Abstract
PI3Kα is a key lipid kinase in the PI3K/Akt pathway. Its frequent oncogenic mutations make it a primary drug target. Calmodulin (CaM) activates PI3Kα independently of extracellular signals, indicating a significant role in oncogenic PI3Kα activation. Here, we reveal the atomic-scale structures of CaM in complexes with the nSH2 and cSH2 domains of the regulatory p85α subunit of PI3Kα, and illustrate how CaM activates PI3Kα by targeting the “soft 1–5–10” CaM-binding motifs in both nSH2 and cSH2 domains. Experiment observed CaM binding cSH2 first, followed by nSH2 binding hours later. CaM typically prefers binding helical peptides. Here we observe that, unlike in cSH2, the CaM-binding motif in nSH2 populates a mixed β-sheet/α-helix/random coil structure. The population shift from a β-sheet toward CaM’s favored α-helical conformation explains why the nSH2 domain needs a longer time for CaM binding in the experiments. The “soft” CaM-binding motifs in both nSH2 and cSH2 domains establish strong CaM–PI3Kα interactions, collectively facilitating PI3Kα activation. This work uncovers the structural basis for CaM-driven PI3Kα activation.
Graphical Abstract

INTRODUCTION
Phosphatidylinositol-4,5-bisphosphate 3-kinase α (PI3Kα) is a lipid kinase that delivers signals in the PI3K/Akt/mTOR pathway by phosphorylating phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-bisphosphate (PIP3) on the membrane, mediating crucial cellular activities including cell growth, proliferation, differentiation, migration, mobility, and apoptosis.1,2 PI3K is frequently mutated in cancers, making it a primary target for cancer therapy.3–5 Statistical data indicate that PIK3CA, that encodes the PI3Kα, and its antagonist PTEN are the second and third most highly mutated genes in cancer.6 PI3Kα oncogenic mutations lead to the abnormal cellular proliferation that results in tumor development.7–9 PI3Kα is an obligate dimer with a p110α catalytic subunit and a p85α regulatory subunit.10,11 Interaction of the p85α subunit stabilizes and inhibits p110α activity.12,13 In response to extracellular signals, activated receptor tyrosine kinases (RTKs) activate PI3Kα. The phosphorylated tyrosine (pTyr) motifs in RTKs constitute high affinity binding sites for the two SH2 domains of p85α, releasing them from the p110α catalytic subunit to activate PI3Kα.14–16 The nSH2 domain in p85α plays a more significant role in inhibiting PI3Kα basal kinase activity, while the cSH2 domain also contributes, but to a lesser extent.14,17,18
Calmodulin (CaM), a calcium (Ca2+) sensor essential for cell life, activates PI3Kα.19–25 CaM-driven PI3Kα activation is independent of the extracellular signals, indicating its significance in unprogrammed tumor growth and division in cancer. The Sacks lab showed that Ca2+-CaM augmented the PI3Kα activity both in vitro and in intact cells, by binding to the two SH2 domains in the p85α subunit.26 The interaction of CaM with the cSH2 domain was readily detected in the experiment, whereas binding to the nSH2 domain required a considerably longer sample exposure time.26 This implies that, despite the similar overall structures, the nSH2 and cSH2 domains in the p85α subunit follow distinct ways to interact with CaM. On the other hand, CaM can be phosphorylated,27 and phosphorylated CaM (pCaM) promotes PI3Kα activation.28,29 Tyrosine phosphorylation of CaM generates a high affinity binding site for interacting with the nSH2 and cSH2 domains. Atomic-scale structures showed that pCaMs interacted with the nSH2 and cSH2 domains in a way similar to the pTyr motifs in RTKs.28 CaM exhibits specificity to KRas, over HRas and NRas.30 Its hydrophobic pockets and negatively charged linker bind strongly to the polybasic hypervariable region (HVR) of KRas with the hydrophobic farnesyl tail.31 The role of CaM in PI3Kα activation, together with CaM’s specificity to KRas, can provide an explanation for how KRas gains potent oncogenic activities in KRas-driven cancers. PI3Kα activation was proposed to be achieved by a ternary structure with KRas (mostly KRas4B) and CaM, in which CaM binds to the cSH2 domain and KRas4B to facilitate the PI3Kα–KRas interactions, and meanwhile binds to and releases the nSH2 domain to activate PI3Kα,19,20,32–34 in line with earlier experiments by Wolfman’s group.35
Despite studies in the past two decades, key questions in CaM-driven PI3Kα activation remain unresolved. First, neither the nSH2 nor the cSH2 domain contains the typical CaM-favored binding surfaces in their native structures. Thus, how the strong CaM–SH2 interactions are established in PI3Kα activation is unclear. Second, given that the release of the nSH2 domain is a dominant event in PI3Kα activation, why CaM needs a longer time to target nSH2 domain versus cSH2 is not understood. Finally, without activated RTKs on the membrane, how CaM promotes PI3Kα membrane localization and how these actions relate to KRas4B on the membrane in PI3Kα activation are elusive.
In this work, we address these questions by revealing the mechanism of the CaM-driven PI3Kα activation from the structural point of view. By modeling and simulations, we solve the atomic-scale structures of CaM–SH2 complexes with the conformational properties that we obtain in line with available experimental data. The nSH2 domain contains one CaM-binding motif, while cSH2 has two. Our results show that although these motifs do not adopt the CaM-favored conformation in their native structures, they are soft and may undergo a structural change into an α-helix conformation to establish strong CaM interactions in PI3Kα activation. Notably, in the complex, CaM is only partially occupied by the SH2 domains, facilitating PI3Kα membrane localization by simultaneously accommodating the KRas4B on the membrane.
METHODS
Generating Initial Configurations of CaM–nSH2 and CaM–cSH2 Complexes.
The initial coordinates of the nSH2 domain (PDB code: 4OVV), cSH2 domain (PDB code: 1H9O), and CaM with extended (PDB code: 1CLL) and collapsed (PDB code: 1CDL) conformations were obtained from the Protein Data Bank (PDB). Rigid dockings were performed for individual nSH2 and cSH2 domains binding to extended and collapsed CaMs. We employed two docking methods here. For the first, we used PatchDock and FireDock online servers36,37 to generate multiple decoys of the complex, in which CaMs with both extended and collapsed conformations individually interact with nSH2 and cSH2 domains. For the second, we established the CaM–cSH2 complex using the HADDOCK server,38 based on NMR data.39 Candidates with an RMSD < 4 Å were classified into clusters. For each combination of the extended and collapsed CaMs in complex with nSH2 and cSH2 domains, four candidates with the highest docking scores were selected, generating a total of 16 configurations, configurations 1–16 (Figure S1 and Table 1). An additional CaM–cSH2 complex (configuration 17) model based on the Haddock/NMR data was also simulated.39
Table 1.
Initial Configurations of the Simulated Systems of CaM–SH2 Complex Generated by PatchDock and FireDock Online Servers
| calmodulin (CaM) | SH2 | ||||
|---|---|---|---|---|---|
| configuration | PDB | conformation | PDB | nSH2/cSH2 | CaM–SH2 complex binding interface on CaM |
| conFigure 1 | 1CDL | collapsed | 4OVV | nSH2 | C-lobe |
| configuration 2 | 1CDL | collapsed | 4OVV | nSH2 | N- and C-lobes |
| conFigure 3 | 1CDL | collapsed | 4OVV | nSH2 | N- and C-lobes |
| conFigure 4 | 1CDL | collapsed | 4OVV | nSH2 | N- and C-lobes |
| conFigure 5 | 1CLL | extended | 4OVV | nSH2 | N-lobe, linker |
| conFigure 6 | 1CLL | extended | 4OVV | nSH2 | C-lobe, linker |
| conFigure 7 | 1CLL | extended | 4OVV | nSH2 | C-lobe |
| conFigure 8 | 1CLL | extended | 4OVV | nSH2 | N-lobe, linker |
| conFigure 9 | 1CDL | collapsed | 1H9O | cSH2 | N-lobe |
| conFigure 10 | 1CDL | collapsed | 1H9O | cSH2 | N- and C-lobes |
| conFigure 11 | 1CDL | collapsed | 1H9O | cSH2 | N- and C-lobes |
| conFigure 12 | 1CDL | collapsed | 1H9O | cSH2 | N- and C-lobes |
| conFigure 13 | 1CLL | extended | 1H9O | cSH2 | N-lobe, linker |
| conFigure 14 | 1CLL | extended | 1H9O | cSH2 | N-lobe, linker |
| conFigure 15 | 1CLL | extended | 1H9O | cSH2 | N-lobe, linker |
| conFigure 16 | 1CLL | extended | 1H9O | cSH2 | N- and C-lobes, linker |
| conFigure 17 | Haddock/NMR model | cSH2 | N- and C-lobes, linker | ||
To facilitate CaM’s interaction with the SH2 domains, the CaM-binding motifs in the nSH2 (residues 389–404) and cSH2 (residues 684–699) domains were converted into full α-helical conformations, generating pseudo-nSH2α and -cSH2α domains with the converted α-helix. To relax the α-helix in the domain, a series of minimization and dynamics cycles were performed for the nSH2α and cSH2α domains using molecular dynamics (MD) simulations. To model the nSH2α and cSH2α domains binding to CaM, the crystal structure of the myosin light chain kinase (MYLK) peptide binding to CaM (PDB code: 1CDL) was used as the template. On the basis of the positions of hydrophobic residues, the superimpositions were performed to individually model the nSH2α and cSH2α domains into the CaM’s N- and C-lobes, generating four initial configurations, configurations α1–α4 (Figure S2 and Table 2). “Rigid-body” minimization and dynamics were performed to optimize the CaM–SH2α interfaces, which avoided large disruptions caused by initial interfacial collapse.
Table 2.
Initial Configurations of the Simulated Systems of CaM–SH2α Complexa
| calmodulin (CaM) | SH2a | ||||
|---|---|---|---|---|---|
| configuration | PDB | conformation | CaM-binding motif | nSH2/cSH2 | CaM–SH2 complex CaM-binding interface |
| configuration α1 | 1CLL | extended | 389–404 | nSH2 | N-lobe |
| configuration α2 | 1CLL | extended | 389–404 | nSH2 | C-lobe |
| configuration α3 | 1CLL | extended | 684–699 | cSH2 | N-lobe |
| configuration α4 | 1CLL | extended | 684–699 | cSH2 | C-lobe |
In the modeled complex, the CaM-binding motif in SH2 was converted to an α-helix.
Molecular Dynamics Simulation Protocols.
A total of 10.5 μs MD simulations were performed for 21 initial configurations. The simulation systems were summarized in Table S2. The all-atom explicit-solvent MD simulations were conducted using the NAMD package40 with the CHARMM all-atom additive force field (version C36).41 The NPT simulations were performed at the temperature of 310 K and pressure of 1 atm. The TIP3 water model was employed to solvate the protein complexes in the cubic periodical unit cell box. Na+ and Cl− ions were used to neutralize the systems and mimic a 150 nM ion concentration. Conjugate gradient energy minimizations were performed to relax the systems. MD simulations were produced with a time step of 2 fs generated by the velocity verlet integration. Covalent bonds with hydrogen atoms were constrained by the RATTLE algorithm. The switch function with the twin-range cutoffs at 12 and 14 Å and the particle mesh Ewald (PME) algorithm were used to describe the short-range van der Waals (vdW) and long-rang electrostatic interactions. The CHARMM, VMD, and TCL scripts were used to analyze the trajectories. We implemented the simulations and analyses following the same protocol as in our previous works.28,31,42
RESULTS
Rigid-Body Binding between CaM and SH2 Domains.
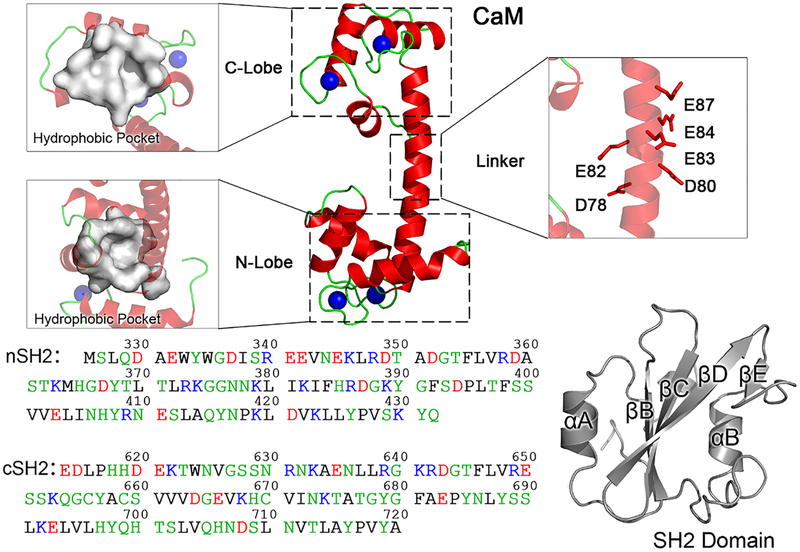
To explore possible binding models between CaM and SH2 (nSH2 and cSH2) domains, docking methods were employed to generate initial conformations. In the docked complexes, CaM and SH2 domains interacted via rigid-body binding without structural changes. CaM is a symmetric molecule, with two lobes connected by a linker (Figure 1). Each lobe (N-terminal lobe, N-lobe; C-terminal lobe, C-lobe) harbors one hydrophobic pocket to accommodate the targets. The linker is flexible, making CaM capable of accommodating numerous targets with distinct conformations. Thus, CaM exhibits (i) an extended conformation with the two lobes apart, and (ii) a collapsed conformation with the two lobes closed when binding to different targets. nSH2 and cSH2 domains share similar overall structures with β-sheet bundles (βB, βC, βD, and βE) surrounded by two α-helices (αA and αB), but have different amino acid sequences (Figure 1).
Figure 1.
Structures and sequences of CaM, nSH2, and cSH2 domains in the p85α subunit. CaM is a symmetric molecule, with two lobes (N- and C-lobe) connected by a flexible linker. The N- and C-lobes each have a hydrophobic pocket, and the linker is populated with residues with negative charges. The nSH2 and cSH2 domains share similar overall structures with a β-sheet bundle surrounded by two α-helixes, but have different amino acid sequences. In the protein sequences, hydrophobic, polar/glycine, positively charged, and negatively charged residues are colored black, green, blue, and red, respectively.
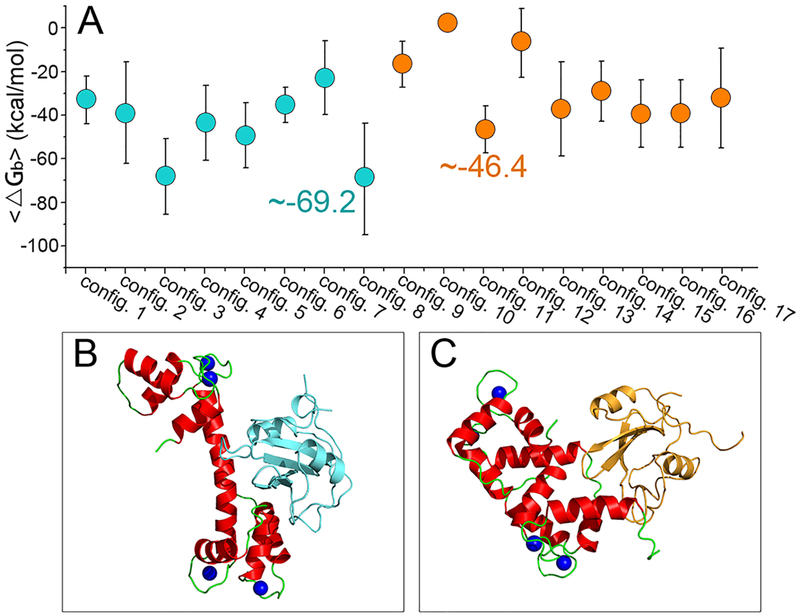
We conducted simulations for 17 configurations of the CaM–SH2 complexes. These complexes presented distinct dynamic properties in the trajectories (Figure S3). To evaluate the binding affinity between CaM and the SH2 domain in the complexes, we used molecular mechanics combined with the generalized Born and surface area continuum solvation (MMGBSA) method to calculate the binding free energies between CaM and the SH2 domains.31 The change in binding free energy was calculated using the same protocol as in our previous studies.31,42,43 In general, the nSH2 domain exhibits stronger binding with CaMs than the cSH2 domain (Figure 2A). Among the models, configuration 8 of CaM–nSH2 complex shows the lowest value of the binding free energy, approximately −69.2 kcal/mol, representing the most stable complex (Figure 2B). For the CaM–cSH2 complex, configuration 11 yields the lowest value of the binding free energy (Figure 2C), approximately −46.4 kcal/mol, which is ~50% higher than the CaM–nSH2 system. The final CaM–SH2 structures show that more salt bridges are formed at CaM–nSH2 interfaces than at CaM–cSH2, in line with the surface analysis on SH2 domains. The charged surface in the nSH2 domain (31.2% positively charged, and 7.1% negatively charged) is 11.2% higher than in the cSH2 domain (17.6% positively charged, and 9.5% negatively charged), contributing to a stronger interfacial electrostatic force. These results suggest that CaM exhibits a higher binding affinity to the nSH2 than to the cSH2 domain through the “rigid-body binding”. However, this is inconsistent with the previous experimental conclusions.26,39 In the experiments, the CaM interactions with the cSH2 domain were established quickly, while those with the nSH2 domain required a longer sample exposure. CaM needs a longer sample exposure time to interact with nSH2 domains. Thus, these results indicate that CaMs do not follow the “rigid-body binding” to target the nSH2/cSH2 domains in PI3Kα activation.
Figure 2.
(A) Binding free energies and most stable structures for (B) CaM–nSH2 and (C) CaM–cSH2 complexes. Color codes: CaM (red), nSH2 domain (cyan), and cSH2 domain (orange).
CaM-Binding Motifs in nSH2 and cSH2 Domains.
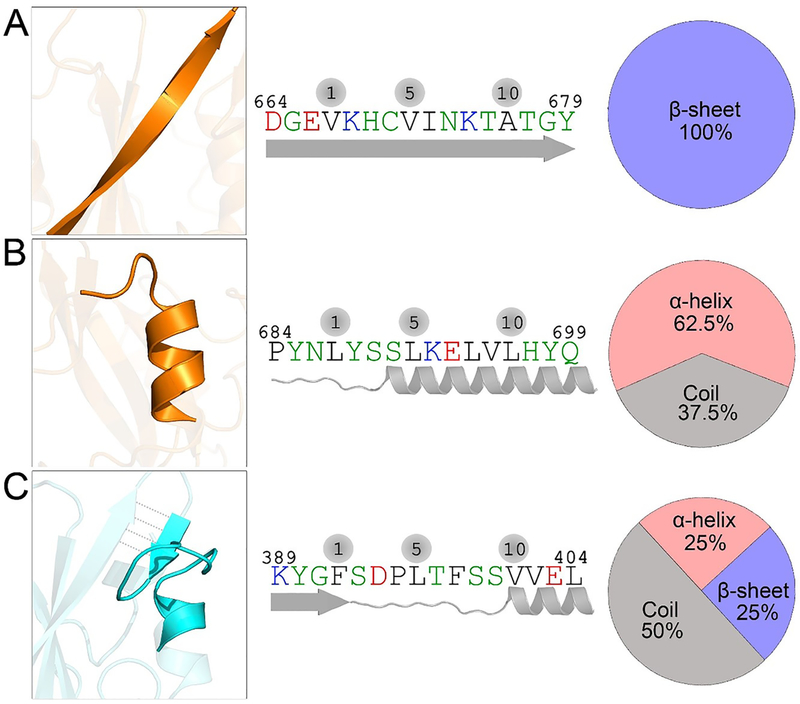
CaM is a calcium sensor essential for cell life.44–46 It has a vast number of targets in vivo.47 These targets share similarities in sequences and structures. Motifs with similar structures and sequences that directly bind to CaM are classified as CaM-binding motifs. In terms of structure, CaM-binding motifs usually adopt the CaM-favored α-helical conformation. In terms of sequence, CaM-binding motifs fall into one of three classes: “1–5–8–14” (ΦXXXΦXXΦXXXXXΦ), “1–8–14” (ΦxrefΦXXXXXΦ), and “1–5–10” (ΦXXXΦXXXXΦ), where Φ denotes the conserved hydrophobic residues (Phe, Ile, Leu, Val, Trp) and X stands for any amino acid. To identify the CaM-binding motifs in nSH2 and cSH2 domains, we analyzed the SH2 sequences. Sequence scanning profiles show that both nSH2 and cSH2 domains contain CaM-binding motifs (Figure S4). For nSH2, it is located at the region of residues 389–404, while there are two CaM-binding motifs in cSH2 spanning residues 664–679 and 684–699 (Figure 3). Only one type of CaM-binding motif, “1–5–10”, was detected in both nSH2 and cSH2 domains; the other two types of CaM-binding motifs (“1–5–8–14” and “1–8–14”) were not observed. We performed secondary structure analysis on the CaM-binding motifs in the nSH2 and cSH2 domains. In the native (crystal) structures, these motifs in the nSH2 and cSH2 domains do not fully adopt the CaM-favored α-helix conformation. The CaM-binding motif in cSH2 at residues 664–679 adopts a 100% β-sheet structure (βD) with strong interactions with adjacent β-sheet bundles (βB, βC, βE) (Figure 3A). Another CaM-binding motif in cSH2 domain at residues 684–699 contains a dominant α-helix conformation (62.5%), and coil structure (37.5%) (Figure 3B). The CaM-binding motif in nSH2 at residues 389–404 contains a mixed coil (50%), α-helix (25%), and β-sheet (25%) conformation (Figure 3C). Except for the CaM-binding motif at residues 664–679 with 100% β-sheet structure, two other CaM-binding motifs in nSH2 (residues 389–404) and cSH2 (residues 684–699) domains are completely solvent-exposed with considerable flexibility. Thus, with certain populations, they may undergo conformational changes into the α-helical conformation, establishing strong CaM interactions, similar to other CaM-binding motifs. However, the CaM-binding motif in the nSH2 domain contains a short β-sheet (βE). To fully convert into an α-helical structure, this short β-sheet has to break hydrogen bonds with adjacent β-sheet bundles and undergo a large conformational change from β-sheet to α-helix. The population shift from a stable, highly populated β-sheet toward the minor α-helix species to retain the equilibrium, where the CaM-favored population is depleted by binding, may be slow, involving a large energy barrier. This slows the CaM interaction with the nSH2 domain, explaining the longer sample exposure time required for CaM–nSH2 interactions in the experiment.26
Figure 3.
CaM-binding motifs in nSH2 and cSH2 domains. (A) One CaM-binding motif in the cSH2 domain at residues 664–679 exhibits a 100% β-sheet structure, and (B) another at residues 684–699 adopts a dominant α-helix conformation. (C) The single CaM-binding motif in the nSH2 domain at residues 389–404 has mixed α-helix, random coil, and β-sheet structures.
CaM Interacts with nSH2/cSH2 Domains via Their CaM-Binding Motifs.
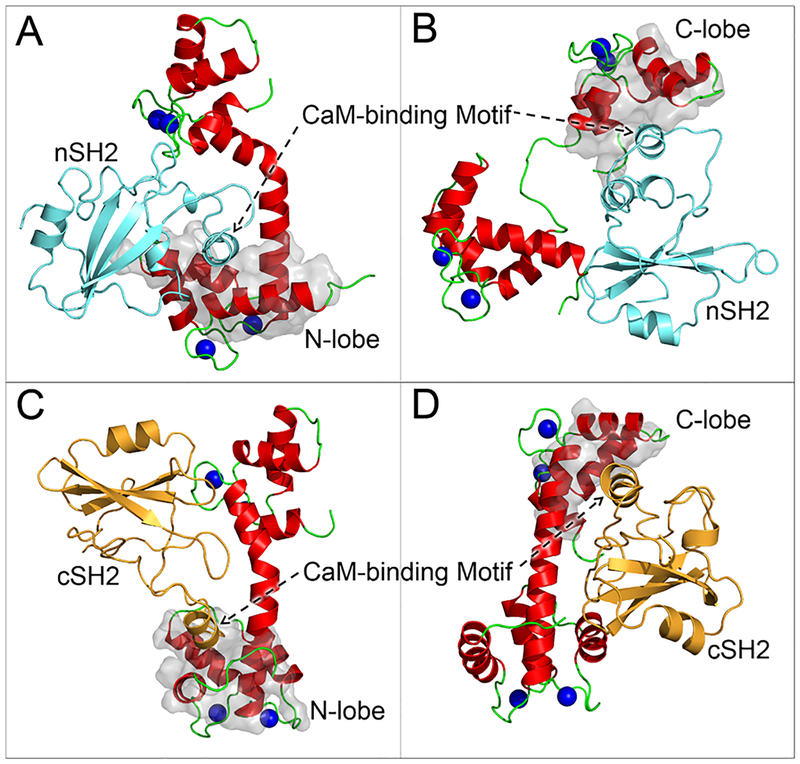
To verify that strong CaM–SH2 interactions were established via the CaM-binding motifs with α-helical conformation and resolve the atomic-scale structures for CaM–SH2 complexes, we modeled the nSH2 and cSH2 domains with CaM-binding motifs adopting the full α-helical conformations (nSH2α and cSH2α) and performed the simulations for CaM–nSH2α and CaM–cSH2α complexes. Myosin light chain kinase (MYLK) is a serine/threonine protein kinase that phosphorylates the regulatory light chain of myosin II. It contains the typical “1–5–10” CaM-binding motif. The crystal structure of the MYLK motif binding to CaM provides a hint for how the “1–5–10” CaM-binding motif interacts with CaM (PDB code: 1CDL).48 We used this crystal structure as the template to model the four CaM–SH2α complexes, configurations α1–α4, in which the CaM-binding motifs in nSH2α (residues 389–404) and cSH2α (residues 684–699) domains individually bind to the N- and C-lobes of CaM (Table 2). The CaM-binding motif in the cSH2 domain at residues 664–679 was not considered and tested here, since with 100% β-sheet structure this region has a much lower probability for a structural change into an α-helical conformation, than the CaM-binding motif at residues 684–699. During the simulations, we observed that these configurations α1–α4 exhibit enhanced stability (Figure 4), as compared to the rigid-body binding between CaM and SH2 (configurations 1–17). The interface between CaM and SH2 appeared to be strong due to the massive interfacial hydrophobic contacts and salt bridges (see below).
Figure 4.
Final structures for (A) configuration α1, (B) configuration α2, (C) configuration α3, and (D) configuration α4 in CaM–SH2α complexes.
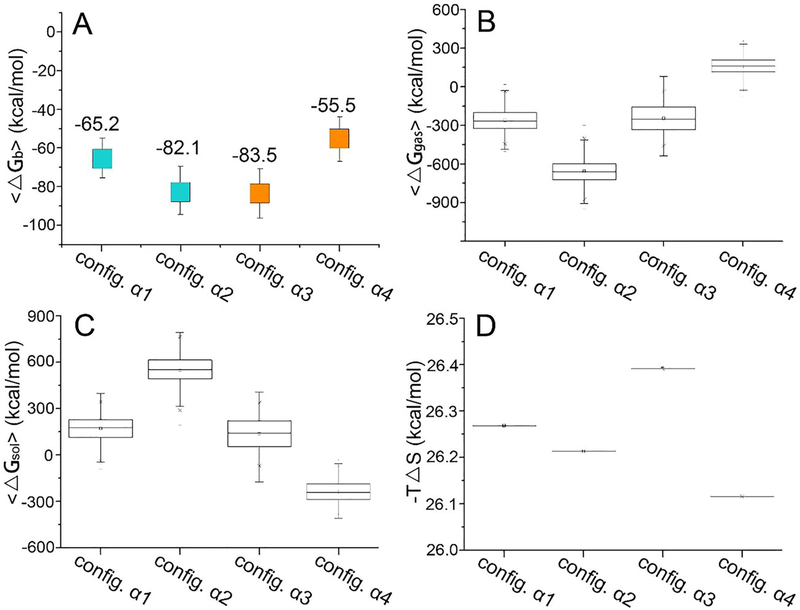
To grade the conformations of the CaM–SH2α complex, we calculated their binding free energies (Figure 5). The binding free energies are approximately −65.2, approximately −82.1, approximately −83.5, and approximately −55.5 kcal/mol for configurations α1–α4, respectively, which are approximately 19–88% lower than the CaM–SH2 systems with rigid-body binding. The strong binding between CaM and SH2α supports that CaM interacts with SH2α via the softened CaM-binding motifs which undergo a structural change into an α-helical conformation, rather than rigid-body binding. SH2α can bind to both N- and C-lobes of CaM. nSH2α displays a preference for the C-lobe, while cSH2α favors CaM’s N-lobe. After CaM-binding motifs change into α-helical conformations, the binding affinities of CaM to nSH2α and cSH2α are highly comparable (−82.1 kcal/mol for nSH2α and −83.5 kcal/mol for cSH2α). This suggests dual targeting of CaM to the nSH2 and cSH2 domains, supporting CaM’s ability to activate PI3Kα.
Figure 5.
Binding free energies for CaM–SH2α complexes. (A) The binding free energies are summarized from the (B) gas phase contribution 〈ΔGb〉, (C) the solvation energy contribution 〈ΔGsol〉, and (D) the entropic contribution, −TΔS.
CaM–SH2α Interfaces.
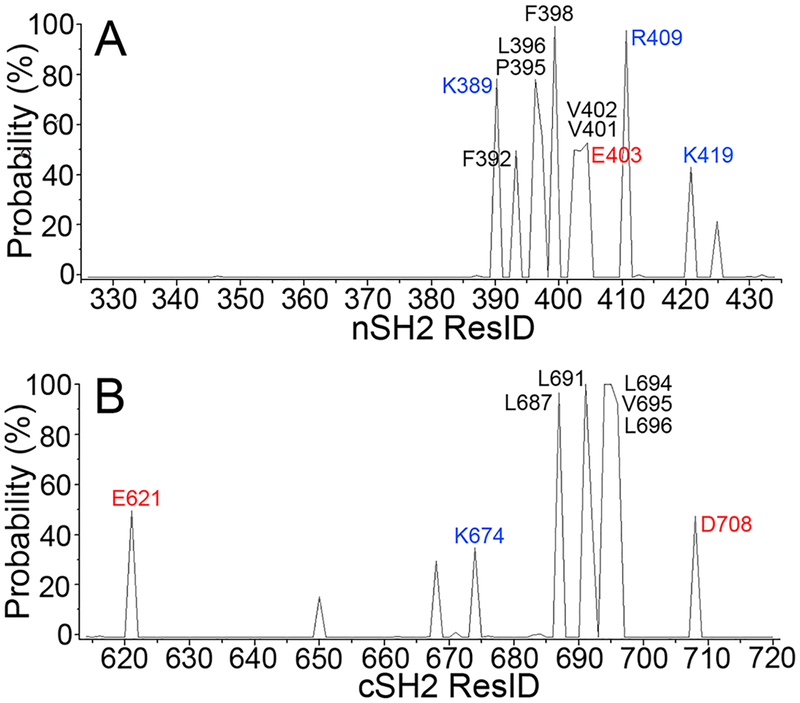
We characterized the interfaces between CaM and SH2α by identifying the key interfacial residues. At the interface, the contact probabilities for interfacial residues that form favorable hydrophilic (hydrogen bonds and salt bridges) and hydrophobic interactions were monitored (Figure 6). Hydrophobic residues play dominant roles in stabilizing the CaM–SH2α interfaces, while some charged residues also contribute. Four charged residues (Lys389, Glu403, Arg409, and Lys419) and six hydrophobic residues (Phe392, Pro395, Leu396, Phe398, Val401, and Val402) in the nSH2 domain, and three charged residues (Glu621, Lys624, and Asp708) and five hydrophobic residues (Leu687, Leu691, Leu694, Val695, and Leu696) in the cSH2 domain, are identified as the interfacial residues with the contact probability >40%.
Figure 6.
Contacting probability for the interfacial residues at CaM–nSH2α and CaM–cSH2α interfaces.
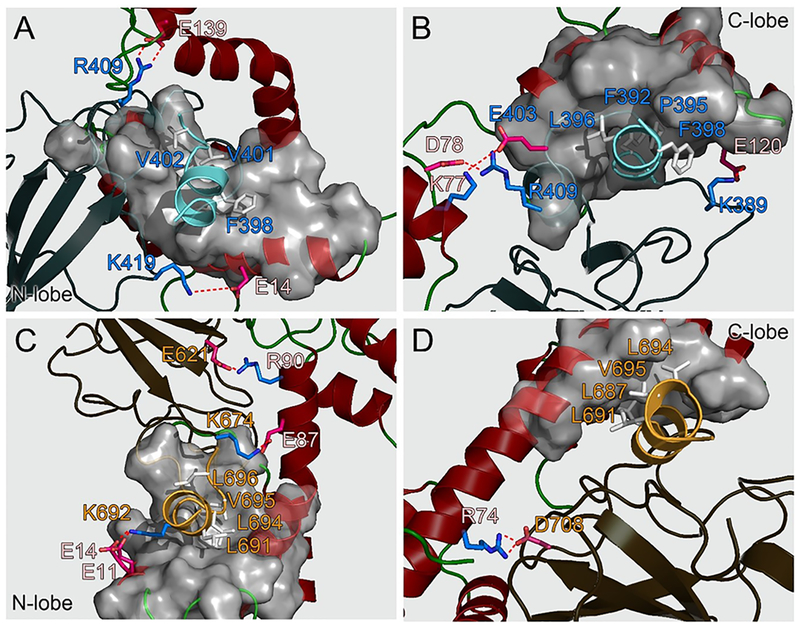
In all CaM–SH2α complexes, the CaM-binding motifs exhibit α-helical conformations, inserting into the hydrophobic pocket and forming strong hydrophobic interactions with CaM (Figure 7). The hydrophobic residues in the CaM-binding motif at positions 1, 5, and 10 (Phe392, Leu396, and Val401 in the nSH2 domain, and Leu687, Leu691, and Leu696 in the cSH2 domain) exert the dominant contributions to the interfacial hydrophobic contacts. While the hydrophobic contacts are strong in all CaM–SH2α complexes, the interfacial hydrophilic contacts mediate the preferences of nSH2 and cSH2 to CaM’s N- and C-lobes. In configuration α1, two interfacial salt bridges (CaMGlu14−nSH2Lys419 and CaMGlu139−nSH2Arg409) were observed, and configuration α2 yielded three interfacial salt bridges (CaMLys77−nSH2Glu403, CaMAsp78−nSH2Arg409, and CaMGlu120−nSH2Lys389). While three salt bridges (CaMGlu11,14−Lys692, CaMGlu87–cSH2Lys674, and CaMArg90−cSH2Glu621) were detected in configuratino α3, only one salt bridge (CaMArg74−cSH2Asp708) was found in configuration α4.
Figure 7.
CaM–SH2α interfaces (A) configuration α1, (B) configuration α2, (C) configuration α3, and (D) configuration α4 with the key residues highlighted.
DISCUSSION
PI3Kα is the key lipid kinase in the PI3K/Akt/mTOR pathway.49 It has frequent oncogenic mutations that lead to its overactivation, a key factor in cell transformation and tumor development.49–52 In response to extracellular signals, PI3Kα is activated by activated RTKs via their pTyr motifs.11,53 The pTyr motifs target the nSH2/cSH2 domains in the p85 subunit, releasing them from the catalytic domain of PI3Kα, thereby activating the kinase.14 The inhibitory effect of p85α on p110α mainly derives from the nSH2 domain. The cSH2 domain also contributes, but less potently.14,17 We have recently elucidated the PI3Kα activation mechanism upon nSH2/cSH2 release (unpublished data). Briefly, the release of the nSH2 domain leads to allosteric motions in the p110α subunit, and, in p85α, iSH2 rotations. These allosteric motions collectively result in the reduced PIP2-ATP distance that facilitates the phosphorylation of PIP2 to PIP3.
How PI3Kα gains excessive activity in cancer in the absence of RTK signals is a significant question that requires comprehension. CaM, a cellular calcium sensor protein, promotes PI3Kα activity in vitro and in intact cells.26,54 It interacts with cSH2, and also nSH2, albeit requiring a longer sample exposure. CaM can be phosphorylated at residue Tyr99.29,55 Phosphorylated CaM (pCaM) generates a binding surface similar to pTyr motifs in RTKs for nSH2 and cSH2 interactions. Our work has shown that the pCaM interactions were stable, with high affinity, indicating the roles of pCaM in promoting the PI3Kα activation.28
Here, we performed modeling and simulations to reveal the structural basis for CaM-driven PI3Kα activation. We determined the atomic-scale structures of CaM–SH2 complexes with properties in line with experimental data, and showed that CaM activates PI3Kα via the “soft” CaM-binding motifs in nSH2 and cSH2 domains. Both nSH2 and cSH2 domains contain the “1–5–10” CaM-binding motifs at the surface. In their populated wild-type state, these CaM-binding motifs do not fully adopt the CaM-favored α-helix structures, and thus cannot follow the “rigid-binding” to interact with CaM. The CaM-binding motif in the cSH2 domain partially adopts the CaM-favored α-helix structure. The CaM-binding motif in the nSH2 domain has a mixed α-helix, a coil, and a short β-sheet structure. To interact with CaM, the CaM-binding motif in nSH2 domain has to populate the CaM-favored α-helix conformation. This process may experience (i) hydrogen bond breaking from the adjacent β-sheet bundle and (ii) a large structural transition from a β-sheet into an α-helix. This process may harbor a large energy barrier, explaining why the nSH2 domain requires a longer time for CaM interactions in PI3Kα activation. The strong and stable interactions between CaM and nSH2α/cSH2α domains were confirmed by the MD simulations. The CaM–SH2α interfaces were dominated by hydrophobic contacts between the CaM-binding motifs of the SH2 domain and the hydrophobic pockets in CaM. The interfacial salt bridges also contribute, mediating the preference of nSH2/cSH2 domains to the N-/C-lobes of CaM. The regions in nSH2/cSH2 domains that CaM binds overlap with those that interact with pTyr motifs, indicating that CaM activates PI3Kα by targeting nSH2/cSH2 domains, in a way that is similar to pTyr motifs in RTKs.15
CaM binds to and releases the nSH2 domain to allosterically activate PI3Kα. However, the role of the interaction of CaM with the cSH2 domain appears to be different. The cSH2 binding surface for CaM and pTyr motifs is solvent-exposed. Thus, the interactions of CaM are not necessary for releasing cSH2 from PI3Kα. In RTK-driven PI3Kα activation, the role of cSH2 interactions with pTyr motifs is likely to bring another pTyr motif in RTKs to the nSH2 domain, facilitating their interactions to release the nSH2 domain. This, however, is not the case for CaM. We observed that, in any CaM–cSH2α complex, only one lobe of CaM is occupied, and the other lobe and the linker are largely free. This raises the possibility that CaM may accommodate other partners at the same time. Our previous data have shown that CaM prefers and binds to the highly variable region (HVR) in KRas4B.31,56–58 KRas4B is a key factor in PI3Kα activation by recruiting it onto the membrane.31,42,59 Different from HRas, NRas (and to a lesser extent KRas4A), the HVR of KRas4B is rich in lysine residues with positive charges, and contains a hydrophobic farnesyl tail.42,60,61 The linker of CaM is negatively charged with many acidic residues (Asp78, Asp80, Glu82, Glu83, Glu84, and Glu87 in Figure 1), which may match the positively charged HVR of KRas4B with the electrostatic force. The free hydrophobic pocket in CaM binds to the hydrophobic farnesyl tail of KRas4B.31 This suggests that the CaM’s interaction with the cSH2 domain may facilitate PI3Kα membrane localization by binding to KRas4B on the membrane (Figure 8).
Figure 8.
Schematic illustration for CaM-driven PI3Kα activation. CaM shifts the equilibrium of the nSH2 and cSH2 domains, making their CaM-binding motifs change into α-helical conformations (nSH2α and cSH2α). Then, CaM forms strong interactions with both nSH2α and cSH2α to activate PI3Kα. CaM binds to and releases the nSH2 domain to allosterically activate PI3Kα, and the CaM interaction with the cSH2 domain facilitates PI3Kα membrane localization by binding of the other lobe of CaM to K-Ras4B on the membrane. Activated PI3Kα phosphorylates PIP2 to PIP3.
CONCLUSIONS
Here, we show a new paradigm for how CaM, the Ca2+ sensor in cells, releases the autoinhibition of PI3Kα lipid kinase by targeting and shifting the equilibrium of CaM-binding motifs in the PI3K inhibitory domains. While the favorable surfaces for CaM binding are not helical in the native conformations of nSH2/cSH2, CaM may shift their equilibrium, making the CaM-binding motifs change into the CaM-favored conformation and form strong CaM–SH2 interactions in PI3Kα activation. This mechanism explains the different scenarios for CaM–nSH2 and CaM–cSH2 interactions in the experiments, and proposes distinct roles for the interaction of CaM with the nSH2 and cSH2 domains. CaM releases the nSH2 domain to allosterically activate PI3K, and CaM’s interaction with cSH2 domain facilitates PI3Kα membrane localization by enhancing PI3Kα–KRas4B interactions on the membrane. Collectively, this work uncovers the structural basis for the CaM-driven PI3Kα activation.
Additionally, it suggests that faster binding observed in the experiment does not necessarily equate with a more stable interaction; it may also imply that the population of the favored species is small, and the population shift to retain the equilibrium to perpetuate the binding reaction is the reason for the experimental observation.
Supplementary Material
ACKNOWLEDGMENTS
This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the high-performance computational facilities of the Biowuilf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.8b05982.
Additional figures including the initial structures of the CaM–SH2 and CaM–SH2α complexes, scanning of CaM-binding motifs in both nSH2 and cSH2 domains, and the final structures of the CaM–SH2 complexes (Figures S1–S4); and an additional table including the summary of the simulation systems (Table S1) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Fruman DA; Chiu H; Hopkins BD; Bagrodia S; Cantley LC; Abraham RT The PI3K pathway in human disease. Cell 2017, 170 (4), 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cantley LC The phosphoinositide 3-kinase pathway. Science 2002, 296 (5573), 1655–1657. [DOI] [PubMed] [Google Scholar]
- (3).Liu P; Cheng H; Roberts TM; Zhao JJ Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discovery 2009, 8 (8), 627–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yap TA; Bjerke L; Clarke PA; Workman P Drugging PI3K in cancer: refining targets and therapeutic strategies. Curr. Opin. Pharmacol 2015, 23, 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nussinov R; Tsai CJ Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci 2014, 35 (5), 256–64. [DOI] [PubMed] [Google Scholar]
- (6).Lawrence MS; Stojanov P; Mermel CH; Robinson JT; Garraway LA; Golub TR; Meyerson M; Gabriel SB; Lander ES; Getz G Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505 (7484), 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Williams R; Berndt A; Miller S; Hon WC; Zhang X Form and flexibility in phosphoinositide 3-kinases. Biochem. Soc. Trans 2009, 37 (4), 615–626. [DOI] [PubMed] [Google Scholar]
- (8).Bader AG; Kang SY; Vogt PK Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (5), 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Burke JE; Perisic O; Masson GR; Vadas O; Williams RL Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc. Natl. Acad. Sci. U. S. A 2012, 109 (38), 15259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Huang CH; Mandelker D; Schmidt-Kittler O; Samuels Y; Velculescu VE; Kinzler KW; Vogelstein B; Gabelli SB; Amzel LM The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 2007, 318 (5857), 1744–8. [DOI] [PubMed] [Google Scholar]
- (11).Kriplani N; Hermida MA; Brown ER; Leslie NR Class I PI 3-kinases: Function and evolution. Adv. Biol. Regul 2015, 59, 53–64. [DOI] [PubMed] [Google Scholar]
- (12).Thorpe LM; Spangle JM; Ohlson CE; Cheng H; Roberts TM; Cantley LC; Zhao JJ PI3K-p110alpha mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85alpha. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (27), 7095–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ito Y; Hart JR; Ueno L; Vogt PK Oncogenic activity of the regulatory subunit p85 beta of phosphatidylinositol 3-kinase (PI3K). Proc. Natl. Acad. Sci. U. S. A 2014, 111 (47), 16826–16829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yu J; Wjasow C; Backer JM Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J. Biol. Chem 1998, 273 (46), 30199–203. [DOI] [PubMed] [Google Scholar]
- (15).Nolte RT; Eck MJ; Schlessinger J; Shoelson SE; Harrison SC Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Mol. Biol 1996, 3 (4), 364–74. [DOI] [PubMed] [Google Scholar]
- (16).Pauptit RA; Dennis CA; Derbyshire DJ; Breeze AL; Weston SA; Rowsell S; Murshudov GN NMR trial models: experiences with the colicin immunity protein Im7 and the p85alpha C-terminal SH2-peptide complex. Acta Crystallogr., Sect. D: Biol. Crystallogr 2001, 57 (10), 1397–1404. [DOI] [PubMed] [Google Scholar]
- (17).Hofmann BT; Jucker M Activation of PI3K/Akt signaling by n-terminal SH2 domain mutants of the p85alpha regulatory subunit of PI3K is enhanced by deletion of its c-terminal SH2 domain. Cell. Signalling 2012, 24 (10), 1950–4. [DOI] [PubMed] [Google Scholar]
- (18).Jucker M; Sudel K; Horn S; Sickel M; Wegner W; Fiedler W; Feldman RA Expression of a mutated form of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase in a Hodgkin’s lymphoma-derived cell line (CO). Leukemia 2002, 16 (5), 894–901. [DOI] [PubMed] [Google Scholar]
- (19).Nussinov R; Muratcioglu S; Tsai CJ; Jang H; Gursoy A; Keskin O K-Ras4B/calmodulin/PI3Kalpha: A promising new adenocarcinoma-specific drug target? Expert Opin. Ther. Targets 2016, 20 (7), 831–42. [DOI] [PubMed] [Google Scholar]
- (20).Nussinov R; Wang G; Tsai CJ; Jang H; Lu S; Banerjee A; Zhang J; Gaponenko V Calmodulin and PI3K signaling in KRAS cancers. Trends Cancer 2017, 3 (3), 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Toufighi K; Yang JS; Luis NM; Aznar Benitah S; Lehner B; Serrano L; Kiel C Dissecting the calcium-induced differentiation of human primary keratinocytes stem cells by integrative and structural network analyses. PLoS Comput. Biol 2015, 11 (5), e1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Orrenius S; Zhivotovsky B; Nicotera P Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell Biol 2003, 4 (7), 552–65. [DOI] [PubMed] [Google Scholar]
- (23).Vergne I; Chua J; Deretic V Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J. Exp. Med 2003, 198 (4), 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Nussinov R; Muratcioglu S; Tsai CJ; Jang H; Gursoy A; Keskin O The key role of calmodulin in KRAS-driven adenocarcinomas. Mol. Cancer Res 2015, 13 (9), 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nussinov R; Zhang M; Tsai CJ; Jang H Calmodulin and IQGAP1 activation of PI3Kalpha and Akt in KRAS, HRAS and NRAS-driven cancers. Biochim. Biophys. Acta, Mol. Basis Dis 2018, 1864 (6 Pt B), 2304–2314. [DOI] [PubMed] [Google Scholar]
- (26).Joyal JL; Burks DJ; Pons S; Matter WF; Vlahos CJ; White MF; Sacks DB Calmodulin activates phosphatidylinositol 3-kinase. J. Biol. Chem 1997, 272 (45), 28183–6. [DOI] [PubMed] [Google Scholar]
- (27).Sacks DB; Fujita-Yamaguchi Y; Gale RD; McDonald JM Tyrosine-specific phosphorylation of calmodulin by the insulin receptor kinase purified from human placenta. Biochem. J 1989, 263 (3), 803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhang M; Jang H; Gaponenko V; Nussinov R Phosphorylated calmodulin promotes PI3K activation by binding to the SH2 domains. Biophys. J 2017, 113 (9), 1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Benaim G; Villalobo A Phosphorylation of calmodulin-Functional implications. Eur. J. Biochem 2002, 269 (15), 3619–3631. [DOI] [PubMed] [Google Scholar]
- (30).Alvarez-Moya B; Barcelo C; Tebar F; Jaumot M; Agell N CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases 2011, 2 (2), 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Jang H; Banerjee A; Chavan T; Gaponenko V; Nussinov R Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling KRas4B membrane extraction. J. Biol. Chem 2017, 292 (30), 12544–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Nussinov R; Tsai CJ; Jang HA New view of pathway-driven drug resistance in tumor proliferation. Trends Pharmacol. Sci 2017, 38 (5), 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nussinov R; Jang H; Tsai CJ; Liao TJ; Li S; Fushman D; Zhang J Intrinsic protein disorder in oncogenic KRAS signaling. Cell. Mol. Life Sci 2017, 74, 3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tsai CJ; Nussinov R The molecular basis of targeting protein kinases in cancer therapeutics. Semin. Cancer Biol 2013, 23 (4), 235–42. [DOI] [PubMed] [Google Scholar]
- (35).Liao J; Planchon SM; Wolfman JC; Wolfman A Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular ras isoforms. J. Biol. Chem 2006, 281 (40), 29730–8. [DOI] [PubMed] [Google Scholar]
- (36).Schneidman-Duhovny D; Inbar Y; Nussinov R; Wolfson HJ PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mashiach E; Schneidman-Duhovny D; Andrusier N; Nussinov R; Wolfson HJ FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Dominguez C; Boelens R; Bonvin AM HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc 2003, 125 (7), 1731–7. [DOI] [PubMed] [Google Scholar]
- (39).Wang G; Zhang M; Jang H; Lu S; Lin S; Chen G; Nussinov R; Zhang J; Gaponenko V Interaction of calmodulin with the cSH2 domain of the p85 regulatory subunit. Biochemistry 2018, 57 (12), 1917–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable molecular dynamics with NAMD. J. Comput. Chem 2005, 26 (16), 1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Brooks BR; Brooks CL; Mackerell AD; Nilsson L; Petrella RJ; Roux B; Won Y; Archontis G; Bartels C; Boresch S; et al. CHARMM: the biomolecular simulation program. J. Comput. Chem 2009, 30 (10), 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Chakrabarti M; Jang H; Nussinov R Comparison of the conformations of KRAS isoforms, K-Ras4A and K-Ras4B, points to similarities and significant differences. J. Phys. Chem. B 2016, 120 (4), 667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Muratcioglu S; Jang H; Gursoy A; Keskin O; Nussinov R PDEdelta binding to Ras isoforms provides a route to proper membrane localization. J. Phys. Chem. B 2017, 121 (24), 5917–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wactawski-Wende J; Kotchen JM; Anderson GL; Assaf AR; Brunner RL; O’Sullivan MJ; Margolis KL; Ockene JK; Phillips L; Pottern L; et al. Women’s health initiative, I., calcium plus vitamin D supplementation and the risk of colorectal cancer. N. Engl. J. Med 2006, 354 (7), 684–96. [DOI] [PubMed] [Google Scholar]
- (45).Schwarz EC; Qu B; Hoth M Calcium, cancer and killing: the role of calcium in killing cancer cells by cytotoxic T lymphocytes and natural killer cells. Biochim. Biophys. Acta, Mol. Cell Res 2013, 1833 (7), 1603–11. [DOI] [PubMed] [Google Scholar]
- (46).Racioppi L; Means AR Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: novel routes for an ancient traveller. Trends Immunol. 2008, 29 (12), 600–607. [DOI] [PubMed] [Google Scholar]
- (47).Berchtold MW; Villalobo A The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta, Mol. Cell Res 2014, 1843 (2), 398–435. [DOI] [PubMed] [Google Scholar]
- (48).Meador WE; Means AR; Quiocho FA Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 1992, 257 (5074), 1251–5. [DOI] [PubMed] [Google Scholar]
- (49).Thorpe LM; Yuzugullu H; Zhao JJ PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15 (1), 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Mandelker D; Gabelli SB; Schmidt-Kittler O; Zhu JX; Cheong I; Huang CH; Kinzler KW; Vogelstein B; Amzel LM A frequent kinase domain mutation that changes the interaction between PI3K alpha and the membrane. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (40), 16996–17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhao L; Vogt PK Helical domain and kinase domain mutations in p110 alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (7), 2652–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Carson JD; Van Aller G; Lehr R; Sinnamon RH; Kirkpatrick RB; Auger KR; Dhanak D; Copeland RA; Gontarek RR; Tummino PJ; et al. Effects of oncogenic p110 alpha subunit mutations on the lipid kinase activity of phosphoinosi-tide 3-kinase. Biochem. J 2008, 409, 519–524. [DOI] [PubMed] [Google Scholar]
- (53).Carpenter CL; Auger KR; Chanudhuri M; Yoakim M; Schaffhausen B; Shoelson S; Cantley LC Phosphoinositide 3-Kinase is activated by phosphopeptides that bind to the Sh2 domains of the 85-Kda subunit. J. Biol. Chem 1993, 268 (13), 9478–9483. [PubMed] [Google Scholar]
- (54).Agamasu C; Ghanam RH; Xu F; Sun Y; Chen YB; Saad JS The interplay between calmodulin and membrane interactions with the pleckstrin homology domain of Akt. J. Biol. Chem 2017, 292 (1), 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Joyal JL; Crimmins DL; Thoma RS; Sacks DB Identification of insulin-stimulated phosphorylation sites on calm-odulin. Biochemistry 1996, 35 (20), 6267–75. [DOI] [PubMed] [Google Scholar]
- (56).Banerjee A; Jang H; Nussinov R; Gaponenko V The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr. Opin. Struct. Biol 2016, 36, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Lu SY; Jang H; Muratcioglu S; Gursoy A; Keskin O; Nussinov R; Zhang J Ras conformational ensembles, allostery, and signaling. Chem. Rev 2016, 116 (11), 6607–6665. [DOI] [PubMed] [Google Scholar]
- (58).Abraham SJ; Nolet RP; Calvert RJ; Anderson LM; Gaponenko V The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry 2009, 48 (32), 7575–7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Gupta S; Ramjaun AR; Haiko P; Wang YH; Warne PH; Nicke B; Nye E; Stamp G; Alitalo K; Downward J Binding of Ras to phosphoinositide 3-kinase p110 alpha is required for Ras-driven tumorigenesis in mice. Cell 2007, 129 (5), 957–968. [DOI] [PubMed] [Google Scholar]
- (60).Nussinov R; Tsai CJ; Chakrabarti M; Jang H A new view of Ras isoforms in cancers. Cancer Res. 2016, 76 (1), 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Nussinov R; Tsai CJ; Jang H; Korcsmaros T; Csermely P Oncogenic KRAS signaling and YAP1/beta-catenin: Similar cell cycle control in tumor initiation. Semin. Cell Dev. Biol 2016, 58, 79–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.