Abstract
Determining the genomic location of DNA binding proteins is essential to understand the function of these factors. Cleavage Under Targets and Release Using Nuclease (CUT&RUN) is a powerful method for mapping protein-DNA interactions in high resolution. In CUT&RUN, recombinant protein A-microccocal nuclease (pA-MN) is recruited by an antibody targeting the chromatin protein of interest, which can be performed on uncrosslinked or crosslinked cells. DNA fragments near sites of antibody binding are released from insoluble bulk chromatin through endonucleolytic cleavage and used to build barcoded DNA sequencing libraries that can be sequenced in pools of at least 30. Therefore, CUT&RUN provides an alternative to ChIP-seq approaches for mapping of chromatin proteins that typically exhibits higher signal-to-noise, using fewer cells, at a lower cost. Here, we describe the method for performing CUT&RUN, generating DNA sequencing libraries, and analyzing the datasets.
Keywords: CUT&RUN, proteinA-MNase, chromatin, transcription factor
INTRODUCTION
Genome-wide profiling of DNA binding proteins is widely used to determine their targets and functions. Chromatin immunoprecipitation (ChIP; Unit 21.19(Raha, Hong et al. 2010)) techniques have been used to map the location of chromatin bound proteins for over three decades (Gilmour 1984, Gilmour 1985, Johnson 2007). Traditional ChIP protocols use formaldehyde crosslinking to fix protein-DNA interactions followed by non-specific shearing of the crosslinked chromatin by sonication. A protein is then immunoprecipitated with an antibody specific for a protein of interest coupled to a magnetic or agarose bead via protein A, protein G, or a secondary antibody. Cleavage Under Targets and Release Using Nuclease (CUT&RUN) is a genome-wide derivative of ChIC (Schmid, Durussel et al. 2004) developed by Steven Henikoff and colleagues that maps native protein-DNA interactions in high resolution (Skene and Henikoff 2017). The CUT&RUN method is diagrammed in Figure 1. Similar to ChIP, chromatin bound proteins are identified using a primary antibody directed against the protein of interest. However, CUT&RUN does not require crosslinking or sonication (although crosslinking can be used (Skene, Henikoff et al. 2018)). Rather, CUT&RUN utilizes a recombinant proteinA-microccocal nuclease (MNase) to specifically digest DNA fragments surrounding the protein of interest. DNA fragments underlying the specific chromatin bound protein of interest are released from bulk chromatin with high specificity due to the recruitment of protein A-MNase to the genomic locations of chromatin proteins through protein A-antibody interactions. Because non-specific association of protein A-MNase with chromatin is relatively low after washing steps, the background signal is often lower than observed using ChIP-seq. Low background is advantageous as it leads to increased enrichment and a decreased requirement for high read coverage to map chromatin proteins genome-wide. CUT&RUN has been used to profile both histone proteins and transcription factors (TFs) from fewer cells than required for ChIP-seq (Skene and Henikoff 2017, Skene, Henikoff et al. 2018).
Figure 1: Cartoon of Workflow.
Shown is a diagram of the CUT&RUN approach.
Here we include all information necessary for performing CUT&RUN from mammalian cells. We describe our implementation of the original CUT&RUN protocol (Skene et al, 2017) that has been used extensively in our labs (Hainer, Boskovic et al. 2018). However, alternative protocols in which cells are permeabilized with digitonin rather than lysed have also been reported to yield excellent results (Janssens, Wu et al. 2018, Skene, Henikoff et al. 2018). Basic Protocol 1 details the steps for performing CUT&RUN from murine embryonic stem (mES) cells. Basic Protocol 2 focuses on building libraries for paired-end Illumina sequencing. Basic Protocol 3 provides steps for analyzing resulting datasets.
BASIC PROTOCOL 1
CUT&RUN of transcription factors in mammalian cells
CUT&RUN is a useful and versatile technique to map the occupancy of DNA binding proteins on chromatin. The following section describes the protocol for CUT&RUN from mES cells. This protocol is amendable to other cell types.
Materials
Mammalian cells
This protocol is written for 500,000 E14 mES cells (RRID:CVCL_C320). Lower cell numbers can be used with a minor modification of the protocol described in Step 9.
Cell counter (such as Biorad TC20 cat. 1450102)
Primary antibody against protein of interest
Recombinant proteinA-MNase (plasmid available from Addgene plasmid #86973)
The proteinA-MNase must be expressed and purified. Details of purification can be found in the original ChIC protocol (Schmid, Durussel et al. 2004).
Phosphate-buffered saline (PBS; Corning cat 21031CV)
Nuclear extraction buffer (see recipe)
Binding buffer (see recipe)
Concanavalian A beads (Polysciences, Inc. cat. 86057)
DynaMag-2 magnetic stand (Life Technologies, cat. 12321D)
Wash buffer (see recipe)
Blocking buffer (see recipe)
2XSTOP buffer (see recipe)
Phenol-chloroform-isoamyl (Fisher cat. BP1752I400)
Phase lock tubes (Quanta Bio cat. 2302830)
Chloroform (Fisher cat. C298500)
20 mg/mL Glycogen (VWR cat. 9005792)
10 mg/mL RNaseA (Thermo Scientific cat. EN0531)
Protease Inhibitors (Life technologies cat. 78439)
10 ng/mL heterologous DNA
This is DNA for a spike in control that should be from a different organism from your sample, for example when performing CUT&RUN on mammalian cells, DNA from S. cerevisiae can be used. DNA can be prepared by crosslinking cells, MNase digesting to mononucleosomes, purifying DNA, and diluting to 10 ng/mL. The spike in can be used for normalization during analysis.
100% Ethanol
100mM CaCl2 (see recipe)
0.1X TE buffer (see recipe)
10% SDS (see recipe)
20 mg/mL Proteinase K (Bioline cat. 37085)
Vortex mixer (such as Fisher cat. 02215414)
Refrigerated (4°C) Rotator (such as VWR cat. 10136084)
Refrigerated centrifuge for 15 mL conical tubes (such as Eppendorf 2231000382)
Refrigerated microcentrifuge (such as Eppendorf cat. 5404000537)
Thermomixer (Eppendorf cat. 2231000574)
Harvest and lyse cells
Chill buffers on ice. Work efficiently and keep tubes on ice unless otherwise indicated.
-
1
Harvest cells and count their density using a cell counter. Add an appropriate number of cells to a 15mL conical tube. The following protocol is written for 500,000 E14 mES cells, but this number can be reduced as necessary.
Our lab routinely uses a BioRad TC10 cell counter according to the manufacturers’ instructions. Other methods of cell counting (e.g., hemocytometer; Phelan and May, 2017) are also fine.
-
2
Centrifuge the cells for 5 minutes 600 X g at 4°C.
-
3
Discard the supernatant and resuspend in 1 mL cold PBS by gently pipetting the cells. Transfer to a 1.5 mL microcentrifuge tube.
-
4
Centrifuge the cells for 5 minutes 600 X g at 4°C.
-
5
Discard the supernatant and resuspend the cells in 1 mL cold Nuclear Extraction (NE) buffer by gently pipetting the cells without introducing bubbles.
-
6
Centrifuge the sample for 5 minutes 600 X g at 4°C.
-
7
Discard the supernatant and resuspend the sample in 600 µL cold NE buffer by gently pipetting the sample without introducing bubbles.
Prepare Concanavalin A beads
Beads can be prepared during the waiting periods in the ‘Harvest and lyse cells’ section.
-
8
Resuspend Concanavalin A beads with gentle vortexing and/or pipetting.
-
9
Transfer 150 µL Concanvalin A bead slurry to a 1.5 mL microcentrifuge tube that contains 850 µL cold Binding buffer.
The amount of bead slurry is dependent upon the number of cells. See Table 1 for example of bead slurry to cell number ratios.
-
10
Place the microcentrifuge tube containing Concanavalin A beads on a magnetic stand for at least 2 minutes, until the solution is clear.
For this and all bead separation steps, confirm the solution is completely clear to avoid loss of beads. After beads accumulate at the magnet-proximal side of the tube, one can invert the tube to capture any beads that may be on the lid and incubate further to allow collection of the remaining beads.
See video 1 for an example of separation using a magnetic stand.
-
11
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
12
Remove the 1.5 mL microcentrifuge tube from the magnetic stand and resuspend the Concanavalin A beads in 1 mL cold Binding buffer by gently pipetting the sample.
-
13
Place the microcentrifuge tube on the magnetic stand for approximately 2 minutes.
-
14
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
15
Remove the 1.5 mL microcentrifuge tube from the magnetic stand and resuspend the Concanavalin A beads in 1 mL cold Binding buffer by gently pipetting the sample.
-
16
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
17
Remove the 1.5 mL microcentrifuge tube from the magnetic stand and resuspend the Concanavalin A beads in 300 µL Binding buffer.
Table 1.
Examples of Concanavalin A bead slurry to cell number ratios.
| Number of cells | Amount of Concanavalin A bead slurry |
|---|---|
| 500,000 | 150 µL |
| 50,000 | 100 µL |
| 5,000 | 75 µL |
Bind nuclei to magnetic beads
-
18
While gently vortexing the nuclei (which were previously resuspended in 600 µL NE buffer in step 7), slowly add the 300 µL of washed and resuspended Concanavalin A beads. During vortexing and bead addition, pipet up and down to assist in complete mixing.
For this and subsequent steps (such as addition of primary antibody and pA-MN), gentle vortexing is crucial. Vortex only vigorously enough to gently mix the beads and be sure the beads mix completely. (Mixing is crucial to evenly distribute beads on nuclei, but harsh vortexing can damage nuclei, potentially leading to higher background or otherwise poor performance.) Set the vortex mixer to setting ~3. See video 2 for an example of gentle vortexing during sample addition.
-
19
Incubate the combined nuclei and beads on a rotating platform for 10 minutes at 4°C.
Block beads
-
20
Place microcentrifuge tube containing bead-bound nuclei on a magnetic stand for approximately 5 minutes.
The amount of time required for bead separation increases once the nuclei are bound. This helps prevent loss of nuclei.
-
21
After complete clearing of the solution, remove and discard the supernatant without disturbing the magnetic beads.
-
22
Remove the microcentrifuge tube from the magnetic stand and resuspend the beads in 1 mL Blocking buffer by gently pipetting the sample.
Gentle pipetting is preferable to vortexing at this step. Take care to minimize introduction of bubbles during this action.
-
23
Incubate the resuspended beads for 5 minutes at room temperature.
Primary antibody
-
24
Place the microcentrifuge tube containing the bead-bound nuclei on a magnetic stand for approximately 5 minutes.
-
25
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
26
Remove microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer with gentle pipetting.
-
27
Place microcentrifuge tube containing bead-bound nuclei on magnetic stand for approximately 5 minutes.
-
28
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
29
Remove microcentrifuge tube from the magnetic stand and resuspend in 250 µL Wash buffer with gentle pipetting.
-
30
Dilute the primary antibody that recognizes against the protein of interest in 250 µL Wash buffer.
The amount of primary antibody used depends on the efficiency of the antibody. A titration of antibody may be necessary (see critical parameters). Typically, a dilution of 1:100 final works well. (5 µL total antibody per 500 µL final binding volume). A master mix of primary antibody should be made for more than one sample.
-
31
While gently vortexing the bead-bound nuclei, slowly add the 250 µL Wash buffer + primary antibody to each sample.
A critical control that should be included in all CUT&RUN experiments is the inclusion of a no primary antibody or non-specific IgG control. This control sample is treated identically to the experimental sample, except that 250 µL Wash buffer alone (no antibody) or with non-specific IgG should be added to this sample in lieu of a primary antibody.
-
32
Incubate the microcentrifuge tube containing the bead-bound nuclei and primary antibody on a rotating platform for 2 hours at 4°C.
-
33
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
After rotation, it is essential to capture all bead-bound nuclei. After initial separation, invert tube to capture any beads on the lid, and allow the microcentrifuge tube to sit on the magnetic stand for additional time.
-
34
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
35
Remove the microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer by gentle pipetting.
-
36
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
-
37
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
38
Remove the microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer by gentle pipetting.
ProteinA-MNase (pA-MN) coupling
pA-MN should be added to both the antibody-containing experimental samples and the no antibody or IgG controls, in order to control for untargeted MNase digestion.
-
39
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
-
40
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
-
41
Remove the microcentrifuge tube from the magnetic stand and resuspend in 250 µL Wash buffer using gentle pipetting.
-
42
Dilute pA-MN in 250 µL Wash buffer.
The amount of pA-MN used depends on the concentration and activity of the purified recombinant protein. A titration of fusion protein must be performed upon each purification to optimize DNA recovery and digestion kinetics, as described (Skene et al, 2017). See critical parameters for a description of pA-MN titration. A master mix of pA-MN should be made for more than one sample.
-
43
While gently vortexing the bead-bound nuclei, slowly add the 250 µL Wash buffer + pA-MN. Pipet up and down during addition to ensure mixing.
-
44
Incubate the microcentrifuge tube on a rotating platform for 1 hour at 4°C.
-
45
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
After rotation, it is essential to capture all bead-bound nuclei. After initial separation, invert tube to capture any beads on the lid, and allow the microcentrifuge tube to sit on the magnetic stand for additional time.
-
46
After complete clearing of the solution, remove and discard the supernatant without disturbing the magnetic beads.
-
47
Remove the microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer by gentle pipetting.
-
48
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
-
49
After complete clearing of the solution, remove and discard the supernatant without disturbing the magnetic beads.
Targeted MNase cleavage
-
50
Place the microcentrifuge tube on a magnetic stand for approximately 5 minutes.
-
51
After complete clearing of the solution, remove and discard the supernatant without disturbing the magnetic beads.
-
52
Remove the microcentrifuge tube from the magnetic stand and resuspend in 150 mL Wash buffer by gentle pipetting.
-
53
Equilibrate to 0°C by incubating tubes for 5 minutes in an ice water bath.
-
54
One tube at a time, briefly remove tubes from ice water and add 3 µL 100mM CaCl2 while gently vortexing. Flick briefly 3-times to quickly mix and return to 0°C ice water.
See video 3 for demonstration. Working rapidly to minimize the time tubes are held outside of the ice water slurry is essential. In addition, complete mixing is essential for proper MNase cleavage.
-
55
Incubate samples on ice water for 30 minutes.
Digestion can be completed in less than 2 minutes. However, additional digestion time allows for higher recovery of DNA fragments with no effect on background (Skene and Henikoff 2017).
-
56
Stop each reaction by addition of 150uL 2XSTOP buffer.
Remember to add the RNaseA, glycogen, and spike-in DNA fresh to the STOP buffer prior to use. See recipe for instructions.
Fragment release and clean up
-
57
Incubate samples for 20 minutes at 37°C on thermomixer (with no shaking) to digest RNA and release cleaved DNA fragments from insoluble nuclear chromatin.
Can incubate at 37°C on a heated block or any instrument that fits 1.5 mL microcentrifuge tubes well.
-
58
Centrifuge 16,000 x g for 5 minutes at 4°C.
-
59
Place samples on a magnetic stand and after separation, immediately transfer supernatants to fresh 1.5 mL tubes.
Supernatant contains the released fragments. Discard beads/pellet. Pellet can be difficult to visualize.
-
60
Add 3 µL 10% SDS and 2.5 µL 20 mg/mL Proteinase K to new 1.5 mL tube containing each sample.
-
61
Invert well and incubate for 10 minutes at 70°C on thermomixer (without shaking).
Can incubate at 70°C on any instrument that fits 1.5 mL microcentrifuge tubes well.
-
62
Add 300 µL Tris-buffered phenol-chloroform-isoamyl solution (25:24:1) to the sample and vortex well.
Solution should become somewhat opaque.
-
63
Transfer to a phase-lock microcentrifuge tube and centrifuge for 5 minutes at 16,000 X g at 4°C.
-
64
Transfer aqueous phase to new (non-phase-lock) microcentrifuge tube and discard the organic phase in an appropriate receptacle.
-
65
Add 300 µL chloroform to the sample and vortex well.
-
66
Centrifuge for 5 minutes at 16,000 X g at 4°C.
-
67
Carefully transfer the aqueous phase to a new 1.5 mL tube and discard the organic phase in an appropriate receptacle.
Minimize contamination of phenol and chloroform, as both can inhibit downstream reactions.
-
68
Add 2 µL of 2 mg/mL glycogen to each sample and mix by vortexing well.
-
69
Add 750 µL 100% ethanol to each sample and mix by vortexing well.
-
70
Incubate samples on ice for approximately 20 minutes.
Sample can also be stored in −20°C for at least several days.
-
71
Centrifuge for 10 minutes at 16,000 X g at 4°C.
-
72
Carefully discard the supernatant without disturbing the glycogen-DNA pellet.
-
73
Wash the pellet with 1 mL 100% ethanol.
Pellet is hardly visible, so be careful when removing the supernatant to not disturb the pellet.
-
74
Centrifuge for 5 minutes at 16,000 X g at 4°C.
-
75
Discard supernatant.
-
76
Briefly centrifuge for 1 minute at 16,000 X g at 4°C to spin down any residual ethanol.
-
77
Discard any residual supernatant.
-
78
Air dry for approximately 5 minutes at room temperature.
Do not overdry samples.
-
79
Dissolve the pellet in 36.5 µL 0.1XTE.
Can store at −20°C or proceed immediately to DNA sequencing library build.
ALTERNATE PROTOCOL 1
Addition of secondary antibody for pA-MN recognition
Some IgG classes bind poorly to protein A. Rabbit IgG is strongly recognized by protein A, as are mouse IgG2a, human IgG1, IgG2, and IgG4. However, antibodies raised in mouse with IgG1, IgG2b, or IgG3 subtypes, Goat IgG, or Sheep IgG are not recognized well by protein A. Therefore, if the primary antibody utilized is not Rabbit IgG or mouse IgG2a subtype, after primary antibody addition and washes (Perform steps 1–38 of Basic Protocol 1) and prior to pA-MN addition (starting at step 39), an additional step of secondary antibody addition can be included. This will allow use of primary antibodies that otherwise would not be strongly bound by protein A.
Materials
Secondary antibody, for example rabbit anti-mouse (Millipore cat. 06–371).
Place microcentrifuge tube on a magnetic stand for approximately 5 minutes.
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
Remove tube from the magnetic stand and resuspend in 250 µL Wash buffer by gently pipetting the sample.
-
Dilute secondary antibody in 250 µL Wash buffer.
Typically, a dilution of 1:100 final works well. (5uL total antibody per reaction). A master mix of secondary antibody should be made for more than one sample. No antibody controls should also include secondary antibody if the experimental sample requires this additional step.
While gently vortexing the bead bound nuclei, slowly add the 250 µL Wash buffer + secondary antibody.
Incubate on a rotating platform for 1 hour at 4°C.
-
Place on a magnetic stand for approximately 5 minutes.
After rotation, it is essential to capture all DNA. After initial separation, invert the tube to capture any beads on the lid, and allow additional time on the magnetic stand.
After complete clearing of the solution, remove and discard the supernatant without disturbing the beads.
Remove the microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer by gently pipetting the sample.
Place tube on a magnetic stand for approximately 5 minutes.
After complete clearing of the solution, remove and discard the supernatant without disturbing magnetic beads.
Remove the microcentrifuge tube from the magnetic stand and resuspend in 1 mL Wash buffer by gentle pipetting.
Proceed to pA-MN addition (starting at step 39 in Basic Protocol 1).
BASIC PROTOCOL 2
CUT&RUN library preparation for paired-end Illumina sequencing
The following section describes the protocol for preparation of CUT&RUN DNA libraries for Illumina paired-end sequencing.
Materials
36.5µL of CUT&RUN enriched DNA (from step 79 of Basic Protocol 1)
10X T4 DNA ligase buffer (NEB cat. B0202S)
10mM dNTPs (NEB cat. N0447S)
10mM ATP (NEB cat. P0756S)
10U/µL T4 PNK (NEB cat. M0201S)
5U/µL T4 DNA polymerase (NEB cat. M0203S)
Taq DNA polymerase (NEB cat. M0273S)
Quick ligation kit (NEB cat. M2200S)
AMPure beads (Beckman Coulter cat. A63881)
10mM Tris-HCl pH 8.0 (see recipe)
80% Ethanol
40% PEG4000 (see recipe)
DynaMag-2 magnetic stand (Life Technologies, cat. 12321D)
100 bp ladder (NEB cat. N3231S)
NEB low molecular weight ladder (NEB cat. N3233S)
5X KAPA buffer (KAPA cat. KK2502)
10mM dNTPs (KAPA cat. KK2502)
KAPA HiFi polymerase (KAPA cat. KK2502)
Thermomixer (Eppendorf cat. 2231000574)
Thermocycler (example Eppendorf 6311000010)
Commercially available paired-end adapters and PCR primers for Illumina TruSeq (or inline PE adapters and primers can be used).
TruSeq adapters and primer sequences are as follows:
TruSeq Universal Adapter
5’AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T
TruSeq Indexed Adapter
5’[Phos]GATCGGAAGAGCACACGTCTGAACTCCAGTCACNNNNNNATCTCGTATGCCGTCTTCTGCTT*G
[Phos] indicates a 5’ phosphate group on the indexed adapter
* Indicates a phosphorothioate bond between the last two bases at the 3’ end
NNNNNN indicates a 6-base barcode for multiplexing samples
PCR primers for library enrichment of TruSeq adapter ligated samples:
P5 PCR Primer 5’ AATGATACGGCGACCACCGA*G
P7 PCR Primer 5’ CAAGCAGAAGACGGCATACGA*G
* Indicates a phosphorothioate bond between the last two bases at the 3’ end
PE inline adapters and primer sequences are as follows:
Adapter Oligo 1: 5’ [Phos]NNNNGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG
Adapter Oligo 2: 5’ ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNN
[Phos] indicates a 5’ phosphate group on the indexed adapter
NNNN indicates a 4-base barcode for multiplexing samples
PE PCR primers for library enrichment:
PE PCR 1.0: 5’ AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
PE PCR 2.0: 5’ CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
PE sequencing primers:
F: 5’ ACACTCTTTCCCTACACGACGCTCTTCCGATCT
R: 5’ CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
End repair, phosphorylation, and A-tailing
-
1
Transfer 36.5 µL CUT&RUN enriched DNA into a 0.5 mL PCR tube.
-
2
Dilute T4 DNA polymerase 1:20 as follows:
17.0 µL nuclease free water
2.0 µL 10X T4 DNA ligase buffer
1.0 µL 5U/µL T4 DNA polymerase
Pipet up and down to mix
-
3
Prepare the end repair/adenylation master mix by combining the following for each sample:
5 µL 10X T4 DNA ligase buffer
2.5 µL 10mM dNTPs
1.25 µL 10mM ATP
3.13 µL 40% PEG 4000
0.63 µL 10 U/µL T4 PNK
0.5 µL diluted T4 DNA polymerase
0.5 µL Taq DNA polymerase
Carefully and gently pipette up and down to mix
-
4
Add 13.5 µL of the master mix to 36.5 µL CUT&RUN enriched DNA in 0.5 mL PCR tube. Mix by carefully pipetting up and down.
Keep samples on ice while adding master mix.
-
5
Incubate as follows in a thermal cycler that was pre-cooled and has no heated lid:
12°C for 15 minutes
37°C for 15 minutes
72°C for 20 minutes
ramp to 4°C
-
6
Immediately remove samples and proceed to adapter ligation.
Proceed immediately to the adapter ligation step.
Adapter ligation
-
7
Place A-tailed library samples on ice.
-
8
Add 5 µL of 1.5 µM annealed adapters (either TruSeq or inline PE Illumina adapters).
-
9
Make a ligation master mix by combining the following (amounts per sample are shown):
55 µL 2X quick ligase buffer
5 µL quick ligase
Gently pipette up and down to mix
-
10
Add 60 µL of the ligation master mix to end prepared DNA + adapter mixture in 0.5 mL PCR tube. Mix by carefully pipetting up and down.
-
11
Incubate at 20°C in a thermocycler that was pre-cooled with no heated lid for 15 minutes.
-
12
Immediately remove samples and proceed to AMPure bead purification.
Over ligating can lead to poor library preparation, so it is important to proceed immediately to the purification step.
AMPure DNA purification
-
13
Transfer the adapter ligated sample to a 1.5 mL microcentrifuge tube.
-
14
Add 38 µL AMPure XP bead solution to each tube containing adapter-ligated DNA.
Bring AMPure beads to room temperature prior to use. Resuspend AMPure beads well by vortexing prior to use.
-
15
Thoroughly resuspend the beads by pipetting up and down gently.
-
16
Incubate the sample for 15 minutes at room temperature to allow DNA to precipitate onto the beads.
-
17
Place sample on a magnetic stand for approximately 5 minutes.
-
18
After complete clearing of the solution, remove and discard the supernatant without disturbing magnetic beads.
-
19
While leaving the microcentrifuge tube on the magnetic stand, add 200 µL 80% ethanol without disturbing the beads.
-
20
Incubate for 30 seconds on the magnetic stand.
-
21
Remove and discard the ethanol without disturbing magnetic beads.
-
22
While leaving the microcentrifuge tube on the magnetic stand, add 200 µL 80% ethanol without disturbing the beads.
-
23
Incubate for 30 seconds on the magnetic stand.
-
24
Remove and discard the ethanol without disturbing magnetic beads.
-
25
Briefly spin the tube (1,000 X g for 5 seconds).
-
26
Place sample on the magnetic stand for approximately 2 minutes.
-
27
Remove any residual ethanol without disturbing the magnetic beads.
-
28
Air dry the beads for approximately 5 minutes at room temperature.
Do not overdry beads. They should be a chocolate-brown, without a gleam from solution.
-
29
Remove the microcentrifuge tube from the magnetic stand and resuspend beads completely with 29 µL 10mM Tris-HCl pH 8.0.
-
30
Incubate for approximately 5 minutes at room temperature to allow DNA to elute from the beads.
-
31
Place the tube on a magnetic stand for approximately 2 minutes.
-
32
Transfer 27.5 µL of eluate to a new 0.5 mL PCR tube.
Be sure to avoid any disruption of the beads by leaving a small volume behind. Eluates can be stored at −20°C after purification, prior to library enrichment.
Library enrichment
-
33
Place samples on ice.
-
34
Prepare a PCR master mix by combining the following (amounts per sample are shown):
10.0 µL 5X KAPA HiFi Fidelity Buffer
1.5 µL 10mM dNTPs
5 µL 20µM P5 or PE PCR 1.0 primer (depending on adapters used)
5 µL 20µM P7 or PE PCR 2.0 primer (depending on adapters used)
1 µL 1U KAPA HiFi HotStart DNA Polymerase
Carefully and gently pipette up and down to mix
-
35
Add 22.5 µL of PCR mix to 27.5 µL purified DNA.
-
36
Pipet up and down to mix well.
-
37Amplify the CUT&RUN libraries using the following PCR conditions:
-
1)98°C 45 seconds
-
2)98°C 15 seconds
-
3)60°C 10 seconds
-
4)Repeat steps 2–3 13 times for a total of 14 PCR cycles
-
5)72°C 1 minute
-
6)ramp to and hold at 4°C
14 PCR cycles may or may not be sufficient, depending on the protein of interest and the antibody used. We recommend performing qPCR following 5 cycles of initial amplification using a procedure analogous to previously described protocols for ATAC-seq (Buenrostro, Wu et al. 2015) to determine the number of cycles required. Furthermore, using lower cell numbers may necessitate additional PCR cycles. Samples can be stored at −20°C after library enrichment.
-
1)
Size selection and purification of library enriched samples
-
7
Pour a 1.5% agarose gel that is preferably 15–20 cm in length.
-
8
Add loading dye and run the entire sample on the agarose gel, using a 100 bp ladder or NEB low molecular weight ladder for comparison. Run the dye front close to the end of the agarose gel to get maximum separation of the library from any adapter dimers.
If necessary, the sample can be precipitated through standard ethanol precipitation and resuspended in a smaller volume prior to running on the agarose gel.
-
9
Cut out a gel slice under long-wave UV light ranging from ~150 bp to ~650 bp in size.
Since adapters on each side of the insert correspond to ~120 bp total, the size range described will include inserts approximately 30–530 bp in size.
-
10
Use a Qiagen gel extraction kit as recommended by the manufacturers. Elute each library in 15 µL nuclease free water.
Samples may be stored at −20°C prior to sequencing.
Quality control of library samples
-
11
Quantify the concentration of the CUT&RUN DNA libraries using dsDNA-specific assay (for example, using a quBit).
-
12
Determine the size distribution of libraries by Fragment Analyzer or equivalent.
Assessing read size for the factor is obtained is the most informative quality control step.
-
13
Optional: Clone and sequence ~10 fragments per library by Sanger sequencing to ensure libraries contain adapters and appropriate inserts.
BASIC PROTOCOL 3
Example analysis pipeline of CUT&RUN datasets
CUT&RUN libraries should be sequenced using paired-end Illumina sequencing (minimum 25 bp each side; longer reads provide minimal additional information). The following protocol provides an example step by step method for analysis of resulting datasets. Alternative programs may be used for analysis.
Materials
Unix-compatible computer
Appropriate software, for example:
Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), to map reads to the genome
Novocode (http://www.novocraft.com/documentation/), to split reads by PE barcode and trim PE barcode
Samtools (http://samtools.sourceforge.net), for making size classes and converting files
Picard (https://broadinstitute.github.io/picard/), to remove duplicates
HOMER (http://homer.ucsd.edu/homer/), for generating genome browser tracks, calling peaks, and averaging data over genomic locations
Sufficient processing power for whole-genome sequence analysis
Split samples by barcode and remove the barcode sequences using Novocode, if inline adapters are used, such as Illumina PE adapters. If Illumina TruSeq adapters are used, this is unnecessary.
Use the awk command in Unix to trim paired-end reads to 21 bases.
Align to annotated genome with Bowtie2 using the -X 1000 parameter.
Remove duplicates using Picard.
Use Samtools to remove low quality reads (MAPQ < 10).
Use the Unix awk command, combined with Samtools, to separate reads into two size classes: < 120 bp for TFs and 150–500 bp for nucleosomes.
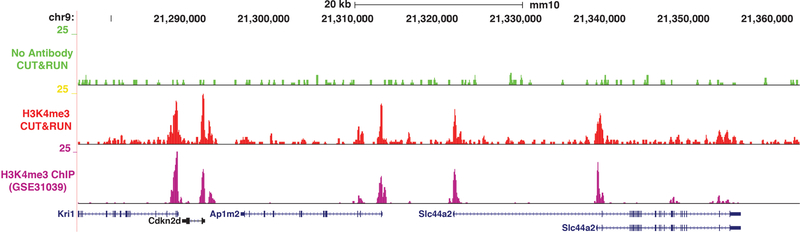
Generate Tag directories and UCSC genome browser tracks using HOMER (Heinz, Benner et al. 2010) using “makeTagDirectories” and “makeUCSCfile” commands (see Figure 4 for an example of a genome browser generated by HOMER).
Align over specific regions of the genome using the “annotatePeaks” command in HOMER.
Call peaks using the “findPeaks” command in HOMER.
Figure 4: Genome browser track for H3K4me3 CUT&RUN compared to available ChIP-seq datasets.
Indicated ChIP-seq datasets from mES cells were downloaded, aligned, and browser tracks were generated. CUT&RUN experiments were performed from 500,000 mES cells. Included are no antibody controls for the matching size distribution.
REAGENTS AND SOLUTIONS
1M HEPES-KOH, pH 7.9
Add 238.3 g HEPES to 800 mL filtered deionized water. Stir at room temperature. Add KOH to reach pH 7.9. Stir at room temperature until dissolved. Bring to final volume of 1 L and filter sterilize. Solution can be stored at room temperature for up to 1 year.
1M KCl
Add 37.275 g potassium chloride to in 400 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 500 mL and filter sterilize. Solution can be stored at room temperature for up to 1 year.
2M spermidine
Add 2.9 g spermidine to 10 mL filtered deionized water. Stir at room temperature until dissolved and filter sterilize. Solution can be stored at room temperature for up to 1 year in a 15 mL conical tube that is covered with foil.
10% Triton X-100
Add 10 g Triton X-100 to 80 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 100 mL and filter sterilize. Solution can be stored at room temperature for up to 1 year.
1M CaCl2
Dissolve 73.51 g CaCl2 in 80 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 100 mL and filter sterilize. Solution can be stored at room temperature for up to 1 year.
1M MnCl2
Dissolve 1.98 g MnCl2 in 10 mL filtered deionized water. Stir at room temperature until dissolved and filter sterilize. Solution can be stored at room temperature for up to 1 year in a 15 mL conical tube that is covered with foil.
1M HEPES-KOH, pH 7.5
Add 238.3 g HEPES to 800 mL filtered deionized water. Stir at room temperature. Add KOH to reach pH 7.5. Stir at room temperature until dissolved. Bring to final volume of 1 L and filter sterilize. Solution can be stored at room temperature for up to 1 year.
5M NaCl
Add 292.2 g NaCl to 800 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 1 L and filter sterilize. Solution can be stored at room temperature for up to 1 year.
30% BSA
Add 3 g BSA to 8 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 10 mL and filter sterilize. Solution can be stored at room temperature for up to 1 year.
0.5M EDTA, pH 8.0
Add 186.1 g disodium EDTA dihydrate to approximately 800 mL filtered deionized water. Stir at room temperature. Add NaOH pellets to reach pH 8.0. Filter sterilize. Solution can be stored at room temperature for up to 1 year.
0.2M EGTA
Add 76.07 g EGTA to approximately 800 mL filtered deionized water. Stir at room temperature. Add NaOH pellets to reach pH 8.0. Filter sterilize. Solution can be stored at room temperature for up to 1 year.
Nuclear extraction (NE) buffer
For 50 mL NE buffer, mix 1 mL 1M HEPES-KOH, pH 7.9 (see recipe), 500 µL 1M KCl (see recipe), 12.5 µL 2M spermidine (see recipe), 500 µL 10% Triton X-100 (see recipe), 12.5 mL 80% glycerol (see recipe), and 35.48 mL pure water. This stock solution can be stored at 4°C for up to 1 year.
On the day of the experiment, add 1X protease inhibitors (Life technologies cat. 78439) to appropriate aliquoted amount of NE buffer.
Binding buffer
For 20 mL Binding buffer, mix 400 µL 1M HEPES-KOH, pH 7.9 (see recipe), 200 µL 1M KCl (see recipe), 20 µL 1M CaCl2 (see recipe), 20 µL 1M MnCl2 (see recipe), and 19.36 mL pure water. This stock solution can be stored at 4°C for up to 1 year.
Wash buffer
For 100 mL Wash buffer, mix 2 mL 1M HEPES-KOH. pH 7.5 (see recipe), 3 mL 5M NaCl (see recipe), 25 µL 2M Spermidine (see recipe), 333 µL 30% BSA (see recipe), and 94.642 mL pure water. This stock solution can be stored at 4°C for up to 1 year.
On the day of the experiment, add 1X protease inhibitors (Life technologies cat. 78439) to appropriate aliquoted amount of Wash buffer.
Blocking buffer
Take 5 mL of prepared wash buffer that contains protease inhibitors (see above) and add 20 µL 0.5M EDTA (see recipe). Invert well to mix.
2XSTOP buffer
For 5mL, mix 200 µL 5M NaCl (see recipe), 200 µL 0.5M ETDA (see recipe), 100 µL 0.2M EGTA (see recipe) and 4.46 mL filtered, deionized water.
On the day of the experiment, add 25 µL 10mg/mL RNaseA (Thermo Scientific cat. EN0531), 10 µL 20mg/mL glycogen (VWR cat. 9005792), and 5 µL 10ng/mL heterologous DNA.
100mM CaCl2
Dilute 1M CaCl2 (see recipe) 1:10 in filtered deionized water. Mix well. Solution can be stored at room temperature for up to 1 year.
10X TE buffer
Add 12.1 g Tris Base and 3.373 g EDTA to 800 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 1 L and filter sterilize. Solution can be stored at room temperature for up to 1 year.
0.1X TE buffer
Dilute 10X TE buffer (see recipe) 1:100 in filtered deionized water. Mix well. Solution can be stored at room temperature for up to 1 year.
10% SDS
Add 100 g sodium dodecyl sulfate to 800 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 1 L and filter sterilize. Solution can be stored at room temperature for up to 1 year.
Wear a mask when working with SDS powder!
40% PEG4000
Add 4 g PEG4000 to 8 mL filtered deionized water. Stir at room temperature until dissolved. Bring to final volume of 10 mL and filter sterilize. Solution can be stored at room temperature for up to 1 year.
1M Tris-HCl, pH 8.0
Dissolve 60.57 g Tris in 400 mL filtered deionized water. Stir at room temperature until dissolved. Bring to a pH of 8.0 by adding concentrated HCl dropwise. Adjust volume to 500 mL with filtered deionized water. Filter sterilize and store at room temperature for up to 1 year.
10mM Tris-HCl, pH 8.0
Dilute 1M Tris-HCl, pH 8.0 buffer (see recipe) 1:100 in filtered deionized water. Mix well. Solution can be stored at room temperature for up to 1 year.
COMMENTARY
Background Information
Transcription factors (TFs) are DNA-binding proteins that recognize short DNA sequence motifs and regulate gene expression through a variety of mechanisms. Histone proteins interact with ~147 bp of DNA to form the basic unit of chromatin, the nucleosome which also modulates gene expression, typically by preventing TF binding through the tight interaction of histones with DNA. Profiling of factor and nucleosome occupancy is essential to understanding DNA templated processes and the activities of these proteins.
Chromatin Immunoprecipitation (ChIP) is the most widely used technique for exploring protein-DNA interactions on chromatin. Traditional ChIP includes crosslinking of protein to DNA using formaldehyde, shearing of crosslinked chromatin with sonication, and immunoprecipitation of the protein of interest (Gilmour 1984, Gilmour 1985). Crosslinking and sonication of chromatin can lead to biases in traditional ChIP techniques including false positives from crosslinking and increased release of fragments in nucleosome depleted regions of the genome from sonication (Meyer and Liu 2014, Baranello, Kouzine et al. 2016).
In addition to potential artifacts from crosslinking and sonication, the resolution of ChIP techniques is limited by chromatin shearing. In crosslinking ChIP, the DNA fragments that are generated are usually 200–400 bp in size. Since many TFs bind sequences only 6–20 bp in size, additional information regarding binding motifs is often necessary to infer the precise location of TFs within broad peaks of ChIP enrichment. The ChIP-exo method increases the resolution of ChIP while maintaining the basic features of ChIP-based methods for enrichment of chromatin (Rhee and Pugh 2011). ChIP-exo involves immunoprecipitation of chromatin fragments from a large number of crosslinked cells, followed by use of an exonuclease to degrade the regions of immunoprecipitated DNA not bound by the chromatin protein. Exonuclease digestion allows for precise localization of TF binding sites to the single nucleotide level. However, the large number of cells required for this technique are not always readily available.
Modifications to traditional ChIP protocols have been developed to increase the sensitivity and therefore reduce the number of cells required. These modifications include ChIPmentation (Schmidl, Rendeiro et al. 2015), carrier-assisted ChIP-seq (Zwart 2013), ULI-NChIP (Brind’Amour, Liu et al. 2015), µChIP (Dahl, Jung et al. 2016), and DROP-ChIP (Rotem, Ram et al. 2015). Several of these techniques enable mapping of abundant histone modifications in fewer than 1,000 cells. However, ChIP-based methods for mapping chromatin occupancy of TFs currently require a minimum of 10,000 cells, and often 2–3 orders of magnitude more.
An orthogonal approach termed DamID utilizes overexpression of bacterial Dam methylase fused to chromatin binding proteins to profile their genomic locations (van Steensel 2000). This approach has been effectively utilized in many systems but requires expression of a Dam-fusion protein for each factor of interest. Furthermore, overexpression of fusion proteins may not be feasible in some settings as it could potentially lead to occupancy at non-physiological locations. Similar to DamID, ChEC (Schmid, Durussel et al. 2004) and ChEC-seq (Zentner, Kasinathan et al. 2015) require fusion of chromatin proteins to MNase.
CUT&RUN is a recently described method for genome-scale profiling that results in high resolution maps of factors (Skene and Henikoff 2017). CUT&RUN was derived from the ChIC technique, developed in 2004 by Ulrich Laemmli and colleagues, which utilized recombinant proteinA-MNase to map chromatin proteins at individual loci (Schmid, Durussel et al. 2004). In 2017, Steven Henikoff and colleagues systematically modified the ChIC approach to accommodate genome-wide profiling of factors (Skene and Henikoff 2017). The authors showed that the background signal for CUT&RUN is extremely low, leading to increased enrichment of factors and a decreased requirement for high read coverage relative to ChIP-seq approaches. The technique itself is rapid and easily performed, with few steps that require optimization for each protein profiled. Consequently, CUT&RUN facilitates profiling of chromatin factors with increased fidelity at lower cost than traditional approaches. In sum, CUT&RUN is an excellent alternative to traditional ChIP-seq techniques, especially when collection of large numbers of cells is unfeasible.
Critical Parameters and Troubleshooting
Harvesting cells and cell lysis
Membrane lysis is essential for proper binding of nuclei to lectin coated beads in subsequent steps. Increasing the incubation time in NE buffer could assist in more efficient lysis for some cell types. When performing CUT&RUN on multiple samples, the same number of cells needs to be used for appropriate comparisons.
Prepare Concanavalin A beads and nuclei binding
Concanavalin A beads must be completely mixed prior to use. Gentle vortexing of nuclei is important to prevent crushing of nuclei.
Block beads
Addition of EDTA at this step prevents MNase from cleaving chromatin prior to activation with CaCl2. Complete resuspension of the beads in blocking buffer allows for all nuclei to be blocked equally.
Primary antibody use
The amount of primary antibody addition is also important to consider. Typically, one starts with a 1:100 final dilution of primary antibody (i.e., 5uL in the 500uL reaction volume). However, since different antibodies exhibit different affinities for their substrates, unequal background binding to chromatin, and other differences, one should typically titrate new antibodies (e.g., 1:50, 1:100, 1:200, 1:400) to identify the optimal dilution for CUT&RUN.
Primary antibody and ProteinA-MNase (pA-MN) coupling
The most critical steps of the CUT&RUN protocol are the proper mixing of nuclei-bound beads during addition of reagents. Incomplete mixing can result in improper and uneven incorporation or spatial differences in the concentrations of primary antibody, pA-MN, or CaCl2. See video 2 for an example of primary antibody and pA-MN addition.
pA-MN titration
The pA-MN must be expressed and purified. Details of purification can be found in the original ChIC protocol (Schmid, Durussel et al. 2004). Once recombinant pA-MN has been generated, the MNase efficiency should be tested. MNase activity should be titrated using standard procedures (Unit 21.1, (Zaret 2005, Skene and Henikoff 2017)). Briefly, cells should be gently lysed and while holding cell numbers constant, a range of pA-MN should be added to lysates (from 0U to 30U). After digestion for a fixed period of time, the reaction is terminated by the addition of EDTA. After purification of DNA, the size distribution should be determined by agarose gel electrophoresis or bioanalyzer analysis. Commercial MNase (such as Clontech 2910A) should be used as a side-by-side control for MNase activity. After endonuclease activity of pA-MN is determined, the amount of pA-MN to be used in a CUT&RUN experiment can be further titrated for CUT&RUN as previously described (Skene and Henikoff 2017). Briefly, a CUT&RUN experiment (Basic Protocol 1) should be performed, but a titration of pA-MN should be used (e.g. 1:50, 1:100, 1:200, 1:400, and 1:800 final dilutions). Libraries should be built (Basic Protocol 2) and fragment enrichment should be assessed and compared between the different pA-MN dilutions (Basic Protocol 3) to identify conditions optimal for mapping of chromatin proteins.
Targeted MNase cleavage
Due to the small volume of CaCl2 added, proper mixing of CaCl2 upon addition is critical. We recommend vortexing and flicking the sample during and immediately following addition, respectively. Furthermore, MNase cleavage is performed at 0°C, which is suboptimal for MNase activity. This is intentional to prevent over-digestion and off-target digestion by pA-MN. Therefore, it is important to act quickly and carefully when performing this step, without heating the sample with one’s hand. We recommend equilibrating the samples to 0°C by incubation in an ice water bath for at least 5 minutes, turning the vortex mixer on, and having 3uL of CaCl2 within your pipette tip prior to removing the sample from the ice water. Also, holding the upper half of the microcentrifuge tube with your fingers prevents heating of the sample from your hand. Finally, by starting a timer upon addition of CaCl2 to the first sample, and stopping the reaction in the same order, appropriate timing is more easily maintained. See video 3 for an example of CaCl2 addition.
Fragment release and clean up
Pre-heating the thermomixer or other incubator to 37°C and 70°C prior to RNase and Proteinase K addition, respectively, assists in rapid clean up. Carefully taking all of and only the aqueous phase during PCI and chloroform purification steps is important to prevent DNA loss and degradation.
Library preparation
Generating master mixes for each step is highly recommended as it helps to control for variation in sample preparation. Continuing with the library build immediately following end repair/ adenylation and adapter ligation is important to promote high quality DNA libraries as over ligation can result in inappropriate ligation intermediates.
Understanding Results
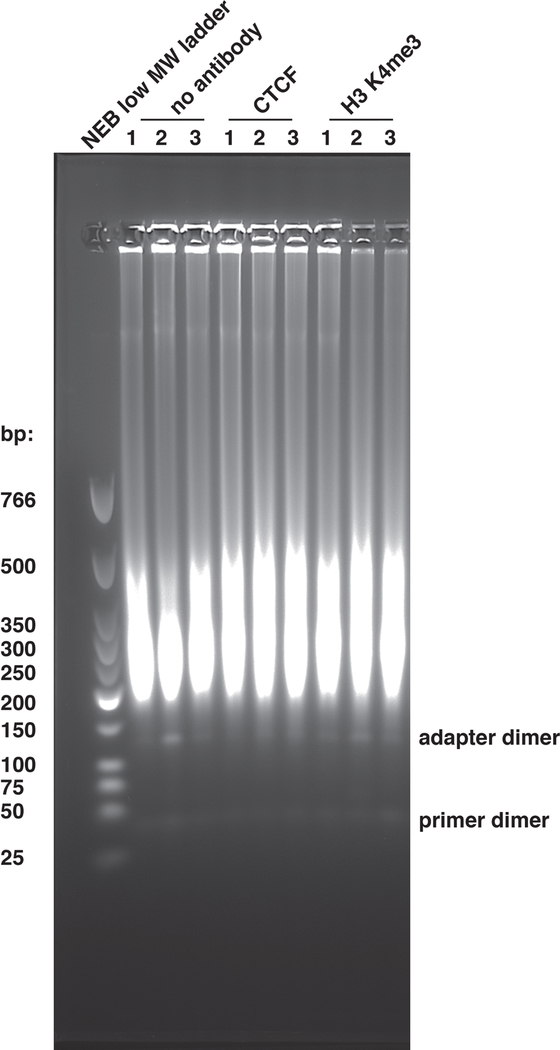
The method described here permits for the mapping of TFs and histone proteins rapidly to high resolution. Prior to gel extraction at the end of the library build, the libraries will be visualized on the agarose gel. Here, a strong “smear” should be observed (Figure 2). Often, adapter dimers and/or PCR dimers are also observed and should not be included in the gel extracted DNA.
Figure 2: Visual example of CUT&RUN libraries on a gel.
Nine CUT&RUN libraries are shown on an agarose gel after experimentation and library preparation, just prior to gel extraction for size selection and purification. The marker included in the leftmost lane is the NEB low molecular weight ladder. Both adapter dimers and PCR dimers are labeled for clarity.
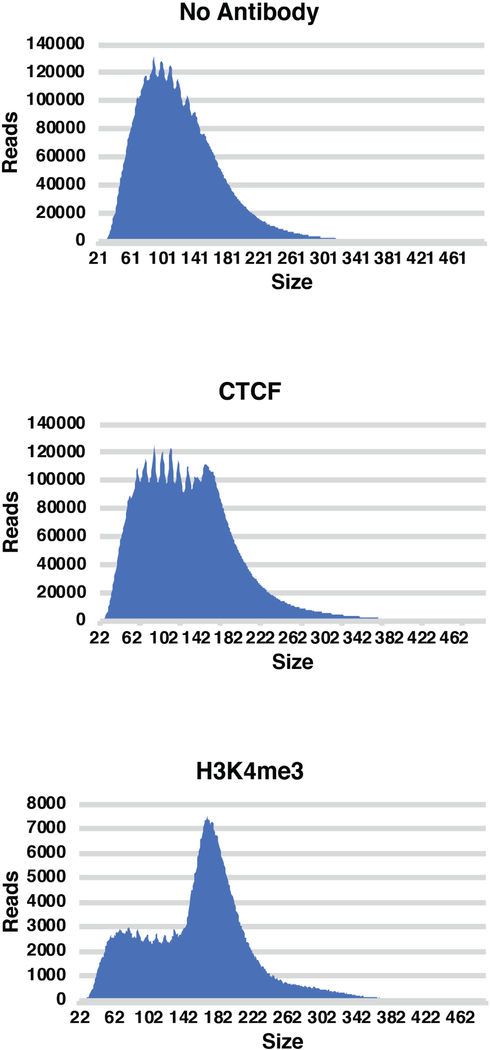
Quality control steps can be taken at the end of the library build. These include quantification of DNA concentration and assessing the size distribution of the library (Figure 3). For profiling of transcription factors, the fragment sizes will be enriched under 120 bp, although large shoulders can occur over 150bp when adjacent nucleosomes are profiled (such as the case with CTCF CUT&RUN). For profiling histone proteins, a large portion of the reads will usually be over 150 bp. Furthermore, prior to deep sequencing, libraries can be cloned, and individual fragments can be sequenced through Sanger sequencing to confirm that adapters are appropriately ligated, the fragments map to multiple locations, and the fragments map to the correct genome of interest.
Figure 3: Example CUT&RUN library size distributions.
After sequencing, the CUT&RUN library size distributions are assessed to confirm enrichment profiles. No primary antibody libraries typically show reduced size enrichment over 150 bp relative to factor-specific libraries. CTCF CUT&RUN libraries (representative of many TF libraries) show a slight right “shoulder” over 150 bp, due to MNase cleavage of DNA on either side of nucleosomes immediately flanking CTCF binding sites. H3K4me3 libraries show a strong enrichment of library fragments over 150 bp, but also include smaller inserts corresponding to open chromatin regions near sites of H3K4me3 enrichment.
Once genome-wide sequencing reads have been obtained and reads have been mapped to the appropriate reference genome, the distribution of recovered fragments can be assessed to confirm appropriate fragment sizes were obtained. Mapping efficiency for 500,000 cells should be approximately 60–70%. Reads can also be mapped to a genome browser (such as UCSC or IGV) to confirm enrichment of the protein of interest at expected locations (Figure 4). Furthermore, if ChIP-seq data are available for the factor profiled, CUT&RUN data can be compared either through visualization on a genome browser track or by aligning the CUT&RUN datasets over the available ChIP-seq peaks. Bound sites can be identified across the genome using peak calling algorithms.
Time Considerations
The time to complete a CUT&RUN experiment and library is significantly less than that of traditional crosslinking ChIP experiments. The steps prior to building the library can be completed in 1 day. The library preparation can be completed in 1 additional day. Detailed time estimates for each step are included below. Quality control and sequencing of the libraries can take as little as one additional day, depending upon available resources. Data analysis depends on the familiarity with the software packages and datasets but is generally not substantially more complex than analyses of ChIP-seq data.
Basic Protocol 1
Buffer preparation: 0.5 hour (0.5 hour bench time)
Lysis and binding to beads: 1 hour (1 hour bench time)
Primary antibody: 2.5 hours (2 hour incubation, 0.5 hour bench time)
pA-MN: 1.5 hours (1 hour incubation, 0.5 hour bench time)
MNase digestion: 1 hour (0.5 hour incubation, 0.5 hour bench time)
Fragmentation and purification: 2 hours (1 hour incubation, 1 hour bench time)
Alternate Protocol 1
Secondary antibody: 1.5 hours (1 hour incubation, 0.5 hour bench time)
Basic Protocol 2
Buffer preparation: 0.5 hour (0.5 hour bench time)
End repair and Adenylation: 1.5 hour (50 minute incubation, 40 minute bench time)
Adapter ligation: 0.5 hour (15 minute incubation, 15 minute bench time)
AMPure Purification: 0.5 hour (20 minute incubations, 10 minute bench time)
Library Enrichment: 40 minutes (20 minute PCR, 20 minute bench time)
Gel extraction: 3 hours (2 hour running time, 1 hour extraction)
Supplementary Material
Separation of sample by magnetic stand. This video demonstrates how to carefully remove supernatant from beads after separation on a magnetic stand, and how to gently resuspend beads during wash steps. By removing the supernatant and immediately resuspending in the Wash buffer, beads do not over-dry.
Addition of sample to bead mixture. This video demonstrates how to gently vortex samples and add master mixes. The technique should be performed when initially adding the Concanavalian A beads to nuclei (Basic protocol 1 step 18), when adding the primary antibody master mix to bead-bound nuclei (Basic protocol 1 step 31), and when adding the pA-MN master mix to bead-bound nuclei (Basic protocol 1 step 43).
Addition of CaCl2 for MNase activation. This video demonstrates a critical step in the protocol (Basic Protocol 1 Step 54). Here, the ice water bath is shown for assistance. After samples are equilibrated to 0°C, the vortex mixer is turned on and 3 µL of 100mM CaCl2 is pipetted. Then the sample is briefly taken out of the ice water, opened, and the 3 µL of CaCl2 is dispensed during gentle vortexing. The 1.5 mL tube is capped, flicked gently three times, and returned to the ice water. To appropriately time the reaction, a timer set to 30 minutes should be started after addition of CaCl2 to the first sample, and then the 2XSTOP buffer can be added to samples in the order that CaCl2 was added.
ACKNOWLEDGEMENT
S.J.H. is a Special Fellow of the Leukemia and Lymphoma Society. T.G.F. is a Leukemia and Lymphoma Society Scholar and supported by NIH grants R01HD072122 and R01HD093783.
Footnotes
INTERNET RESOURCES
Information on IgG affinities: https://www.neb.com/tools-and-resources/selection-charts/affinity-of-protein-ag-for-igg-types-from-different-species
Picard website: http://broadinstitute.github.io/picard/
HOMER website: http://homer.ucsd.edu/homer/
Samtools website: http://samtools.sourceforge.net
Bowtie2 website: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
LITERATURE CITED
- Baranello L, Kouzine F, Sanford S and Levens D (2016). “ChIP bias as a function of cross-linking time.” Chromosome Res 24(2): 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind’Amour J, Liu S, Hudson M, Chen C, Karimi MM and Lorincz MC (2015). “An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations.” Nat Commun 6: 6033. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY and Greenleaf WJ (2015). “ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide.” Curr Protoc Mol Biol 109: 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, Preissl S, Jermstad I, Haugen MH, Suganthan R, Bjoras M, Hansen K, Dalen KT, Fedorcsak P, Ren B and Klungland A (2016). “Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition.” Nature 537(7621): 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DSLJT, (1984). “Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes.” PNAS 81: 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DSLJT, (1985). “In vivo interactions of RNA Polymerase II with genes of Drosophila melanogaster.” Molecular and Cellular Biology: 2009–2018. [DOI] [PMC free article] [PubMed]
- Hainer SJ, Boskovic A, Rando OJ and Fazzio TG (2018). “Profiling of pluripotency factors in individual stem cells and early embryos.” bioRxiv 286351(March 21, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H and Glass CK (2010). “Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities.” Mol Cell 38(4): 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens DH, Wu SJ, Sarthy JF, Meers MP, Myers CH, Olson JM, Ahmad K and Henikoff S (2018). “Automated in situ profiling of chromatin modifications resolves cell types and gene regulatory programs.” bioRxiv 418681(September 16, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DSMA; Myers RM,; Wold B (2007). “Genome-wide mapping of in vivo protein–DNA interactions.” Science 316: 1497–1502. [DOI] [PubMed] [Google Scholar]
- Meyer CA and Liu XS (2014). “Identifying and mitigating bias in next-generation sequencing methods for chromatin biology.” Nat Rev Genet 15(11): 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K and May KM (2017). Mammalian Cell Tissue Culture Techniques. Curr. Protoc. Molec. Bio 10.1002/cpmb.31 [DOI] [PubMed] [Google Scholar]
- Raha D, Hong M and Snyder M (2010). “ChIP-Seq: a method for global identification of regulatory elements in the genome.” Curr Protoc Mol Biol Chapter 21: Unit 21 19 21–14. [DOI] [PubMed] [Google Scholar]
- Rhee HS and Pugh BF (2011). “Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution.” Cell 147(6): 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA and Bernstein BE (2015). “Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state.” Nat Biotechnol 33(11): 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Durussel T and Laemmli UK (2004). “ChIC and ChEC; genomic mapping of chromatin proteins.” Mol Cell 16(1): 147–157.This paper developed recombinant proteinA-MNase and described its use in their ChIC technique to map the chromatin binding protein Gbd in yeast.
- Schmidl C, Rendeiro AF, Sheffield NC and Bock C (2015). “ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors.” Nat Methods 12(10): 963–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG and Henikoff S (2018). “Targeted in situ genome-wide profiling with high efficiency for low cell numbers.” Nat Protoc 13(5): 1006–1019. [DOI] [PubMed] [Google Scholar]
- Skene PJ and Henikoff S (2017). “An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites.” Elife 6.This paper developed CUT&RUN as a novel genome-wide mapping technique that leads to high resolution mapping of TFs and histone proteins in yeast and human K562 cells.
- van Steensel BHS, (2000). “Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase.” Nature Biotechnology 18(4): 424–428. [DOI] [PubMed] [Google Scholar]
- Zaret K (2005). “Micrococcal Nuclease Analysis of Chromatin Structure.” Curr Protoc Mol Biol Chapter 21. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Kasinathan S, Xin B, Rohs R and Henikoff S (2015). “ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo.” Nat Commun 6: 8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart WK,R; Wesseling J; Rutgers E; Linn S; Carroll JS (2013). “A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples.” BMC Genomics 14: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Separation of sample by magnetic stand. This video demonstrates how to carefully remove supernatant from beads after separation on a magnetic stand, and how to gently resuspend beads during wash steps. By removing the supernatant and immediately resuspending in the Wash buffer, beads do not over-dry.
Addition of sample to bead mixture. This video demonstrates how to gently vortex samples and add master mixes. The technique should be performed when initially adding the Concanavalian A beads to nuclei (Basic protocol 1 step 18), when adding the primary antibody master mix to bead-bound nuclei (Basic protocol 1 step 31), and when adding the pA-MN master mix to bead-bound nuclei (Basic protocol 1 step 43).
Addition of CaCl2 for MNase activation. This video demonstrates a critical step in the protocol (Basic Protocol 1 Step 54). Here, the ice water bath is shown for assistance. After samples are equilibrated to 0°C, the vortex mixer is turned on and 3 µL of 100mM CaCl2 is pipetted. Then the sample is briefly taken out of the ice water, opened, and the 3 µL of CaCl2 is dispensed during gentle vortexing. The 1.5 mL tube is capped, flicked gently three times, and returned to the ice water. To appropriately time the reaction, a timer set to 30 minutes should be started after addition of CaCl2 to the first sample, and then the 2XSTOP buffer can be added to samples in the order that CaCl2 was added.