Abstract
Flow cytometry approaches combined with a genetically-encoded targeted fluorescent biosensor are used to determine the subcellular compartmental availability of the oxidized form of nicotinamide adenine dinucleotide (NAD+). The availability of free NAD+ can affect the activities of NAD+-consuming enzymes such as Sirtuin, PARP/ARTD and cyclic ADPR-hydrolase family members. Many methods for measuring the NAD+ available to these enzymes are limited because they cannot determine free NAD+ as it exists in various subcellular compartments distinctly from bound NAD+ or NADH. Here, we outline an approach to express the sensor in mammalian cells, monitor NAD+-dependent fluorescence intensity changes using flow cytometry approaches, and analyze the data obtained. The benefit of flow cytometry approaches with the NAD+ sensor is the ability to monitor compartmentalized free NAD+ fluctuations simultaneously within many cells, which greatly facilitates analyses and calibration.
Keywords: NAD+, metabolite, nicotinamide adenine dinucleotide, genetically-encoded biosensor, fluorescent sensor, circularly-permutated fluorescent protein, sirtuin, PARP, ARTD, CD-38
Introduction
In eukaryotes the intermediary metabolite nicotinamide adenine dinucleotide predominantly exists as its oxidized form (NAD+), serving two major cellular roles that are evolutionarily conserved (Verdin, 2015; Yang et al., 2016). One role for NAD+ is in oxidoreductive (redox) reactions, and a key example includes NAD+ fueling the Krebs/citric acid cycle to ultimately drive ATP production. The NAD+/NADH ratio controls flux through these pathways, and as several of the steps are reversible, the concentration of NAD+ may contribute to the directionality of these reactions as well. The majority of intracellular NAD+ is engaged in redox reactions and is thus tightly associated with protein (Holzer et al., 1956; Bucher et al., 1958; Williamson et al., 1967). Equally important for cellular fitness is the distinct role of NAD+ as a required substrate for a broad class of enzymes, termed NAD+-consuming enzymes. In mammals, these include 7 Sirtuin deacylase family members, 17 ADP-ribose transferases (PARP/ARTD family members), and at least three cyclic ADP-ribose hydrolases (Houtkooper et al., 2010; Gerdts et al., 2015). These enzymes cleave the glycosidic linkage between the nicotinamide and ribose moieties, resulting in the consumption of an NAD+ molecule for each enzymatic cycle. As such, NAD+-consuming enzymes depend on the local availability of free NAD+ molecules.
Many human pathologies—including neurodegeneration, cardiovascular disease, metabolic syndrome, and cancer—are linked to misregulated NAD+-consuming enzymes (Houtkooper et al., 2010; Verdin, 2015; Yang et al., 2016). The local concentration of NAD+ therefore has been proposed to contribute to these diseases by being limiting for specific NAD+-consuming enzymes, many of which have KM(NAD+) values that approximate local physiological concentrations of NAD+ (Houtkooper et al., 2010). The challenge, nevertheless, is being able to measure the amount of free NAD+ in cells that is available to NAD+-consuming enzymes in biologically relevant contexts. The abundance of the protein-bound NAD+ creates a challenge when assessing the amount of free NAD+ available as substrate. The protein-bound NAD+ in redox reactions does not turn over, thus steady-state levels of the bound fraction minimally fluctuate under normal physiology. As such, the abundance of the protein-bound fraction may overwhelm any detection of free NAD+ fluctuations in total measurements. The free NAD+ fraction, in contrast, is susceptible to steady-state changes through the balance between its biosynthetic and consumption pathways (Fig. 1).
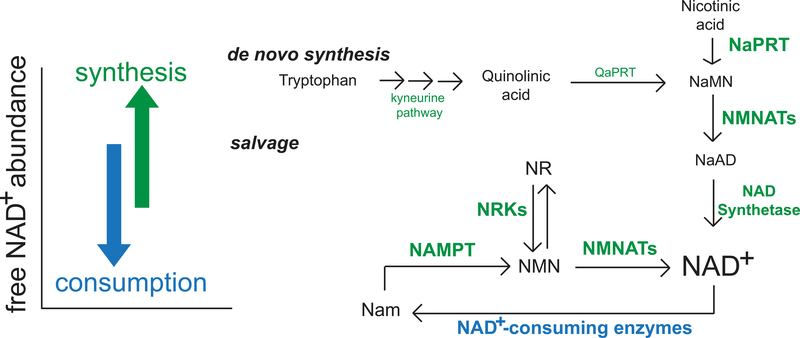
Fig. 1. Regulation of NAD+ steady-state concentrations.
Free intracellular NAD+ pools are regulated predominantly by biosynthetic (green) and consumption (blue) pathways, which synthesize or turn over NAD+ molecules, respectively. In mammalian cells, NAD+ synthesis largely depends on the salvage pathway. Thus, the specific targeting of the mammalian salvage pathway through inhibition of NAMPT results in robust depletion of NAD+ abundance in the subcellular compartments tested here because resident consuming mechanisms are left intact. QaPRT, quinolinate phosphoribosyltransferase; NaMN, nicotinic acid mononucleotide; NMNAT, nicotinamide mononucleotide adenylyltransferase; NaAD, nicotinic acid adenine dinucleotide; NR, nicotinamide riboside; NMN, nicotinamide mononucleotide; NRK, nicotinamide riboside kinase; Nam, nicotinamide.
Local concentrations of NAD+ can differ in different biological compartments, and subcellular concentrations of NAD+ can be regulated independently through biosynthetic pathways or depleted by the local consuming enzymes. Understanding this would provide (1) an important framework for understanding NAD+ regulation; (2) information about where, when, how and the extent to which local free NAD+ concentrations may be limiting in disease, and (3) help identify treatments or approaches that may be able to influence local free NAD+ availability. Towards this goal, we have developed a genetically-encoded fluorescent sensor for free NAD+ that can be localized to specific subcellular compartments for direct measurements (Fig. 2) (Cambronne et al., 2016). We have generated three versions of the NAD+ biosensor targeted to the nucleus, cytoplasm or mitochondrial matrix and describe here an approach to monitor the steady-state levels of free NAD+ in these specific subcellular compartments. Details about the sensor’s design can be found in Cambronne et al., 2016.
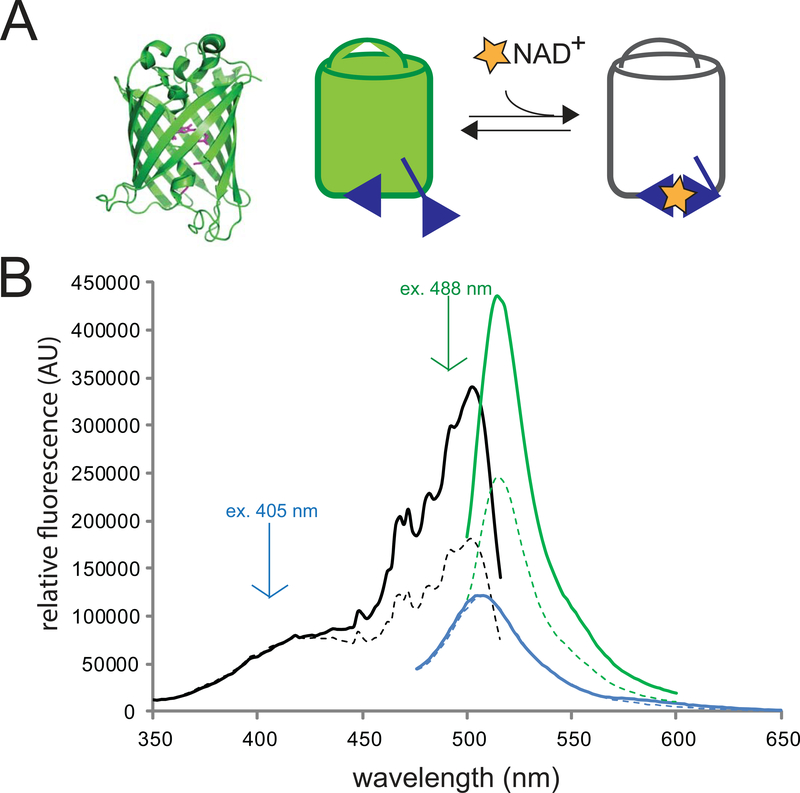
Fig. 2. Characteristics of the NAD+ sensor.
a) (left) Structural depiction of a circularly-permuted fluorescent protein similar to cpVenus (modified PDB: 3EVP). Highlighted in magenta are the new N- and C- termini (formally residues 145 and 146, respectively), which have been relocated closer to residues that constitute the chromophore centered in the beta-barrel. (right) When the bi-partite NAD+-binding pocket of the sensor (blue) binds NAD+ (yellow star) the sensor’s fluorescence decreases.
b) The NAD+ sensor harbors a primary peak in its excitation spectrum monitored at 530 nm (black), which can be excited at 488 nm to produce fluorescence ~520 nm (green). A secondary excitation peak is excitable at 405 nm to yield fluorescence ~510 nm (blue). The presence of NAD+—shown here by addition of 500 μM NAD+ (dotted line)—decreases the fluorescence following 488 nm excitation but has minimal effect on the fluorescence from excitation at 405 nm.
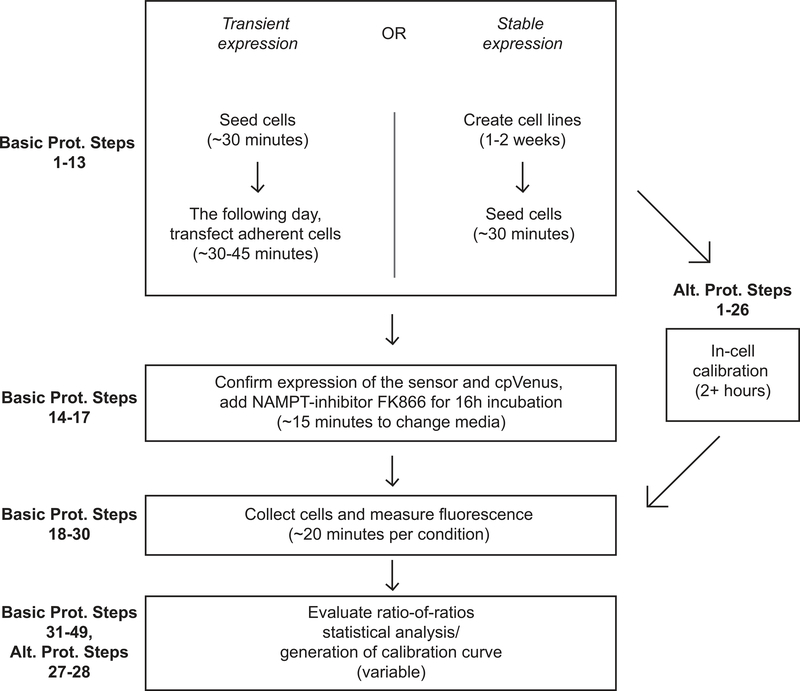
In this protocol we describe two methods for measuring free NAD+ with the sensor using flow cytometry approaches in mammalian cells (Fig. 3). The first protocol describes a transient approach for expressing the sensor from a plasmid (Basic Protocol steps 1–17) and obtaining relative measurements of NAD+ fluctuations (Basic Protocol steps 18–39). We additionally cover how to perform statistical analyses to evaluate the significance of measured changes from experimental replicates (Basic Protocol steps 40–50). This transient transfection protocol works best with flow cytometry measurements where it is relatively easy to obtain a large population of fluorescent cells for evaluation. The alternate protocol (Alternate Protocol 1) will describe the initial experimental preparation required to calibrate the sensor in cells using flow cytometry, including its stable expression and monitoring of the sensor in stable mammalian cell lines. Alternate Protocol Steps 16–28 then cover how to create an in-cell calibration curve for interpolating cytoplasmic measurements obtained from flow cytometry. This method may be more difficult for labs not experienced with cell culture or labs who do not wish to spend time creating the stably expressing line, but it does offer the ability to measure quantitative values of NAD+ concentration. Figure 3 shows a diagram of the work-flow for each method.
Fig. 3. Flow chart outlining protocol steps.
Basic Protocol 1: Transient Expression of the NAD+ Sensor and Analysis via Flow Cytometry
Measurement of subcellular localization of free NAD+ is critical to understanding changes in function and regulation of NAD+ consuming enzymes within eukaryotic systems. We developed a genetically-encoded fluorescent biosensor capable of accurately detecting fluctuations in free NAD+ within targeted subcellular compartments that can be analyzed through flow cytometric methods. Here, we describe how to transiently transfect a cell line with the NAD+ sensor. Then we describe how to treat these cells with FK866 {N-[4-(1-benzoyl-4-piperidinyl)butyl]-3-(3-pyrindyl)-2Epropenamide}. FK866 is a highly specific small molecule inhibitor for nicotinamide phosphoribosyltransferase (NAMPT). By inhibiting the enzyme responsible for a rate-limiting step in mammalian NAD+ biosynthesis (Revollo et al., 2004) we can effectively lower NAD+ concentrations through its depletion across cellular compartments and measure this reduction as an increase in fluorescence intensity with the sensor and flow cytometry (Cambronne et al., 2016). We describe how to evaluate these fluorescence changes using flow cytometry. We also show how to use FlowJo software to perform a ratiometric analysis of the flow cytometry data and how to use STATA-14 software to evaluate the statistical significance of the relative changes within the experimental replicates.
Materials
Dulbecco’s Modified Eagle Medium (DMEM) + 4.5 g/L D-Glucose, + 110 mg/L Sodium Pyruvate (Thermo Fisher Cat# 11995–065); store at 4°C.
Fetal Bovine Serum (FBS) (Thermo Fisher Cat# 10437028); store at −20°C until use, then keep at 4°C.
1M HEPES Buffered Solution pH 7.4 (Thermo Fisher Cat# 15630080)
Dulbecco’s Phosphate Buffered Saline (DPBS) without Calcium Chloride, without Magnesium Chloride (Thermo Fisher Cat# 14190–144)
0.05% vol/vol EDTA-Trypsin (Thermo Fisher Cat# 25300–062), store at −20°C.
Lipofectamine 2000 (Thermo Fisher Cat# 11668–019); store at 4°C.
NAD+ Sensor Purified Plasmid DNA (2 μg for 35 mm dish. 20 μg for 10 cm dish.)
cpVenus Purified Plasmid DNA (2 μg for 35 mm dish. 20 μg for 10 cm dish.)
HeLa cells: Healthy, proliferating and maintained in a 10 cm dish or similar.
Cells lines are to be regularly checked to ensure authenticity and lack of mycoplasma infection via PCR approaches.
HEK293T cells: Healthy, proliferating and maintained in a 10 cm dish or similar.
Cells lines are to be regularly checked to ensure authenticity and lack of mycoplasma infection via PCR approaches.
Serum-free Opti-MEM Media +HEPES, +2.4 g/L Sodium Bicarbonate, +L-Glutamine (Thermo Fisher Cat# 31985–070); store at 4°C.
FK866 (Cayman Chemical, cat# 13287); store stock at 50 mM at −20°C: Serially dilute fresh to final 10 nM concentration before use on cells.
Dimethyl Sulfoxide (DMSO) (Fisher Chemicals 67–68-5)
β-Nicotinamide adenine dinucleotide hydrate (NAD+), (Sigma N1636, >99% pure, 25mg); store powder at −20°C with desiccant.
1N Sodium Hydroxide (NaOH) (Fisher Scientific cat#SS2661): Used to adjust pH, can be diluted to 0.1N.
5% wt/vol Digitonin (Thermo Fisher cat# BN2006); store at 4°C. Heat at 95°C for 30 seconds to 2 minutes and vortex to re-dissolve prior to use.
Propidium Iodide (Sigma P4170, 668.39 g/mL): make up as 10 mg/mL in PBS. Aliquot and store in dark at −20°C. Dilute immediately prior to use.
MColorpHast pH-indicator strip 5.0–10.0 (Millipore109533)
Mitotracker Red CMXRos (Thermo Fisher M7512)
Hoechst 33342 (Thermo Fisher H3570)
Biosafety Cabinet
Eppendorf New Brunswick Galaxy 170S CO2 Cell Culture Incubator (or similar) kept at 37°C with 5% CO2
6-well TC treated plastic plates (Corning Cat# 353406)
Serological pipet tips:
2mL Aspirating Pipet (Fisher# 13–678-11C)
5mL Pipet (Fisher# 13–678-11D)
10mL Pipet (Fisher# 13–678-11E)
25mL Pipet (Fisher# 13–678-11)
Portable Pipet-Aid (Corning Cat# 4099) or similar.
Eppendorf Research Plus Micropipette in sizes P10, P20, P200, P1000 or similar.
Barrier Tips:
10uL (USA Scientific 1121–2710)
20 uL (USA Scientific 1123–1710)
200 uL (USA Scientific 1120–8710)
1000 uL (USA Scientific 1122–1730)
5mL polystyrene round-bottom tubes (Corning Cat# 352235)
Hemocytometer (Fisher Scientific Cat# 0267110)
Ice
Flow Cytometer: BD LSR II, BD Fortessa (or similar)
Filer Set: 405–2, Laser: 405nm; Detector 525 ± 25 nm
Filter Set: 488–1, Laser: 488nm, Detector 530 ± 15 nm
Filter Set: 561–3, Laser: 561nm, Detector: 670 ± 15 nm
FlowJo Data Analysis Software Version 10-Microsoft (or similar)
Inverted wide-field light and fluorescent microscope with Dapi, GFP, and DsRed compatible filter sets (Sole SM II light engine, or similar)
STATA-14 statistical analysis software (or similar)
Seeding Cells
-
1Calculate the number of experimental conditions needed.
- Before starting, ensure HeLa cells are healthy, free of mycoplasma contamination, and have been proliferating steadily.
-
2Aspirate the media from HeLa cells in 10 cm plate.
- Do not leave cells to dry out without media or PBS.
- All steps involved with seeding cells should be performed in a biosafety cabinet with sterile technique to prevent contamination of cells.
-
3Wash cells with 10 mL PBS and remove. Add 1 mL of 0.05% vol/vol EDTA-trypsin onto cells and incubate at 37°C and 5% CO2 until all cells are detached.
- To keep cells healthy do not trypsinize cells longer than 5 minutes.
- Verify that cells have completely detached using an inverted wide-field light microscope.
- Cells should be rounded up or free-floating before continuing.
-
4
Quench trypsin by adding 10 mL of complete cell culture media (DMEM + 10% vol/vol FBS + 25 mM HEPES) to the dish. Triturate cells into a single-cell suspension.
-
5
Use the hemocytometer or digital cell counter to count and calculate the number of cells per mL. You may need to dilute cells to accurately count.
-
6
Determine the number of cells needed for the total experiment. We suggest starting at ~250 000 HeLa cells per well of a 6-well plate; each well holds 2 mL of volume. Dilute stock of cells appropriately in complete growth media and seed HeLa cells in a 6-well plate at a concentration such that they will reach ~50% confluency in 24 hours.
Transfection
-
7
Twenty-four hours after seeding, HeLa cells are ready for transfection with expression plasmids for the sensor or cpVenus controls. Confirm that cells are ~50% confluent using the light microscope.
-
8In a sterile tissue culture hood, prepare a master mix containing Lipofectamine 2000 transfection reagent diluted i n serum-free Opti-MEM media. For each well, the ratio is 5 μL of Lipofectamine 2000 in 200 μL of Opti- MEM media for 2 μg of total DNA transfected; e.g. for 4 transfections, prepare 20 uL of Lipofectamine 2000 in 800 μL of Opti- MEM media. Vortex to mix.
- Lipofectamine 2000 reagent should be kept cold on ice until use.
- Serum free Opti-MEM media should be a peach color to indicate correct pH
-
9
Prepare tubes containing either the expression plasmid DNA for the sensor or cpVenus diluted in Opti-MEM media. For each 6-well, dilute 2 μg of DNA in 200 μL Opti-MEM; e.g. for 2 transfections with the sensor plasmid representing treated and untreated conditions, dilute 4 μg sensor plasmid in 400 μL Opti-MEM media. Vortex to mix well.
-
10
Add 400 μL of the Lipofectamine 2000 transfection master mix to the tubes containing either the diluted sensor or cpVenus plasmids. Vortex to mix well and incubate at room temperature for 20 minutes.
-
11
After incubation, add 400 μL per well of the Lipofectamine-DNA transfection mixture dropwise onto the already seeded HeLa cells.
-
12
Return the cells to the incubator for 2 hours.
-
13After 2 hours, remove media containing the transfection mixture and replace with new complete growth media.
- Increased incubation with the transfection mixture may be toxic for this cell type.
-
14Confirmation of a successful transfection can be observed with fluorescence microscopy in the green channel by the next day. Transfected cells should fluoresce green and localize to expected subcellular compartments.
- Do not leave cells out of the incubator for over 10 minutes as this may negatively affect their health.
- It is important to confirm that the sensors are localized to expected subcellular compartments via microscopy. Broad illumination with a GFP-compatible filter is sufficient to visualize the sensor. Either prior to the experiment or in parallel, the subcellular localization of the sensor in live cells can be correlated with nuclear Hoechst 33342 staining and mitochondrial staining with red mitotracker; both dyes are cell permeable and amenable to use in live cells. Subcellular localization of the sensor in live cells can also be estimated by comparison of the sensor’s expression pattern relative to brightfield images.
Treatment with small molecule NAMPT inhibitor FK866.
-
15
Twenty-four hours post-transfection, cells can be treated with freshly diluted 10 nM FK866. We always freshly dilute our FK866 from the DMSO stock in media with serial dilutions from 50 mM down to 10 nM. The 0 nM FK866 treatment should include serially diluted DMSO similar to the 10 nM FK866 treatment to serve as a control.
-
16
Aspirate media from transfected HeLa cells and gently replace with either 0 or 10 nM FK866 complete growth media as indicated in Table 1.
-
17Return cells to incubator for 16 hours.
- Depending on the biological compartment and cell type, we have observed differences in the rate of NAD+ depletion (Cambronne et al, 2016). We suggest 16 hours as a reasonable starting point for these experiments, where we have consistently seen depletion in most compartments. When we incubate HeLa cells with FK866 for over 18 hours we begin to see cell morphological changes in the population that indicate compromised cellular health (Fig. 4). To ensure that we are evaluating relatively healthy cells and to prevent misinterpretation by secondary effects, we limit our FK866 treatments to 16 hours or less.
Table 1. Minimal experimental conditions required for a single assay.
This table outlines the minimum experimental conditions required to evaluate NAD+ changes in a single subcellular compartment. To evaluate multiple sensors that target to distinct subcellular localizations, an equivalently-targeted cpVenus control is required and a similar set of conditions are needed for each sensor.
| Treatment: | To be transfected: | ||
|---|---|---|---|
| cpVenus control | NAD+ Sensor | No transfection | |
| 0 nM FK866 | Condition 1 | Condition 2 | Condition 5* |
| 10 nM FK866 | Condition 3 | Condition 4 | (optional) |
This control will be used to set up cytometry gates and distinguish the cells expressing the sensor compared to untransfected cells during fluorescence measurements.
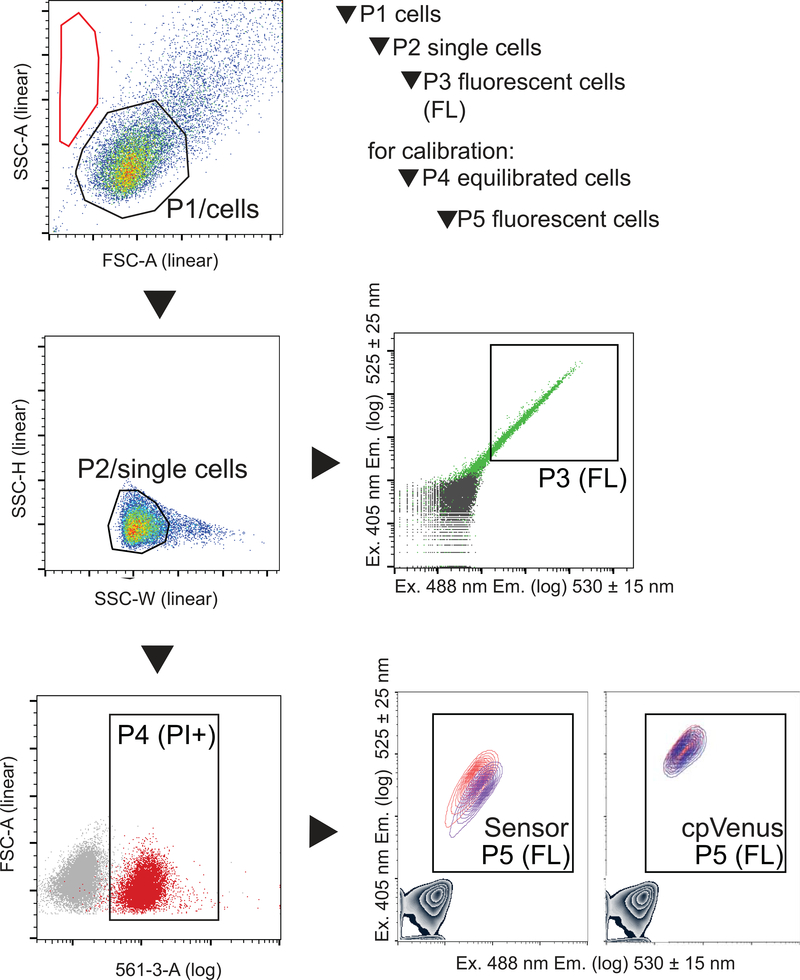
Fig. 4. Hierarchical gates to evaluate fluorescent cells.
Gates are applied identically to all samples. The P1 gate outlines the uniform population of HeLa cells for evaluation (typically >75% of total events). Under these specific conditions, events in the red region represent cellular debris or mechanically disrupted cells; if >50% of events are in this region it indicates an overall unhealthy sample. From the P1/cells population, single cells are identified by the P2 gate using their size distribution (typically > 90% of P1). The P3/fluorescent population is derived from the P2/single cell population. The P3 fluorescent cell population is defined by a gate that excludes non-fluorescent cells in the untransfected sample (gray) and includes transfected cells (green, typically >50% of P2). There should be ~10 000 events in P3 for an accurate evaluation. To calibrate the sensor in cells, an additional gate is required to identify the permeabilized and equilibrated cells. This P4 gate (typically >85% of P3) is derived from uniform, single cells in P2, and is delineated by equilibrated intracellular propidium iodide (PI). Fluorescence is then evaluated in P5, which is derived from P4. In this example, a gray density contour plot in the lower left quadrant represents cells that do no express either sensor or cpVenus. Cells that express either Sensor or cpVenus populate P5, and the fluorescence of the populations treated with either equilibrated buffer (red) or 500 μM NAD+ (indigo) are overlaid. Data was collected on a BD Fortessa flow cytometer and analyzed on FlowJo V10 software.
Preparation of cells for flow cytometry evaluation.
-
18
Prepare and label flow cytometry sample tubes on ice.
-
19Remove growth media from cells and gently wash 1X with PBS.
- This preparation must be within 5–10 minutes from start to finish due to the analysis being performed on live cells out of the incubator. If you have more samples than can be handled in this time, trypsinize, collect cells, and analyze in small batches (i.e. one 6-well plate at a time). From here onward, contamination of the cell culture is no longer a consideration and cells can be prepared for analysis outside of the biosafety cabinet if needed.
-
20
Add 100 μL of 0.05% EDTA-trypsin to each well of cells and incubate at 37°C until you can easily see cells come off the plate.
-
21Add 400uL freshly-prepared complete growth media to each well to quench the trypsin.
- It is critical to ensure that the media used to resuspend cells (complete growth media including HEPES) is within near-neutral pH. If it contains phenol red, the medium should be orange-red in color without any traces of pink or magenta.
- HEPES is necessary in the media to buffer the cell suspension during flow analysis, which is typically performed on the bench outside of an incubator.
-
22Triturate with a 1mL pipette tip to obtain single cell suspension and immediately transfer to sample tube on ice.
- Including serum in the media and keeping the cells on ice helps to prevent cells from clumping.
Flow cytometry measurements
-
23
Refer to Table 2 for instrumental guidelines and data parameters to collect.
-
24
Ensure that the lasers have been warmed up and are stabilized, and that lines have been cleaned from the previous user with bleach, detergent, and water. Ideally, your template is pre-loaded on the instrument so that they can be immediately processed. If not, save a new template from your analysis when finished. When choosing the speed to perform the cytometry analysis, rapid evaluation of fluorescence across samples is preferable, so it is recommended to run samples on “high” if possible.
-
25
(Optional) It is important that the cells are sufficiently separated as a single-cell suspension otherwise the flow can become clogged. The cell suspension can be filtered through a 40–70 μm nylon mesh to ensure that no significant clumps are present in the sample.
-
26The first data should be collected from untransfected HeLa cells (condition 5, Table 1) to ensure the gates have been accurately drawn around the three populations.
- For the HeLa sample, confirm that cells appear in P1 and P2 but not in P3 (Fig. 4, gray population. Gates P1-P3 are utilized for this protocol. Gates P1-P5 are utilized for Alternate Protocol.)
-
27
The population of cells is evaluated in a hierarchy to obtain fluorescent measurements from healthy, single, and transfected cells (Fig. 4). Voltages of each laser should be adjusted per instrument and experiment, based on untransfected HeLa cells (condition 5, Table 1). All cell gates should be centered within the plot. The P1 gate identifies the healthy cell population with relatively uniform forward and side scatter; x-axis is set to FSC-A (linear) and the y-axis to SSC-A (linear) (Fig 4).
-
28
The P2 gate defines single cells and is derived from the P1 population in a new plot (Fig. 4). To perform a doublet-exclusion, select the P1 population for viewing and set the x-axis to SSC-W (linear) and the y-axis to SSC-H (linear). Draw the gate (P2) around the population on the left. This gate represents individual cells, as opposed to multiple cells stuck together that will appear as a group with a greater width.
-
29
The P3 gate is derived from the P2 population. P3 identifies cells that have been transfected and are fluorescent. The P3 gate is defined by exclusion of the untransfected, non-fluorescent cells in condition 5 (Fig. 4). Viewing the cells in P2 in a new plot, set the x-axis to log display for excitation at 405 nm and capture emission by filter set 525 ± 25 nm. The y-axis should be set in log display to excite at 488 nm and capture emission with filter set 530 ± 15 nm. Draw a gate to the right and above the cell population in the HeLa sample, ensuring that you do not include any events in this gate (Fig. 4). This gate will represent the fluorescent, sensor or cpVenus-expressing cell populations, which will be unpopulated for untransfected HeLa cells.
-
30The order of the remainder of the samples is not critical but should be completed within 15–20 minute sessions. It is necessary to collect at least 10 000 cells in P3 to rigorously evaluate the fluorescence of each sensor or cpVenus population. Save data from all events.
- Pause point. Once data has been collected, data can be analyzed at the user’s convenience.
Table 2. Measurements needed for flow cytometry analysis.
The voltage guidelines are provided here only as aides for setting up a template. They must be adjusted by the user such that all measurements are on-scale for the instrument’s specific lasers and set-up. Different instruments may have different voltage settings or may not have voltage controls.
| Name | Scale | Approximate voltage guidelines for HeLa cells |
|---|---|---|
| SSC-A | Linear | ~275 |
| FSC-A | Linear | ~220 |
| SSC-H | Linear | No Recommendation |
| SSC-W | Linear | No Recommendation |
| Filter Set: Ex. 488 nm, Em. 530 ± 15 nm | Log | ~275 |
| Filter Set: Ex. 405 nm, Em. 525 ± 25 nm | Log | ~250 |
Analyzing fluorescence using FlowJo Software
-
31
Data can be exported in FCS file format for analysis with a post-capture software such as FlowJo to determine the 488/405 nm fluorescence ratio per cell.
-
32
Open a new workspace and drag all FCS files (each representing a different experimental condition) into the workspace.
-
33
Double click the HeLa untransfected sample (condition 5, Table 1), which opens the data in a new window. Similar gates, as used to collect data in steps 26–28, will need to be re-defined here. We suggest viewing the data as a density plot in pseudocolor (Fig. 4).
-
34Once initial gates are defined, apply these gates to all samples to ensure that identical gating is performed on all samples. Confirm by checking for each condition that the gate is transferred and appropriately drawn such that it includes the correct cell population. If necessary, the gate can be redrawn and replaced in all samples with a larger gate.
- Confirm that a relatively equal number of cells (~10 000 cells) are being evaluated in the P3 gate across all experimental conditions, excluding the HeLa control sample.
-
3535. Determine fluorescence ratio of 488nm/405nm per cell. In the FlowJo software, select “derive parameters” under the “Tools” tab. You will enter the “formula” by clicking “Insert Reference” to select the 488 nm measurement, click the ÷ key, and “Insert Reference” to select the 405 nm measurement.
- Obtaining the 488/405 nm measurement per cell is distinct from taking the average 488 nm fluorescence of the population and dividing by the average 405 nm fluorescence of the same population of cells. This number describes the level of fluorescence change due to NAD+ (488nm) compared to the measurement of the amount of sensor expressed (405nm).
-
36
Apply this “Derived” value to “All Samples”. A histogram of the derived value should result in a defined peak. It is appropriate to use the geometric mean of this derived ratio as the fluorescence value for each experimental condition because fluorescence measurements are log-amplified. These ratiometric values can be exported into a table using the Table Editor function.
Analyzing Sensor Data to determine the “ratio of ratios”
-
37
The “ratio of ratios” value represents the ratiometric measurement of the sensor with respect to its cpVenus control or relative to untreated controls. Divide the ratiometric 488/405 nm fluorescence value for the sensor by the corresponding value for the cpVenus control subjected to the same condition, e.g. the measurements for sensor and cpVenus both treated with FK866. This normalization accounts for any NAD+ independent fluorescence changes (Table 3).
-
38
To discern whether relative NAD+ changes have occurred, divide the ratio of ratios value determined in Basic Protocol step 37 for treated cells by analogous ratio of ratio value for untreated cells. With FK866 treatment, the sensor’s fluorescence is expected to increase in relative brightness. A relative increase in fluorescence indicates less free NAD+, and is denoted by a relative value greater than 1; correspondingly, more NAD+ is indicated by a relative value less than 1 (Tables 3–6).
-
39
To relate the measurement to an intracellular concentration, interpolate this ratio-of-ratio value to a standard curve (Alternate Protocol).
Table 3. Example of how to compare ratiometric measurements.
The ratio of ratios provides a relative comparison to determine whether an NAD+-dependent fluorescence change occurred. This table contains a normalization step first, then a ratio is being determined from the normalized values. The statistical significance of the change is evaluated with experimental replicates, described in Basic Protocol steps 40–49. Because the comparison utilizes division, the order of the comparisons (either option 1, relative to cpVenus or option 2, relative to untreated control) does not matter.
| Geometric mean 488/405 nm fluorescence values of each population | ||||||
| nuclear cpVenus | nuclear sensor | cytoplasmic cpVenus | cytoplasmic sensor | mitochondrial cpVenus | mitochondrial sensor | |
| 0 nM FK866 | 6.83 | 2.26 | 7.79 | 2.51 | 22.1 | 5.37 |
| 10 nM FK866 | 6.85 | 3.34 | 7.46 | 3.78 | 22.8 | 7.30 |
| Option 1: Compared to cpVenus | ||||||
| 0 nM FK866 | 2.26/6.83 = 0.33 | 2.51/7.79 = 0.32 | 5.37/22.1 = 0.24 | |||
| 10 nM FK866 | 3.34/6.85 = 0.49 | 3.78/7.46 = 0.51 | 7.3/22.8 = 0.32 | |||
| Fluorescence change | Nuclear | Cytoplasmic | Mitochondrial | |||
| Ratio of ratios (treated/untreated) | 0.49/0.33 = 1.48 | 0.51/0.32 = 1.57* | 0.32/0.24 = 1.32* | |||
| Option 2: Compared to untreated | ||||||
| 6.85/6.83 = 1.00 | 3.34/2.26 = 1.48 | 7.46/7.79 = 0.96 | 3.78/2.51 = 1.51 | 22.8/22.1 = 1.04 | 7.3/5.37 = 1.36 | |
| Fluorescence change | Nuclear | Cytoplasmic | Mitochondrial | |||
| Ratio of ratios (treated/untreated) | 1.48/1.00 = 1.48 | 1.51/0.96 = 1.57 | 1.36/1.04 = 1.32* | |||
values based on non-rounded numbers.
Table 6. Data and statistical analysis from mitochondrial sensor replicates.
Top. Data represents biological replicates using transiently transfected mitochondrial sensor and mitochondrial cpVenus in HeLa cells. Data are arranged for input into STATA14 statistical software. “0” represents cells treated with 0 nM FK866. “1’ represents cells treated with 10 nM FK866. The fl values represent the geometric mean from 488/405 nm fluorescence ratio of the healthy, single HeLa cells measured by cytometry. “experiment”, biological replicate; “fl”, 488/405 nm geometric mean fluorescent value; “treatment”, +/− 10 nM FK866; “sensor”, +/− mitochondrial NAD+ sensor.
Bottom. Calculated statistical interaction table from STATA14 using REML. The p-value for the ratio of ratios across replicates (bold red) is reported under the statistical interaction “_IsenXtre_1_1”. This represents the fluorescence change with treatment in the mitochondrial sensor, relative to changes in cpVenus. Calculating the exponential of the coefficient value, “Coef.”, represents the mean fold-change in fluorescence for the ratio of ratios across replicates. i.e. e^(0.25) = 1.28, 95% CI (1.05 to 1.57), p= 0.016.
The fluorescence change upon treatment in nuclear cpVenus across replicates is represented by “_Itreatment_1”: e^(coef) = e^(0.04) = 1.04, p=0.574.
To calculate the average fluorescent change in the mitochondrial sensor independent of cpVenus, combine the “treatment” and “sensor X treatment interaction” using the lincom command (step 50). The combined values (blue) represents the change in the sensor: e^(coef) = e^(0.29) = 1.34, 95% CI (1.16 to 1.54), p<0.001.
| experiment | fl | treatment | sensor |
|---|---|---|---|
| 1 | 24.9 | 0 | 0 |
| 1 | 23.4 | 1 | 0 |
| 1 | 7.01 | 0 | 1 |
| 1 | 8.41 | 1 | 1 |
| 2 | 22.1 | 0 | 0 |
| 2 | 22.8 | 1 | 0 |
| 2 | 5.37 | 0 | 1 |
| 2 | 7.30 | 1 | 1 |
| 3 | 20.4 | 0 | 0 |
| 3 | 23.8 | 1 | 0 |
| 3 | 4.61 | 0 | 1 |
| 3 | 6.76 | 1 | 1 |
| log_fl | Coef. | Std. Err. | z | P> |z| | [95% Conf. | Interval] |
|---|---|---|---|---|---|---|
| _Isensor_1 | −1.39 | 0.07 | −19 | 0 | −1.53 | −1.25 |
| _Itreatment_1 | 0.04 | 0.07 | 0.56 | 0.574 | −0.10 | 0.18 |
| _IsenXtre_1_1 | 0.25 | 0.10 | 2.41 | 0.016 | 0.05 | 0.45 |
| _cons | 3.11 | 0.08 | 41.36 | 0 | 2.96 | 3.26 |
| _IsenXtre_1_1 + _Itreatment_1 | 0.29 | 0.07 | 3.97 | 0 | 0.15 | 0.43 |
Statistical analysis using STATA14 to evaluate significance of relative changes from experimental replicates.
-
40
Organize data as shown in Tables 4–6. We suggest using a comma-delimited file, e.g. filename.csv, as we have experienced few issues with this format. Experimental replicates are grouped and indicated by numbers 1 to n. Input geometric mean fluorescence values under “fl.” Treatment is designated with either “0” indicating untreated, or “1” indicating FK866-treated. cpVenus is designated with “0” and Sensor is designated with “1”.
-
41
Import the data file under “text data” in the STATA14 software. You may have to browse to find the location where you had saved the file. When uploaded correctly, the program will indicate it found 4 variables (columns) and x observations (number of rows). The uploaded data can also be verified by checking under “data editor.”
-
42Perform a log transformation of the geometric mean fluorescence values to help control for variability. This can be accomplished by entering the following command “generate log_fl=log(fl)”
- Use commands that reflect the exact case and format of the headings in your data file.
-
43
To denote that the data represents repeated measures (replicates), enter the following command: “xtset experiment”.
-
44
To apply the statistical model, enter the following command:
Table 4. Data and statistical analysis from nuclear sensor replicates.
Top. Data represents biological replicates using transiently transfected nuclear sensor and nuclear cpVenus in HeLa cells. Data are arranged for input into STATA14 statistical software. “0” represents cells treated with 0 nM FK866. “1” represents cells treated with 10 nM FK866. The fl values represent the geometric mean from 488/405 nm fluorescence ratio of the healthy, single HeLa cells measured by cytometry. “experiment”, biological replicate; “fl”, 488/405 nm geometric mean fluorescent value; “treatment”, +/− 10 nM FK866; “sensor”, +/− nuclear NAD+ sensor.
Bottom. Calculated statistical interaction table from STATA14 using REML. The p-value for the ratio of ratios across replicates (bold red) is reported under the statistical interaction “_IsenXtre_1_1”. This represents the fluorescence change following FK866 treatment in the nuclear sensor, relative to changes in cpVenus. Calculating the exponential of the coefficient value, “Coef.”, represents the mean fold-change in fluorescence for the ratio of ratios across replicates. i.e. e^(0.42) = 1.52, 95% CI (e^0.25 – e^0.59) = 95% CI (1.28 to 1.80), p<0.001.
The fluorescence change following FK866 treatment in nuclear cpVenus across replicates is represented by “_Itreatment_1”: e^(coef) = e^(0.03) = 1.03, p=0.629.
To calculate the average fluorescent change in the nuclear sensor independent of cpVenus, combine the “treatment” and “sensor X treatment interaction” using the lincom command (step 50). The combined values (blue) represents the change in the sensor: e^(coef) = e^(0.45) = 1.57, 95% CI (1.39 to 1.77), p<0.001.
| experiment | fl | treatment | sensor |
|---|---|---|---|
| 1 | 9.97 | 0 | 0 |
| 1 | 10.00 | 1 | 0 |
| 1 | 3.78 | 0 | 1 |
| 1 | 6.45 | 1 | 1 |
| 2 | 6.83 | 0 | 0 |
| 2 | 6.85 | 1 | 0 |
| 2 | 2.26 | 0 | 1 |
| 2 | 3.34 | 1 | 1 |
| 3 | 6.70 | 0 | 0 |
| 3 | 7.28 | 1 | 0 |
| 3 | 2.30 | 0 | 1 |
| 3 | 3.56 | 1 | 1 |
| log_fl | Coef. | Std. Err. | z | P> |z| | [95% Conf. | Interval] |
|---|---|---|---|---|---|---|
| _Isensor_1 | −1.05 | 0.06 | −17.09 | 0 | −1.17 | −0.93 |
| _Itreatment_1 | 0.03 | 0.06 | 0.48 | 0.629 | −0.09 | 0.15 |
| _IsenXtre_1_1 | 0.42 | 0.09 | 4.89 | 0 | 0.25 | 0.59 |
| _cons | 2.04 | 0.16 | 12.73 | 0 | 1.73 | 2.36 |
| _IsenXtre_1_1 + _Itreatment_1 | 0.45 | 0.061337 | 7.40 | 0 | 0.33 | 0.57 |
“xi:xtmixed log_fl i.sensor*i.treatment || experiment:, reml”
-
45
A summary of the model’s estimated results will display in a format similar to what is shown in Tables 4–6. The values shown in these tables will be present as a readout from the program, including the coefficient (Coef.) and the 95% confidence (95% Conf.) and the p-value described in later steps.
-
46
The estimate for the ratio of ratios across multiple experiments is represented by the third row, “IsenXtre_1_1”, also known as the statistical interaction (Tables 4–6). The p-value from this row represents the probability that—after controlling for any changes in the fluorescence of cpVenus—any observed difference in sensor fluorescence is indistinguishable from variation due to random sampling error. Thus, a small p-value represents situations where the “null” model is a poor explanation of the data. In these experiments the p-values are less than 0.05.
-
47
To determine the mean fold change in fluorescence for the ratio of ratios across replicates, calculate the exponential of the coefficient (Coef.).
-
48To examine the changes in the sensor and the changes in cpVenus independently, the estimates can be found as follows:
- For cpVenus, the estimate for the fluorescence change across replicates is represented by the row named “_Itreatment_1” (Tables 4–6). In these experiments, the changes in cpVenus are not significantly different with FK866 treatment. The fold change can be calculated by taking the exponential of the coefficient in this row, and the 95% confidence interval can be similarly calculated by taking the exponential of the values in the column labeled “Coef.”.
-
49
To calculate the average fluorescent change in the sensor independent of cpVenus, combine the “treatment” and “sensor X treatment interaction” using the following command: “lincom _IsenXtre_1_1 + _Itreatment_1”. This generates a new table, which you can use to determine the reported p-value for this condition, calculate the fold-change by taking the exponential of the coefficient, and calculate the 95% confidence interval by taking the exponential of the values in the column labeled “Coef.”.
Alternate Protocol 1: Calibration of sensor fluorescence to intracellular NAD+ concentrations.
Apart from evaluating relative changes in steady-state NAD+ levels between control and experimental conditions, the sensor can be used to ascribe quantitative measurements to these changes. To accomplish this, the fluorescence of the sensor needs to be calibrated with known NAD+ concentrations to generate a standard curve. Experimental fluorescence readings (488/405 nm) are then interpolated from the reference curve to obtain NAD+ measurements. For accuracy, a standard curve will need to be generated for each instrument setup and should be repeated if there are changes in instrumentation. As NAD+ is intrinsically acidic, this approach requires a buffered NAD+ stock solution, which is described in steps 1–4.
The approach for calibration described here involves acutely permeabilizing cells using digitonin such that internal stores of NAD+ equilibrate with externally provided concentrations of NAD+ (Zhao et al., 2011, Cambronne et al., 2016). Equilibration of cells is monitored by internalization of the molecular dye, propidium iodide (PI), which has a similar molecular weight to NAD+. When PI is excited at 561 nm, it emits a fluorescence that can be monitored with a 670 ± 15 nm filter. This excitation and filter combination permits PI fluorescence to be simultaneously monitored with the sensor’s fluorescence in the same cell. Measurements of the sensor’s 488/405 nm fluorescence in PI-positive cells can be then correlated with applied NAD+ concentrations to generate a calibration curve. Sensor measurements used to generate an in-cell calibration curve can be obtained from 10 cm plates of cells transiently transfected the previous day with cytoplasmically localized cpVenus or Sensor. It may be technically easier and more cost-effective, nevertheless, to use stably-expressing cell lines of the cytoplasmic cpVenus or Sensor (Fig. 3).
Generation of buffered 50 mM NAD+ stock at pH 7.4.
-
1
Resuspend 25 mg of NAD+ (sigma N1636, >99% pure) in 500 μL of buffer (100 mM Tris pH 7.4 + 150mM NaCl). The buffer composition is important, as NAD+ is unstable in phosphate buffers (Anderson & Anderson, 1963), and addition of salt mimics physiological concentrations.
-
2
Initially increase the pH with ~25 μL of 1N NaOH to approximately pH 6.5. Then slowly add 0.1N NaOH (~10 μL) to increase pH ~7.5. Mix well and use pH strips to confirm pH.
-
3
Total volume is now approximately 535 μL. Add remaining volume of buffer up to 753.66 uL total for a 50 mM stock. Mix well.
-
4
Aliquot and store at −20°C or −80°C for up to 1 year.
Determine experimental conditions and digitonin concentration.
-
5The first step is to determine the appropriate concentration of digitonin for permeabilization. Trypsinize and collect HEK293T cells from an 80–90% confluent 10 cm plate into 5 mL of growth media. Count cells and adjust volume so that cells are resuspended at a concentration of 1 million cells per mL in growth media.
- A more concentrated or less concentrated cell suspension can be used, but this will affect the required amount to digitonin. We suggest 1 million HEK293T cells per mL as an appropriate starting concentration that can be permeabilized by 0.001% vol/vol digitonin in 15 minutes at room temperature.
-
6Set aside 500 μL of cell suspension so that these cells will not receive any PI dye. This will serve as the “no-PI” control.
- This control is required to ascertain the difference between +/− PI internalization.
-
7
Withdraw 5 mL of the cell suspension. To the 5 mL cell suspension, add 10 μL of a 15 mM PI stock (~10 mg/mL), such that final concentration of PI is 30 μM. Vortex to mix well.
-
8
Prepare and label 9 polystyrene round-bottom tubes with the following indicated digitonin concentrations: 0%, 0.00010%, 0.00025%, 0.00050%, 0.00075%, 0.00100%, 0.00125%, 0.00150%, and 0.00175% vol/vol.
-
9
9. Aliquot 500 μL of the cell/PI mixture into each of the pre-labeled polystyrene round-bottom tubes and set these aside.
-
10
Analyze the “no-PI” control cells on flow cytometer to set gates for live cells of uniform side and forward scatter (P1) and single cells (P2, from P1). From the single-cell population (P2), select the y-axis to read the FSC-A (linear) and the x-axis to monitor PI fluorescence following from excitation at 561 nm and using emission filter 670 ± 15 nm (log). Using the “no-PI” control as a guide, draw a gate that excludes that population and only includes PI-positive cells (P4). (Fig. 4)
-
11
Next, analyze the 0% digitonin cell/PI mixture. If any PI-staining is observed in this sample, it indicates membrane-compromised and likely unhealthy cells. Unhealthy cells are characterized by strong PI-staining and should be excluded from the P4 gate. In contrast, when cells are permeabilized and equilibrated with PI (as will be observed in subsequent steps) the staining is less intense and is only present in digitonin-permeabilized samples. Use this sample to define the P4 gate to also exclude the unhealthy “high-PI” cells (Fig. 4).
-
12Using a 0.05% vol/vol digitonin stock—which has been serially diluted from the 5% wt/vol digitonin stock in 100 mM Tris pH.7.4 + 150 mM NaCl—calculate the appropriate volume of digitonin for each percentage to be tested. Add this volume to the appropriate 500 μL cell/PI aliquot. Vortex to mix and incubate at room temperature for 15 minutes.
- It is critical to incubate each sample for exactly the same amount of time, so addition of digitonin is staggered such that analysis can be performed at the 15 minute mark. We empirically determined that fifteen minutes was sufficiently long enough such that PI could internally equilibrate while cells remained viable.
-
13
After 15 minutes, analyze each sample. Collect 10 000 cells in the P2 gate, record the percentage of P2 cells in the P4 gate (PI+).
-
14
Plot a semi-log graph with the percentage of digitonin on the x-axis and the percentage of PI-positive cells on the y-axis. Examples of this are available as seen in Cambronne et al. 2016., Figure S11 and Zhao et al., Cell Metabolism, 2011., Figure 2E. The lowest amount of digitonin required to equilibrate the population is the amount to use to generate the calibration curve. We have previously found that 0.001% vol/vol is typically an appropriate amount to permeabilize HEK293T cells at a concentration of 1 million cells per mL.
To monitor the sensor’s fluorescence in the PI-equilibrated population.
-
15
Prior to monitoring the sensor’s and cpVenus’ responses to NAD+, it is important to define the P5 gate using untransfected or parental cells that do not express any fluorescent protein. This can be accomplished as an extension of Alternate Protocol steps 5–14, utilizing the same samples that were used to determine an appropriate digitonin concentration. The P5 gate defines the fluorescent population and is derived from the P4 (PI+) population. It is defined by exclusion of cells in the non-fluorescent sample, and inclusion of cells wherein cpVenus or sensor proteins are expressed (Fig. 4). To define P5, analyze the P4 population (PI+) on a new plot. Set the x-axis display to log format with excitation at 405 nm and the emission filter set 525 ± 25 nm. Set the y-axis display to log format with excitation at 488 nm and the emission filter set 530 ± 15 nm. Ratiometric fluorescent measurements of the Sensor and cpVenus (488/405 nm) will be obtained from the P5 population.
Calibrating in-cell sensor fluorescence to NAD+ concentrations.
-
16
This calibration requires 10 cm dishes of cells each expressing either cytoplasmic localized cpVenus or Sensor. It is possible to use 10 cm dishes of cells that have been previously transiently transfected for either cytoplasmic cpVenus or Sensor, or use stably-expressing lines (Fig. 3). To generate stably-expressing lines, refer to the protocol provided in Lai et al., 2013. These expression plasmids are compatible for lenti-viral production using 2nd generation viral helpers. This calibration is best suited for cytoplasmic measurements.
-
17
Label polystyrene flow cytometry round-bottom tubes with the range of NAD+ concentrations to be tested. We recommend starting concentrations of 10 μM, 30 μM, 100 μM, 300 μM, 1 mM, 3 mM, 10 mM.
-
18
Dilute the NAD+ stock in 100mM Tris pH 7.4 + 150 mM NaCl to represent the 5X experimental values to test. E.g. to measure effects of 100 μM NAD+, dilute the NAD+ to a 5X stock of 500 μM NAD+. Aliquot 100 μL of the 5X stocks into the appropriate, pre-labeled tubes. Set aside these tube for the time being. They will eventually be mixed with 400 μL of a cell/PI/digitonin suspension.
-
19
Trypsinize with 1 mL of 0.05% trypsin for 5 minutes and collect cells from a 10 cm dish that is 80–90% confluent. Count cells and resuspend them at a concentration of ~ 1 million per mL in growth media. A minimum of 5.5 mL is required for 9 data points; to account for pipetting error prepare 6 mL. Collect and analyze one cell line at a time, either Sensor or cpVenus, to facilitate and maintain consistent timing.
-
20
Remove 500 μL of the cell suspension as a control condition that will not receive PI or digitonin. Use this control to set and confirm P1 and P2 gates for uniform and single cell populations, respectively.
-
21
Add PI to the remaining 5.5 mL of cell suspension for a final concentration of 30 μM.
-
22
Remove 500 μL of cell/PI suspension as a control condition (PI, no digitonin). Analyze this sample to define P4 as previously outlined in Alternate Protocol steps 10–11 (Fig. 4).
-
23To the remainder of the cells (5 mL), add 12.5 μL of a 0.05% vol/vol digitonin stock, such that the final percentage is 0.00125% vol/vol, or 1.25X the required digitonin concentration that was empirically determined. When 400 μL of this cell/PI/digitonin suspension is mixed with 100 μL of the diluted NAD+, the final concentration of digitonin will be 0.001% vol/vol. Once the digitonin has been added, immediately vortex and distribute 400 μL of the cells/PI/digitonin suspension to the each pre-labelled tube that already contains 100 μL of 5X NAD+.
- Staggered preparation of sample is critical for ensuring accurate timing for each sample.
-
24
Vortex the cell/PI/digitonin/NAD+ mixture and incubate 15min.
-
25
Analyze samples and collect 10 000 events from P5.
-
26
It is recommended to complete the flow cytometer analysis of all samples from each cell line within 5–10 min from each other for accurate measurements.
-
27
Pause point. On a semi-log graph, plot the log NAD+ concentrations on the x-axis and the ratio of ratio Sensor/cpVenus (488/405 nm) values on the y-axis. Examples of expected changes can be found in Cambronne et al., 2016, Fig S12.
-
28
Biological replicates can be fit to a sigmoidal regression model and 95% confidence intervals calculated. Ratio-of-ratio y measurements can be interpolated onto this curve to obtain values for x.
Reagents and Solutions:
Cell culture growth media:
DMEM + 10% vol/vol FBS + 25 mM HEPES
Under sterile conditions in a tissue culture hood, add 50 mL of FBS to 500 mL DMEM media. Add 12.5 mL HEPES Buffer pH 7.4 to media. Mix. Store at 4°C up to a month if unopened.
50 mM FK866 in DMSO
To make a stock of 50 mM FK866, add 255 μL DMSO to 5 mg FK866 powder. Vortex to mix.
Store at −20°C in 50 μL aliquots up to one year.
Serially dilute in growth media to 10 nM working concentration immediately before use. Do not store aqueous dilutions for over 24 hours.
50 mM NAD+ stock
Resuspend 25 mg of NAD+ in 500 μL of buffer (100 mM Tris pH 7.4 + 150mM NaCl). Add ~25 μL of 1N NaOH at approximately pH 6.5, then slowly add 0.1N NaOH (~10 μL) to increase pH ~7.5. Mix well and use pH strips to confirm pH. Add remaining volume of buffer up to 753.66 μL total for a 50 mM stock. Mix well, aliquot and freeze for up to 1 year.
Dilution Buffer (100 mM Tris pH 7.4, 150 mM NaCl)
Used for dilution of NAD+ and digitonin during calibration. Dilute 5 mL of 1M Tris pH 7.4 and 1.5 mL of 5M NaCl with 43.5 mL of ultrapure water (total volume 50 mL). Filter sterilize to remove dust and particulates. Can be stored at room temperature (20–25°C) for up to 2 years.
Commentary:
Background Information
In the late-1990s, the Tsein lab found that GFP-like proteins could maintain fluorescence despite permutation of the primary sequence and even tolerate large peptide insertions, as long as its beta-barrel structure was intact to protect the fluorophore (Baird et al., 1999). By inserting calmodulin into eYFP, Baird et al. pioneered the first single-FP (fluorescent protein) sensor for calcium (Baird et al., 1999). This was further developed into a ratiometric intracellular Ca2+ sensor by Miyawaki, Nagai, and colleagues (Nagai et al., 2001), which set the groundwork for the current GECO and GCaMP sensor series commonly used today (Tian et al., 2009; Zhao et al., 2011; Akerboom et al., 2012; Chen et al., 2013; Wu et al., 2013), and also represents the strategy that has been subsequently utilized in many other single fluorescent-protein biosensor designs, including those used for the NADH:NAD+ sensors (Hung et al., 2011; Zhao et al., 2011; Bilan et al., 2014; Zhao et al., 2015).
The NAD+ sensor combines a circularly-permuted fluorescent protein (cpVenus) with a binding pocket to the target of interest, in this case NAD+ (Fig. 2a). We developed a unique bi-partite NAD+ binding pocket modeled after the NAD+-binding domain from bacterial DNA ligase, which specifically binds NAD+ and undergoes a conformational change upon NAD+ binding (Gajiwala et al., 2004; Lahiri et al., 2012). Attachment of the NAD+-binding pocket to cpVenus physically links the fluorescence of the cpVenus chromophore to local NAD+ availability. In other words, binding of free NAD+, representing its local availability, in turn affects fluorescence from the chromophore of the sensor resulting in decreased fluorescence. We further engineered the binding pocket to ensure reversible and non-destructive binding of NAD+, and showed that it was specific for NAD+, uniquely bound the free fraction, and could measure NAD+ within its estimated physiological range. For this NAD+ sensor, increasing NAD+ availability decreases the fluorescence intensity of the sensor (Fig.2). Thus, there is an inverse relationship between the sensor’s fluorescence intensity and free NAD+ concentration that we can reliably detect between ~30 μM and 1 mM. Because the sensor is genetically encoded, we can incorporate subcellular localization sequences to target the sensor to specific subcellular compartments to determine local NAD+ measurements (Cambronne et al., 2016).
Methods to measure free NAD+ are needed because unlike NADH—whose free fraction can be distinguished from protein-bound NADH in cells due to distinct differences in intrinsic fluorescence lifetimes (Patterson et al., 2000; Zhang et al., 2002)—the oxidized NAD+ molecule has no intrinsic fluorescence. Moreover, estimates of free NAD+, using free NADH or NADH/NAD+ measurements are unlikely to accurately reflect free NAD+ concentrations because of a high NAD+/NADH ratio in most subcellular compartments. In mammalian cells, the ratio of free NAD+ to NADH is estimated at ~700:1 in the nucleocytoplasm, and ~7:1 in mitochondria (Williamson et al., 1967). Due to their disproportionate ratio and because the concentration of free NAD+ exceeds that of free NADH, current ratiometric NADH:NAD+ sensors are prone to saturation for NAD+ measurements when used in cells (Hung et al., 2011; Zhao et al. 2011; Bilan et al., 2014; Zhao et al., 2015), and monitoring changes in free NADH can overestimate the resulting changes in NAD+ concentrations.
Approaches to directly detect the free NAD+ relevant for cellular signaling pathway would additionally need to be able to inform about its subcellular measurements. NAD+ is highly compartmentalized and its levels are maintained at distinct steady-state concentrations in different subcellular compartments (Yang et al., 2007; Pittelli et al., 2010; Cambronne et al., 2016). Regulation of these subcellular pools is complex due to the multiple and paralogous biosynthetic and consuming enzymes that are differentially expressed according to cell or tissue type (Felici et al., 2013; Mori et al., 2014; Cambronne et al., 2016), as well as differentially localized within subcellular compartments (Raffaelli et al., 2002; Berger et al., 2005; Nikiforov et al., 2011; Mori et al., 2014). Moreover, the rates of individual enzymes can be controlled independently (Raffaelli et al., 2002; Berger et al., 2005; Sorci et al., 2007; Nikiforov et al., 2011). Thus, local concentrations of NAD+ can differ in different biological compartments and subcellular concentrations of NAD+ can be regulated independently through biosynthetic pathways or depleted by the previously mentioned local consuming enzymes (Fig. 1). Methods that rely on preparation of cellular extracts—such as liquid-chromatography mass spectrometry, or colorimetric or fluorogenic assays—often compromise spatial information and it is usually unclear what proportion of the collected pool represents the free NAD+ fraction. Magnetic resonance scanning can distinguish NAD+ from NADH and has been used for patient diagnosis (Zhu et al., 2015), but it cannot distinguish bound from free fractions and is limited for subcellular interrogations. Lastly, while both indirect readouts such as the PARAPLAY assay (Dölle et al., 2010) or direct NAD+ measurements using a semisynthetic sensor (Sallin et al., 2018) have been extremely informative, our emphasis has been to develop a completely genetically-encoded method to directly detect free intracellular NAD+.
Critical Parameters
There are two important controls required when using the NAD+ sensor. Distinct from the brighter fluorescence that follows excitation at 488 nm and represents NAD+-dependent changes, we recommend also taking a secondary measurement of fluorescence between ~500–525 nm following excitation at 405 nm (Fig. 2b). Fluorescence after excitation at 405 nm proportionally tracks in vitro with the abundance of the sensor or cpVenus, and thus its intensity can be used for normalization of sensor expression levels in cells (Cambronne et al., 2016). The second important control is cpVenus alone (without the NAD+ binding pocket), whose fluorescence is not affected by NAD+ (Cambronne et al., 2016). We found that pH influences the fluorescence of the sensor and cpVenus control similarly between pH conditions 6.6 to 8.0 (Cambronne et al., 2016). Thus, parallel analysis of fluorescent changes in cpVenus can be used to normalize for pH effects and other non-specific changes in fluorescence within this range (Bilan et al, 2014, Cambronne et al., 2016). Most intracellular compartments fall within this pH range, including the nucleus, cytosol, mitochondria, peroxisomes, endoplasmic reticulum, and the cis-golgi network (Casey et al,. 2010). Relatedly, complete growth media used to resuspend cells prior to analysis must be made fresh such that it is within near-neutral pH. Media that is not near neutral pH is unable to sufficiently buffer the cell suspension for the duration of the flow analysis. If media contains phenol red, it should be orange-red in color without any traces of pink or magenta and is ideally opened and supplemented immediately before analysis. Phenol red will not interfere with the flow analysis because cells are diluted in sheath buffer during the analysis. To confirm experimental and instrumental setups for sensor measurements, we recommend utilizing FK866 to deplete intracellular NAD+.
Similar to other FP sensors, this NAD+ sensor is sensitive to the pH of its environment, and thus incompatible with cell compartments pH < 6.5, including the trans-golgi network, secretory granules, endosomes, and lysosomes (Casey et al., 2010). In cases when the pH of a compartment is low, overall fluorescence is eliminated both from cpVenus and the sensor (Cambronne et al., 2016). Thus, a general limitation of the sensor is that it cannot monitor NAD+ under experimental conditions that would eliminate overall fluorescence, e.g. pH less than 6.5 or exogenous H2O2. NAD+-dependent changes only alter and do not completely eliminate fluorescence, so a lack of fluorescence from both cpVenus and the sensor would indicate experimental conditions where the sensor cannot measure NAD+. This point highlights the necessity of the cpVenus parallel control.
We also do not currently know the response time of the sensor to respond to NAD+ fluctuations. This has not been a limitation in our measurements, but the time lag has not been formally defined. The sensor also has a limited sensitivity when measuring NAD+ levels outside the range of 30 μM to 1 mM. The dynamic range of fluorescence for the sensor is ~50%, corresponding to a ~30% increase or ~20% decrease in fluorescence (Cambronne et al., 2016). Measurements of modest changes can be improved by reproducibility in measurements of a large sample size, facilitated by flow cytometry approaches.
The NAD+ sensor is compatible with common instrumentation setups and has unique and advantageous properties over previously determined methods.
At extreme supra-physiological concentrations, NAD+ precursor molecules NR and NMN can influence the sensor. To our knowledge, the extent to which steady-state intracellular NMN or NR increases intracellularly upon exogenous treatments is currently still unknown. Notwithstanding, we found that 25 μM NMN (25X supra-physiological) and 100 μM NR (>6000x supra-physiological) only minimally affected the NAD+ sensor in vitro, and so we recommend using these concentrations to exogenously treat cells when sensor measurements are taken. It is also critical to make fresh media for the experiment as listed in the Reagents and Solutions section.
This version of the sensor may have limited sensitivity to reliably detect modest NAD+ changes, and the apparent sensitivity of the sensor also depends on how well the particular instrumentation can detect fluorescence. As standard practice when using flow cytometry, we recommend analyzing 10 000 fluorescent cells per condition. An analysis using a power of 90% and data obtained on our cytometer indicated that this was sufficient to distinguish changes in all examined compartments. We have provided a list of recommended filter sets to use for experimental measurements. We provide a straightforward approach using the small molecule FK866, an NAMPT inhibitor, to check instrument capabilities. We highly recommend using this positive test before moving in to experimental conditions.
Troubleshooting
FK866 Treatment:
- Treated cells are unhealthy (Fig. 4) or have aberrant morphology.
-
FK866 concentration may be too highRe-confirm calculation of serial dilution. Dilute fresh in media. Can test different concentrations.
-
-
FK866 incubation exceeded 18 hours
Ensure that evaluation is performed within 18 hours.
- Treatment has no effect.
-
Incorrect FK866 concentration or incubation timeRe-confirm calculation of serial dilution. Dilute in fresh media right before use.
-
-
Incorrect FK866 concentration because too much growth media was left in well
Remove growth media completely and replace with 10 nM FK866 final concentration.
FK866 was not properly stored
Remake stock in DMSO, aliquot and store at −20°C. Can confirm efficacy of FK866 treatment by whole cell chromatography, which should be able to detect depletion of total cellular NAD+.
-
Insufficient buffering of the samples during analysis
Use freshly made media that contains HEPES to collect cells and confirm that media is orange-red in color.
Minimize the time it takes to analyze samples by performing small batches and having the instrument ready ahead of time.
In-cell calibration
- Fluorescence of cpVenus is altered upon equilibration with high concentrations of NAD+
-
NAD+ stock solution may not be sufficiently buffered and remains slightly acidicRemake NAD+ stock solution and ensure that the final stock solution containing Tris and NaCl is buffered accurately to pH 7.4. When further diluted to make the 5X stock, acidity should remain at pH 7.4.
-
- Sensor or cpVenus cells lose fluorescence upon permeabilization but without the addition of NAD+
-
Cells are unhealthy or dyingTry equilibrating with less digitonin. Re-establish the minimal amount of digitonin required to permeabilize the cell type. It may be also necessary to shorten the incubation time before analysis. (see below, cells dying)
-
-
Inadvertent introduction of quencher
Ensure that none of the buffers, instrument buffers, or reagents contain any compounds that can quench fluorescence.
- Cells dying before measurements can be obtained
-
Too much digitoninReestablish and use the minimal concentration of digitonin required to allow for PI-equilibration within 15 minutes for each cell type.
-
-
Impure digitonin stock
Use only highly pure digitonin from source that has been recrystallized or from a stock that is >99% pure. Digitonin should readily go into aqueous solution at 5% and should be clear.
Digitonin can be recrystallized to improve its purity. Digitonin is first dissolved in ethanol at 75°C, precipitated in ice water for 20 minutes, and pelleted by centrifugation. Repetition of these steps 2x can result in ~60% recovery that is isolated by vacuum drying. Weighed material is now pure enough for use.
-
Too long of an incubation
Once cells are equilibrated, consistently maintain the incubation time for all samples to ~15 minutes. This can be achieved by staggering the addition of digitonin and analysis among the samples.
-
Precipitated digitonin resulting in inconsistent permeabilization
Warm up digitonin solution in 90–95°C water bath/heat block and vortex to re-solubilize the digitonin into a clear solution. This should ensure accurate application of digitonin.
- No observation of PI-equilibrated cells
-
Insufficient digitonin concentration for cell typeEstablish minimum effective digitonin concentration to equilibrate intracellular PI within 15 minutes for specific cell type (Alternate Protocol, step 23).
-
-
Incorrect laser excitation or fluorescent filter set
Confirm that an appropriate excitation laser and filter set is being used.
-
Accidental omission of PI dye
Confirm that PI has been added to sample.
-
May have missed the proper timepoint
Establish with timepoints the incubation time required for the specific cell type.
- Measurements are off-scale of calibration curve
-
Lower scale measurements that require substantial NAD+ diffusion out of the cell are a limitation for this in-cell calibration method.A bead-based calibration of the instrumentation can be used for this low range (Cohen et al. Methods for Using a Genetically Encoded Fluorescent Biosensor to Monitor Nuclear NAD+. Methods Mol Biol. 1813:391–414. (2018).).
-
-
Instrument has been changed
Determine whether there have been any adjustments or calibration of laser settings, voltages, or alignments in between experimental replicates. Calibration can be repeated for new instrumental settings. Replicates should be all obtained under the same configuration that will also be used for data collection.
- Insufficient number of fluorescent cells
-
Transfection efficiency was too lowUse a stably expressing line to ensure that all cells that are PI-equilibrated in P4 will be fluorescent in P5.
-
- Large standard deviations in replicates when generating calibration curve
-
Experimental inconsistencies that may include incubation timing, NAD+ concentrations, permeabilization of cells.Ensure that the 15 minute incubation time with NAD+ and digitonin is always consistent among samples. This can be achieved by staggering the addition of digitonin and NAD+. Confirm that NAD+ stock concentrations are consistent across experiments and that the NAD+ has not degraded. Using freshly diluted 5X stocks aids in consistent pipetting. Confirm that the digitonin stock is pure and has not precipitated.
-
Statistical analysis
- Not statistically significant
-
Too much variability among replicatesUse freshly made media to collect cells each time.Minimize the time it takes to analyze samples by performing small batches and having the instrument ready ahead of time.Because the absolute values of the measurements depend on the specific instrument and lasers, ensure that replicates are performed on the same instrument with exactly the same settings.
-
- Fluorescence from cpVenus is changing.
-
pH across samples is not sufficiently stabilized. Phenol-red indicator in media will turn from orange to pink.Confirm that collection media contains HEPES.Use freshly made orange-red media (but not yellow-red) to collect cells each time.Minimize the time it takes to analyze samples by performing small batches, run on high, have high transfection efficiency, and having the instrument ready ahead of time.Can overlay a thin layer of mineral oil atop each sample to minimize sample exposure to air.
-
- Command not recognized
-
Data was not uploaded correctly or with different headingsConfirm that correct data was uploaded in data editor and headings are identical to what is used in command line.
-
Flow cytometry
- Cannot see cells.
-
Incorrect instrument setupMake sure instrument set-up is correct and lasers are on. Adjust voltages as needed. Can first view on log scale to find cells before changing to linear scale.
-
-
Clogged instrument
Potentially, unclog instrument by triturating cells and passing through a 0.45 μm nylon mesh to remove clumps
-
No/few cells are detected as fluorescent
Poor transfection (see Transfection).
Flow analysis
- Cannot see events or gates
-
Events are off-scaleAdjust axis similarly to Fig. 4., both filter component and scale.
-
- Gates or derived parameters are not applied to all samples
-
Did not apply to “all samples”Drag desired gate hierarchy or evaluation to “all samples” heading on top.
-
- Events do not look like example
-
Gates may not be organized as hierarchyMake sure that gate P3 is defined from P2, and P2 is defined from P1.
-
- Cannot click on derive parameter option
-
When defining a new parameter, a specific subpopulation cannot be actively selected.Move cursor to deselect specific subpopulation.
-
Trypsinization:
- Cells will not detach from plate.
-
Media may not have been complete washed from dishTry gently rinsing cells once more with PBS, aspirate completely before adding new trypsin.
-
Transfection:
- Poor transfection. Few fluorescent cells.
-
Low quality DNAConfirm with spectrophotometry a distinct and clear 260 nm peak, that the 260/280 nm ratio is ~1.8, and that the 260/230 nm ratio is ~2. Confirm by resolving the DNA on an agarose gel that the majority of the prep is supercoiled. Re-purify DNA using a maxiprep column and repeat transfection.
-
-
Incorrect DNA plasmid
Restriction digest analysis and sequencing can determine whether the plasmids are correct. Re-purify DNA as needed and repeat transfection.
-
Insufficient number of healthy cells
Ensure cells are at least 50% confluent and healthy before transfection and repeat transfection. Thaw new cells if needed.
-
Did not incubate DNA and lipofectamine mixture for 20–30 minutes before adding to cells
Too little or too much incubation time of the DNA and Lipofectamine 2000 can affect transfection efficiency. Repeat transfection and keep track of incubation time.
-
Changed media too soon after transfection
Incubate HeLa cells with transfection mixture for 2 hours.
- Cells did not survive transfection
-
Low quality DNAConfirm with spectrophotometry a distinct and clear 260 nm peak, that the 260/280 nm ratio is ~1.8, and that the 260/230 nm ratio is ~2. Confirm by resolving the DNA on an agarose gel that the majority of the prep is supercoiled. Re-purify DNA using a maxiprep column and repeat transfection.
-
-
Forgot to change the media 2 hours after transfection
Prepare new cells and transfection mixture and incubate HeLa cells with transfection mixture for 2 hours.
-
Forgot to use complete media
Check that complete medium including serum was used. Start new transfection.
- Incorrect subcellular localization
-
Incorrect DNA plasmidRestriction digest analysis and sequencing can determine whether the plasmids are correct. Re-purify DNA as needed and repeat transfection.
-
-
Ectopic expression is too high, overwhelming endogenous targeting mechanisms
Re-transfect with half the amount of DNA and half the volume of lipofectamine 2000.
Statistical Analyses
To evaluate the likelihood that any observed fluorescence changes occurred by chance, we analyze the data using a mixed-effects model (McCulloch, Searle, Neuhaus, 2008) in which the experimental replicate is regarded as a random factor and the selective conditions of treatment and either sensor or cpVenus expression are both considered as fixed factors. Compared to other statistical evaluations, e.g. t-tests, this approach is most appropriate for estimating variance components of ratiometric measurements (Wulff, 2008). To help stabilize variance and limit the impact of outliers, we first log transform the geometric mean fluorescence intensity value for the ratiometric 488/405 nm measurement. The requirement for this transformation is based on our observations that the untransformed data violates the model’s assumptions of normality of the error distribution and homogeneity of error variances; log-transformed data showed no such violations. We recommend estimating variance by performing a Residual Maximum Likelihood (REML) analysis (Wulff, 2008; Swallow, Monahan, 1984). It is performed using the 488nm/405nm geometric mean values for each experimental condition, and a minimum of three biological replicates is required for sufficient power in the statistical analysis. A paired two-way ANOVA may also be appropriate, and this classical analysis is incorporated into many commonly-used statistical programs. However, advantages of REML include that its estimates of variance components are approximately unbiased even in small-sample settings, and that it separately fits the fixed and random effects and thus does not require a balanced experimental design for consistency of estimation (Wulff, 2008). We present an example here of how to perform the REML statistical analysis using the commonly used STATA14 software (Basic Protocol steps 40–49).
Understanding Results.
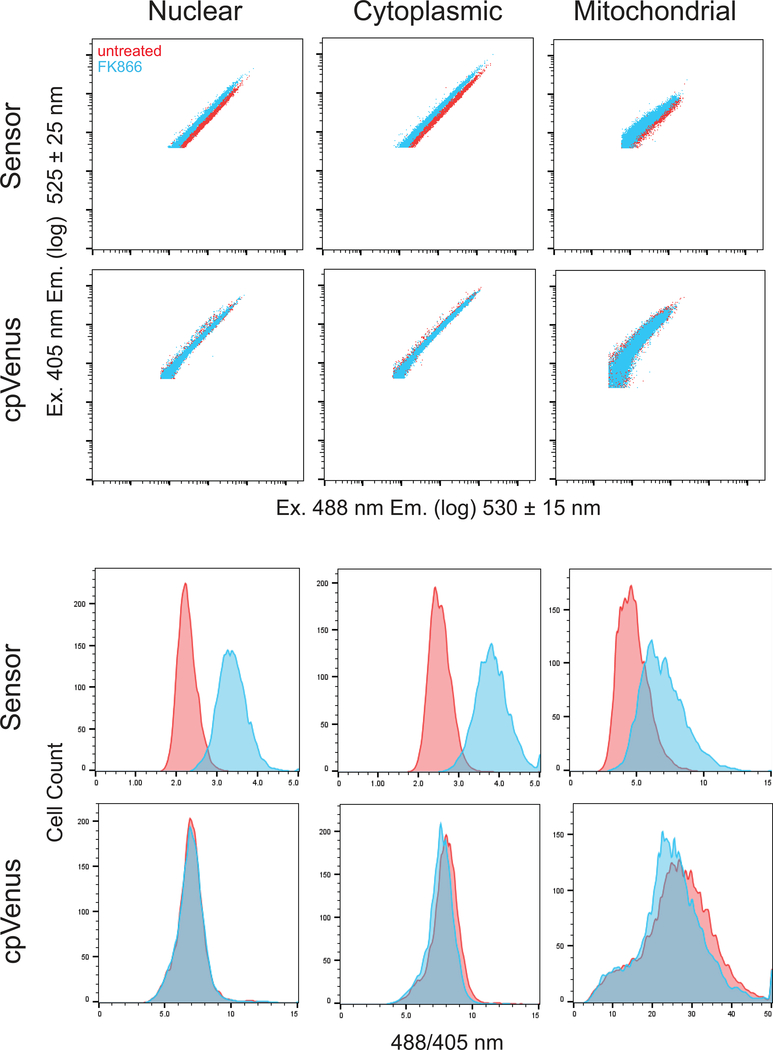
Cells treated with FK866 are unable to sufficiently replenish the NAD+ that is turned over by NAD+-consuming enzymes, and free intracellular pools are depleted over time (Pittelli et al., 2010; Cambronne et al., 2016). We have found that 16 hours of FK866 treatment resulted in robust depletion of free NAD+ in the subcellular compartments examined (Cambronne et al., 2016). Decreased NAD+ concentrations result in brighter fluorescence intensity of the sensor after excitation at 488 nm, and minimally affect fluorescence after excitation at 405 nm (Cambronne et al., 2016). Thus, the brighter 488/405 nm fluorescence when measured per cell (derived parameter) in FK866-treated samples, compared to untreated samples, represents differences in NAD+-availability that has been normalized for the expression of the sensor (Figure. 5 and Tables 4–6). Accordingly, the broad range in fluorescent intensities spanning the log scale that is due to transient transfection and varying plasmid copy number, collapses after normalization into a relatively tight peak on a linear scale representing the 488/405 nm fluorescence of that experimental sample (Fig. 5). NAD+ concentrations do not affect fluorescence of the cpVenus protein (Cambronne et al., 2016), thus we can thus further normalize our observed fluorescent changes to that of cpVenus subjected to the same experimental conditions (Fig. 5 and Tables 4–6). This allows researchers to discern NAD+-specific changes to fluorescence from non-specific factors influencing fluorescence. A detailed explanation of how to calculate and statistically evaluate the ratio of ratios is found in Basic Protocol steps 31–49. Measurements can further be compared to a standard curve after the sensor is calibrated for a specific experimental set-up. Calibration of the sensor is performed in parallel and described in Alternate Protocol. By targeting the sensor to a specific subcellular compartment, the protocol described here for determining the fluorescent ratio of ratios provides a reliable method for monitoring changes in free NAD+ concentrations in specific subcellular compartments.
Fig. 5. Representative data from cytometry measurements.
(Top) Logarithmic dot-plot fluorescent measurements of ~10 000 cells in P3 that have been transfected with plasmids expressing either the sensor or cpVenus, as indicated. Each column represents data determined from the indicated subcellular compartment. x-axis, fluorescence following 405 nm excitation; y-axis, fluorescence following 488 nm excitation. (Bottom) Histograms showing the distribution of the same P3 population as a function of the 488/405 nm ratio per cell. Data was collected on a BD LSRII flow cytometer and analyzed on FlowJo V10 software.
Time Considerations (Fig. 3)
Steps 1–6, seed cells, 30 minutes (day 1)
Steps 7–13, transfection, 2–3 hours (day 2)
Step 14, confirm expression and subcellular localization in stable cell lines or following transfection, 10 minutes (day 3)
Steps 15–17, start 16 h FK866 treatment, 15 minutes (day 3)
Steps 18–30, define gates and collection of flow cytometry measurements, 20–30 minutes (day 4)
Pause point
Steps 31–49, evaluate ratio-of-ratios, statistical analysis (variable)
Pause point
Alternate Protocol Steps 1–4, buffering NAD+ stock, 30–45 minutes
Pause point
Alternate Protocol Steps 5–15, determining experimental conditions and digitonin concentration (0.5–2 hours, variable)
Pause point
Alternate Protocol Steps 16–26, calibration of cpVenus and sensor-expressing cells with respect to equilibrated NAD+ concentrations. (~1 hour per replicate analyzing both sensor- and cpVenus-expressing cells)
Pause point
Alternate Protocol Steps 27–28, evaluation of ratio-of-ratios and generation of calibration curve (variable)
Table 5. Data and statistical analysis from cytoplasmic sensor replicates.
Top. Data representing biological replicates using transiently transfected cytoplasmic sensor and cytoplasmic cpVenus in HeLa cells. Data are arranged for input into STATA14 statistical software. “0” represents cells treated with 0 nM FK866. “1” represents cells treated with 10 nM FK866. The fl values represent the geometric mean from 488/405 nm fluorescence ratio of the healthy, single HeLa cells measured by cytometry. “experiment”, biological replicate; “fl”, 488/405 nm geometric mean fluorescent value; “treatment”, +/− 10 nM FK866; “sensor”, +/− cytoplasmic NAD+ sensor.
Bottom. Calculated statistical interaction table from STATA14 using REML. The p-value for the ratio of ratios across replicates (bold red) is reported under the statistical interaction “_IsenXtre_1_1”. This represents the fluorescence change following FK866 treatment in the cytoplasmic sensor, relative to changes in cpVenus. Calculating the exponential of the coefficient value, “Coef.”, represents the mean fold-change in fluorescence for the ratio of ratios, across replicates, i.e. e^(0.40) = 1.49, 95% CI (1.38 to 1.60), p<0.001.
The fluorescence change in cytoplasmic cpVenus with treatment across replicates is represented by the “_Itreatment_1” row: e^(coef) = e^(0.00) = 1.00, p=0.982. To calculate the fluorescent change in cytoplasmic sensor independent of cpVenus, combine the “treatment” and “sensor X treatment interaction” using the lincom command (step 50). The combined values (blue) represents the change in the sensor: e^(coef) = e^(0.40) = 1.49, 95% CI (1.42 to 1.57), p<0.001.
| experiment | fl | treatment | sensor |
|---|---|---|---|
| 1 | 9.65 | 0 | 0 |
| 1 | 9.56 | 1 | 0 |
| 1 | 3.06 | 0 | 1 |
| 1 | 4.35 | 1 | 1 |
| 2 | 7.79 | 0 | 0 |
| 2 | 7.46 | 1 | 0 |
| 2 | 2.51 | 0 | 1 |
| 2 | 3.78 | 1 | 1 |
| 3 | 7.33 | 0 | 0 |
| 3 | 7.74 | 1 | 0 |
| 3 | 2.35 | 0 | 1 |
| 3 | 3.62 | 1 | 1 |
| log_fl | Coef. | Std. Err. | z | P> |z| | [95% Conf. | Interval] |
|---|---|---|---|---|---|---|
| _Isensor_1 | −1.14 | 0.03 | −43.51 | 0 | −1.19 | −1.09 |
| _Itreatment_1 | 0.00 | 0.03 | 0.02 | 0.982 | −0.05 | 0.05 |
| _IsenXtre_1_1 | 0.40 | 0.04 | 10.72 | 0 | 0.32 | 0.47 |
| _cons | 2.10 | 0.07 | 28.18 | 0 | 1.96 | 2.25 |
| _IsenXtre_1_1 + _Itreatment_1 | .40 | .0261934 | 15.19 | 0.000 | .35 | .45 |
Significance Statement.
NAD+ serves as a substrate for NAD+-consuming enzymes, which play central roles in cell regulation and gene expression. The free NAD+ available to these enzymes is thus a critical measurement for understanding any NAD+-consuming process. Here we describe a method using flow cytometry for measuring free intracellular NAD+ with a genetically-encoded fluorescent sensor. This approach can provide direct and dynamic measurements of free NAD+ in targeted physiological compartments. It can distinguish free NAD+ from the bound NAD+ used in redox biochemistry, measure NAD+ concentrations in specific physiological compartments, and is specific for NAD+ in relation to NADH. Other methods are either limited to indirect measurements of NAD+ consumption or rely on cell-permeable dyes.
Acknowledgements
We thank Mike Lasarev for his initial help guiding the statistical analysis and DongHo Kim for technical assistance in developing the sensor. Work was supported by the Hillcrest Committee, Pew Charitable Trust and the NIH (NS088629, UL1TR000128, P30CA069533, DP2GM126897). We apologize to all researchers whose primary works have not been cited owing to space constraints.
Footnotes
Conflicts of Interest
NAD+ sensors are available from Oregon Health & Science University (OHSU) under a material transfer agreement with the authors. MLS, MSC, RHG, and XAC are inventors on patent application PCT/US15/62003 for the NAD+ sensor.
Literature Cited
- Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderón NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, … Looger LL (2012). Optimization of a GCaMP calcium indicator for neural activity imaging. The Journal of neuroscience : the official journal of the Society for Neuroscience, 32(40), 13819–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B & Anderson C (1963). The Effect of Buffers on Nicotinamide Adenine Dinucleotide Hydrolysis. J Biol Chem 238, 1475–1478. [PubMed] [Google Scholar]
- Baird GS, Zacharias DA & Tsien RY (1999). Circular permutation and receptor insertion within green fluorescent proteins. PNAS 96, 11241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M & Ziegler M (2005). Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280, 36334–36341. [DOI] [PubMed] [Google Scholar]
- Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G, & Belousov VV (2013). Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochimica et biophysica acta, 1840(3), 951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher Th. & Klingenberg M (1958). Wege des Wasserstoffs in der lebendigen Organisation. Angew. Chem 70, 552–570. [Google Scholar]
- Cambronne XA, Stewart M, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen Michael S., Goodman RH (2016). Biosensor reveals multiple sources for mitochondrial NAD⁺. Science 352, 1474–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Grinstein S & Orlowski J (2010). Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 11, 50–61. [DOI] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, … Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Stewart ML, Goodman RH, & Cambronne XA (2018). Methods for Using a Genetically Encoded Fluorescent Biosensor to Monitor Nuclear NAD+ In Chang P (Ed.), ADP-ribosylation and NAD+ Utilizing Enzymes: Methods and Protocols (pp. 391–414). New York, NY: Springer New York; 10.1007/978-1-4939-8588-3_26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölle C, Niere M, Lohndal E & Ziegler M (2010). Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation. Cell Mol Life Sci 67, 433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici R, Lapucci A, Ramazzotti M & Chiarugi A (2013). Insight into molecular and functional properties of NMNAT3 reveals new hints of NAD homeostasis within human mitochondria. PLoS One. 8, e76938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala KS & Pinko C (2004). Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure 12, 1449–59. [DOI] [PubMed] [Google Scholar]
- Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, & Milbrandt J (2015). SARM1 activation triggers axon degeneration locally via NAD⁺ destruction. Science (New York, N.Y.), 348(6233), 453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer H, Lynen F & Schultz G (1956). Determination of diphosphopyridine nucleotide/reduced diphosphopyridine nucleotide quotient in living yeast cells by analysis of constant alcohol and acetaldehyde concentrations. Biochem Z. 328, 252–63. [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ & Auwerx J (2010). The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 31, 194–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, & Yellen G (2011). Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell metabolism, 14(4), 545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T; Yang Y; Ng SK (2013). Advances in mammalian cell line development technologies for recombinant protein production. Pharamceuticals. 6(5), 579–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri SD, Gu R-F, Gao N, Karantzeni I, Walkup GK, & Mills SD (2012). Structure Guided Understanding of NAD+ Recognition in Bacterial DNA Ligases. ACS Chemical Biology, 7(3), 571–580. 10.1021/cb200392g. [DOI] [PubMed] [Google Scholar]
- McCulloch C, Searle S, Neuhaus J (2008). Generalized, Linear, and Mixed Models, 2e Wiley [Google Scholar]
- Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G, Ruggieri S, Raffaelli N, … Orsomando G (2014). Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PloS one, 9(11), e113939. doi: 10.1371/journal.pone.0113939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES & Miyawaki A (2001). Circularly permuted green fluorescent proteins engineered to sense Ca2+. PNAS 98, 3197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov A, Dölle C, Niere M & Ziegler M (2011). Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem 286, 21767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Arkhammar P, Thastrup O & Piston DW (2000). Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci USA. 97, 5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A (2010). Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. The Journal of biological chemistry, 285(44), 34106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelli N, Sorci L, Amici A, Emanuelli M, Mazzola F, & Magni G (2002). Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochemical and Biophysical Research Communications, 297(4), 835–840. 10.1016/S0006-291X(02)02285-4 [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA & Imai S (2004). The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 279, 50754–63. [DOI] [PubMed] [Google Scholar]
- Sallin O, Reymond L, Gondrand C, Raith F, Koch B, & Johnsson K (2018). Semisynthetic biosensors for mapping cellular concentrations of nicotinamide adenine dinucleotides. eLife, 7, e32638. doi: 10.7554/eLife.32638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci L, Cimadamore F, Scotti S, Petrelli R, Cappellacci L, Franchetti P, … Magni G (2007). Initial-Rate Kinetics of Human NMN-Adenylyltransferases: Substrate and Metal Ion Specificity, Inhibition by Products and Multisubstrate Analogues, and Isozyme Contributions to NAD+ Biosynthesis. Biochemistry, 46(16), 4912–4922. 10.1021/bi6023379 [DOI] [PubMed] [Google Scholar]
- Swallow W, Monahan J (1984). Monte Carlo Comparison of ANOVA, MIVQUE, REML, and ML Estimators of Variance Components. Technometrics 28:47–57 [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, … Looger LL (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods, 6(12), 875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E (2015). NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–13. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P & Krebs HA. (1967). The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 103, 514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Prole DL, Shen Y, Lin Z, Gnanasekaran A, Liu Y, Chen L, Zhou H, Chen SR, Usachev YM, Taylor CW, Campbell RE (2014). Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. The Biochemical journal, 464(1), 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff SS (2008). The equality of REML and ANOVA estimators of variance components in unbalanced normal classification models. Statistics & Probability Letters 78, 405–411. [Google Scholar]
- Yang Y & Sauve AA (2016). NAD+ metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta. 1864, 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA (2007). Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell, 130(6), 1095–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, & Goodman RH (2002). Regulation of corepressor function by nuclear NADH. Science 295, 1895–7. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, Hu H, Chen X, Zhou HM, Huang X, Yang K, Zhu Q, Wang X, Yi J, Zhu L, Qian X, Chen L, Tang Y, Loscalzo J, … Yang Y (2015). SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell metabolism, 21(5), 777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE (2011). An expanded palette of genetically encoded Ca2+ indicators. Science (New York, N.Y.), 333(6051), 1888–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 14, 555–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XH, Lu M, Lee BY, Ugurbil K & Chen W (2015). In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci USA. 112, 2876–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Stewart M, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen Michael S., Goodman RH (2016). Biosensor reveals multiple sources for mitochondrial NAD⁺. Science 352, 1474–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The NAD+ sensor described in this protocol was originally published in Cambronne, X.A., et al. in Science in 2016. [Google Scholar]