Abstract
Aptameric sensors can bind molecular targets and produce output signals; a phenomenon that is used in bioassays. In some cases, it is important to distinguish between monomeric and oligomeric forms of a target. Here we propose a strategy on how to convert a monomer/oligomer non-selective sensor to an oligomer-selective sensor. We designed an aptazyme that produced a high fluorescent output in the presence of oligomeric alpha-synuclein (a molecular marker of Parkinson’s disease), but not with its monomeric form. The strategy is potentially useful in the design of point-of-care tests for diagnosis of neurodegenerative diseases.
Keywords: aptamers, aptazymes, deoxyribozymes, Parkinson’s disease, biosensors, split probes
Graphical Abstract
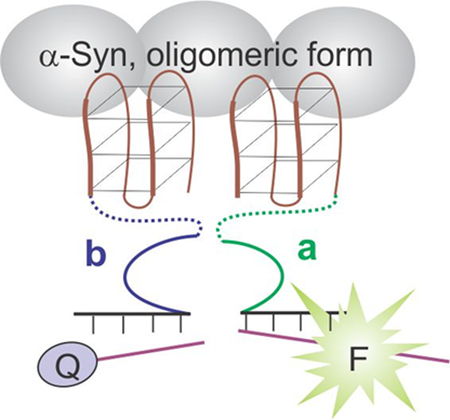
FaptaSyme: Each DNA strand a and b consists of a full α-synuclein (α-syn)-binding aptamer and a half of the catalytic core of an RNA cleaving deoxyribozyme, which is able to fluorescently report only the oligomeric form of α-syn, but not its monomeric form.
Aptamers are short single-stranded DNA, RNA or peptide molecules that, like antibodies, bind selective targets ranging from small molecules to whole cells.[1] A great variety of aptamer-based sensors have been developed so far and used in analytical assays.[2] For example, module aptameric sensor strategy uses chimeric constructs that recognize analytes and trigger conformational changes form a binding site for a non-fluorescent dye and make it fluorescent.[3] Another strategy converts aptamers into aptazymes,[3] chimeras of aptamers and catalytic RNA or DNA sequences, in which the catalytic function is suppressed.[4] A ligand bound to the aptameric portion of the aptazyme triggers conformational changes of the structure, which activates the catalytic function followed by production of an output signal. In the split approach, suppression of the catalytic function is achieved due to splitting of the aptazyme into two separate portions. The two aptazyme strands re-unite in the presence of a target and form a catalytic RNA or DNA structure, which can be registered by a number of methods.[5]
In this study, we attempted to design an aptazyme sensor differentiating between monomeric and oligomeric forms of a practically significant target protein, α-synuclein (α-syn), which is one of the amyloidogenic proteins undergoing polymerization in case of neurodegenerative diseases. The split approach resulted in the aptazyme sensor that responded to both forms of α-syn. On the other hand, the discrimination of the aggregated species among a pool of structurally similar monomeric molecules is crucial for the development of therapeutic and diagnostic strategies.[6] In order to convert the monomer/oligomer non-selective to oligomer-selective sensor, we proposed a strategy, coined Full-Aptamer-Split-DNAzyme (FaptaSyme), as detailed below. The strategy might be useful for the design of sensors that selectively recognize protein oligomers over their monomeric forms, and thus can be used in the diagnosis of neurodegenerative diseases.
The second most common neurodegenerative disease after Alzheimer’s disease is Parkinson’s disease (PD) with 500,000 individuals currently living with PD and nearly 60,000 new cases each year in the US.[7] PD’s progressive neurodegenerative pathology has been associated with self-aggregation of a native monomeric α-syn ultimately resulting in the formation of durable and cytotoxic oligomeric species. The oligomeric species impart marked neurotoxicity in dopaminergic neurons and illustrate migratory potential to neighboring healthy neurons where the process iterates. The current golden standard for PD diagnostics is a qualitative system with accuracy that is contingent upon physician experience.[8] This, along with a lack of standardized clinical testing procedures, lends to a 25% misdiagnosis rate.[9] Since oligomeric α-syn can be found in patients’ blood and cerebral spinal fluid (CSF) at early stages of PD development, it can thereby serve as a biomarker for a quantitative PD diagnostic system; which to date does not exist due, in part, to the difficulty with differentiating between monomeric and oligomeric species of α-syn.[10] Detection of oligomeric α-syn, therefore, serves as a differentiating factor between non-pathogenic (monomeric) and pathogenic (oligomeric) α-syn species for PD diagnosis. Therefore, oligomer-selective detection of α-syn has been a focus of a number of studies.[10]
Initially, we designed a split aptazyme sensor inspired by the work of Yamamoto et al.[5a,d] and Yoshida et al.[5c] We used RNA-cleaving deoxyribozymes (DZ) 10–23,[11] a catalytic DNA that cleaves RNA phosphodiester bonds (Figure 1A). DZ 10–23 can cleave a fluorophore- and a quencher-labeled reporter substrate (F_sub in Figure 1A), thereby generating a fluorescent output signal. Moreover, it was shown that DZ catalytic cores can be deactivated by splitting into two subunits, which can be then reactivated due to the analyte-dependent re-association of the subunits.[12] The strategy was shown to be efficient for the detection of DNA and RNA analytes, due to the catalytic amplification of the output signal by DZ activity,[13] as well as simplicity of the assay optimization.[14]
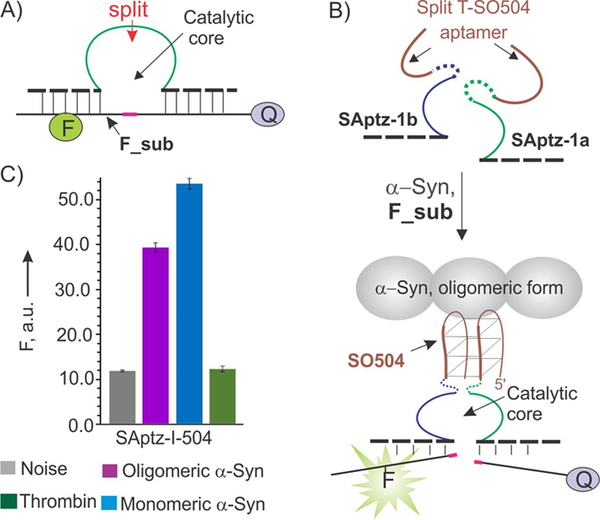
Figure 1.
Design of split aptazyme (SAptaz) sensor for the detection of α-synuclein (α-syn). A) A catalytic DNA oligonucleotide, DZ-10–23, can cleave a fluorophore- and a quencher-labeled fluorogenic substrate (F_sub). A position for splitting the DZ as discovered by Mokany et al.[10] is indicated by the red arrow. B) Split aptazyme design strategy: both DZ and the aptamer DNA strands are split into two parts. Each half of the aptamer is linked to one-half of DZ to form SAptz-1a and SAptz-1b strands, which form the catalytic DZ core when aptameric portions bind α-syn (see Figure S1 for more details). C) Selectivity of α-syn recognition by the split SAptaz-1 sensors. All samples contained 200 nM F_sub, 2 nM SAptz-1a, 50 nM SAptz-1b, and either no protein target (Noise) or 100 nM either oligomeric or monomeric α-syn, or 26 μM of thrombin (Negative control).
A series of α-syn-binding aptamers were isolated recently by Tsukakoshi et al.[15] One of the aptamers, namely T-SO504 (5’-CAG GGG TGG GCA AAG GGC GGT GGT G) has a G-rich sequence and presumably forms a G-quadruplex structure[16] when bound to its target. This was used for splitting the aptamer sequence into two fragments between the G-rich regions according to the strategy introduced by us earlier.[17] The two DNA strands (SAptz-1a and SAptz-1b, in Figure 1B) bind α-syn by re-forming the full aptamer structure, which brings the two DZ portions in close proximity, thus forming the 10–23 catalytic core. The concentrations of SAptz-1a and SAptz-1b strands were optimized to achieve the highest signal-to-background (S/B) ratio (similar to that shown in Figure S2), where signal is the fluorescence of the sensor in the presence of oligomeric α-syn, while the background is the florescence of the sensor in the absence of the target (grey bar in Figure 1C). The sensor did not respond to 26 μM of the protein thrombin (green bars in Figure 1), which is known to bind some G quadruplex-containing DNA structures[18] and is abundant in patient blood.[19] Interestingly, this system resulted in a high signal in the presence of both monomeric and oligomeric forms of α-syn (magenta and blue bars in Figure 1C), which makes this sensor suitable for the detection of either oligomeric or monomeric α-syn forms. At the same time, diagnosis of PD requires selective recognition of the oligomeric α-syn.
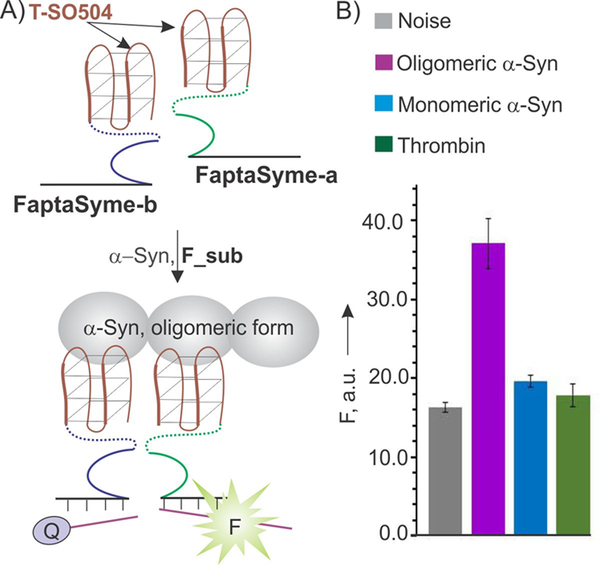
In order to enable the aptazyme sensor to differentiate between oligomeric and monomeric forms of α-syn, we proposed the FaptaSyme design shown in Figure 2A. In this design, full T-SO504 aptamers were attached to each of the two parts of the split DZ 10–23 via long flexible oligoethylene glycol linkers to enable high flexibility (strands FaptaSyme-a and FaptaSyme-b). The two aptamers, when bound to two neighboring oligomer-forming α-syn units and giving sufficient flexibility, should bring the two 10–23 halves in close proximity thus enabling catalytic core formation followed by fluorescent signaling. Indeed, the sensor was found to detect only the oligomeric form of α-syn (Figure 2B). Importantly, the sensor’s performance was not significantly affected by the presence of thrombin even at 260-fold excess (Figure S3). Expectedly, FaptaSyme’s response is time dependent demonstrating about 12-fold increase in sensitivity upon increase of the incubation time from 2 to 18 hours (Figure S4).
Figure 2.
Full-Aptamer-Split-DNAzyme (FaptaSyme) sensor design and performance. A) Design strategy: only the DZ sequence is split in half; each segment is linked to a full aptamer sequence. FaptaSyme-a and FaptaSyme-b strands bind α-syn by their aptameric portions and form a DZ catalytic core followed by fluorescent signaling. In both cases, DZ and aptameric portions were linked by oligoethylene glycol linkers (dashed lines). B) Selectivity of α-syn recognition by the FaptaSyme sensor. All samples contained 200 nM F_sub, 2 nM FaptaSyme-a, 50 nM FaptaSyme-b, and either no protein target (Noise) or 100 nM oligomeric or monomeric α-syn, or 26 μM of thrombin (Negative control). The concentrations of FaptaSyme were optimized, as detailed in Figure S2.
It was reported that aptamer-protein binding affinity can be increased by using a conjugate of two aptamers against the same target.[20] In FaptaSyme design, the resultant signal-producing complex (Figure 2A, bottom) possesses two aptameric units bound to the target. This design, therefore, could potentially increase affinity as well as lead to the improved limit of detection (LOD) in comparison with monodentate aptameric sensors. Therefore, we measured LODs for the two designs. LODs were found to be 13.4 and 5.6 nM for SAptz-1 and FaptaSyme designs, respectively (Figure S5), thus indicating minor improvement in LOD for the FaptaSyme design. The linear dynamic range for SAptz-1 was estimated to be 1–100 nM. For FaptaSyme design, however, the concentration-dependence curve deviated from linear at the concentrations above 40 nM (Figure S5B). We explain this observation by the excess amount of the target α-syn over sensor strands (highest concentration was 50 nM FaptaSyme-b), a set up that disfavors the formation of 10–23 catalytic core due to increased probability of FaptaSyme-a and FaptaSyme-b to be bound to different α-syn molecules. In practice, this fact should be taken into account if a linear concentration-dependent sensor’s response is required. A limiting factor for achieving lower LODs is the Kd of the original aptamer, T-SO504, which was presumably in the range of 60–70 nM, similar to the Kds measured for other aptamers.[15] The observed LODs below the Kd value can be attributed to the catalytic signal amplification of the DZ sensor, which is a common advantage of the catalytic sensors. Further improvements in LOD depends on selection of aptamers with lower Kd.
The originally selected aptamer, T-SO504, was reported to be oligomer-selective based on nitrocellulose filter-binding experiments.[15] However, in this study we detected monomer-binding activity of the SAptz sensor for the solution assays (Figure 1C). While the observed discrepancy in selectivity requires additional experimental studies, we hypothesize that an aptamer -(monomeric) α-syn complex with a high koff (and high kon) would not survive washing stages required for the filter-based assay.[15] At the same time, our solution-based assay took place under equilibrium conditions, which were dependent on koff /kon ratio, but not absolute koff or kon values. In other words, it is possible that T-SO504 possessed monomeric α-syn binding activity, which was not detected by the filter-binnding assay.
There are multiple examples of amyloidogenic proteins that are able to aggregate, including islet amyloid polypeptide and Aβ,[21] Tau protein,[22] as well as insulin and transthyretin.[23] State of the art techniques for detection of α-syn, as well as protein markers of other amyloidogenic diseases include antibody-based detection, dot blots and/or ELISA’s.[24] Limitations with the antibody-based methods include high production cost, variations in binding properties, and selectivity between lots. Moreover, antibodies themselves do not allow for signal accumulation without the addition of secondary compounds, and immunosorbent assays require large quantities of reagents or sample, which increases expenditures. Alternatively, oligomer-selective aptamers can be derived by in vitro selection using a stage of counter-selection with the monomeric form of a protein.[15] Here, we introduced an approach to design an oligomer-selective aptameric sensor that does not require sophisticated oligomer-selective selection schemes. We demonstrated that an aptamer that binds both monomeric and oligomeric forms can be redesigned according to the FaptaSyme strategy to be used to exclusively detect oligomeric forms of a target protein. Moreover, due to the catalytic signal amplification, FaptaSyme achieves lower than expected LODs without compromising selectivity and reliability of the assay. It was demonstrated, that α-syn–binding aptamers can bind other amyloidogenic oligomers including amyloid β, which is involved in the development of Alzheimer’s disease.[15] Therefore, we expect that the proposed approach will potentially enable analysis of other amyloidosis-related proteins.
In conclusion, we proposed a strategy that enables high selectivity towards the oligomeric form of a protein target using a monomer/oligomer-nonselective aptamer. We have designed a FaptaSyme sensor that enables selective detection of α-syn protein species with limits of detection about 6 nM. The sensor is inexpensive, stable during storage, and produces a fluorescent signal - characteristics that makes this technology suitable for translational and clinical applications in the future. This proposed approach potentiates the design of point-of-care diagnostics for neurodegenerative diseases.
Supplementary Material
Acknowledgements
Financial support from the NSF CCF (1423219) and National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Grant number 1R15AI103880–01A1) is gratefully acknowledged. D. M. K. was supported by the ITMO University Fellowship and Professorship Program.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].(a) Ellington AD and Szostak JW, Nature 1990, 346, 818–22; [DOI] [PubMed] [Google Scholar]; (b) Mercier MC, Dontenwill M and Choulier L, Cancers (Basel) 2017, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sefah K, Phillips JA, Xiong X, Meng L, Van Simaeys D, Chen H, Martin J and Tan W, Analyst 2009, 134, 1765–1775; [DOI] [PubMed] [Google Scholar]; (d) Pfeiffer F, Rosenthal M, Siegl J, Ewers J and Mayer G, Curr Opin Biotechnol 2017, 48, 111–118; [DOI] [PubMed] [Google Scholar]; (e) Reverdatto S, Burz DS and Shekhtman A, Curr Top Med Chem 2015, 15, 1082–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].(a) Thean D, Ebo JS, Luxton T, Lee XC, Yuen TY, Ferrer FJ, Johannes CW, Lane DP and Brown CJ, Sci Rep 2017, 7, 1763; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ding F, Gao Y and He X, Bioorg Med Chem Lett 2017, 27, 4256–4269; [DOI] [PubMed] [Google Scholar]; (c) Farzin L, Shamsipur M and Sheibani S, Talanta 2017, 174, 619–627; [DOI] [PubMed] [Google Scholar]; (d) Alizadeh N, Memar MY, Moaddab SR and Kafil HS, Biomed. Pharmacother 2017, 93, 737–745; [DOI] [PubMed] [Google Scholar]; (e) Tang J, Huang N, Zhang X, Zhou T, Tan Y, Pi J, Pi L, Cheng S, Zheng H and Cheng Y, Int. J. Nanomedicine 2017, 12, 3899–3911; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Cho EJ, Lee JW and Ellington AD, Annu. Rev. Anal. Chem. (Palo Alto Calif) 2009, 2, 241–264; [DOI] [PubMed] [Google Scholar]; (g) Labib M and Berezovski MV, Adv. Biochem. Eng. Biotechnol 2014, 140, 155–181. [DOI] [PubMed] [Google Scholar]
- [3].(a) Stojanovic MN, Kolpashchikov DM. J. Am. Chem. Soc 2004, 126, 9266–9270; [DOI] [PubMed] [Google Scholar]; (b) Pei R, Rothman J, Xie Y, Stojanovic MN Nucleic Acids Res. 2009, 37, e59; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Strack RL, Song W, Jaffrey Nat SR. Protoc. 2014, 9, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].(a) Soukup GA and Breaker RR, Proc. Natl. Acad. Sci. U. S. A 1999, 96, 3584–3589; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vinkenborg JL, Karnowski N and Famulok M, Nat. Chem. Biol 2011, 7, 519–527; [DOI] [PubMed] [Google Scholar]; (c) Wu D, Gao T, Lei L, Yang D, Mao X and Li G, Anal Chim Acta 2016, 942, 68–73. [DOI] [PubMed] [Google Scholar]
- [5].(a) Yamamoto R, Baba T and Kumar PK, Genes Cells 2000, 5, 389–396; [DOI] [PubMed] [Google Scholar]; (b) Yamamoto-Fujita R and Kumar PK, Anal. Chem 2005, 77, 5460–5466; [DOI] [PubMed] [Google Scholar]; (c) Yoshida W, Sode K and Ikebukuro K, Biotechnol. Lett 2008, 30, 421–425; [DOI] [PubMed] [Google Scholar]; (d) Xu W and Lu Y, Anal Chem 2010, 82, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Sosic A, Meneghello A, Cretaio E and Gatto B, Sensors (Basel) 2011, 11, 9426–9441; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lin Z, Chen L, Zhu X, Qiu B and Chen G, Chem Commun (Camb) 2010, 46, 5563–5565; [DOI] [PubMed] [Google Scholar]; (g) Yu H, Canoura J, Guntupalli B, Lou X and Xiao Y, Chem. Sci 2017, 8, 131–141; [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Yuan B, Zhou Y, Guo Q, Wang K, Yang X, Meng X, Wan J, Tan Y, Huang Z, Xie Q and Zhao X, Chem. Commun 2016, 52, 1590–1593. [DOI] [PubMed] [Google Scholar]
- [6].(a) Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC and Tycko R, Cell 2013, 154, 1257–1268; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sarell CJ, Karamanos TK, White SJ, Bunka DH, Kalverda AP, Thompson GS, Barker AM, Stockley PG and Radford SE, J Biol Chem 2014, 289, 26859–26871; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].(a) Guhathakurta S, Bok E, Evangelista BA and Kim YS, Prog. Neurobiol 2017, 154, 21–36; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nussbaum RL and Ellis CE, N. Engl. J. Med 2003, 348, 1356–1364. [DOI] [PubMed] [Google Scholar]
- [8].Charlesworth G, Gandhi S, Bras JM, Barker RA, Burn DJ, Chinnery PF, Gentleman SM, Guerreiro R, Hardy J, Holton JL, Lees A, Morrison K, Sheerin UM, Williams N, Morris H, Revesz T and Wood NW, Neurobiol. Aging 2012, 33, 838 e837–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schrag A, Ben-Shlomo Y and Quinn N, J. Neurol. Neurosurg Psychiatry 2002, 73, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Massano J and Bhatia KP, Cold Spring Harb Perspect Med. 2012, 2, a008870; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun K, Xia N, Zhao L, Liu K, Hou W, Liu L. Sensors and Actuators 2017, 245, 87–94. [Google Scholar]
- [11].Santoro SW and Joyce GF, Biochemistry 1998, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- [12].(a) Kolpashchikov DM Chembiochem 2009, 10, 1443–1445; [DOI] [PubMed] [Google Scholar]; (b) Gerasimova YV, Cornett E, Kolpashchikov DM, Chembiochem 2010, 11, 811–817; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mokany E, Bone SM, Young PE, Doan TB and Todd AV, J Am. Chem. Soc 2010, 132, 1051–1059; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mokany E, Tan YL, Bone SM, Fuery CJ, Todd AV, Clin Chem. 2013, 59, 419–426; [DOI] [PubMed] [Google Scholar]; (e) Mokany E, Todd AV, Methods Mol Biol. 2013, 1039, 31–49; [DOI] [PubMed] [Google Scholar]; (f) Gerasimova YV, Kolpashchikov DM, Chem. Biol 2010, 17, 104–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].(a) Zagorovsky K, Chan WCW, Angew. Chem. Int. Ed 2013, 52, 3168–3171; [DOI] [PubMed] [Google Scholar]; j) Gerasimova YV and Kolpashchikov DM, Angew. Chem. Int. Ed. Engl 2013, 52, 10586–10588; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gerasimova YV, Cornett EM, Edwards E, Su X, Rohde KH, Kolpashchikov DM, Chembiochem 2013, 14, 2087–2090; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gerasimova YV, Yakovchuk P, Dedkova LM, Hecht SM, Kolpashchikov DM, RNA 2015, 21, 1834–1843; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cox AJ, Bengtson HN, Gerasimova YV, Rohde KH, Kolpashchikov DM, Chembiochem 2016, 21, 2038–2041; [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Cox AJ, Bengtson HN, Rohde KH, Kolpashchikov DM, Chem. Commun 2016, 52, 14318–14321; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Kamar O, Sun SC, Lin CH, Chung WY, Lee MS, Liao YC, Kolpashchikov, M. C. DM Chuang Chem. Commun 2017, 53, 10592–10595; [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Bakshi SF, Guz N, Zakharchenko A, Deng H, Tumanov AV, Woodworth CD, Minko S and Kolpashchikov DM, E. Katz J. Am. Chem. Soc 2017, 139, 12117–12120; [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Bengtson HN, Homolka S, Niemann S, Reis AJ, da Silva PE, Gerasimova YV, Kolpashchikov DM, Rohde Biosens KH. Bioelectron. 2017, 94, 176–183; [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Bakshi SF, Guz N, Zakharchenko A, Deng H, Tumanov AV, Woodworth CD, Minko S, Kolpashchikov DM, Katz E Nanoscale 2018, 10, 1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smith AL, Kolpashchikov DM ChemistrySelect, 2017, 2, 5427–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tsukakoshi K, Abe K, Sode K and Ikebukuro K, Anal. Chem 2012, 84, 5542–5547. [DOI] [PubMed] [Google Scholar]
- [16].(a) Kwok CK and Merrick CJ, Trends Biotechnol 2017, 35, 997–1013; [DOI] [PubMed] [Google Scholar]; (b) Ma DL, Wu C, Dong ZZ, Tam WS, Wong SW, Yang C, Li G and Leung CH, Chem. Asian. J 2017, 12, 1851–1860. [DOI] [PubMed] [Google Scholar]
- [17].Kolpashchikov DM, J. Am. Chem. Soc 2008, 130, 2934–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zavyalova E, Tagiltsev G, Reshetnikov R, Arutyunyan A and Kopylov A, Nucleic Acid Ther 2016, 26, 299–308. [DOI] [PubMed] [Google Scholar]
- [19].Wolberg AS and Campbell RA, Transfus Apher Sci. 2008, 38, 15–23; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].(a) Kim Y, Cao Z and Tan W, Proc. Natl. Acad. Sci. U. S. A 2008, 105, 5664–5669; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vorobyeva M, Vorobjev P and Venyaminova A, Molecules 2016, 21; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bhatia S, Camacho LC and Haag R, J. Am. Chem. Soc 2016, 138, 8654–8666. [DOI] [PubMed] [Google Scholar]
- [21].(a) O’Nuallain B, Williams AD, Westermark P and Wetzel R, J. Biol. Chem 2004, 279, 17490–17499; [DOI] [PubMed] [Google Scholar]; (b) Seeliger J, Evers F, Jeworrek C, Kapoor S, Weise K, Andreetto E, Tolan M, Kapurniotu A and Winter R, Angew. Chem. Int. Ed. Engl 2012, 51, 679–683. [DOI] [PubMed] [Google Scholar]
- [22].Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ and Lee VM, Science 2003, 300, 636–640. [DOI] [PubMed] [Google Scholar]
- [23].Udomprasert A, Bongiovanni MN, Sha R, Sherman WB, Wang T, Arora PS, Canary JW, Gras SL and Seeman NC, Nat Nanotechnol 2014, 9, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Covell D, Robinson J, Akhtar R, Grossman M, Weintraub D, Bucklin H, Pitkin R, Riddle D, Yousef A, Trojanowski J and Lee V, Neuropathol. Appl. Neurobiol 2017, 43, 604–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.