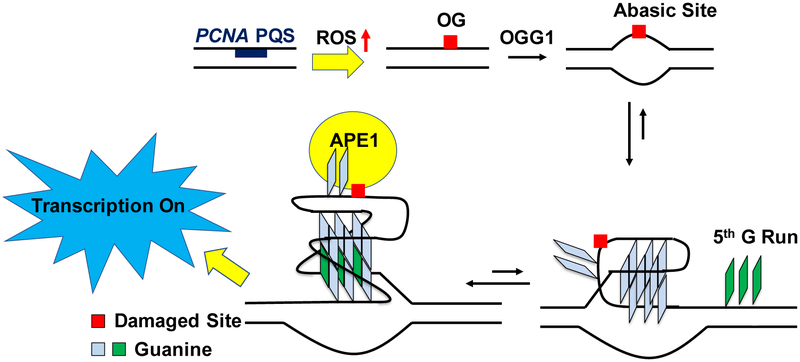
Abstract
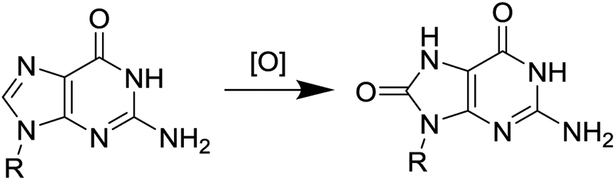
Due to its low redox potential, guanine (G) is the most frequent site of oxidation in the genome. Metabolic processes generate reactive oxygen species (ROS) that can oxidize G to yield 8-oxo-7,8-dihydroguanine (OG) as a key two-electron oxidation product. In a genome, G-rich sites including many gene promoters are sensitive to oxidative modification, and some of these regions have the propensity to form G-quadruplexes (G4s). Recently, OG formation in G-rich gene promoters was demonstrated to regulate mRNA expression via the base excision repair (BER) pathway. The proliferating cell nuclear antigen (PCNA) gene was previously found to be activated by metabolic ROS, and the gene has a five G-track potential G4 in the coding strand of its promoter. Herein, we demonstrated the ability for four G runs of the PCNA promoter sequence to adopt a parallel-stranded G4. Next, we identified G nucleotides in the PCNA G4 sequence sensitive to oxidative modification. The G oxidation product OG and its initial BER product an abasic site were synthetically incorporated into the four- and five-track PCNA sequences at the sensitive sites followed by interrogation of G4 folding by five methods. We found the modifications impacted the G4 folds with positional dependency. Additionally, the fifth G track maintained the stability of the modified G4s by extrusion of the oxidatively modified G run. Finally, we synthetically inserted a portion of the promoter into a reporter plasmid with OG at select oxidation prone positions to monitor expression in human glioblastoma cells. Our results demonstrate that OG formation in the context of the PCNA G4 can lead to increased gene expression consistent with the previous studies identifying metabolic ROS activates transcription of the gene. This study provides another example of a G4 with the potential to serve as a regulatory agent for gene expression upon G oxidation.
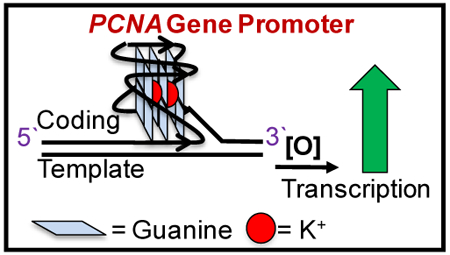
Graphical Abstract
Introduction
Mutations in DNA caused by reactive oxygen species (ROS) are linked to a myriad of diseases such as cancer, Alzheimer’s disease, and other neurological disorders.1–3 The DNA base guanine (G), having the lowest redox potential, is a frequent site of oxidation in the genome.4 Metabolic processes generate the ROS O2•- that reacts in the cell to yield the moderately stable products ONOO- or H2O2.5, 6 These intermediate products can diffuse through the cell and generate either CO3•- or HO• as the active oxidants in the highest yield. Metabolically derived oxidants target G to yield the two-electron oxidation product 8-oxo-7,8-dihydroguanine (OG) as one of the major products.5–7 This oxidatively modified G base is suspected to induce G→T transversion mutations found in genomes in many diseases resulting from ROS damage (Figure 1).8, 9 These observations led to the long-held belief that oxidation to DNA is inherently mutagenic; however, more recent studies support the proposal that OG formation in gene promoters may also function in a gene regulatory role (i.e., epigenetic-like).10–13 Several examples exist in which OG formation in a gene promoter increases transcriptional activity. For instance, genes coding for the vascular endothelial growth factor (VEGF), tumor necrosis factor alpha (TNFα), B-cell lymphoma 2 (BCL2), Kirsten rat sarcoma viral oncogene homolog (KRAS), and sirtuin 1 (SIRT1) all show a positive correlation between upregulation and possible OG formation in the gene promoter.14–18 Central to the proposal of OG as a regulatory DNA base modification is the intertwining of DNA repair and transcriptional induction.19
Figure 1.
The two-electron oxidation of G yields OG.
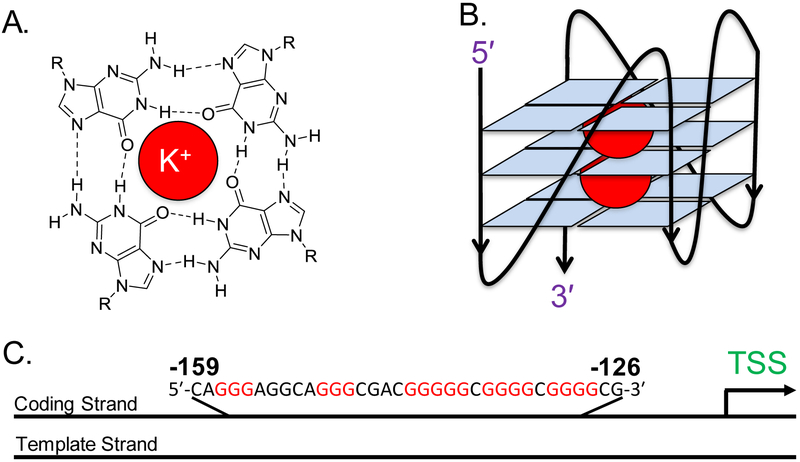
Likely sites of guanine oxidation include G-rich regions of the human genome, and some of these regions have the propensity to form G-quadruplexes (G4s). A G4 secondary structure in DNA is composed of three stacked guanine tetrads, each consisting of four guanine bases associated with one another through Hoogsteen hydrogen bonding (Figures 2A and2B). The classical sequence motif typically observed for a G-quadruplex is 5′-G≥3NXG≥3NXG≥3NXG≥3-3′ (x = 1–7 nucleotides), and use of this sequence pattern has identified via bioinformatics over 350,000 of these potential G-quadruplex forming sequences (PQS) in the human genome.20 Experimental determination of the number of sequences capable of adopting a G4 in the human genome by G4-Seq found >700,000, and G4-ChIP-Seq found ~10,000 sequences that fold within human skin cells to regulate transcription.21, 22
Figure 2.
(A) The structure of a G-tetrad, in which four guanine bases associate via Hoogsteen pairing around a K+ ion, represented by the red circle. (B) A G4 representation, in which three tetrads are stacked on top of one another and stabilized by two K+ ions to adopt a parallel-stranded G4 fold. (C) The PCNA PQS showing its position in the coding strand between −126 and −159 relative to the transcription start site (TSS) of the gene.
The VEGF gene is an example in which oxidative stress, increased gene expression, oxidation of a promoter PQS, and involvement of DNA repair in the process have been identified. There exists a positive correlation between oxidative stress and VEGF transcription levels in mammalian cells under oxidative stress conditions.14 Additional work revealed that VEGF transcriptional activation is dependent on the DNA repair enzyme 8-oxoguanine glycosylase 1 (OGG1), which is responsible for recognizing and excising OG from the genome.14, 23 The VEGF promoter contains one of the best structurally characterized and accepted G4s in the coding strand for regulation of the expression of this gene that was previously demonstrated.24, 25 Our prior work on the subject found G oxidation in the VEGF G4 sequence is incompatible with G4 folding unless that G is not required in a tetrad. However, stable folding can occur by a second mechanism in which a 5th G track allows extrusion of the damaged G run, maintaining the G4 fold by incorporation of the 5th G track as the “spare tire.”26 All of these observations begged the question, does G oxidation to OG in the G rich VEGF G4 induce transcription? Our laboratory addressed this question using chemical tools to identify oxidative modification of the VEGF G4 regulating a reporter gene could induce transcription via a possible G4 fold and the BER pathway.27, 28
The PCNA gene, which codes for the proliferating cell nuclear antigen, is another gene with a PQS in the coding strand of the promoter.29 This PQS is located between positions −126 and −159 on the coding strand, and it has five G tracks (Figure 2C). This sequence is bound by three equivalents of the SP1 transcription factor that is typical of promoter PQSs.30 More importantly, this sequence was identified to adopt a G4 fold in human cells by utilizing G4-ChIP-Seq and ChIP-Seq for the G4-specific helicases XPB and XPD.22, 31 These observations provide compelling data to support the proposal that the PCNA G-rich sequence can fold within the cellular context.
An additional interesting observation is that PCNA expression is activated by UV-induced oxidative stress, as increases in gene activity were shown at the mRNA and protein levels by Chang, et al.32 In a second study, PCNA was significantly upregulated in human colorectal cancer cells after exposure to H2O2.33 The authors also found that PCNA induction is not dependent on nucleotide excision repair (NER) or repair of double-strand breaks; however, these studies did not evaluate whether the BER pathway was important in gene activation.32 Further, they observed that inhibition of the AP-1 transcription factor that interacts with apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) or inhibition of G oxidation by the antioxidant N-acetylcysteine led to a reduction in PCNA activity.34 These results support a strong possibility that G oxidation occurred in the PCNA promoter to induce expression.32 Lastly, PCNA over-expression has been detected in malignancies ranging from colorectal cancer to breast cancer, diseases that also show dramatic increases in metabolically derived oxidative stress.35–39 Considering that the PCNA gene promoter contains a PQS found to fold in human cells, the gene is activated by oxidative stress, and BER was indirectly implicated in gene induction, the question was proposed whether oxidative modification of a G nucleotide in the PCNA promoter PQS induces gene expression via BER activation similarly to VEGF activation under oxidative stress that we previously reported on.27, 28 This study first sought to characterize the G4 formed in the PCNA promoter, and then to determine where this sequence is prone to oxidation. Once the oxidation sites were identified we synthetically installed the G oxidation product OG or the initial repair product of OG an abasic site at select hotspot sites to understand how the modifications impact G4 folding in the four- and five-track PCNA sequence contexts. Finally, OG was synthesized in the PCNA PQS context within a promoter driving expression of a luciferase gene on a plasmid that was transfected into human glioblastoma cells to understand the impact on transcription. When OG was present in the PCNA PQS context induction of transcription of the reporter gene was observed, and this finding is consistent with the previous cell-based studies that found PCNA gene was activated under oxidative stress conditions.32, 33
Materials and Methods
Preparation and Purification of Oligodeoxynucleotides
The oligodeoxynucleotides (ODNs) were synthesized at the DNA/Peptide Core Facility at the University of Utah using commercially available phosphoramidites, and the OG-containing ODNs were processed following a previously reported protocol.26 The cleaved and deprotected ODNs were purified using an anion-exchange HPLC column (DNAPac PA100 250 × 4 mm, 5 µm) running mobile phase compositions that were A = 1 M LiCl and 20 mM LiOAc (pH 7) in 9:1 ddH2O:MeCN, and B = 9:1 ddH2O:MeCN. To elute the ODNs, mobile phase flowed through the column at a rate of 3 mL/min, beginning at 5% B and increasing linearly to 100% B over 35 minutes. The ODNs were tracked by monitoring the elution profile by the absorbance at 260 nm. The HPLC-purified ODNs were lyophilized to decrease the volume and then dialyzed against ddH2O for 36 h to remove the purification salts. The dialyzed samples were lyophilized to dryness and resuspended in ddH2O followed by determination of the stock concentrations via determination of the A260 nm using a Nanodrop UV-vis spectrometer. The extinction coefficients for the ODNs were determined by the nearest-neighbor approximation, and G was substituted for OG for the calculation of the modified strands.
Characterization of the G4 Folds
To induce G4 formation, the ODNs were annealed by heating them to 90 ˚C and then gradually cooling the samples to room temperature in a water bath, before being chilled overnight at 4 ˚C. To collect the 1H-NMR spectra, the ODNs were annealed in a solvent containing 20 mM KPi, 50 mM KCl, with 10% D2O (pH 7). The 1H-NMR spectra were recorded on an 800-MHz NMR spectrometer using the Watergate solvent suppression pulse sequencing. To perform the circular dichroism (CD) analysis, thermal melt (Tm), and thioflavin-T experiments, the samples were annealed in 20 mM KPi, 120 mM KCl, 12 mM NaCl (pH 7.4), at ODN concentrations of 10 μM, 2.5 μM and 1 μM, respectively. The CD profiles were recorded at room temperature and plots of molar ellipticity were achieved using ε = 263,000 L•mol−1•cm−1 for the PCNA 4-track sequence and ε = 332,000 L•mol−1•cm−1 for the PCNA 5-track sequence. Thermal melt curves were performed by monitoring the ODN absorbance at 295 nm as the temperature was varied from 20 to 100 °C at a ramp rate of 1 °C/min with thermal equilibration of 1 min at each step in the temperature ramp, and the absorbance readings at each 1 °C temperature change were recorded. The fluorescence assays were measured using 0.5 μM Thioflavin-T, with the λex = 425 and λem scanned from 440–700 nm. The increase in fluorescence of the G4 samples was compared to the emission of control DNA sequences (i.e., established G4 fold, c-MYC as a positive control, and single-stranded DNA as a negative control).
Gel Electrophoresis
The ODNs were radiolabeled at the 5′-end with 32P using T4 polynucleotide kinase to radiolabel the ODN with [γ−32P]-ATP following a literature protocol.40 To induce G4 folds, the radiolabeled ODNs were annealed in the same manner as described above. To determine the reactive sites, 50-μL portions of the G4 samples were oxidized using 50 mM riboflavin and 350-nm light for time intervals ranging 0–12 minutes following a protocol previously reported by our laboratory.41 After the oxidation reaction, the samples were dialyzed against ddH2O for 12 h and then lyophilized to dryness. To determine the sites of oxidation, the sample was resuspended in 1 M freshly prepared aqueous piperidine and heated at 90 ˚C for 1 h following a previously reported protocol determined to efficiently induce strand scission at known G oxidation products.42 After the piperidine reaction, the samples were lyophilized to remove the piperidine. Next, the dry samples were resuspended in 10 μl of loading dye (30% glycerol, 0.25% bromophenol blue, and 0.25% xylene cyanol), followed by loading 5 μL of the sample onto a 20% denaturing polyacrylamide gel (PAGE) and electrophoresed at 75 W for >1.5 h in 1x TBE electrolyte. The oxidation sites were visualized using storage-phosphor autoradiography and a phosphorimager.
For native gel electrophoresis, the ODNs were labeled with 32P and annealed to induce G4 folds both of which were described above. The samples were dried and resuspended in 40 µL of loading dye and loaded onto a 20% native PAGE gel that included 0.1 M KOAc in the gel mix and 1x TBE electrolyte solution (pH ~8). The native gel samples were studied at pH 8; thus, the poly-dC ladder used was not capable of adopting i-motif folds rendering these strands as a suitable ssDNA control.43
Plasmid Construction
Primers containing the PCNA PQS and flanked by recognition sites for the Nt. BspQ1 nicking endonuclease were designed and inserted into the psi-CHECK-2 plasmid (Promega) following a method previously outlined in our laboratory that harnessed conditions to avoid non-specific oxidation of G nucleotides in the sequence.27 The plasmid was equipped with coding sequences for both the Renilla (Rluc) and firefly luciferase (luc) genes. The Rluc gene is driven by the SV40 promoter that was modified with the PCNA sequence, while luc is driven by the HSV-TK promoter and this luciferase served as an internal control for the measurements. Expression from these two luciferase genes was determined by the dual-glo luciferase assay. Following PCR amplification, the plasmid was transformed into NEB-5-alpha competent E. coli cells according to the High Efficiency Transformation protocol (NEB). The sample was plated and grown overnight (~16 h) at 37 ˚C before the colonies were picked and incubated in growth media overnight. Plasmid DNA was extracted using a miniprep kit (Qiagen) before being submitted to the University of Utah DNA sequencing core facility for Sanger sequencing to confirm the proper insertion of the PCNA PQS in to the plasmid (refer to Figure S1 for an example Sanger chromatrogram showing G4 incorporation).
To insert site-specific modifications into the plasmid, 5 μg of the modified plasmid with Nt.BspQ1 recognition sites was added to 50 μL 1x Nt.BspQ1 reaction buffer with 5 units of Nt.BspQ1 (NEB). The nicking endonuclease reaction proceeded for 60 min at 50 °C, followed by quenching the reaction for 20 min at 80 °C. Next, 1 nmol (~1,000-fold excess) of 5′-phosphorylated oligomer with the site-specific modification was added to the reaction mixture, and then the mixture was thermally cycled between an 80 °C water bath for 2 min and ice for 2 min, which was repeated 4 times. Next, 7 μL of 10x ligase buffer and 800 units of T4 DNA ligase were added to the thermally cycled mixture, and it was left to react for 6 h at 20 °C. The modified plasmids were purified from protein extracts using an Ultra Clean PCR Cleanup kit (MoBio) according to the manufacturer’s protocol, and finally, the concentrations were determined using a Nanodrop UV-vis spectrometer.
Cell Culture
Glioblastoma cells (U87 MG) purchased from ATCC were grown in Dulbecco’s Modified Eagle Medium (DMEM) and supplemented with 10% FBS, gentamicin (20 µg/mL), glutamax (1x), and nonessential amino acids (1x). Transfection experiments were performed in white 96-well plates using X-tremeGene HP DNA transfection agent (Roche) with 400 ng plasmid. The dual-glo luciferase assay (Promega) was performed according to manufacturer′s protocol to measure the Rluc expression relative to luc. Each experimental group included 8 replicates.
Results and Discussion
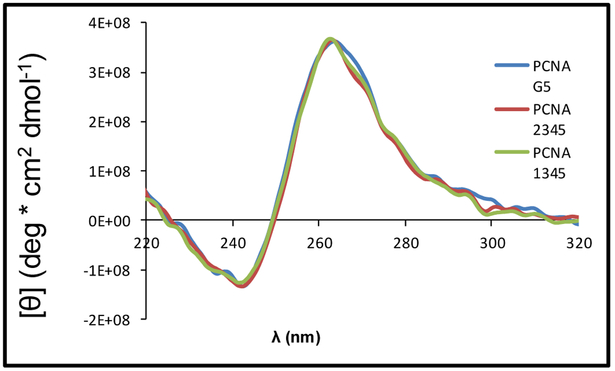
The aim of the initial characterizations was to determine which of the five G runs in the PCNA sequence adopt the principle G4 fold in solution. To address this question, we followed a similar method as was used to understand the same question asked of the c-MYC G4 sequence.44 First, three sequences were synthesized: a wild-type strand with all five G runs (G5), a strand with the first G run substituted with thymine bases (PCNA 2345), and a strand with the second G run (PCNA 1345) substituted with thymine bases. Substitutions with T were not conducted on the 3′ side of the PQS because where possible, G4 folds generally prefer shorter loop lengths in which the 3`-most G runs would provide the shortest loops.45 The sequences were then subjected to thermal melting (Tm) analysis to identify the stability of the sequences. The Tm values were determined by following denaturation of the folded G4 via the decrease in the absorbance at 295 nm as the temperature was increased from 20 to 100 °C. The wild-type G4 with five G runs was found to have a low thermal stability, followed by the strand with the second G run mutated to Ts (Table 1). In contrast, the complete sequence with the 5′ most G run mutated to Ts and the truncated four track sequence comprised of the four 3′ most G runs had high thermal stabilities. This final observation supports the predominant structure in solution to be one containing a G4 consisting of the four 3′-most G runs. The CD spectra recorded for the five-track sequences showed a λmax at 262 nm and a λmin at 243 nm, indicating the G4s to be parallel stranded on the basis of comparison to literature data (Figure 3).46 These initial data support the wild-type PCNA sequence is a parallel-stranded G4 that folds using the four 3′ most G runs. We would like to point out that the five G-track PCNA sequences were extremely challenging to work with. The synthetic yields were poor, and the sequences consistently precipitated out of solution; thus, the data for the five G-track sequences were challenging to obtain and in a few instances the data were unobtainable.
Table 1.
The PCNA G4 sequences studied and their Tm values.
| Strand | Sequence | Tm (°C) |
|---|---|---|
| PCNA G5 | 5′-CAGGGAGGCAGGGCGACGGGGGCGGGGCGGGGCG-3′ | 70.6 +/− 0.6 |
| PCNA 2345 | 5′-CATTTAGGCAGGGCGACGGGGGCGGGGCGGGGCG-3′ | 74.6 +/− 1.4 |
| PCNA 1345 | 5′-CAGGGAGGCATTTCGACGGGGGCGGGGCGGGGCG-3′ | 70.2 +/− 2.5 |
| PCNA G4 | 5′-CAGGGCGACGGGGGCGGGGCGGGGCG- 3′ | 74.1 +/− 0.6 |
Figure 3.
The CD spectra of the folded five-track PCNA sequences that include the unmodified PCNA G5, PCNA with the first G-run mutated to Ts (PCNA 2345), and the PCNA with the second G-run mutated to T nucleotides (PCNA 1345). The spectra were recorded in 20 mM KPi (pH 7.4) buffer with 120 mM KCl, 12 mM NaCl at 20 °C.
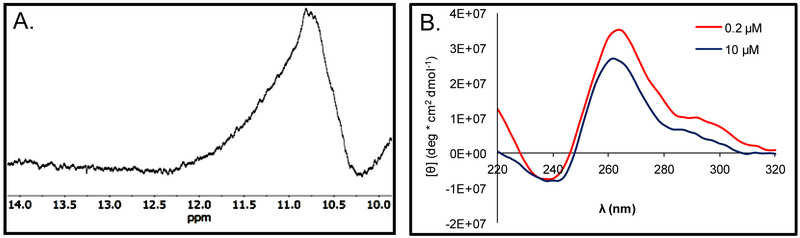
Because the five G-track PCNA sequence can adopt many possible G4 folds, we chose to truncate the sequence to the four 3′-most G runs found to be the most stable in solution to minimize the number of folds it could adopt. This truncated PCNA sequence (PCNA G4) was studied in greater detail to gain a better understanding of the G4 fold. Next, the 1H-NMR spectra for the wild-type sequence was recorded at ~300 µM to identify imino proton shifts diagnostic of G:G Hoogsteen base pairs in a G4 fold (10–12 ppm), following a prior literature report as a guide.47 Figure 4A shows the 1H-NMR spectrum collected for the PCNA 4-track sequence in which a broad peak centered at 11 ppm was found that supports G:G Hoogsteen base pairing in a G-tetrad. The broadness of the peak suggests that the G4 is polymorphic and several folds exist in solution. This observation was expected on the basis of the G4 having three G runs with more than four G nucleotides that could adopt up to 12 possible structures. The next analysis of the wild-type sequence was recording of the CD spectra at 20 °C in buffer with K+ and Na+ concentrations designed to match physiological conditions with the G4 concentrations at 0.2 or 10 µM. The CD studies were conducted with lower DNA concentrations (0.2 or 10 µM) than the NMR experiments. The spectra at both DNA concentrations showed a λmax of 262 nm, indicating the PCNA PQS adopts a parallel-stranded G4 with no dependency on the strand concentration (Figure 4B), on the basis of comparison to a literature report.46
Figure 4.
Characterization of the 4-track PCNA PQS by spectroscopic methods. (A) The 1H-NMR shows a chemical shift diagnostic of imino protons in G:G Hoogsteen base pairs in a G-tetrad between 10–12 ppm. (B) The CD spectra has a λmax of 262 nm for the folded sequence at varying concentrations.
The concentration-dependency in the CD spectra was studied as the first experiment to support the PCNA sequence adopts an intramolecular G4 fold. The CD spectra at a high and low concentration did not change suggesting the structure remains the same (Figure 4B). The intramolecularity of this PQS was further supported by a non-denaturing PAGE examination that was conducted at 1 and 10 µM (Figure S2). The data from the PAGE analysis suggest intramolecular folding of the wild-type PCNA sequences because quadruplex-DNA folds are compact and migrate at a greater rate than the single-stranded DNA ladder band of similar length used for comparison48 that was observed for the folded PCNA strands. Additional support for intramolecular folding of the wild-type PCNA G4 was derived from a concentration-dependent Tm study. We found the Tm of the four-track fold did not change as the concentration was increased from 2 to 20 µM, which suggests intramolecular folding at these concentrations (Figure S3). The next sets of studies were conducted at low concentrations (<10 µM), at which intramolecular folds were assumed.
In the following study a Thioflavin-T (ThT) assay was performed that utilizes a light-up probe repurposed by Mergny and co-workers for detecting the presence of folded G4s because ThT is G4 selective and shows minimal interaction with ss- or dsDNA.49 The established c-MYC G4 was used as a positive control and the C-rich complementary strand for the PCNA PQS was harnessed as a negative control. Under the conditions of the analysis, the C-rich complement did not adopt an i-motif fold that would interfere with the analysis, on the basis of a pH-dependent CD study (Figure S4). Application of the ThT assay to the controls provided the expected results: high fluorescence for c-MYC (120-fold increase above background) and low fluorescence for the ssDNA (<10-fold increase above background). The ThT assay applied to the four G-track PCNA sequence found an 80-fold increase in fluorescence at 490 nm that further supports G4 formation (Figure S4).
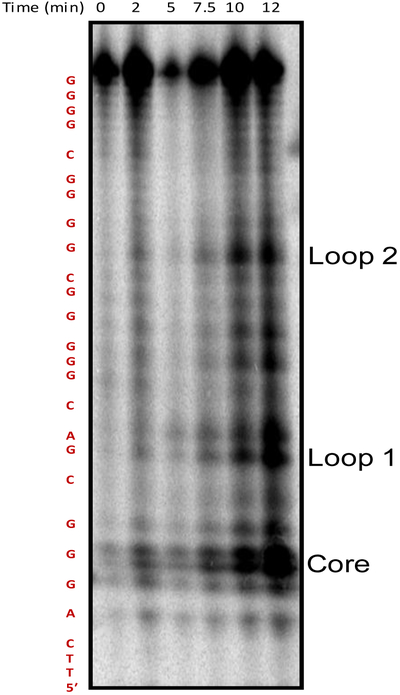
Following the initial structural analysis of the wild-type PCNA sequence, the 4-track PCNA sequence was folded at 10 µM concentration in buffer with physiologically relevant amounts of K+ and Na+ ions (140 and 12 mM, respectively), and the DNA was subjected to G-specific oxidizing conditions to identify the G nucleotides most prone to oxidation. The one-electron photooxidant riboflavin was used for the oxidation studies because the reaction pathway and products are well established.5 Post oxidation mapping of the oxidation sites was achieved via hot piperidine-induced cleavage following a method demonstrated to cleave all products of riboflavin-mediated oxidation of G detected (2Ih, Sp, Gh, and Iz/Z), with the exception of OG.42 Visualization of the cleaved sites by PAGE separation and storage-phosphor autoradiography analysis revealed the most reactive sites to be positions −149 (core), −145 (loop 1), and −136 (loop 2) relative to the transcription start site (TSS, Figures 5 and S5). The core or loop designations will be used for the remainder of the text. In the context of the PCNA PQS, position −149 is found in a run of three Gs, suggesting that this particular guanine must participate in G-tetrad formation, and therefore, exists in a G4 core position. The reason the G nucleotide at position −145 in a loop was highly reactive is because this position resides in the loop of the folded structure, in which previous studies found loop G nucleotides to be hyperreactive toward oxidation.50 Oxidation products of G are not compatible with the formation of G-tetrads;51–53 therefore, oxidation in the core position is expected to interrupt G4 formation when the four G-track sequence is considered. Meanwhile, the guanine labeled loop 1 is not a part of a G run, and therefore, does not participate in G4 tetrad formation. Lastly, oxidation to the G labeled loop 2 is part of a run of four G nucleotides and is likely forced into a loop position following oxidation allowing the other three G nucleotides to form a stable G4 fold as a result of the incompatibility of OG for tetrad formation. Now that we have identified sites prone to oxidation in the PCNA PQS when folded as a G4, studies were then conducted to understand the impact of oxidation at each of these sites on the G4 fold.
Figure 5.
Denaturing PAGE showing PCNA PQS to have oxidation hotspots at positions −149 (core), −145 (loop 1), and −136 (loop 2) relative to the TSS. The PCNA G4 studied with the reactive sites underlined is 5′-TTCA GGG CGAC GGGGG C GGGG C GGGG CT, there were two T nucleotides added to the 5′ end to facilitate observation of the cleaved G closest to the 5′ end. See also Fig. S5.
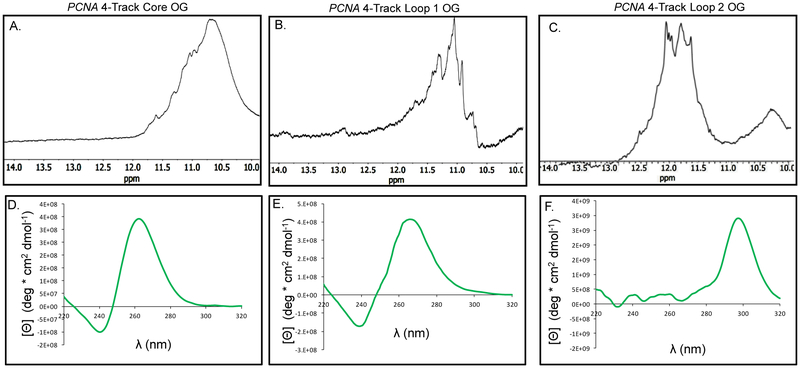
We synthesized OG into the PCNA strand at each of the positions found in the oxidation studies using a standard solid-phase synthesis protocol. Selection of OG for these studies is based on previous work that found OG was a product of G oxidation in the G4 context under riboflavin-mediated oxidations in the presence of physiological amounts of reducing agent.41 The positions studied include the site labeled as the core position, the loop 1 site, and the site labeled loop 2. First, 1H-NMR spectra were collected for the OG-containing strands at ~300 µM to identify imino shifts between 10–12 ppm, indicative of G:G Hoogsteen bonding in a G-tetrad (Figures 6A-C).
Figure 6.
Characterization of the four G-track PCNA PQS with OG synthesized at sites sensitive to oxidation in the sequence. (A–C) The 1H-NMR spectra for the PCNA PQSs with OG at the three different positions studied (D–F) The CD spectra recorded for the PCNA PQS with OG in the core, loop 1, and loop 2 positions.
Before conducting the next set of experiments, the chemically modified G4s were analyzed by native-PAGE analysis to determine whether the molecularity changed when folded at 1 or 10 µM. The analysis found the modified G4s migrated on the gel consistent with intramolecular folds (Figure S2). Next, the CD spectra for the OG-containing G4s provide additional insights to the topologies of the folds. When OG was located in the core position, the λmax was at 265 nm. This change in the λmax in the CD spectrum54 is consistent with previous studies identifying OG causing the structure to unravel to an unstable fold,51–53 discussed more below. When OG was placed in the first loop position, the CD spectrum had a λmax at 262 nm that is consistent with a parallel-stranded fold similar to the wild-type PCNA sequence. Lastly, placement of OG in loop 2 provided a CD spectrum with a λmax at 290 nm (Figure 6 D-F). This final spectrum was significantly different than the others measured and is consistent with the sequence adopting an antiparallel-stranded G4 fold.
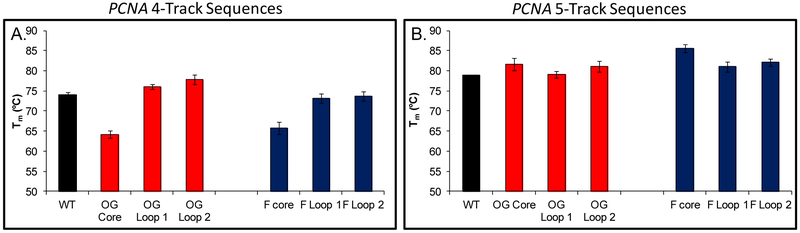
Previous work from our laboratory demonstrated that PQSs with more than four G runs can accommodate OG in a core position by replacing the damaged run with a fifth G run close to the principle G4 fold.26 Because the PCNA PQS has a fifth G run, we determined whether a similar mechanism to tolerate modifications to the nucleic acid bases in the context of a G4 fold occurred. We synthesized OG and its proposed initial repair product, an abasic site,55 that was studied as the THF (F) analog at the same locations as analyzed in the four G-track sequence. First, thermal melting assays were performed on the ODNs in both the G4 and G5 contexts. The thermal stabilities of the damaged five-track strands were similar to the wild-type sequence, which stands in contrast to the modified four-track strand at the core position that was found to be significantly less stable than the wild-type sequence. These differences suggest that the fifth domain becomes incorporated into the G-quadruplex fold when damage to a core G run has occurred (Figure 7).
Figure 7.
Thermal melting assays performed on the damage-containing G4 and G5 strands showing that the fifth domain is engaged in the G5 context. (A) Thermal melting profiles for the PCNA G4 show a decreased thermal stability when damage occurs in the core position. (B) Thermal stability for the PCNA G5 remains high when the modifications occur in the core position, in which the stability is maintained by the “spare tire,” or 5th domain, in the native PCNA sequence. The error bars represent the standard deviation determined from triplicate analysis of each sample.
To provide further evidence for these observations, we returned to the ThT assay in which the OG- and F-containing 4-track sequences exhibited a decrease in FI/FIo relative to the wild-type sequence, while the FI/FIo of the OG- and F-containing five-track sequences remained high (Figure S6). Lastly, CD analysis was performed on the sequences, confirming that the modification-containing G5 sequences adopt a parallel-stranded G-quadruplex (Figure S7).
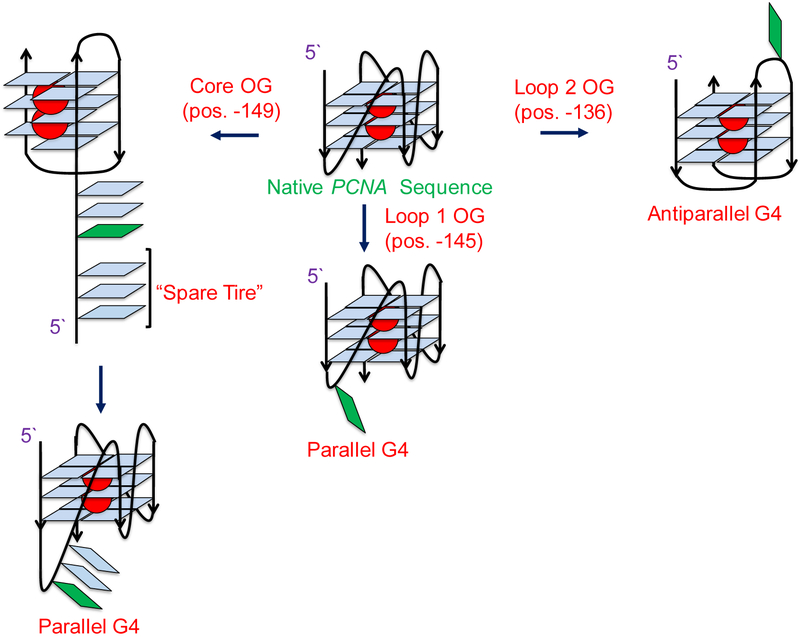
To summarize these findings, introduction of the oxidation product OG at the core G nucleotide in the PCNA gene disrupts G-tetrad formation and results in a low stability fold (Figure 7A). This observation is consistent with previous studies leading to a similar observation.51–53 In contrast, when the oxidation product OG was positioned in loop 1, no impact on the G4 fold was observed relative to the wild-type sequence (Figure 6E and7A). Finally, the guanine in loop 2 when modified is likely looped out as is made evident by the conformational change that results when the G4 shifts from parallel to anti-parallel, on the basis of the CD signature (Figure 6F). These observations are summarized in the pathway outlined in Figure 8.
Figure 8.
Summary of the effects of G oxidation on the PCNA G4. Oxidation to position −149 (core) destabilizes the quadruplex and leads to triplex formation, suggesting the G to be in a definitive core position. Oxidation at position −145 (loop 1) does not affect the topology of the fold, confirming it to be in a loop position, while oxidation to position −136 (loop 2) results in a conformational change, shifting the G4 from parallel to anti-parallel topology. This indicates that position −136 (loop 2) can exist in either a loop or a core position.
In the final cell-based study, the PCNA PQS with OG incorporated at specific locations was cloned into the promoter region of a luciferase gene in a plasmid, and the vector was then transfected into human glioblastoma cells (U87-MG). The plasmid studied also contains the firefly luciferase gene that was not modified and was used as an internal standard for quantitative analysis of the impact of OG on gene expression. The time required prior to measuring the luciferase expression is based on our previous studies that found 48 h to provide a maximum level of expression change when OG was present.28 Our experiments focused on the native 5-track PCNA sequence. When OG was present in the core position of the PCNA PQS, the observed expression level was similar to that of the unmodified PCNA G4 sequence (Figure 9). Next, synthesis of OG at either of the loop positions previously found to be sensitive to oxidation in the PCNA PQS was demonstrated to be consequential for gene expression, as roughly a four-fold increase in Rluc expression was observed relative to the wild-type PCNA sequence (Figure 9). These studies identified that when OG is present in the context of the PCNA promoter in a loop position of the PQS on the coding strand, gene expression is enhanced. A reason why OG placed at the core position failed to induce transcription is not clear. A possibility is that the G4 fold with OG at a core site generates a structure that is not compatible with the proposed mechanism, as described below.
Figure 9.
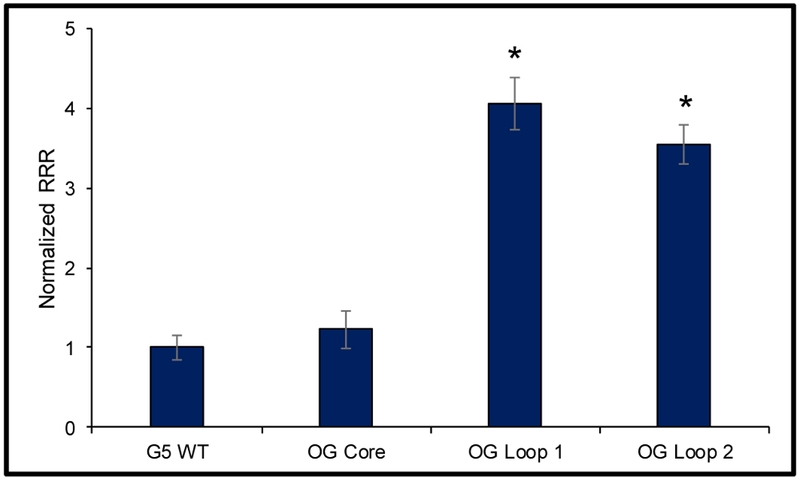
Incorporation of OG in the PCNA PQS led to a statistically significant increase in Rluc expression. *The results are significant at the 95% confidence interval on the basis of the Student’s t test.
The observation that guanine oxidation in the PCNA PQS turns transcription on is consistent with previous studies that found oxidation of a PQS in the coding strand of the promoter close to the TSS can turn on gene expression through the BER pathway.28 There exist further examples in the literature of oxidative stress leading to an increase in PCNA expression at the mRNA and protein levels.32, 56, 57 Further, this gene induction occurs independently of NER or repair of double-strand breaks.32 The present results support the general hypothesis that the interplay of possible G4 formation and oxidative stress can act as an on/off switch for transcription, as OG concentrations are increased in combination with some DNA repair, cell cycle, and stress response genes during inflammation.7, 27, 28, 58 Lastly, we hypothesize that incorporation of the fifth domain yields a structural conformation when the G4 is modified that is incompatible with APE1 cleavage of the strand, but allows APE1 to bind and stall on the DNA for recruitment of activating proteins. The structural transition is supported by the increased Tm values found when the fifth domain was present (Figure 7B), and the inability of APE1 to process an abasic site in the G4 context was previously reported,59 consistent with our proposal.
The studies in our laboratory28 and the present paper have not explored the role of the complementary strand that can adopt a i-motif fold in the gene regulatory process. This question arises because work on other sequences capable of G4 and/or i-motif formation have suggested the i-motif fold plays a significant role in the regulatory process.60 Although, we note that ROS derived from metabolic processes is strongly biased for G oxidation and as a result of the G richness of the PQS strand, oxidations will occur in this strand. By focusing the oxidation on the PQS strand, the BER proteins that are essential for gene induction will target this strand leading to the repair-mediated gene activation observed, on the basis of our previous studies.28 Thus, the G-rich strand is important for the induction processes but the full scope of the i-motif strand is not known.
The results to the VEGF inquiry led our laboratory to propose a mechanism of how oxidative damage, G4 formation, and transcriptional regulation are all interconnected (Figure 10).27 The synthetic reporters were transfected into mammalian cells to map the gene induction process when OG was located in the VEGF promoter PQS. The studies found OG was released by the glycosylase OGG1 to form a duplex-destabilizing abasic site (Figure 10). The destabilization allowed the structure to possibly adopt the now thermodynamically preferred G4 fold by placing the abasic site in a loop that did not interfere with the G4 structure. This was achievable because the VEGF PQS possesses a fifth G track (i.e., spare tire) that allows looping out the damaged track and replacing it with the non-damaged G track to avoid destabilizing the structure (Figure 10). The Tm studies comparing modifications in the PCNA four-vs. five-track sequences (Figure 7) are similar to our previous results on VEGF,26 supporting a similar folding process of the G4 strand. Last, the abasic site was then bound in the G4 context, but poorly cleaved, by apurinic/apyrimidinic endonuclease I (APE1). A key feature of APE1 is that it interacts with activating transcription factors such as HIF-1α and AP-1 for gene induction while the binding was attenuated on the abasic site in the G4 context (Figure 10).51, 59, 61 To date, we have yet to complete mapping of the protein players in the proposed pathway. The present work on the PCNA PQS, in tandem with our previous studies on the VEGF PQS,27 adds support for a hypothesis that G oxidation to OG focuses the BER process to unmask a G4 fold for gene regulation. Lastly, this study has identified another candidate gene with a promoter PQS that can function as a redox switch for gene regulation during oxidative stress.
Figure 10.
Proposed mechanism for how OG provides the drive for G4 formation in the PCNA promoter. First, OG removal by OGG1 yields a duplex-destabilizing abasic site that allows a structural switch to the G4 fold. Next, APE1 binds the abasic site when it is looped out in the G4 fold to recruit activating transcription factors for gene induction. The proposal is made on the basis of comparisons to our previous work regarding VEGF activation by a similar process.27
Conclusion
We have demonstrated that the PCNA promoter contains a G-rich region on the coding strand that is capable of forming a parallel-stranded G-quadruplex (Figure 3). The initial in vitro studies identified the G4 sequence to be prone to oxidation at specific sites (Figure 5). Next, studies to understand the impact of the G oxidation product OG on the G4 fold found positional dependency in the G4 topology relative to the site OG was synthetically incorporated (Figure 8). In a final study, the promoter of a luciferase gene was modified to include the PCNA PQS with and without OG present near the TSS in the coding strand. When the plasmid containing OG at one of three different sites studied was transfected into human glioblastoma cells, those with OG in a loop position of the PCNA PQS gave nearly four-fold greater expression than the plasmid without OG (Figure 9); however, placement of OG at the core position did not lead to significant enhancement of transcription. The present findings add additional support for understanding how guanine oxidation in the context of a promoter PQS may function as a key regulatory modification that impacts gene expression.10, 27–29 Furthermore, these results provide molecular details to a possible mechanism by which the PCNA gene is activated during oxidative stress conditions as previously found.32, 33 In the proposed mechanism, the G-rich PQS promoter element of the PCNA gene is subject to G oxidation to yield OG. This oxidation event directs DNA repair to the promoter for initiating events that ultimately induce transcription of the gene. In contrast to our prior work on this topic in the VEGF,27, 28 NTHL1,27 and RAD1762 promoter PQS contexts, the PCNA PQS contains longer G runs that result in this sequencing being more sensitive to oxidative stress while also causing the G4 formed to adopt many different folds, as evident by the 1H-NMR spectra (Figures 4 and 6A-C). Interestingly, this sequence can shift the structural fold upon oxidation (Figure 8) that was only observed for the RAD17 sequence in which 5′ and 3′ nucleotides were found to be a contributing factor in the structural shift.62 The present study adds additional support to growing interests in understanding whether oxidative modifications to the guanine heterocycle serve a biological function other than causing mutations to the genome.
Supplementary Material
Acknowledgment
We thank the National Cancer Institute for financial support of the project (R01 CA090689). The oligonucleotides and Sanger sequencing were provided by the University of Utah Health Sciences Core facilities that are supported in part by a National Cancer Institute Cancer Center Support grant (P30 CA042014). The authors thank Yun Ding and Judy Zhu (University of Utah) for their assistance with the cell studies.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: XYZ.
Sanger sequencing chromatogram, thermal melting curves, CD spectra, and NMR spectra
Conflict of Interest
The authors are not in conflict of interest in this work.
References:
- [1].Lonkar P, and Dedon PC (2011) Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer 128, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Federico A, Morgillo F, Tuccillo C, Ciardiello F, and Loguercio C (2007) Chronic inflammation and oxidative stress in human carcinogenesis. Int. J. Cancer 121, 2381–2386. [DOI] [PubMed] [Google Scholar]
- [3].Finkel T, and Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- [4].Steenken S, and Jovanovic SV (1997) How easily oxidizable Is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 119, 617–618. [Google Scholar]
- [5].Fleming AM, and Burrows CJ (2017) Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med 107, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cadet J, Wagner JR, Shafirovich V, and Geacintov NE (2014) One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol 90, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, and Dedon PC (2012) Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Nat. Acad. Sci. U. S. A. 109, E1820–E1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shibutani S, Takeshita M, and Grollman AP (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434. [DOI] [PubMed] [Google Scholar]
- [9].Pfeifer GP, and Besaratinia A (2009) Mutational spectra of human cancer. Human. Genet. 125, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fleming AM, and Burrows CJ (2017) 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA repair 56, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Seifermann M, and Epe B (2017) Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark? Free Radic. Biol. Med 107, 258–265. [DOI] [PubMed] [Google Scholar]
- [12].Antoniali G, Malfatti MC, and Tell G (2017) Unveiling the non-repair face of the Base Excision Repair pathway in RNA processing: A missing link between DNA repair and gene expression? DNA Repair (Amst.) 56, 65–74. [DOI] [PubMed] [Google Scholar]
- [13].Wang R, Hao W, Pan L, Boldogh I, and Ba X (2018) The roles of base excision repair enzyme OGG1 in gene expression. Cell Mol. Life Sci 75, 3741–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, and Gillespie MN (2015) An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol 309, L1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, and Boldogh I (2016) Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 291, 25553–25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Antoniali G, Lirussi L, D’Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, and Tell G (2014) SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell. 25, 532–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cogoi S, Ferino A, Miglietta G, Pedersen EB, and Xodo LE (2018) The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 46, 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, and Avvedimento EV (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science (New York, N.Y.) 319, 202–206. [DOI] [PubMed] [Google Scholar]
- [19].Fong YW, Cattoglio C, and Tjian R (2013) The intertwined roles of transcription and repair proteins. Mol. Cell. 52, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huppert JL, and Balasubramanian S (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, and Balasubramanian S (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881. [DOI] [PubMed] [Google Scholar]
- [22].Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, and Balasubramanian S (2016) G-quadruplex structures mark human regulatory chromatin. Nat. Genet 48, 1267–1272. [DOI] [PubMed] [Google Scholar]
- [23].Nishimura S (2002) Involvement of mammalian OGG1(MMH) in excision of the 8-hydroxyguanine residue in DNA. Free Radic. Biol. Med 32, 813–821. [DOI] [PubMed] [Google Scholar]
- [24].Agrawal P, Hatzakis E, Guo K, Carver M, and Yang D (2013) Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 41, 10584–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun D, Liu W-J, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, and Hurley LH (2008) The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex–interactive agents. Mol. Cancer Ther 7, 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fleming AM, Zhou J, Wallace SS, and Burrows CJ (2015) A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: do these “spare tires” have an evolved function? ACS Cent. Sci 1, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fleming AM, Ding Y, and Burrows CJ (2017) Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Nat. Acad. Sci. U.S.A. 114, 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fleming AM, Zhu J, Ding Y, and Burrows CJ (2017) 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on–off switch for transcription. ACS Chem. Biol 12, 2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fleming AM, Zhu J, Ding Y, Visser JA, Zhu J, and Burrows CJ (2018) Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5′-untranslated regions. Biochemistry 57, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, and Hocker M (2003) Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem 278, 8190–8198. [DOI] [PubMed] [Google Scholar]
- [31].Gray LT, Vallur AC, Eddy J, and Maizels N (2014) G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol 10, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chang YC, Chang HW, Liao CB, and Liu YC (2002) The roles of p53, DNA repair, and oxidative stress in ultraviolet C induction of proliferating cell nuclear antigen expression. Ann. N.Y. Acad. Sci 973, 384–391. [DOI] [PubMed] [Google Scholar]
- [33].Zhong J, Ji L, Chen H, Li X, Zhang J, Wang X, Wu W, Xu Y, Huang F, Cai W, and Sun ZS (2017) Acetylation of hMOF modulates H4K16ac to regulate DNA repair genes in response to oxidative stress. Int. J. Biol. Sci 13, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bhakat KK, Mantha AK, and Mitra S (2009) Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 11, 621–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yonemura Y, Fonseca L, Tsugawa K, Ninomiya I, Matsumoto H, Sugiyama K, Ohoyama S, Fushida S, Kimura H, and Miyazaki I (1994) Prediction of lymph node metastasis and prognosis from the assay of the expression of proliferating cell nuclear antigen and DNA ploidy in gastric cancer. Oncology 51, 251–257. [DOI] [PubMed] [Google Scholar]
- [36].Kunihiro M, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G, and Shimamoto F (1998) Combined expression of HLA-DR antigen and proliferating cell nuclear antigen correlate with colorectal cancer prognosis. Oncology 55, 326–333. [DOI] [PubMed] [Google Scholar]
- [37].Malkas LH, Herbert BS, Abdel-Aziz W, Dobrolecki LE, Liu Y, Agarwal B, Hoelz D, Badve S, Schnaper L, Arnold RJ, Mechref Y, Novotny MV, Loehrer P, Goulet RJ, and Hickey RJ (2006) A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proc. Nat. Acad. Sci. U.S.A. 103, 19472–19477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tang X (2013) Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 332, 3–10. [DOI] [PubMed] [Google Scholar]
- [39].Yonemura Y, Kimura H, Fushida S, Tugawa K, Nakai Y, Kaji M, Fonseca L, Yamaguchi A, and Miyazaki I (1993) Analysis of proliferative activity using anti-proliferating cell nuclear antigen antibody in gastric cancer tissue specimens obtained by endoscopic biopsy. Cancer 71, 2448–2453. [DOI] [PubMed] [Google Scholar]
- [40].Fleming AM, Muller JG, Dlouhy AC, and Burrows CJ (2012) Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J. Am. Chem. Soc 134, 15091–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fleming AM, and Burrows CJ (2013) G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol 26, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fleming AM, Alshykhly O, Zhu J, Muller JG, and Burrows CJ (2015) Rates of chemical cleavage of DNA and RNA oligomers containing guanine oxidation products. Chem. Res. Tox 28, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fleming AM, Ding Y, Rogers RA, Zhu J, Burton AD, Carlisle CB, and Burrows CJ (2017) 4n-1 Is a “sweet spot” in DNA i-motif folding of 2’-deoxycytidine homopolymers. J. Am. Chem. Soc 139, 4682–4689. [DOI] [PubMed] [Google Scholar]
- [44].Brooks TA, Kendrick S, and Hurley L (2010) Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 277, 3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guédin A, Gros J, Alberti P, and Mergny J-L (2010) How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, and Webba da Silva M (2011) Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. Engl. 50, 10645–10648. [DOI] [PubMed] [Google Scholar]
- [47].Adrian M, Heddi B, and Phan AT (2012) NMR spectroscopy of G-quadruplexes. Methods 57, 11–24. [DOI] [PubMed] [Google Scholar]
- [48].Sun D, and Hurley LH (2010) Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods in Mol. Biol. (Clifton, N.J.) 608, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Renaud de la Faverie A, Guédin A, Bedrat A, Yatsunyk LA, and Mergny J-L (2014) Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 42, e65–e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Burrows CJ, and Muller JG (1998) Oxidative nucleobase modifications leading to strand scission. Chem. Rev 98, 1109–1152. [DOI] [PubMed] [Google Scholar]
- [51].Zhou J, Fleming AM, Averill AM, Burrows CJ, and Wallace SS (2015) The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lech CJ, Cheow Lim JK, Wen Lim JM, Amrane S, Heddi B, and Phan AT (2011) Effects of site-specific guanine C8-modifications on an intramolecular DNA G-quadruplex. Biophys J. 101, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vorlickova M, Tomasko M, Sagi AJ, Bednarova K, and Sagi J (2012) 8-oxoguanine in a quadruplex of the human telomere DNA sequence. Febs J. 279, 29–39. [DOI] [PubMed] [Google Scholar]
- [54].Koirala D, Mashimo T, Sannohe Y, Yu Z, Mao H, and Sugiyama H (2012) Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. 48, 2006–2008. [DOI] [PubMed] [Google Scholar]
- [55].Allgayer J, Kitsera N, Bartelt S, Epe B, and Khobta A (2016) Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 44, 7267–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rodrigues CE, Capcha JMC, de Bragança AC, Sanches TR, Gouveia PQ, de Oliveira PAF, Malheiros DMAC, Volpini RA, Santinho MAR, Santana BAA, Calado R. d. T., Noronha I. d. L., and Andrade L (2017) Human umbilical cord-derived mesenchymal stromal cells protect against premature renal senescence resulting from oxidative stress in rats with acute kidney injury. Stem Cell Res. Ther 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Guo R, Li W, Liu B, Li S, Zhang B, and Xu Y (2014) Resveratrol protects vascular smooth muscle cells against high glucose-induced oxidative stress and cell proliferation in vitro. Med. Sci. Monit. Bas. Res 20, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bochman ML, Paeschke K, and Zakian VA (2012) DNA secondary structures: stability and function of G-quadruplex structures. Genetics 13, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Broxson C, Hayner JN, Beckett J, Bloom LB, and Tornaletti S (2014) Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 42, 7708–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Abou Assi H, Garavis M, Gonzalez C, and Damha MJ (2018) i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 46, 8038–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bhakat KK, Mantha AK, and Mitra S (2009) Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxidants & redox signaling 11, 621–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhu J, Fleming AM, and Burrows CJ (2018) The RAD17 promoter sequence contains a potential tail-dependent G-quadruplex that downregulates gene expression upon oxidative modification. ACS Chem. Biol 13, doi: 10.1021/acschembio.1028b00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.