Abstract
Functional connectomes computed from fMRI provide a means to characterize individual differences in the patterns of BOLD synchronization across regions of the entire brain. Using four resting-state fMRI datasets with a wide range of ages, we show that individual differences of the functional connectome are stable across 3 months to 1–2 years (and even detectable at above-chance levels across 3 years). Medial frontal and frontoparietal networks appear to be both unique and stable, resulting in high ID rates, as did a combination of these two networks. We conduct analyses demonstrating that these results are not driven by head motion. We also show that edges contributing the most to a successful ID tend to connect nodes in the frontal and parietal cortices, while edges contributing the least tend to connect cross-hemispheric homologs. Our results demonstrate that the functional connectome is stable across years and that high ID rates are not an idiosyncratic aspect of a specific dataset, but rather reflect stable individual differences in the functional connectivity of the brain.
Introduction
Using fMRI and functional connectivity analyses, it is possible to establish a functional connectome for an individual. It has previously been shown that young adults’ functional connectomes are unique and stable across multiple days to one week (Finn et al., 2015; Miranda-Dominguez et al., 2014). In this context, the term unique means that an individual’s functional connectivity matrix has characteristics that are specific to an individual. The term stability, on the other hand, means that these characteristics are not a moment-to-moment attribute of the data, but rather reflect features that are detectable across time. This uniqueness and stability can be measured using an identification (ID) test, which identifies an individual from a pool of other individuals. To determine the ID rate of a dataset requires at least two scans from an individual, in which one is designated as a reference scan and the other a target scan. Using this framework, the ability to identify individuals via their functional connectomes from data acquired in sessions separated by multiple days has been replicated many times in adult subjects (Amico and Goni, 2018; Biazoli et al., 2017; Finn et al., 2017; Finn et al., 2015; Horien et al., 2018; Kaufmann et al., 2018; Noble et al., 2017; Vanderwal et al., 2017; Waller et al., 2017) and in children and adolescents (Kaufmann et al., 2018; Kaufmann et al., 2017). In addition, these studies have consistently shown that regions in the frontal and parietal cortices (i.e. medial frontal and frontoparietal networks) tend to be important for defining individual uniqueness in functional connectivity data (Amico and Goni, 2018; Finn et al., 2017; Finn et al., 2015; Kaufmann et al., 2017; Miranda-Dominguez et al., 2014; Vanderwal et al., 2017; Waller et al., 2017).
Nevertheless, questions remain as to the stability of the functional connectome over longer periods of time and the extent to which unique networks remain important in discriminating among individuals over longer time frames, especially given that these networks are comprised of regions that undergo changes throughout the lifespan. For example, it is known that children and adolescents experience marked changes in numerous structural and functional measures, especially in frontal and parietal regions (Giedd et al., 1999; Gogtay et al., 2004; Gu et al., 2015; Kaufmann et al., 2017; Vasa et al., 2018). Older adults have been shown to exhibit changes in functional connectivity measurements in these regions as well (Chan et al., 2014; Damoiseaux, 2017; Ng et al., 2016). Hence, these populations provide an intriguing opportunity to test if individual differences in the connectome are stable despite these changes and to determine if unique networks across days remain unique at longer time frames. In fact, a preliminary report using a single dataset suggested that younger subjects (7–15 year olds) do seem to exhibit unique and stable whole-brain connectomes over 1–2 years (Miranda-Dominguez et al., 2018).
In addition, there are other reasons to investigate the stability of the connectome over longer time intervals. Having knowledge about the stability of the connectome over longer time scales can inform prediction-based studies that might prove useful in a clinical context. This predictive framework, however, necessitates that unique patterns of functional connectivity (ultimately underlying the association with behavior) remain unique across longer time-scales. That is, if subject-specific patterns in connectivity data apparent at shorter time-scales disappear at longer intervals, there is little hope in using connectivity data in long-term prediction studies. Hence, knowledge about how the connectome changes over multiple years can inform prediction-based work in clincal scenarios.
Here, using four independent longitudinal fMRI datasets, from four different sites, we confirm the uniqueness of the connectome over a period of years. We extend these findings by characterizing the relative importance of specific networks in discriminating among individuals, and determine the anatomical locations of the most and least informative edges across years. The findings reveal that connectomes are indeed stable across time spans of 1–2 years, individual differences are detectable up to 3 years between scans at above chance levels, and the networks driving detection of individual differences across days (i.e. medial frontal and frontoparietal networks) also drive detection across longer time frames.
Methods
Overview
We have two main goals in this paper: 1) determine connectome stability using connectome-based identification over the course of years during periods of large-scale changes in the brain and 2) determine the importance of specific networks and anatomical locations in maintaining subject uniqueness (Figure 1). We do this by analyzing four independent resting-state datasets (three comprised of adolescents/younger adults and one comprised of older adults) with multiple scan sessions (i.e. up to three separate time points years apart) and some with multiple test-retest scans per session (i.e. two rest runs per scan session). To aid interpretation we mainly present results from session 1 – session 2 in the main text, but provide results from the other sessions and test-retest scans in the Supplementary Material.
Figure 1.
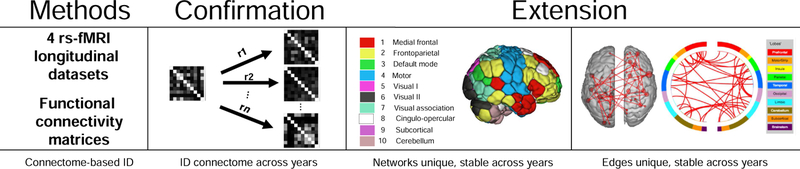
Overview of the goals of this paper. To test connectome-stability, we used connectome-based identification to determine if individual differences in functional connectivity data are detectable across years. The stability and uniqueness of networks across years is also characterized, in addition to determining the locations of the most and least informative edges.
Description of datasets
We utilized four longitudinal rs-fMRI datasets of neurotypical subjects. The University of Pittsburgh School of Medicine dataset, the University of Utah dataset, and the University of McGill dataset (hereafter referred to as Pitt, Utah, and UM, respectively) were downloaded from the Consortium for Reliability and Reproducibility (CoRR; http://fcon_1000.projects.nitrc.org/indi/CoRR/html/samples.html; Zuo et al., 2014); the Southwest University Longitudinal Imaging Multimodal dataset (hereafter referred to as SLIM; Liu et al., 2017) was downloaded through the International Data-sharing Initiative (INDI; http://fcon_1000.projects.nitrc.org/). All datasets were collected in accordance with the institutional review board or research ethics committee at each site. All resting-state scans were acquired on Siemens 3-T Trio scanners. Relevant demographic characteristics and imaging parameters are described in Table 1; full details of the Pitt, SLIM, and UM datasets can be found elsewhere (Hwang et al., 2013; Liu et al., 2017; Orban et al., 2015). To provide a fuller picture of the demographic data, we provide histograms of the number of years between scans and age at the time of the first scan for all datasets (Supplemental Figure 1 and 2). We note that in the Utah data set, two resting-state scans were provided for session 2 (hereafter session2a and session 2b), and in the UM data set two resting-state scans were provided for session 1 (hereafter session1a and session1b) and for session 2 (hereafter session2a and session2b).
Table 1.
Demographic and imaging characteristics of datasets used in this study.
| SLIM | Pitt | UM | Utah | |
|---|---|---|---|---|
| Number of sessions | 3 | 3 | 2a | 2a |
| Subjects in session 1 and 2 (females) | 105 (49) | 93 (45) | 79 (58) | 26 (0) |
| Subjects in session 3 (females) | 105 (49) | 30 (16)b | - | - |
| Low-motion subjects session 1- session 2 | 47 | 44 | 27 (Session 1a – session 2a) | 16 (Session 1 – session 2a) |
| Low-motion subjects session 1- session 3 | 38 | 15 | 27 (Session 1a – session 2b) | 14 (Session 1 – session 2b) |
| Low-motion subjects session 2- session 3 | 40 | 16 | 25 (Session 1b – session 2a) | - |
| Low-motion subjects session 1b –session 2b | - | - | 25 (Session 1b – session 2b) | - |
| Age at scan 1 (mean+ SD)c | 19.67 + 0.96 years | 15.23 + 2.83 years | 65.3 + 6.28 years | 20.23 + 8.28 years |
| Years between scan 1 and 2 (mean+ SD) | 0.84 + 0.29 years | 1.76 + 0.41 years | 0.305 + 0.067 years | 2.54 + 0.29 years |
| Years between scan 2 and 3 (mean+ SD) | 1.532+ 0.18 years | 1.53 + 0.30 years | - | - |
| Years between scan 1 and 3 (mean+ SD) | 2.37 + 0.28 years | 3.16 + 0.26 years | - | - |
| Scan duration in minutes (volumes) | 8 (242) | 5 (200) | 5 min 45 sec (150) | 8 (240) |
| TR in seconds | 2 | 1.5 | 2 | 2 |
Both UM and Utah had 2 scans sessions. However, the UM subjects had 2 resting-state runs per session (4 scans total); the Utah subjects had 2 resting-state runs in session 2 (3 scans total). See ‘Description of datasets’ in the Methods for details about how these sessions were named.
Scan session 2 and 3 occurred on the same day in one female subject in this session; we removed it when conducting analyses that required us to incorporate the number of years between scans 2 and 3.
All datasets provided age in years at scan session 1. For SLIM and Utah, the ages were reported as integers in years only, while the time between scan sessions was reported in days. To standardize the reporting of age among all four datasets, we only report age at session 1 here.
Note that because of numerous differences between the datasets (eyes open/closed during rest, resting-state scan collected after other functional runs, differences in the number of years between scans, etc.) we did not compare ID rates between datasets; rather, our focus was on defining the upper bounds of identifiability within each dataset. In addition, identification success is influenced by sample size (Waller et al., 2017; Horien et al., 2018), so care must be taken to consider the number of subjects when interpreting ID rates. It should also be noted that given a large enough sample size, a successful identification might not be possible.
Preprocessing
The preprocessing strategy used has been described in detail elsewhere (Greene et al., 2018). All preprocessing steps were performed using BioImage Suite (Joshi et al., 2011) unless otherwise indicated. We note that we only preprocessed the Pitt, Utah, and UM subjects, as the SLIM data was preprocessed beforehand as in Liu et al. (2017); for SLIM, we downloaded (http://fcon_1000.projects.nitrc.org/indi/retro/southwestuni_qiu_index.html) pre-calculated connectivity matrices based on the 268-node functional atlas described below. The preprocessing steps included: skull-stripping the 3D magnetization prepared rapid gradient echo (MPRAGE) images using optiBET (Lutkenhoff et al., 2014) and performing linear and non-linear transformations to warp a 268-node functional atlas from MNI space to single subject space using BioImage Suite as in Greene et al., (2018). In the UM dataset, no subjects were deemed to have major atrophy precluding successful registration using the MNI template. Functional images were slice-time and motion corrected using SPM8 (https://www.fil.ion.ucl.ac.uk/spm/software/spm8/). Covariates of no interest were regressed from the data, including linear, quadratic, and cubic drift, a 24 parameter model of motion (Satterthwaite et al., 2013) mean cerebral-spinal fluid signal, mean white matter signal, and the overall global signal. Data were temporally smoothed with a zero-mean unit-variance low-pass Gaussian filter (approximate cutoff frequency of 0.12 Hz). The results of skull-stripping, nonlinear, and linear registrations were inspected visually after each step by the corresponding author (CH).
Node and network definition
We used a 268-node functional atlas described previously (Finn et al., 2015). For each subject the average time-course of each region of interest (“node” in graph theoretic terminology) was calculated, and the Pearson correlation coefficient was calculated between every other node to achieve a symmetric 268 × 268 matrix of correlation values representing edges (connections between nodes) in graph theoretic terminology. We subsequently normalized the matrix to z-scores via a Fisher transformation and only considered the upper triangle of the matrix, yielding 35,778 unique edges for whole-brain analyses. These nodes were then grouped into 10 functional networks as in previous work (Finn et al., 2015; Noble et al., 2017). Network names are listed in Figure 5a.
Figure 5.
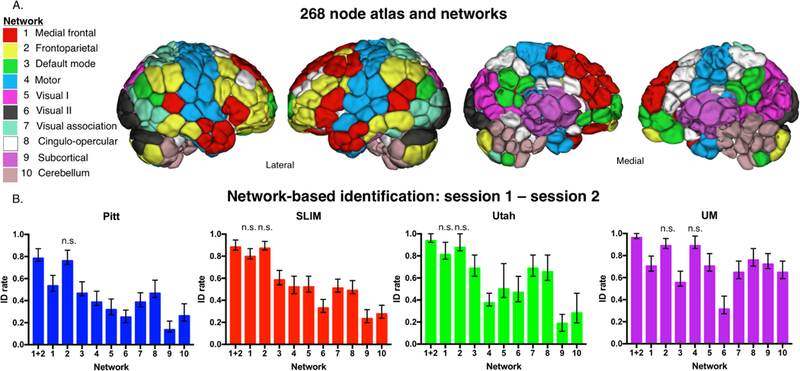
Network-based identification of low-motion subjects. Panel A: Node and network labels. We utilized a 268-node functional atlas. Nodes were further grouped into the 10 functional networks indicated here. Network names are to the left; anatomic locations are shown on the brains to the right. Panel B: We performed identification using only within-network edges (networks 1 – 10; indicated below the x-axis of each graph). We also combined networks 1 and 2 and performed identification with these edges (indicated as ‘1+2’ below the x-axis). Dataset name is indicated at the top of the graph. Networks that do not have a statistically lower ID rate (P > 0.05) than combined network 1 and 2 are labelled ‘n.s.’ above the corresponding network; all other networks that are unlabeled have an ID rate that is lower than combined network 1 and 2 (P < 0.05; i.e. these networks do have ‘n.s’ above the bar). Error bars correspond to 95 percent confidence intervals obtained via bootstrapping. Note that we considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2.
To ensure our results were not being driven by the use of a specific atlas, we repeated the whole-brain ID analyses using a 368-node functional atlas. The 368-node brain atlas was created by combining three different delineations of brain regions. For the cortex, we applied a groupwise parcellation algorithm (Shen et al., 2010) and obtained 164 nodes in the left hemisphere and 163 nodes in the right hemisphere. This functional parcellation of the cortex was computed based on resting-state BOLD data from 120 subjects collected at Yale (unpublished data). For the subcortical area, we adopted anatomic definitions of subcortical structures (Lacadie et al., 2008). There are seven nodes in each of the left and right subcortical regions. For the cerebellum, we adapted the 17 network definition proposed by Buckner et al. (2011), eliminating some nodes of small size, leaving 13 nodes in each of the left and right cerebellum. We also included one node for the brain stem area. Using this atlas for identification, the results verified that we were able to attain high ID rates using a different atlas.
Standardizing nodes and edges in a dataset
In the Pitt and SLIM dataset, it was necessary to eliminate some subjects and edges due to incomplete functional scan coverage. We describe the process for both datasets here.
In the Pitt dataset, we started with 100 unique subjects with session 1 and session 2 data; 30 subjects had all 3 scans. From the session 1 – session 2 scan pairs, we excluded one subject (subject ID 25316) that was missing an anatomical scan from session 1. To deal with subjects missing edges due to incomplete functional coverage in the remaining subjects, we plotted the number of edges missing per subject for each session: this revealed most subjects had most edges (data not shown), though 6 subjects were missing > 6,500 of the 35,778 unique edges in the full connectivity matrix. These subjects were removed from further analyses.
In the SLIM dataset, we started with 553 subjects in session 1, 239 subjects in session 2, and 227 subjects in session 3. We required that subjects have all imaging data in all three sessions; this reduced the number of subjects to 121. As before, we plotted the number of edges missing per subject for each session: this again revealed most subjects had most edges (data not shown), though 16 subjects were missing > 5,000 of the 35,778 unique edges in the full connectivity matrix. These subjects were removed from further analysis.
From here, if any remaining subject was missing an edge in Pitt or SLIM, we removed that edge from all subjects. Of the 35,778 edges in the whole-brain functional connectome, 29,646 edges/subject remained in the Pitt dataset; 31,626 edges/subject remained in the SLIM dataset. All 35,778 edges were covered for all subjects in the Utah and UM datasets (after removing 1 subject missing ~1000 edges from all scans in UM). The number of edges missing in each network, differences between networks in terms of missing edges, and differences in each network among the datasets must be kept in mind when interpreting ID rates, especially in the network-based ID results. See Supplemental Table 1 for the final list of the subject IDs used in this study. See Supplemental Figure 3 for the proportion of edges remaining in network pairs for SLIM and Pitt.
Identification procedure
The identification procedure has been described in detail previously (Finn et al., 2015). Briefly, a database is first created consisting of all subjects’ connectivity matrices from a particular session for a specific dataset. In an iterative process, a connectivity matrix from a subject is then selected from a different session and denoted as the target. Pearson correlation coefficients are calculated between the target connectivity matrix and all the matrices in the database. If the highest Pearson correlation coefficient is between the target subject in one session and the same subject in the second session, this would be recorded as a correct identification. We repeat the identification test such that each subject serves as the target subject once. This process is repeated until identifications have been performed for all subjects, sessions, and database-target combinations. We then averaged both database-target pairs (because these can be reversed) for a dataset to achieve an average ID rate for a given scan pair. This approach was used for whole-brain ID as well as network-based ID, in which connectivity matrices are formed using only the edges from a particular network of interest. To determine if ID rates were achieved at above-chance levels, we used permutation testing to generate a null distribution. Specifically, subject identities were randomly shuffled and ID was performed with these shuffled labels. ID rates obtained using the correct labels were then compared to this null distribution to determine significance. To compare ID rates between pairs of networks in a dataset we performed bootstrapping: we randomly subsampled ~80% of the subjects in each iteration and performed identification. The process was repeated 1000 times to generate 95% confidence intervals and determine significance.
Controlling for motion and artificial elevations in ID rate
To avoid confounds due to motion, we performed identification on low-motion subjects (i.e. a mean frame-frame displacement (FFD) threshold of < 0.1mm for both scans) as described previously (Horien et al., 2018); the number of low-motion subjects in each dataset is provided in Table 1. Imposing a threshold like this has previously been shown to limit confounds due to motion in different samples (Greene et al., 2018).
To further determine that identification was not due to idiosyncratic head movements specific to an individual across sessions, we conducted identification analyses using estimates of head movement parameters, which has been described in detail elsewhere (Finn et al., 2015). Briefly, we calculated discrete motion distribution vectors for each subject based on FFD over an entire scan. We computed the mean and standard deviations of the FFD across all subjects for each dataset. Based on the number of volumes collected, we then specified 15 bins for the Utah and SLIM subjects, 10 bins for the Pitt subjects, and 8 bins for the UM subjects to span the grand mean +/−3 standard deviations, and motion distribution vectors were subsequently calculated. These vectors were then submitted to the identification procedure. The results verified that motion was not a major factor in identification.
In addition, to determine if there was a relationship between motion and identifiability, we correlated the mean FFD with self-correlation scores (i.e. the correlation across edges between time 1 and time 2 for each subject) and also variance of FFD with self-correlation scores. Specifically, we calculated the mean/variance of FFD for a subject across a session, then calculated the average across the scan pairs involved in the identification for that subject. Across all subjects, we then correlated the averaged mean/variance of FFD (i.e. a single value for mean/variance of FFD across the two sessions) with self-correlation scores. The results again confirmed that motion was not driving identifiability.
We also tested to see if the ID results were driven by an artificially reduced sample size. To do so, we performed ID using all subjects in a sample (i.e. both the high and low-motion subjects).
Edge-based and correlation analyses
To determine the role of specific edges in the identification process, we quantified highly unique and highly consistent edges via the differential power (DP) measure and the group consistency measure described in detail elsewhere (Finn et al., 2015). We provide a brief overview of the intuition here; a formal mathematical description is provided in the Supplemental Methods. DP provides an estimate, for each given edge, of the likelihood that within-subject similarity is higher than that between subjects. To calculate the DP, the product of an edge value from time 1 and time 2 from the same subject is compared to the product of the same edge value from time 1 and time 2 from unmatched subjects (note that this a “paired” edge—for example, the value for edge 1 from time 1 is multiplied by the value for edge 1 from time 2). If the within-subject product is higher than the between-subject product across all subjects in a sample, this corresponds to a high DP value, and the edge is helpful in identification.
To calculate the group consistency measure, we multiply a given edge value from time 1 and time 2 within a subject (again, note that this a paired edge). We do this across all edges for all subjects and calculate the mean for each edge. Edges with high values in this measure are therefore high across all individuals in the group and are not helpful in identification. In the course of these analyses, we also determined the network(s) to which any given pair of edges belong (within-and between-networks); to account for differences in network sizes in both of these analyses, we divided the number of significant edges in a network pair by the number of total edges in the network.
To investigate the relationship between self-correlation scores (i.e. for a single subject, a self-correlation score is the correlation across all edges from time 1 and time 2 for that subject), age, and time between scans, we performed correlation (Pearson) analyses between self-correlation and the number of years between scans; we also correlated self-correlation scores with age. We further performed partial correlation analyses in which we assessed the relationship between self-correlation scores and age while controlling for time between scans and vice-versa.
Code availability
Analyses were conducted in Matlab; code is available online at https://www.nitrc.org/frs/?group_id=51. Questions regarding using or adapting these scripts should be directed to the corresponding author (CH). The corresponding author assumes all responsibility for accuracy/integrity of all data and code.
Results
Whole-brain identification results
Analyses using the whole-brain connectivity matrix and low-motion subjects (mean FFD < 0.1 mm for both sessions) yielded identification rates well above chance using all four datasets (P < 0.0001 obtained via permutation testing; Figure 2A). To aid interpretation of how ID rates using the connectivity matrices compare to a null distribution, we show the results of permutation testing in Figure 2A and Figure 2B. To determine if ID rates within a dataset were artificially elevated by the reduced sample sizes introduced by the motion threshold, we performed identification again with all subjects in a given dataset. Note that this increases the difficulty of the ID procedure in two ways: by increasing the number of subjects a participant might incorrectly match to, and by introducing harder to identify subjects into the ID pipeline, as high-motion subjects tend to look less similar from scan-to-scan (Amico and Goni 2018), which might decrease ID rates independent of sample size (Horien et al., 2018). Nevertheless, when we included all subjects in the ID pool, we observed ID rates again highly above chance (P < 0.0001 obtained via permutation testing; Figure 2B). To ensure results were not specific to the 268-node atlas, we repeated the whole-brain ID analyses shown in Figure 2A and 2B using a 368-node atlas. We achieved high ID rates in both the low-motion subjects (Supplemental Figure 4A) and when all subjects in a sample were included (Supplemental Figure 4B), increasing confidence that high ID rates are not specific to the 268-node atlas. Finally, we performed ID on subjects grouped by sex, as well as on the high-motion subjects only and observed that ID rates were similarly high (Supplemental Figure 5).
Figure 2.
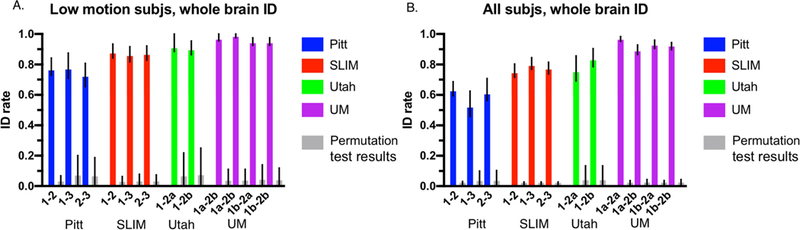
Identification results using whole brain data. Panel A: results using only low-motion subjects—only the subjects with a mean FFD < 0.1 mm were included in this analysis. Panel B: results using all subjects from a dataset—both high and low-motion subjects were included. Both panels: Each dataset is indicated below the x-axis, along with the sessions that were involved in identification. ID rate is indicated on the y-axis. Error bars correspond to 95% confidence intervals obtained via bootstrapping. Note that for a given scan pair two bars are shown: the actual ID rate obtained is shown to the left (see legend for the colors of different datasets); directly to the right of this are the results from permutation testing from that scan pair. Specifically, we are showing the mean permutation ID rate obtained after 1000 iterations with 95% confidence intervals.
We next assessed if there was an association between FFD and within-subject correlation scores. Specifically, we computed the mean FFD of a subject over a run, calculated the average across the two scans used in the ID process for that subject, and, across all subjects, correlated the average mean FFD with self-correlation scores. We used a similar process to correlate variance of FFD with self-correlation scores. For the low-motion subjects, we found that only 1/12 possible scan pairs across all datasets showed a significant positive correlation between mean FFD and self-correlation (Utah session 1-session 2b; r = 0.583, P = 0.0286; Supplemental Table 2). No other significant positive associations emerged, indicating that increased motion does not tend to lead to increased self-correlation scores. We found no significant positive correlations between variance of FFD with self-correlation scores (though we did observe one significant negative correlation (Pitt session 1-session 2; r = −0.3244, P = 0.0317; Supplemental Table 2). When we repeated these analyses with all subjects in a dataset, we found multiple occurrences of a significant negative correlation—that is, the higher the mean FFD or variance of FFD, the lower the self-correlation scores (Supplemental Table 2). Hence, these results suggest that across a sample, high in-scanner motion actually leads to decreased identifiability, increasing confidence that the high ID rates we obtained in Figure 2A and 2B are not being driven by motion.
Finally, we assessed if subject-specific movement patterns in the scanner were driving the high ID rates. We calculated a motion distribution vector (see Methods for details) capturing patterns of movement for each subject in a scan and used this for identification. In the low-motion subjects, the highest ID rate we obtained in Pitt was 2.27% (P = 0.5490) and in Utah was 3.57% (P = 0.8380); the ID rates for the other scan pairs in these datasets were all 0 (Table 2). In SLIM, we found 2/3 of the scan pairs had head-motion ID rates lower than chance levels (indicating that motion estimates could not be used to ID individuals). However, SLIM session 2 – session 3 had a motion ID rate of 11.25%, which was greater than chance (P = 0.004; Table 2). A similar pattern emerged in UM: 3/4 scan pairs had chance-level ID rates; UM session 1a – session 2a had an ID rate of 18.52% (P = 0.001; Table 2).
Table 2.
Results of using motion-based vectors to ID participants when only low-motion subjects are included in the analysis. The dataset and scan pair used is in the leftmost column; the head motion-based ID rate is in the middle column (0–1 scale; 0 = no successful identifications; 1 = all subjects successfully identified); the associated P-value (obtained through permutation testing) is in the rightmost column.
| Dataset (scan pair) | Head motion-based ID rate | p-value |
|---|---|---|
| Pitt (se1-se2) | 0.0227 | 0.5490 |
| Pitt (se1-se3) | 0 | 0.9999 |
| Pitt (se2-se3) | 0 | 0.9999 |
| SLIM (se1-se2) | 0.0106 | 0.7860 |
| SLIM (se1-se3) | 0.0263 | 0.5490 |
| SLIM (se2-se3) | 0.1125 | 0.0040 |
| Utah (se1-se2a) | 0 | 0.9999 |
| Utah (se1-se2b) | 0.0357 | 0.8380 |
| UM (se1a-se2a) | 0.1852 | 0.0010 |
| UM (se1a-se2b) | 0.0741 | 0.1580 |
| UM (se1b-se2a) | 0.0600 | 0.3310 |
| UM (se1b-se2b) | 0.0600 | 0.3200 |
When we repeated these analyses using all subjects in a dataset, we found numerous ID rates greater than chance (although the rates are lower than those obtained using the connectivity matrix; Supplemental Table 3). Hence, we cannot rule out that subject-specific movements may contribute somewhat to a successful ID. However, given the apparent lack of a positive correlation between mean/variance of FFD and self-correlation (especially in the low-motion subjects; Supplemental Table 2), and the low head motion-based ID rates we obtained (relative to connectivity-based ID), the overall contribution appears to be small.
Within-subject correlation scores, age, and time between scan results
To further investigate connectome stability, we next examined the relationship between self-correlation scores, age, and years in between scans by performing correlation analyses. To control for motion effects, we imposed the same 0.1 mm mean FFD threshold as above. We observed that there appears to be a negative correlation between self-correlation and time between scans, though, in general, the relationship was not statistically significant (all P > 0.05 with the exception of SLIM session1-session2, r = −0.3105, P = 0.0337; and UM session1b-session2a, r = −0.5528, P = 0.0042; Figure 3). There was no consistent relationship between self-correlation and age, with the only significant result coming from Utah session1-session2a (r = −0.6205, P = 0.0103; all other P-values > 0.05; Figure 4).
Figure 3.
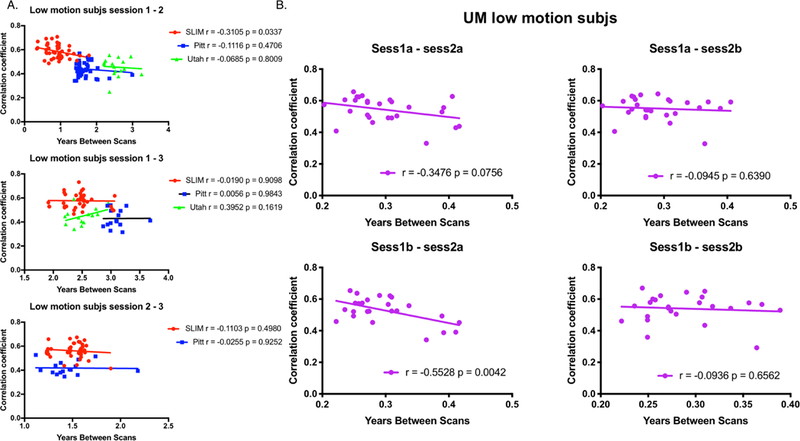
Plotting self-correlation against years in between scans for low-motion subjects. Panel A: Results for SLIM, Pitt, and Utah. SLIM is represented by red circles; Pitt is represented by blue squares; Utah is represented by green triangles. Panel B: Results for UM. Both panels: On the x-axis in both panel A and B is the number of years between scans. On the y-axis is the within-subject correlation coefficient (see Methods for details about calculating the within-subject correlation coefficient). The scan pair of interest (e.g. the target scan and the database scan) used in the identification process is indicated above each plot. Each plot also contains the results of correlation analyses for each dataset (r = Pearson correlation coefficient; p = P-value). Note that in Panel A for Utah we considered session 2a as session 2 and session 2b as session 3.
Figure 4.
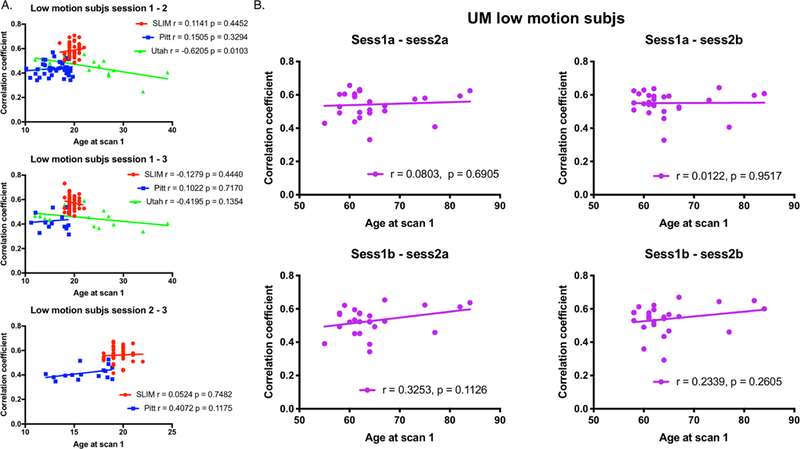
Plotting self-correlation against age at scan 1 for low-motion subjects. Panel A: Results for SLIM, Pitt, and Utah. SLIM is represented by red circles; Pitt is represented by blue squares; Utah is represented by green triangles. Panel B: Results for UM. Both panels: On the x-axis in both panel A and B is the age of a subject at scan 1. On the y-axis is the within-subject correlation coefficient (see Methods for details about calculating the within-subject correlation coefficient). The scan pair of interest (e.g. the target scan and the database scan) used in the identification process is indicated above each plot. Each plot also contains the results of correlation analyses for each dataset (r = Pearson correlation coefficient; p = P-value). Note that in Panel A for Utah we considered session 2a as session 2 and session 2b as session 3.
We also performed analyses with all subjects in a given dataset, as well as subjects grouped by gender, and with high-motion subjects only (Supplemental Figures 6–13). We observed largely consistent results, in that there is an (expected) negative correlation between self-correlation and time between scans, though the relationship is weak; there is no clear relationship between age and self-correlation score in the datasets studied here; and motion does not appear to be driving the results (Supplemental Figures 6–9). Further, no clear trends emerged due to gender (Supplemental Figures 10–13).
In addition, we performed partial correlation analyses of the low-motion subjects to explore the relationship between self-correlation and years between scans while controlling for age (and vice-versa). No clear patterns emerged (Supplemental Table 4).
Taken together, these data reinforce that self-correlation scores tend to be stable between scans and there is no clear relationship between self-correlation and age. In addition, the results do not seem to be driven by motion.
Network-based identification results
Having determined whole-brain connectivity data is stable across years, we next tested the contributions of specific networks to this stability. We grouped the connectivity data for each subject into 10 functional networks (Figure 5A), and subsequently performed identification analyses using only the edges from a given network. Given the performance of the medial frontal and frontoparietal networks in previous work (Finn et al., 2015; Waller et al., 2017; Kaufmann et al., 2017), we also combined the edges from these networks (hereafter ‘combined network 1+2’) and performed identification.
Focusing on the low-motion subjects from session 1 – session 2, we observed that edges in networks 1 and 2 tended to lead to the highest ID rates, as well as combined network 1+2 (Figure 5B), consistent with the original fingerprinting work of Finn et al. (2015). For example, using the results of bootstrapping analyses and an alpha of 0.05, combined network 1+2 had higher ID rates than all other networks in Pitt (except compared to individual network 2); all other networks in SLIM (except compared to individual networks 1 and 2); all other networks in Utah (except compared to individual networks 1 and 2); and all other networks in UM (except compared to individual networks 2 and 4). We observed similar patterns of performance when we repeated identification on all subjects in a dataset: networks 1 and 2 resulted in the highest ID rates (Supplemental Figure 14).
To further assess the importance of combined network 1+2 in defining individual uniqueness, we next assessed ID performance using between-network pairs of edges in the low-motion subjects of session 1 – session 2 (Figure 6). Using the results of bootstrapping analyses and an alpha of 0.05, we observed that combined network 1+2 resulted in higher ID rates than 43/45 between-network edges in SLIM (non-significant network pairs: 1–2, 5–6); 42/45 between-network edges in Utah (non-significant network pairs: 1–2, 2–7, 2–8); 42/45 between-network edges in Pitt (non-significant network pairs: 1–2, 4–5, 5–6); and 42/45 between-network edges in UM (non-significant network pairs: 1–2, 4–5, 5–6). These results echo the original fingerprinting work of Finn et al. (2015), emphasizing the importance of the medial frontal and frontoparietal network. Performing between-network ID on all subjects in a dataset resulted in a similar pattern: the highest ID rates tended to result from edges connecting to networks 1 and 2 (Supplemental Figure 14).
Figure 6.
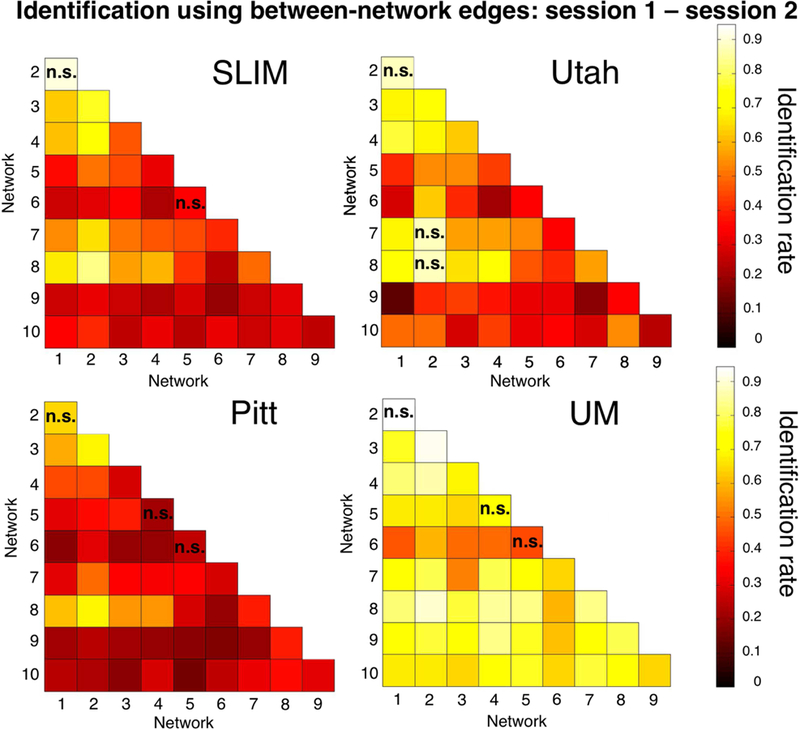
Between-network identification of low-motion subjects. We repeated the identification analyses using only the between-network edges. Shown are the results from session 1-session 2. Dataset name is indicated above the appropriate matrix. The scale of the color bars is the same for all matrices. We considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2. Note also that the diagonal of the matrix (which is not shown) consists of the within-network edges; these data are shown in Figure 5B. Networks in the matrix with an ‘n.s.’ do not have a statistically lower ID rate than combined network 1+2 (P > 0.05); combined network 1+2 had a statistically higher ID rate than all other networks (P < 0.05; i.e. these networks do not have an ‘n.s’). Note that statistical significance is assessed by comparing the ID distributions for each network obtained through bootstrapping; thus, it is possible for a network with a low ID rate in the Figure (i.e. network pair 5–6 in SLIM) to not be significantly different than combined network 1+2 (despite having a much lower ID rate).
Edge-based analyses results
Having established that the medial frontal and frontoparietal networks tend to be stable across years in individual subjects, we lastly assessed the importance of specific edges to subject uniqueness in the low-motion subjects of session 1 – session 2. Using the DP measure, which calculates how characteristic an edge tends to be, we determined which edges were important in the identification process. For visualization purposes we show the anatomical locations of DP edges in the 99.9 percentile (Figure 7). Significant edges tended to be distributed across the entire brain, with particular representation in prefrontal, temporal, and parietal cortices. Less stringent thresholds resulted in similar overall patterns (albeit with more edges; Supplemental Figure 15). Quantifying network representation revealed that networks 1 and 2 tended to harbor significant edges (Figure 8): approximately 78% of edges were within or connected to networks 1 and 2 in Utah; approximately 60% in SLIM; approximately 58% in Pitt; and approximately 48% in UM. The overrepresentation of significant edges in networks 1 and 2 held at a range of thresholds (Supplemental Table 5). DP results were stable when we re-performed the analyses on all subjects in each dataset (Supplemental Figure 16 and 17).
Figure 7.
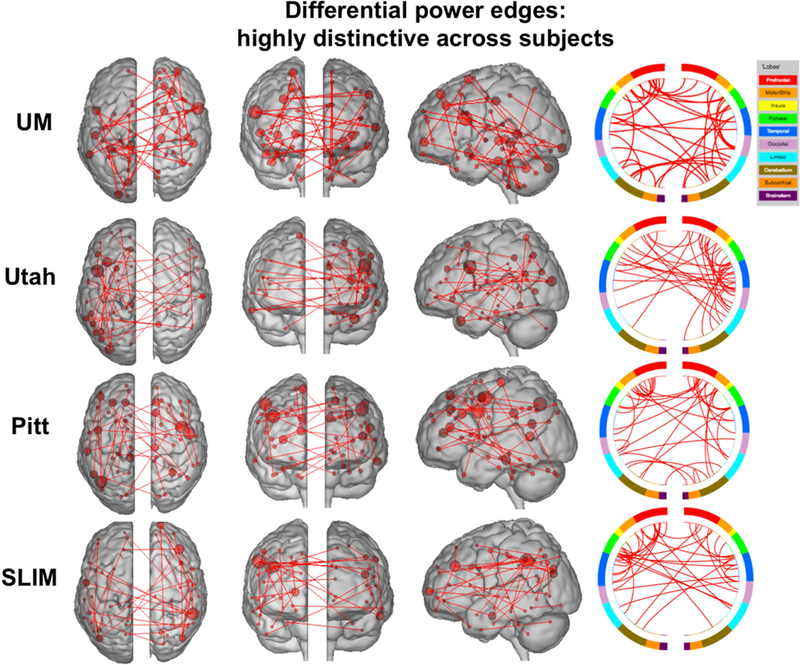
Results of edge-based analyses – differential power (highly discriminative) edges in low-motion subjects. The edges shown here were in the top 99.9 percentile of highly unique edges among subjects – these edges were most helpful in identification. Each row corresponds to results from a different dataset. For a given row, in the three leftmost images of the brain, the red lines indicate edges connecting the red spheres, representing nodes. Nodes are sized according to degree, the number of edges connected to that node. For a given row, on the right, the same nodes and edges are visualized on a circle plot, in which nodes are grouped according to anatomic location. The top of the circle represents anterior; the bottom, posterior. The left half of the circle plot corresponds to the left hemisphere of the brain. A legend indicating the approximate anatomic ‘lobe’ is shown to the top right of the figure. Results are shown for session 1 – session 2. Note that we considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2.
Figure 8.
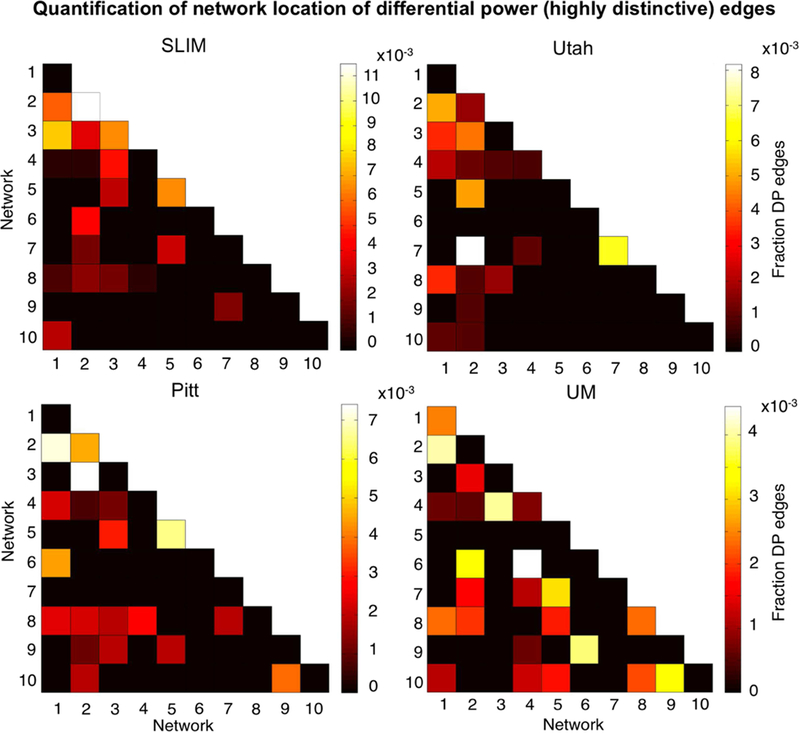
Network representation of differential power (highly unique) edges in low-motion subjects. Here we are quantifying the network location of the data shown in Figure 7 (i.e. these are the 99.9 percentile most unique edges). Hotter colors indicate more edges are in the network pair; if an element in the matrix is black it indicates no edges were in this network pair. Results are normalized by network size. Data are shown for session 1 – session 2. Note that the color bars are not necessarily the same among all datasets. Note also that we considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2.
To quantify the extent to which individual edges do not contribute to subject uniqueness, we calculated the group consistency measure, which quantifies edges that are highly consistent within a single subject and across all subjects in a dataset. Because they are highly consistent for all subjects in a dataset, such edges do not discriminate among individuals. For visualization we show the anatomical locations of group consistency edges in the 99.9 percentile; these edges tended to link cross-hemispheric homologs (Figure 9). This pattern of connecting to cross-hemispheric homologs was stable across a range of thresholds (Supplemental Figure 15). Quantifying network representation revealed that the majority of these edges tended to be located in visual networks (networks 5 and 6), as well as the cerebellum in UM (network 10; Figure 10). Group consistency results were stable when this analysis was repeated on all subjects in each dataset (Supplemental Figure 16 and 17).
Figure 9.
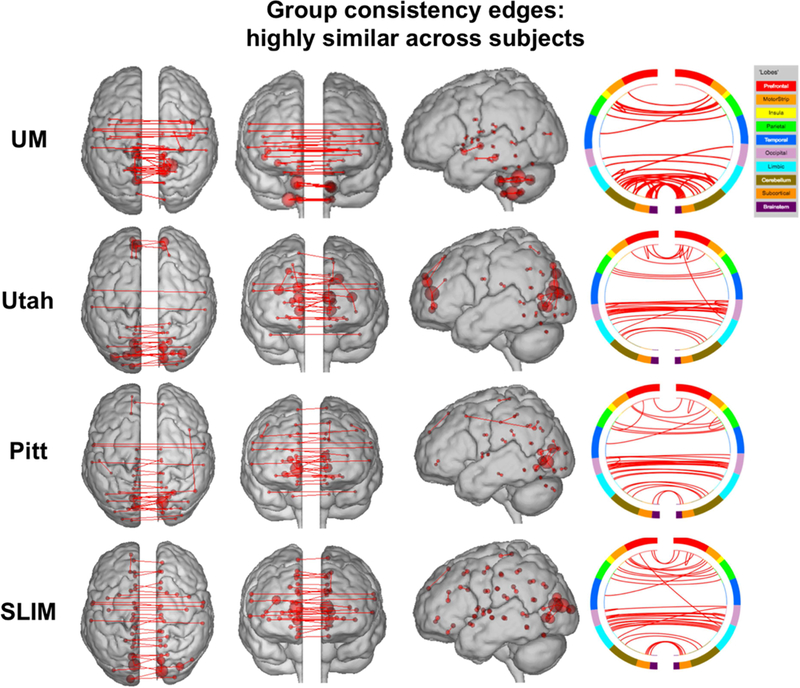
Results of edge-based analyses – group consistency (highly similar) edges in low-motion subjects. The edges shown here were in the top 99.9 percentile of highly similar edges among subjects – these edges were least helpful in identification. Each row corresponds to results from a different dataset. For a given row, in the three leftmost images of the brain, the red lines indicate edges connecting the red spheres, representing nodes. Nodes are sized according to degree, the number of edges connected to that node. For a given row, on the right, the same nodes and edges are visualized on a circle plot, in which nodes are grouped according to anatomic location. The top of the circle represents anterior; the bottom, posterior. The left half of the circle plot corresponds to the left hemisphere of the brain. A legend indicating the approximate anatomic ‘lobe’ is shown to the top right of the figure. Results are shown for session 1 – session 2. Note that we considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2.
Figure 10.
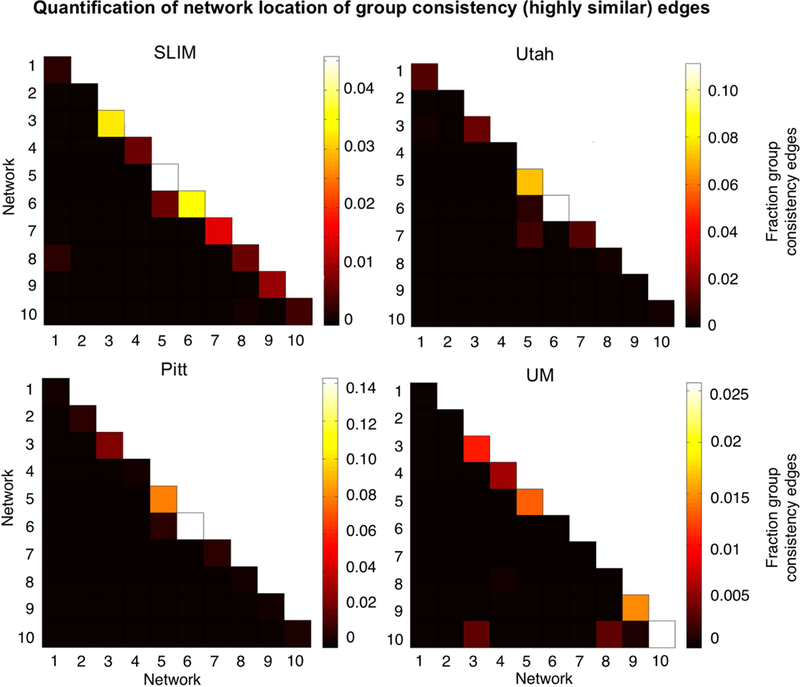
Network representation of group consistency (highly similar) edges in low-motion subjects. Here we are quantifying the network location of the data shown in Figure 9 (i.e. these are the 99.9 percentile most similar edges). Hotter colors indicate more edges are in the network pair; if an element in the matrix is black it indicates no edges were in this network pair. Results are shown for session 1 – session 2. Note that the color bars are not necessarily the same for all datasets. Note also that we considered Utah session 2a as session 2; for UM we considered session1a as session 1 and session 2a as session 2.
Discussion
In this work, we set out to assess the stability of the functional connectome over longer periods of time, as well as the extent to which networks found to be unique over the course of days (i.e. medial frontal and frontal parietal networks; Finn et al., 2015) remain unique across months to years. Utilizing connectome-based identification in four longitudinal resting-state fMRI datasets, we determined that individual differences in the functional connectome are indeed stable across 1–2 years and that individual differences are detectable up to 3 years between scans at above chance levels. Further, the networks driving individual uniqueness across days also drive uniqueness across months to years. By utilizing samples comprised of younger participants (children and adolescents) as well as older adults, our results also demonstrate that even in periods of significant brain changes, individuals retain a unique signature of functional connectivity patterns. Hence, consistent with other recent work (Finn et al., 2015; Gordon et al., 2017b; Gratton et al., 2018; Laumann et al., 2015; Poldrack et al., 2015), we find that individual differences are a stable feature of brain organization revealed by functional connectivity analyses.
Replication and open science
An important aspect of the scientific process is reliability and reproducibility, and practical steps have been proposed in the context of fMRI (Nichols et al., 2017; Poldrack et al., 2017). Especially in a field like neuroimaging, which has largely failed to make an impact on clinical practice, studies are needed that assess the replicability and generalizability of findings (Nichols et al., 2017; Poldrack et al., 2017). Given the advent of the open data movement in neuroimaging (Poldrack et al., 2017; Poldrack et al., 2013; Poldrack and Gorgolewski, 2014; Zuo et al., 2014), there are multiple open source datasets readily available to assess the reproducibility of findings. Here, we leveraged these resources to use four different datasets, with different sample characteristics, to address the stability of functional connectivity patterns. Given the need to utilize test-retest datasets, we would not have been able to conduct the analyses reported here if it were not for open source data. Using these resources allowed us to replicate the findings of previous reports (Amico and Goni, 2018; Biazoli et al., 2017; Finn et al., 2017; Finn et al., 2015; Horien et al., 2017; Kaufmann et al., 2018; Kaufmann et al., 2017; Miranda-Dominguez et al., 2014; Noble et al., 2017; Vanderwal et al., 2017; Waller et al., 2017) and add to the growing body of literature in connectome-based identification studies by extending this work, specifically focusing on identification across longer time frames. Thus, our overall approach speaks to the power of open data in neuroimaging, both in confirming and advancing initial results.
How can a network be “stable” but undergo developmental changes?
It has previously been shown that networks comprised of nodes in frontal and parietal association cortices are important for defining individual uniqueness across a shorter time frame (Finn et al., 2017; Finn et al., 2015; Kaufmann et al., 2017; Miranda-Dominguez et al., 2014; Vanderwal et al., 2017; Waller et al., 2017). Our new findings extend these results by showing that these same networks are stable over longer time scales. While our findings are consistent with other work highlighting the degree to which these regions exhibit differences between subjects (Finn et al., 2015; Gordon et al., 2017a; Gratton et al., 2018; Miranda-Dominguez et al., 2014; Mueller et al., 2013), our results are intriguing given that these same regions undergo changes in children and adolescents (Giedd et al., 1999; Gogtay et al., 2004; Gu et al., 2015; Kaufmann et al., 2017; Vasa et al., 2018) as well as in older adults (Chan et al., 2014; Damoiseaux, 2017; Ng et al., 2016). One possible explanation lies in the identification approach used here. The connectome-based ID approach does not require a participant to have precisely the same connectivity matrix from scan-to-scan; all that is needed for a correct ID is that a subject must look more like her/himself than anyone else. In this framework, single edges that change across test-retest scans (Noble et al., 2017; Pannunzi et al., 2017) and are expected to change across development might not be enough to overcome the large-scale, subject-specific patterns in FC that allow a successful identification. Hence, the high ID rates obtained here add a layer of nuance to the development literature: children and adolescents (and older adults) undergo neurodevelopmental changes leading to differences in functional connectivity measures due to age, but these changes occur in an individual-specific way. That is, despite brain changes, individuals still tend to look like themselves across development.
Motion considerations
Given the effect of in-scanner movement on estimates of functional connectivity (Power et al., 2015; Satterthwaite et al., 2012), motion is an important variable to consider. Motion has also been demonstrated to exhibit high test-retest reliability (Zuo and Xing, 2014), potentially complicating identification. We therefore undertook numerous steps to address movement in our preprocessing and analysis pipelines, and we observed that identification rates were high independent of motion. We did find that motion might be playing a small role in a successful ID— for example, we obtained above-chance motion-based ID rates in 1/4 scan pairs in UM and 1/3 scan pairs in SLIM. However, we also found that the higher the movement in a scan, the lower the subject’s self-correlation score. This result is consistent with previous work suggesting that motion hinders a correct ID (Amico and Goni, 2018; Finn et al., 2017; Finn et al., 2015; Horien et al., 2017; Vanderwal et al., 2017). Hence, while we cannot rule out that motion might play a small role in our findings, the high connectivity-based ID rates obtained here do not appear to be driven exclusively by in-scanner motion and seem to depend largely on differences in functional connectivity.
Individual differences and individual parcellations
We applied a group parcellation to our resting-state data, as have all connectome-based identification studies to this point. Recent work has indicated that individuals exhibit meaningful differences in the spatial topography of networks (e.g. differences in spatial topography are predictive of phenotypic measures; Kong et al., 2018), and that these differences can confound estimates of connectivity when a group parcellation is used (Bijsterbosch et al., 2018). Thus, it is possible that individual differences in spatial topography are driving subject identification (along with, or in place of, differences in functional connectivity). A potential way to circumvent this confound is using individualized parcellations (Braga and Buckner, 2017; Gordon et al., 2017b; Kong et al., 2018; Laumann et al., 2015; Salehi et al., 2017; Wang et al., 2015). Given that our goal was confirming and extending work using group parcellations, we did not individualize parcellations in this study. Further, given that substantial amounts of data have sometimes been required to establish accurate individual differences (Laumann et al., 2015; Gordon et al., 2017; Braga and Buckner, 2017), how best to use and validate these methods with “typical” scan times (e.g., the resting-state scans used here with 5–8 minutes of data per scan) is unclear, especially considering that little individualized parcellation work has been conducted in younger children and older adults, a majority of the participants in this study. Applying individualized parcellation techniques in these populations could be a fruitful area of future research, as could examining the stability of network topography more generally.
Limitations and future considerations
We only examined resting-state data here; hence, the extent to which task-based connectivity patterns remain stable is unclear. Other studies have shown that identification rates are highest across days when subjects are completing a task in the scanner (possibly by increasing subject-specific signal-to-noise and augmenting unique patterns of functional connectivity; Finn et al., 2017; Vanderwal et al., 2017), so identifiability could be investigated across longer time frames when subjects are completing task-based scans to see if a similar pattern holds. Task-based connectivity has recently been shown to better reveal brain-behavior associations than rest-based connectivity, again emphasizing how individual differences in connectivity patterns can be amplified by performance of a task (Greene et al., 2018). Another limitation is that we were unable to compare ID rates across samples given the numerous differences among the datasets. Thus, questions about how identifiability varies due to age in longitudinal samples await further investigation. Also, we have previously shown that in-scanner motion and the length of resting-state scans affect ID rates, while spatiotemporal resolution does not (Horien et al. 2018). The impact of other variables (length of time in between scans, the impact of eyes open versus eye closed resting-state runs, etc.) on identifiability requires additional data in order to be explored.
We observed that edges arising from the medial frontal and frontoparietal networks tended to result in the highest ID rates. These networks are comprised mostly of cortical nodes, which have been noted to be the most reliable in the brain (Noble et al., 2017; Shah et al., 2016) and which probably underlies the success of these networks here. One possible explanation is that these networks could be less prone, relative to networks comprised of mostly non-cortical nodes, to distortions of the data from susceptibility artifacts (Noble et al., 2017; Raj et al., 2001).
A number of other questions (Finn et al., 2015; Finn and Constable, 2016) remain open for study. For example, is an individual’s functional connectome unique from birth (or even in utero), or is the distinctiveness something that develops at a later age? If so, what is the developmental trajectory? The stability of the connectome across even longer time frames (i.e. across decades) is unknown and could be investigated. It is also unclear if an individual’s connectome ever becomes unidentifiable from scan-to-scan. Does identifiability degrade at a certain age? If so, how does this relate to measures of behavior and cognition? Finally, can we exploit the fact that the connectome is unique and stable in clinical scenarios?
Conclusions
In sum, we have shown that subjects have unique and stable functional connectomes over the span of years and that medial frontal and frontoparietal networks remain important in defining individual uniqueness over these long time scales. Leveraging the stability of functional connectome data to generate meaningful models related to phenotypic or clinical variables remains an important goal for this field.
Supplementary Material
Research highlights.
Used 4 longitudinal datasets to generate functional connectivity matrices
Whole-brain matrices are unique and stable across months to years
Medial frontal and frontoparietal networks tended to be both unique and stable
Edges in the frontal and parietal cortices tended to be most discriminative
Acknowledgements
This work was supported by a Medical Scientist Training Program training grant (NIH/NIGMS T32GM007205; C.H.). The authors thank Abigail Greene for helpful comments on a draft of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amico E, Goni J, 2018. The quest for identifiability in human functional connectomes. Sci Rep 8, 8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazoli CE Jr., Salum GA, Pan PM, Zugman A, Amaro E Jr., Rohde LA, Miguel EC, Jackowski AP, Bressan RA, Sato JR, 2017. Commentary: Functional connectome fingerprint: identifying individuals using patterns of brain connectivity. Front Hum Neurosci 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijsterbosch JD, Woolrich MW, Glasser MF, Robinson EC, Beckmann CF, Van Essen DC, Harrison SJ, Smith SM, 2018. The relationship between spatial configuration and functional connectivity of brain regions. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga RM, Buckner RL, 2017. Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron 95, 457–471 e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT, 2011. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol 106, 2322–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS, 2014. Decreased segregation of brain systems across the healthy adult lifespan. Proc Natl Acad Sci U S A 111, E4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, 2017. Effects of aging on functional and structural brain connectivity. Neuroimage 160, 32–40. [DOI] [PubMed] [Google Scholar]
- Finn ES, Scheinost D, Finn DM, Shen X, Papademetris X, Constable RT, 2017. Can brain state be manipulated to emphasize individual differences in functional connectivity? Neuroimage [DOI] [PMC free article] [PubMed]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 18, 1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Todd Constable R., 2016. Individual variation in functional brain connectivity: implications for personalized approaches to psychiatric disease. Dialogues Clin Neurosci 18, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL, 1999. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci 2, 861–863. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF 3rd, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM, 2004. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A 101, 8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Petersen SE, 2017a. Individual Variability of the System-Level Organization of the Human Brain. Cereb Cortex 27, 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt-Drazen C, Gratton C, Sun H, Hampton JM, Coalson RS, Nguyen AL, McDermott KB, Shimony JS, Snyder AZ, Schlaggar BL, Petersen SE, Nelson SM, Dosenbach NUF, 2017b. Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807 e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Laumann TO, Nielsen AN, Greene DJ, Gordon EM, Gilmore AW, Nelson SM, Coalson RS, Snyder AZ, Schlaggar BL, Dosenbach NUF, Petersen SE, 2018. Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Variation. Neuron 98, 439–452 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AS, Gao S, Scheinost D, Constable RT, 2018. Task-induced brain state manipulation improves prediction of individual traits. Nat Commun 9, 2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Satterthwaite TD, Medaglia JD, Yang M, Gur RE, Gur RC, Bassett DS, 2015. Emergence of system roles in normative neurodevelopment. Proc Natl Acad Sci U S A 112, 13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C, Noble S, Finn ES, Shen X, Scheinost D, Constable RT, 2018. Considering factors affecting the connectome-based identification process: Comment on Waller et al. Neuroimage 169, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K, Hallquist MN, Luna B, 2013. The development of hub architecture in the human functional brain network. Cereb Cortex 23, 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Scheinost D, Okuda H, Belhachemi D, Murphy I, Staib LH, Papademetris X, 2011. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics 9, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T, Alnaes D, Brandt CL, Bettella F, Djurovic S, Andreassen OA, Westlye LT, 2018. Stability of the Brain Functional Connectome Fingerprint in Individuals With Schizophrenia. JAMA Psychiatry 75, 749–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T, Alnaes D, Doan NT, Brandt CL, Andreassen OA, Westlye LT, 2017. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat Neurosci 20, 513–515. [DOI] [PubMed] [Google Scholar]
- Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, Sun N, Zuo XN, Holmes AJ, Eickhoff SB, Yeo BTT, 2018. Spatial Topography of Individual-Specific Cortical Networks Predicts Human Cognition, Personality, and Emotion. Cereb Cortex [DOI] [PMC free article] [PubMed]
- Lacadie C, F.R., Arora J, Constable RT, Papademetris X, 2008. Brodmann areas defined in MNI space using a new Tracing Tool in Bioimage Suite. Hum Brain Mapp, conference abstract
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, Schlaggar BL, Mumford JA, Poldrack RA, Petersen SE, 2015. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wei D, Chen Q, Yang W, Meng J, Wu G, Bi T, Zhang Q, Zuo XN, Qiu J, 2017. Longitudinal test-retest neuroimaging data from healthy young adults in southwest China. Sci Data 4, 170017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhoff ES, Rosenberg M, Chiang J, Zhang K, Pickard JD, Owen AM, Monti MM, 2014. Optimized brain extraction for pathological brains (optiBET). PLoS One 9, e115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, Fair DA, 2014. Connectotyping: model based fingerprinting of the functional connectome. PLoS One 9, e111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA, 2018. Heritability of the human connectome: A connectotyping study. Network Neuroscience 2, 10.1162/netn_a_00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H, 2013. Individual variability in functional connectivity architecture of the human brain. Neuron 77, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KK, Lo JC, Lim JKW, Chee MWL, Zhou J, 2016. Reduced functional segregation between the default mode network and the executive control network in healthy older adults: A longitudinal study. Neuroimage 133, 321–330. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Das S, Eickhoff SB, Evans AC, Glatard T, Hanke M, Kriegeskorte N, Milham MP, Poldrack RA, Poline JB, Proal E, Thirion B, Van Essen DC, White T, Yeo BT, 2017. Best practices in data analysis and sharing in neuroimaging using MRI. Nat Neurosci 20, 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D, 2017. Influences on the Test-Retest Reliability of Functional Connectivity MRI and its Relationship with Behavioral Utility. Cereb Cortex 27, 5415–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban P, Madjar C, Savard M, Dansereau C, Tam A, Das S, Evans AC, Rosa-Neto P, Breitner JC, Bellec P, Group, P.-A.R., 2015. Test-retest resting-state fMRI in healthy elderly persons with a family history of Alzheimer’s disease. Sci Data 2, 150043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzi M, Hindriks R, Bettinardi RG, Wenger E, Lisofsky N, Martensson J, Butler O, Filevich E, Becker M, Lochstet M, Kuhn S, Deco G, 2017. Resting-state fMRI correlations: From link-wise unreliability to whole brain stability. Neuroimage 157, 250–262. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafo MR, Nichols TE, Poline JB, Vul E, Yarkoni T, 2017. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat Rev Neurosci 18, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Barch DM, Mitchell JP, Wager TD, Wagner AD, Devlin JT, Cumba C, Koyejo O, Milham MP, 2013. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinform 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Gorgolewski KJ, 2014. Making big data open: data sharing in neuroimaging. Nat Neurosci 17, 1510–1517. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, Hunicke-Smith S, Simpson ZB, Caven T, Sochat V, Shine JM, Gordon E, Snyder AZ, Adeyemo B, Petersen SE, Glahn DC, Reese Mckay D., Curran JE, Goring HH, Carless MA, Blangero J, Dougherty R, Leemans A, Handwerker DA, Frick L, Marcotte EM, Mumford JA, 2015. Long-term neural and physiological phenotyping of a single human. Nat Commun 6, 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE, 2015. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage 105, 536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj D, Anderson AW, Gore JC, 2001. Respiratory effects in human functional magnetic resonance imaging due to bulk susceptibility changes. Phys Med Biol 46, 3331–3340. [DOI] [PubMed] [Google Scholar]
- Salehi M, Karbasi A, Shen X, Scheinost D, Constable RT, 2017. An exemplar-based approach to individualized parcellation reveals the need for sex specific functional networks. Neuroimage [DOI] [PMC free article] [PubMed]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH, 2013. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64, 240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE, 2012. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage 60, 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah LM, Cramer JA, Ferguson MA, Birn RM, Anderson JS, 2016. Reliability and reproducibility of individual differences in functional connectivity acquired during task and resting state. Brain Behav 6, e00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Papademetris X, Constable RT, 2010. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. Neuroimage 50, 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwal T, Eilbott J, Finn ES, Craddock RC, Turnbull A, Castellanos FX, 2017. Individual differences in functional connectivity during naturalistic viewing conditions. Neuroimage 157, 521–530. [DOI] [PubMed] [Google Scholar]
- Vasa F, Seidlitz J, Romero-Garcia R, Whitaker KJ, Rosenthal G, Vertes PE, Shinn M, Alexander-Bloch A, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, consortium N, Sporns O, Bullmore ET, 2018. Adolescent Tuning of Association Cortex in Human Structural Brain Networks. Cereb Cortex 28, 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller L, Walter H, Kruschwitz JD, Reuter L, Muller S, Erk S, Veer IM, 2017. Evaluating the replicability, specificity, and generalizability of connectome fingerprints. Neuroimage 158, 371–377. [DOI] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, Baker JT, Stufflebeam SM, Wang K, Wang X, Hong B, Liu H, 2015. Parcellating cortical functional networks in individuals. Nat Neurosci 18, 1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner JC, Buckner RL, Calhoun VD, Castellanos FX, Chen A, Chen B, Chen J, Chen X, Colcombe SJ, Courtney W, Craddock RC, Di Martino A, Dong HM, Fu X, Gong Q, Gorgolewski KJ, Han Y, He Y, He Y, Ho E, Holmes A, Hou XH, Huckins J, Jiang T, Jiang Y, Kelley W, Kelly C, King M, LaConte SM, Lainhart JE, Lei X, Li HJ, Li K, Li K, Lin Q, Liu D, Liu J, Liu X, Liu Y, Lu G, Lu J, Luna B, Luo J, Lurie D, Mao Y, Margulies DS, Mayer AR, Meindl T, Meyerand ME, Nan W, Nielsen JA, O’Connor D, Paulsen D, Prabhakaran V, Qi Z, Qiu J, Shao C, Shehzad Z, Tang W, Villringer A, Wang H, Wang K, Wei D, Wei GX, Weng XC, Wu X, Xu T, Yang N, Yang Z, Zang YF, Zhang L, Zhang Q, Zhang Z, Zhang Z, Zhao K, Zhen Z, Zhou Y, Zhu XT, Milham MP, 2014. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data 1, 140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Xing XX, 2014. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci Biobehav Rev 45, 100–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


