membrane that typically occludes molecules of certain molecular weight from moving between the two chambers. For this reason, equilibrium dialysis is mostly suited for protein interaction with low molecular weight signaling nucleotides. Once the sample reaches equilibrium, the amount of radioactivity is measured from each chamber. When there is no binding interaction, the ratio of signal of the protein chamber over the buffer control is one. A ratio greater than one implies binding, and the magnitude of the value is indicative of the affinity between the protein and ligand. While equilibrium dialysis is technically a straightforward assay, it is difficult to adapt for high-throughput applications.
A third assay is the electromobility shift assay (EMSA). In EMSA, the unbound nucleic acid is separated from the protein-nucleic acid complex by running the complex through a polyacrylamide gel. While this technique is also straightforward, there are a few issues due to the passage of the sample through the gel. Since separation by electrophoresis takes time, the protein-nucleic acid complex can separate, leaving a smearing of the nucleic acid on the gel. For small signaling nucleotides, there is a possibility of the free nucleotide running at the dye front and even off the gel. The advantage of EMSA is that the difference in size between the nucleic acid and the protein-nucleic acid complex can be easily resolved.
Interactions between fluorescently labeled nucleic acids and proteins can be detected using a number of assays. Fluorescently labeled nucleic acids can be used in equilibrium dialysis and EMSA in place of a radiolabeled ligand. In addition, fluorescently labeled nucleic acids can be detected by fluorescence anisotropy. The principle of fluorescence anisotropy is based on the ability of the fluorophore to absorb and fluoresce in solution. A free fluorescently labeled nucleic acid that has a lower molecular weight will rotate faster and have a smaller anisotropy. When bound to a larger molecular weight protein, the time required for rotation will be increased and thus allow more fluorescence in response to polarized light. Fluorescence anisotropy allows for detection of both binding affinity and changes in the size and shape of the labeled nucleic acid. Fluorescence anisotropy is particularly useful for high throughput identification of small molecule inhibitors that interfere with protein nucleic acid interactions. While powerful, fluorescently labeled molecules can take on different chemical properties imparted by a covalently linked fluorophore. This issue is particularly significant as the size of the nucleic acid of interest is reduced.
While all of the above assays are valid methods for detecting protein-nucleic acid interactions, DRaCALA provides an easy method to rapidly characterize these interactions. In addition, DRaCALA is amenable for high-throughput screens for small molecule inhibitors as well as novel binding proteins. On the other hand, DRaCALA requires the use of radio-labeled ligands, so proper radiation safety techniques need to be followed.
Critical Parameters
Notes have been provided throughout Basic Protocols 1–4 for specific issues that may arise. High-throughput screens described in Basic Protocol 5 and 6 have issues associated with processing large number of samples. In particular, care should be taken to prevent air bubbles from being present in the lysate, the pin tool wash solution, or the pin tool. The presence of air bubbles will interfere with capillary action.
Troubleshooting
If no binding is observed for a positive control, verify the protein concentration is above Kd.
For Basic Protocol 6, a result in which all wells show binding suggests that there are problems with the underlying conditions used for the screen. Check that the correct concentration of ligand and lysate are used. For some ligands (for example ATP), cell lysates will have an abundance of proteins that bind the ligand. Such ligands are not compatible for screening of cell lysates.
If possible, purified proteins should be stored in the binding buffer to reduce artifacts that arise when combining different buffers.
Highly concentrated protein samples create viscous mixtures that may lead to deformed spots and hinder analysis by DRaCALA. Dilution of the protein will resolve these irregular migration patterns.
Detergents in the binding buffer can cause irregular migration patterns. Reduce detergent concentration to resolve these issues.
Nitrocellulose must be kept dry. Prolonged exposure of nitrocellulose to humid air will lead to non-uniform migration patterns.
Enzymes that can degrade the nucleic acid will lead to false negative results. If hydrolysis is suspected, try using buffer without a divalent cation or a different divalent cation that does not promote enzymatic activity (for example, switching Mg++ to Ca++).
Statistical Analyses
Appropriate statistical analyses have been indicated for each protocol.
Understanding Results
Calculating the Kd requires a known initial concentration of the protein stock. This can be determined by a number of assays. A recommended method for calculating protein concentration for proteins that are >90% pure is to use Beer’s law using absorbance measured at 280 nm (Abs280) and the extinction coefficient (ε) calculated from the protein sequence using the online ProtParam tool.
Time Considerations
Time required to perform DRaCALA consists of mixing reagents, applying mixtures onto nitrocellulose, capillary action time (~10–20 seconds) and drying time (5–10 minutes). So, while the time for executing the experiment is short, preparation of all of the protein samples, labeled nucleic acids, reagents, and equipment requires additional time and is of upmost importance. Plan for additional time for exposing the radioactivity on phosphorimager screen and/or film. Depending on the specific activity and amount of radiolabeled ligand used, prolonged exposure times may be required (up to 48 hours).
ACKNOWLEDGEMENTS
NIH AI110740, NIH AI133670
LITERATURE CITED
- Alfthan K (1998). Surface plasmon resonance biosensors as a tool in antibody engineering. Biosens Bioelectron, 13(6), 653–663. doi:S0956-5663(98)00020-7 [pii] [DOI] [PubMed] [Google Scholar]
- Brand L, & Johnson ML (2008). Preface. Methods in Enzymology (fluorescence 2008). Methods Enzymol, 450, xv–xvi. doi: 10.1016/S0076-6879(08)03419-8 [DOI] [PubMed] [Google Scholar]
- Cashel M (1969). The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem, 244(12), 3133–3141. [PubMed] [Google Scholar]
- Cobbs WH, Barkdoll AE 3rd, & Pugh EN Jr. (1985). Cyclic GMP increases photocurrent and light sensitivity of retinal cones. Nature, 317(6032), 64–66. [DOI] [PubMed] [Google Scholar]
- Connors KA (1987). Binding constants : the measurement of molecular complex stability. New York: Wiley. [Google Scholar]
- Corrigan RM, Bellows LE, Wood A, & Gründling A (2016). ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci U S A, 113(12), E1710–1719. doi: 10.1073/pnas.1522179113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, & Gründling A (2013). Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A, 110(22), 9084–9089. doi:1300595110 [pii] 10.1073/pnas.1300595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Roelofs KG, Luo Y, Sintim HO, & Lee VT (2011). A rapid assay for affinity and kinetics of molecular interactions with nucleic acids. Nucleic Acids Res. doi:gkr1299 [pii] 10.1093/nar/gkr1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff MR Jr., Grubbs J, & Howell EE (2011). Isothermal titration calorimetry for measuring macromolecule-ligand affinity. J Vis Exp(55). doi: 10.3791/2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer M, deCrombrugghe B, Pastan I, & Perlman R (1970). Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc Natl Acad Sci U S A, 66(2), 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erneux C, Couchie D, Dumont JE, Baraniak J, Stec WJ, Abbad EG, . . . Jastorff B (1981). Specificity of cyclic GMP activation of a multi-substrate cyclic nucleotide phosphodiesterase from rat liver. Eur J Biochem, 115(3), 503–510. [DOI] [PubMed] [Google Scholar]
- Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, . . . Gomelsky M (2014). GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol, 93(3), 439–452. doi: 10.1111/mmi.12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M, & Crothers DM (1981). Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res, 9(23), 6505–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostell A, Vinterback L, & Sjobom H (2013). Protein-Ligand Interactions Using SPR Systems. Methods Mol Biol, 1008, 139–165. doi: 10.1007/978-1-62703-398-5_6 [DOI] [PubMed] [Google Scholar]
- Haynes L, & Yau KW (1985). Cyclic GMP-sensitive conductance in outer segment membrane of catfish cones. Nature, 317(6032), 61–64. [DOI] [PubMed] [Google Scholar]
- Kuo JF, & Greengard P (1970). Cyclic nucleotide-dependent protein kinases. VI. Isolation and partial purification of a protein kinase activated by guanosine 3’,5’-monophosphate. J Biol Chem, 245(10), 2493–2498. [PubMed] [Google Scholar]
- Lakowicz JR (2006). Principles of Fluorescence Spectroscopy: Springer; US. [Google Scholar]
- Lieberman OJ, Orr MW, Wang Y, & Lee VT (2014). High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem. Biol, 9(1), 183–192. doi: 10.1021/cb400485k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, & Gold GH (1987). A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature, 325(6103), 442–444. doi: 10.1038/325442a0 [DOI] [PubMed] [Google Scholar]
- Nesper J, Reinders A, Glatter T, Schmidt A, & Jenal U (2012). A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics, 75(15), 4874–4878. doi: 10.1016/j.jprot.2012.05.033 [DOI] [PubMed] [Google Scholar]
- Nirenberg M, & Leder P (1964). Rna Codewords and Protein Synthesis. The Effect of Trinucleotides Upon the Binding of Srna to Ribosomes. Science, 145, 1399–1407. [DOI] [PubMed] [Google Scholar]
- Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, & Lee VT (2015). Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A, 112(36), E5048–5057. doi: 10.1073/pnas.1507245112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, & Lee VT (2017). Differential Radial Capillary Action of Ligand Assay (DRaCALA) for High-Throughput Detection of Protein-Metabolite Interactions in Bacteria. Methods Mol Biol, 1535, 25–41. doi: 10.1007/978-1-4939-6673-8_3 [DOI] [PubMed] [Google Scholar]
- Patel DK, Gebbie MP, & Lee VT (2014). Assessing RNA interactions with proteins by DRaCALA. Methods Enzymol, 549, 489–512. doi: 10.1016/B978-0-12-801122-5.00021-0 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, & Gourse RL (2004). DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell, 118(3), 311–322. doi: 10.1016/j.cell.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, . . . Lee VT (2015). Systematic Identification of Cyclic-di-GMP Binding Proteins in Vibrio cholerae Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS Pathog ., 11(10), e1005232. doi: 10.1371/journal.ppat.1005232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, & Lee VT (2011). Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U. S. A, 108(37), 15528–15533. doi:1018949108 [pii] 10.1073/pnas.1018949108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Ohana P, Mayer R, . . . Benziman M (1987). Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature, 325, 279–281. [DOI] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, & Gourse RL (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell, 50(3), 420–429. doi: 10.1016/j.molcel.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant JM (1974). Some applications of calorimetry in biochemistry and biology. Annu Rev Biophys Bioeng, 3(0), 35–51. doi: 10.1146/annurev.bb.03.060174.000343 [DOI] [PubMed] [Google Scholar]
- Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, . . . Woodward JJ (2014). The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell, 158(6), 1389–1401. doi: 10.1016/j.cell.2014.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, & Lin LN (1989). Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem, 179(1), 131–137. doi:0003-2697(89)90213-3 [pii] [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zbornikova E, Rejman D, & Gerdes K (2018). Novel (p)ppGpp Binding and Metabolizing Proteins of Escherichia coli. MBio, 9(2). doi: 10.1128/mBio.02188-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou E, Seminara AB, Kim SK, Hall CL, Wang Y, & Lee VT (2017). Thiol-benzo-triazolo-quinazolinone Inhibits Alg44 Binding to c-di-GMP and Reduces Alginate Production by Pseudomonas aeruginosa. ACS Chem Biol, 12(12), 3076–3085. doi: 10.1021/acschembio.7b00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G, Schwartz D, & Beckwith J (1970). Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci U S A, 66(1), 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]