Abstract
Mycoviruses that induce hypovirulence in phytopathogenic fungi are interesting because their potential use as biological control agents of the plant diseases caused by their fungal hosts. The recently identified chrysovirus Fusarium oxysporum f. sp. dianthi virus 1 (FodV1) has been associated to the induction of hypovirulence in Fusarium oxysporum f. sp. dianthi, the forma specialis of F. oxysporum that causes vascular wilt in carnation (Dianthus caryophyllus). In this work, we have used confocal laser scanner microscopy and two isogenic GFP-labeled strains of F. oxysporum f. sp. dianthi infected (V+) and not infected (V−) with the Fusarium oxysporum f. sp. dianthi virus 1, respectively, to analyze the effect of mycovirus FodV1 on the plant colonization pattern of its fungal host. Results demonstrate that FodV1-viral infection affects the speed and spatial distribution of fungal colonization into the plant. Initial stages of external root colonization were similar for both strains, but the virus-free strain colonized the internal plant tissues faster than the virus-infected strain. In addition, other differences related to the specific zone colonized and the density of colonization were observed between both F. oxysporum f. sp. dianthi strains. The hyphae of both V− and V+ strains progressed up through the xylem vessels but differences in the number of vessels colonized and of hyphae inside them were found. Moreover, as colonization progressed, V− and V+ hyphae propagated horizontally reaching the central medulla but, while the virus-free strain V− densely colonized the interior of the medulla cells, the virus-infected strain V+ appeared mainly in the intercellular spaces and with a lower density of colonization. Finally, the incidence of FodV1-viral infections in a collection of 221 isolates sampled between 2008 and 2012 in the geographic area where the originally infected isolate was obtained has been also analyzed. The very low (<2%) incidence of viral infections is discussed here. To the best of our knowledge, this work provides the first microscopic evidence about the effect of a hypovirulence-inducing mycovirus on the pattern of plant colonization by its fungal host.
Keywords: Fusarium oxysporum, carnation, chrysovirus FodV1, hypovirulence, CLSM microscopy, mycovirus-dissemination
Introduction
Fungal viruses (mycoviruses) are found infecting the main taxonomic groups of fungi, including phytopathogenic species. Although there are a few recent evidences that extracellular transmission may occur (Yu et al., 2013; Liu et al., 2016; Marzano et al., 2016), mycoviruses are mainly transferred between strains by hyphal anastomosis (horizontal transmission), and during the production of spores (vertical transmission). Most of the viral infections in fungi are cryptic, i.e., no appreciable changes are observed in the phenotype of the host, but in some cases an alteration of particular phenotypic traits occurs as a consequence of the viral infection. Mycoviruses inducing hypovirulence in phytopathogenic species, i.e., that reduce the virulence of the fungus against its host plant, are the most interesting to study, because the possibility of using them as biological control agents of the diseases caused by their fungal hosts (reviewed in: Ghabrial and Suzuki, 2009; Pearson et al., 2009; Xie and Jiang, 2014; Ghabrial et al., 2015). With the exception of a circular single stranded (ss) DNA mycovirus from the plant pathogenic fungus Sclerotinia sclerotiorum (Yu et al., 2010), mycoviruses inducing hypovirulence have all single or double-stranded (ds) RNA genomes, and are included in the Totiviridae, Chrysoviridae, Partitiviridae, Hypoviridae, Narnaviridae, and Reoviridae families.
The species Fusarium oxysporum contains important plant-pathogens that cause a wide range of plant diseases, mostly involving a vascular wilt syndrome. Strains of F. oxysporum are classified in formae speciales on the basis of the host plant affected. F. oxysporum f. sp. dianthi is the forma specialis of F. oxysporum that infects carnation (Dianthus caryophillus), causing the most devastating carnation disease worldwide (Garibaldi and Gullino, 1987; Baayen, 1988). A few mycoviruses have been described infecting different F. oxysporum strains, but none of them could be associated with the induction of hypovirulence in its fungal host (Kilic and Griffin, 1998; Sharzei et al., 2007). Recently, we have described Fusarium oxysporum f. sp. dianthi virus 1 (FodV1), the first hypovirulence-inducing mycovirus described in the species F. oxysporum. FodV1 was detected infecting isolate Fod 116 of F. oxysporum f. sp. dianthi (Lemus-Minor et al., 2015). FodV1 consists of four dsRNA segments of 3,555 bp (dsRNA1), 2,809 bp (dsRNA2), 2,794 bp (dsRNA3), and 2,646 bp (dsRNA4), respectively. FodV1 dsRNA1 and dsRNA3 encode a RNA-dependent RNA polymerase (RdRp) and a coat protein, respectively; dsRNA2 and dsRNA4 encode hypothetical proteins with unknown functions (Lemus-Minor et al., 2015). Phylogenetic analysis with the aminoacid sequences identifies FodV1 as a member of the Chrysoviridae family, and places it in the same branch as the so-called “chryso-like” mycoviruses, all of them related with the alteration of phenotypic traits in their hosts (Darissa et al., 2012; Lee et al., 2014; Lemus-Minor et al., 2015). The effect of FodV1 on the phenotype of the fungus has been analyzed using two versions of isolate Fod 116: the original one infected with a very high titer of the virus (116V+), and another one with a residual titer obtained by single conidia selection (116V−). Results obtained by comparison of both versions evidenced a significant effect of FodV1 on all the phenotypic traits analyzed (Lemus-Minor et al., 2018). Presence of a high titer of FodV1 in isolate Fod 116V+ modified the morphology and decreased the radial growth of the colony on solid medium, reduced the conidiation in liquid medium, and lowered the virulence against carnation of its fungal host. It has also been demonstrated that FodV1 can be transferred to another isolate in the laboratory by hyphal anastomosis (Lemus-Minor et al., 2019). To prove this, we have used the originally infected strain Fod 116V+ as a donor, and strain Fod 77, a vegetatively compatible virus-free strain that had previously been transformed with a hygromycin resistance gene (Fod 77HygR), as a recipient. Results obtained showed that FodV1 not only was transferred to the recipient isolate, but also accumulated in it at a similarly high level. Comparative analysis of isolates Fod 77HygR (virus-free) and Fod 77HygRV+ (virus-infected) proved that FodV1 induces similar phenotypic alterations in the new infected isolate that those described in the original infected isolate Fod 116V+ (Lemus-Minor et al., 2019).
Previous fungal isolation assays using carnation plants inoculated with V+ and V− versions of isolate Fod 116 suggested that hypovirulence caused by mycovirus FodV1 could be associated to a delay in the progress of plant colonization by the virus-infected isolate (Lemus-Minor et al., 2018), but this point has not yet been experimentally supported. Using confocal laser scanner microscopy (CLSM) and GFP-labeled strains of F. oxysporum f. sp. dianthi infected and not infected with the Fusarium oxysporum f. sp. dianthi virus 1 (FodV1), respectively, we have investigated the effect of mycovirus FodV1 on the ability of the fungal isolate to colonize the plant. In addition, as the success in using mycoviruses for the control of phytopathogenic fungi depends on their efficient transmission among isolates in the natural fungal populations, we have analyzed the efficiency of FodV1-viral dispersion by examining the incidence of FodV1 viral-infections in a wide collection of F. oxysporum f. sp. dianthi isolates from the same geographic area as the original infected isolate 116V+.
Materials and Methods
Fungal Isolates and Culture Conditions
Mycovirus FodV1 was first identified in isolate Fod 116 (116V+), a race 2 strain of F. oxysporum f. sp. dianthi obtained in 2008 from a diseased carnation plant collected in the carnation growing area of Chipiona (Cádiz province, Spain) (Gómez-Lama Cabanás et al., 2012; Lemus-Minor et al., 2015). To study the prevalence of FodV1-viral infections, a collection of 221 F. oxysporum f. sp. dianthi isolates obtained between 2008 and 2012 from plants and soils in the geographic area of Cádiz and Seville provinces has been analyzed (Table 1). To perform the histologic analyses by CLSM, isogenic virus-free (V−) and virus-infected (V+) GFP-labeled strains of isolate Fod 77 were obtained (see below). All strains were stored as conidial suspensions at −80°C in glycerol, and propagated in potato dextrose broth (PDB) liquid medium with agitation (125 rpm) at 25°C in the dark.
Table 1.
Fusarium oxysporum f. sp. dianthi isolates analyzed for the presence of FodV1 viral infection.
| Isolate (s)a | Geographic originb | Source | Year | Race group assignation by PCR patternc |
|---|---|---|---|---|
| Fod 108, 110, 111, 120, 121 | Chipiona(Ca) | Plant | 2008 | R1t |
| Fod 112, 113, 114, 117 | Chipiona(Ca) | Plant | 2008 | R2I |
| Fod 118, 119 | Chipiona(Ca) | Plant | 2008 | R2II |
| Fod 115 | Chipiona(Ca) | Plant | 2008 | – |
| Fod 124, 127, 132, 134, 136, 138 | Chipiona(Ca) | Plant | 2009 | R2I |
| Fod 122, 128, 130, 140, 142 | Chipiona(Ca) | Plant | 2009 | R1t |
| Fod 183, 185, 187 | Chipiona(Ca) | Plant | 2010 | R2I |
| Fod 181, 189, 191 | Chipiona(Ca) | Plant | 2010 | R1t |
| Fod 179, 195, | Chipiona(Ca) | Soil | 2010 | R2II |
| Fod 197 | Chipiona(Ca) | Soil | 2010 | R1t |
| Fod 144, 146, 148, 150, 158, 160, 162, 164 | La Colonia(Se) | Plant | 2010 | R2I |
| Fod 152, 154 | Lebrija(Se) | Plant | 2010 | R1t |
| Fod 156 | Lebrija(Se) | Plant | 2010 | R2II |
| Fod 223, 225, 227, 229, 231, 233, 235, 237, 239, 241, 245, 247, 249, 251, 253 | Chipiona(Ca) | Plant | 2011 | R2I |
| Fod 200, 201, 203, 205 | Chipiona(Ca) | Plant | 2011 | R1t |
| Fod 207 | Chipiona(Ca) | Soil | 2011 | R2I |
| Fod 256, 258, 259, 260, 261, 264, 265, 267, 269, 272, 275, 281 | Chipiona(Ca) | Soil | 2011 | R2II |
| Fod 270, 280 | Chipiona(Ca) | Soil | 2011 | R1t |
| Fod 211 | Lebrija(Se) | Plant | 2011 | R2I |
| Fod 207, 210, 213, 215, 217, 219, 221 | Lebrija(Se) | Plant | 2011 | R2I |
| Fod 327, 328, 329, 330, 331, 332, 333, 334, 335, 336, 341.1, 357, 358, 359, 360, 361, 364, 368, 373.2, 433, 434, 435, 453 | Chipiona(Ca) | Plant | 2012 | R2I |
| Fod 365, 452 | Chipiona(Ca) | Plant | 2012 | R2II |
| Fod 341.2, 342, 343, 344, 345, 346, 347, 348, 349, 350, 351, 363, 366, 367, 371, 372, 373.1, 471 | Chipiona(Ca) | Plant | 2012 | R1t |
| Fod 337, 338, 339, 340, 369, 370, 375, 437, 438, 439, 442, 443, 444, 445, 446, 451, 469 | Chipiona(Ca) | Soil | 2012 | R2II |
| Fod 362, 376, 440, 441, 447, 448, 450, 454, 459, 463, 476 | Chipiona(Ca) | Soil | 2012 | R2II |
| Fod 352, 353, 354, 355, 356, 374, 449, 455, 456, 457, 458, 460, 461, 462, 464, 465, 466, 467, 468, 470, 472, 473, 474, 475, 477, 478, 479, 480, 481, 482, 483, 484, 485, 486, 487 | Chipiona(Ca) | Soil | 2012 | R1t |
| Fod 383, 384, 385, 386, 387, 388 | Lebrija(Se) | Plant | 2012 | R2I |
| Fod 377, 378, 379, 380, 381, 382, 391, 392, 393, 394 | Lebrija(Se) | Plant | 2012 | R1t |
| Fod 400, 402 | Lebrija(Se) | Soil | 2012 | R2I |
| Fod 389, 390, 395, 403, 404, 405 | Lebrija(Se) | Soil | 2012 | R2II |
| Fod 396, 397, 398, 399, 401 | Lebrija(Se) | Soil | 2012 | R1t |
Isolates had been previously characterized to race-group by specific-PCR amplifications, as described in Gómez-Lama Cabanás et al. (2012).
dsRNA from each isolate was purified by cellulose column chromatography and analyzed by RT-PCR using specific primers FodV1f/FodV1r directed to the RdRp sequence of mycovirus FodV1 (Lemus-Minor et al., 2018).
Ca, Cádiz province; Se, Sevilla province.
Race-group assignation by molecular markers. R1t, race 1molecular group type; R2I, race 2 molecular group I; R2II, race 2 molecular group II.
(–) Unknown data.
dsRNA Extraction and FodV1 Detection
Infection by mycovirus FodV1 was determined in dsRNA extracts. To obtain the dsRNA-enriched extracts, 3 mg of mycelia were ground in liquid nitrogen, suspended in 2 x STE buffer (50 mM Tris-HCl, pH 7.0, 0.1 M NaCl, 1 mM EDTA) and 10% SDS, and the total nucleic acids were extracted with phenol. The extracts were then purified by cellulose column chromatography (Valverde et al., 1990), and visualized by electrophoresis on 1% agarose gels stained with RedSafe Nucleic Acid Staining Solution (iNtRON Biotechnology, Seongnam-si Gyeonggi-do, Korea). Presence of FodV1-dsRNA in the extracts was detected by retrotranscription (RT) followed by a polymerase chain reaction (PCR) using primers directed to the RNA-dependent RNA polymerase (RdRp) sequence of FodV1. Primers FodV1RT (5′-GGGTGGAGACTTGCGATCAT-3′) and FodV1F/FodV1R (5′-GGCCTGCTGACCCCCGACATAG-3′/5′-GACCCGAGGCAGCTGAACCCAATA-3′) were used for the RT and the PCR, respectively. RT reactions were performed from 2 μL of dsRNA extract using the enzyme NZY M-MuLV reverse transcriptase (NZYTech). PCR amplifications were performed using 2 μL of the cDNA synthesized, and the enzyme GoTaq® DNA polymerase (Promega Corporation, Madison, WI USA).
RT and PCR amplification conditions were as described in Lemus-Minor et al. (2018). The products of the RT-PCR reactions were analyzed by electrophoresis on 1.5% agarose gels, purified from the gel and sequenced, and the sequences were analyzed by homology with the RdRp sequence of FodV1 using the MegAling program (DNASTAR Lasergene 15 software package).
Obtention of a GFP-Expressing Version of Isolate Fod 77 and Transference of Mycovirus FodV1
To generate a version of strain Fod 77 constitutively expressing a green fluorescent gene (GFP), an Agrobacterium tumefaciens-mediated transformation (ATMT) method was used. To obtain a conidial suspension, strain Fod 77 was grown in YEPS (1% yeast extract, 2% bactopeptone, 2% sucrose) for 4 days at 25°C under 200 rpm shaking. Conidia were harvested by filtration through sterile Miracloth assembled in funnels and centrifuged at 4,000 × g for 10 min. The conidia concentration was determined using a hemocytometer and diluted to a final concentration of 107 conidia/ml. A binary vector containing a hygR marker and a green fluorescent protein (GFP) cassette (Sarmiento-Villamil et al., 2018) was generated using a variant of the OSCAR method (Paz et al., 2011; Gold et al., 2017), and introduced into the AGL-1 Agrobacterium tumefaciens strain by electroporation. ATMT of the conidial suspension of Fod 77 was performed as described by Dobinson et al. (2004) with minor modifications. Transformants were selected on PDA containing hygromycin B (75 μg/ml). To confirm the integration of the selective marker in the genome, a PCR analysis of the transformants was performed with primers Hyg-F (5′-AAAGCCTGAACTCACCGCGACG-3') and Hyg-R (5′-AGCGCGTCTGCTGCTCCATAC-3′), which amplified a 736 bp region of the hygR gene.
To generate the mycovirus-infected GFP-tagged strain, FodV1 was transferred from isolate Fod 116V+ to isolate Fod 77-GFP by hyphal anastomosis. For that, mycelial plugs of the mycovirus donor and recipient strains were transferred to a PDA plate, placing them about 1–1.5 cm apart. Plates were then incubated for 10 days at 25°C to allow strains come into contact and exchange genetic material. Mycelial plugs were then taken from different points of the area of contact of both strains, and transferred to fresh PDA plates containing hygromicin B (75 μg/ml) to selectively grow the recipient strain. Selection plates were incubated for 7 days and then flasks with 100 ml of PDB were inoculated with 2–3 fragments of fungal tissue from each plate. After 4 days of growth, mycelium was collected from the liquid cultures by filtration, and ground to fine powder under liquid nitrogen. Samples were then subjected to chromatography on cellulose to detect presence of dsRNA viral molecules as described previously.
Colonization and Virulence Bioassays
One-month cuttings of susceptible carnation cultivar Candela were inoculated by root-deeping in a suspension of conidia (106 conidia/ml) of the GFP-tagged isolate Fod 77V+ or isolate Fod 77V−. Candela cuttings treated with water (not inoculated) were used as controls. Obtention of fungal inoculum, inoculation of carnation cuttings, and greenhouse conditions, were as described in Gómez-Lama Cabanás et al. (2012). Two bioassays were carried out simultaneously, using 35 plants (replicates) per treatment for each bioassay. A set of 8 plants per treatment were maintained throughout a total period of 40 days to evaluate the progress in the disease severity symptoms. Fusarium wilt symptoms were evaluated every 2 days using a scale of disease from 0 (no symptoms) to 5 (dead plant). Disease severity values were used to calculate the percentage of the standardized area under the disease progress curve (sAUDPC). Analysis of variance (ANOVA) was used to analyze sAUDPC, and significant differences among means for disease severity values with each isolate were determined using the Fisher's least significant difference (LSD P ≤ 0.05). ANOVA analyses were performed using the GraphPad Prism 6 program.
One week before ending the bioassay, a plant inoculated with each V− and V+ Fod 77 strain was used to isolate the fungus, as described in Lemus-Minor et al. (2018). Mycelia from representatives of the fungal colonies obtained were analyzed by multiplex PCR (Gómez-Lama Cabanás et al., 2012) to confirm the forma specialis and race-group of the fungus, and by cellulose column chromatography to confirm presence or absence of FodV1-dsRNA.
Microscopic Monitoring of Pathogen Progression in Carnation Plants
Plants inoculated with the GFP-labeled isolates Fod 77 V− or Fod 77V+ were examined using confocal laser scanner microscopy (CLSM) every 24–48 h during 40 days after inoculation. Two plants per treatment were analyzed each time for CLSM analysis. Plants were carefully uprooted from the pots and the roots were dipped in tap water to be slightly washed. During the first week, root samples were taken daily and observed directly under the confocal microscope. From day 8, samples from the root crown and the internodes of the stem were also exhaustively analyzed by confocal microscopy. Around 1 cm long of stem and root crown tissue were sectioned using a Vibratome Series 1000 plus (TAAB Laboratories Equipment Ltd, Aldermarston, UK) under distilled water as described previously (Prieto et al., 2007). If colonization was observed in an internode, serial cuts of the next upper internode were also prepared to stage the colonization progress of the fungal isolate through the vascular bundles. Single confocal optical sections were thoroughly analyzed from all these vibratome sections using an Axioskop 2 MOT microscope (Carl Zeiss, Jena GmbH, Germany), which has a Krypton and an Argon lasers, controlled by LSM5 PASCAL software (Carl Zeiss). Both GFP-tagged F. oxysporum f. sp. dianthi strains infected and not infected with mycovirus FodV1, respectively, were detected using the 488 nm Argon laser emission light (detection at 500–520 nm). The recorded images were then transferred to Zeiss LSM Image Browser version 4.0 (Carl Zeiss) for an exhaustive study. Finally, confocal stacks were composed and deeply analyzed to assess colonization of Candela root and stem tissues by F oxysporum f. sp. dianthi strains. Brightness and contrast were slightly modified in final figures using PhotoShop 10.0 software (Adobe Systems, San Jose, CA, USA).
Results
Tranformation of Isolate Fod 77 With the gfp Gene and Transference of Mycovirus FodV1
To obtain a F. oxysporum f. sp. dianthi strain expressing the green fluorescent protein (GFP), a binary vector containing a hygR cassette next to a sGFP gene under the control of the Aspergillus nidulans Pgdp promoter between the right and left border of the T-DNA (Sarmiento-Villamil et al., 2018) was used. This vector was used for Agrobacterium tumefaciens-mediated transformation of parental virus-free isolate Fod 77. Three of the transformants obtained following two rounds of selection on PDA plates containing hygromycin B (75 μg/ml) were selected for detailed analysis. The presence of the transgene in the selected transformants was confirmed by performing PCR amplification of a 736 bp amplicon using the primer pair specific for the selectable gene hyg, while no amplification was observed in wild type Fod 77 isolate (data not shown). Comparison of growth rates and colony morphology on PDA plates of the three transformants and the wild type Fod 77 showed no significant differences.
One of the three transformants was selected as virus-free version of the GFP-tagged Fod 77, which was named Fod 77-GFP. Before transferring mycovirus FodV1 to Fod 77-GFP, we performed a pathogenicity test to discard that the integration of the T-DNA carrying the gfp gene had any effect on the virulence of the fungal isolate. To do that, we inoculated the wild type strain Fod 77 and the GFP expressing strain Fod 77-GFP on susceptible carnation cultivars Candela and Pink Bijou, and compared the progress of disease severity symptoms. Results obtained confirmed that Fod 77-GFP was as virulent as the parental wild type (Supplementary Table 1).
To generate isogenic lines with and without viral infection, the mycovirus FodV1 was transferred from isolate Fod 116V+ to the compatible isolate Fod 77-GFP by hyphal anastomosis. FodV1 donor strain Fod 116 and recipient strain Fod 77-GFP were co-cultivated on PDA plates. All the samples taken from different points of the area in contact between both strains and subcultured on PDA with hygromicin were found to contain the virus. One of these samples was chosen for further work as the FodV1-infected version of the GFP-tagged Fod 77.
Mycovirus FodV1 Decreases the Speed and Modifies the Spatial Distribution of the Plant Colonization by Its Fungal Host
A detailed microscopic study of Candela root and stem tissues by CLSM was carried out to reveal the progress of vascular colonization by the GFP-labeled strains Fod 77-virus free (V−) and Fod 77-virus infected (V+). Candela carnation plants were inoculated with both Fod 77 strains, respectively, and samples were taken every 24–48 h and used for CLSM analyses. A set of the inoculated carnation plants were maintained throughout the whole bioassay and used to register the progress of external disease severity symptoms.
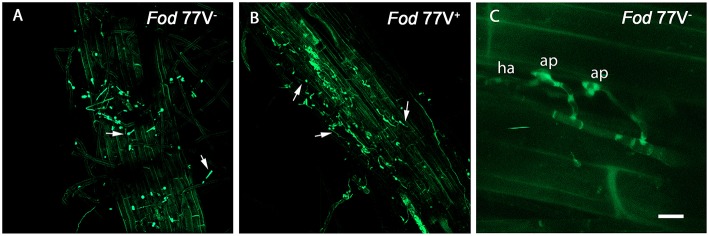
Weak autofluorescence from cell walls was always observed using CLSM, but did not interfere with detection of the GFP-tagged pathogen, even helping to image plant tissues and cells morphology and avoiding extra tissue staining. No autofluorescent native microorganisms were detected on/in Candela tissues at any time. Observation of intact roots between 1 and 7 days post inoculation (dpi) for both treatments showed that initial stages of carnation root colonization were similar in plants inoculated with the virus-free (V−) or the virus-infected (V+) Fod 77-GFP isolate. F. oxysporum f. sp. dianti conidia were readily detected on the root surface just 1 dpi, and most of these conidia had begun to germinate at 2 dpi (Figures 1A,B). After 2–4 more days, a hank of hyphae was observed wrapping the root surface. In these early days we were even able to visualize some events of fungal penetration, identifying the formation of apressoria over the surface of the root epidermal cells and haustoria intracellularly (Figure 1C). Since penetration of epidermal root cells was detected, cross-sections of the root crown zone were examined every 48 h to detect the presence of hyphal internal colonization.
Figure 1.
External colonization of carnation roots by the virus-free (V−) and the virus-infected (V+) GFP-strains of Fusarium oxysporum f. sp. dianthi isolate 77. Images were taken using a confocal laser scanning microscope and intact roots sampled at two (A,B) and three (C) days after inoculation with the virus-free strain V− (A,C) or the virus-infected strain V+ (B). (A,B) Superficial colonization of adventitious roots by the virus-free (A) and the virus-infected (B) strains showing single and germinating conidia (arrowed). (C) Event of penetration of the root epidermis cells showing the formation of apressoria (ap) on the cell surface, and haustoria (ha) inside the epidermal cell. Scale bar = 50 μm (A,B) and 5 μm (C).
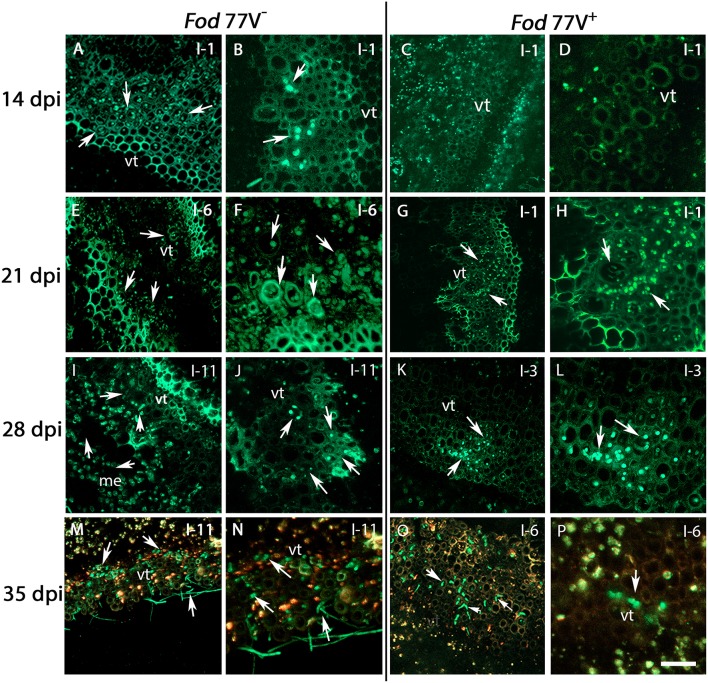
Root crown sections obtained at 6, 8, 10, and 12 dpi from plants inoculated with either strain Fod 77V+ or Fod 77V− were all free of fungal colonization. Colonization in this zone was first detected at 14 dpi only in crown sections from carnation plants inoculated with the virus-free strain (Figures 2A,C). At this same sampling time some hyphae of the V− strain were already slightly reaching the vascular tissue in the first internode of the stem (Figures 3A,B). In contrast, no fungal signals were detected in plants inoculated with the strain carrying the virus (V+) at this stage (Figures 3C,D). In fact, colonization of the root crown of plants inoculated with the strain carrying the virus was firstly detected at 21 dpi (this means 7 days later than with the strain without the virus) (Figures 2B,D). In addition, our observations also suggested that, although hyphae of both fungal strains (V− and V+) colonized the vascular tissue in the root crown zone, the number of vessels colonized and the hyphae density inside them seemed to be always markedly lower in the plants inoculated with the strain carrying the virus (V+) (Figure 2).
Figure 2.
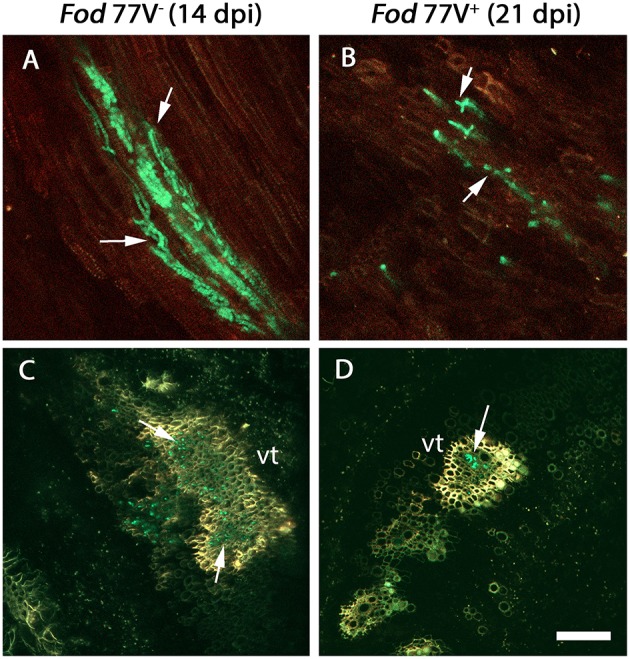
Colonization of the root crown by the virus-free (V−) and the virus-infected (V+) GFP- strains of F. oxysporum f. sp. dianthi isolate 77. (A) Longitudinal and (C) transversal root crown sections of plants inoculated with the virus-free strain V− (green) 14 days after inoculation. (B) Longitudinal and (D) transversal root crown sections of plants inoculated with the virus-infected strain V+ (green) 21 days after inoculation. Both, the virus free and the virus-infected strain, colonized the plant vascular tissue (arrowed), but the number of vessels colonized as well as the number of hyphae inside them were lower in plants inoculated with the strain carrying the virus. vt, vascular tissue. Scale bar = 50 μm (A,B) and 100 μm (C,D).
Figure 3.
Internal colonization of stem internodes after inoculation of carnation plants with the virus-free (V−) and the virus-infected (V+) GFP-strains of F. oxysporum f. sp. dianthi isolate 77. Transversal sections of the internodes (I) were obtained at 14, 21, 28, and 35 days post inoculation (dpi), and were analyzed by confocal laser scanner microscopy. (A,B) Hyphae of the virus-free strain were detected in the vascular tissue of the first internode (I-1) at 14 dpi. (C,D) At the same sampling time (14 dpi), hyphae of the virus–infected strain were not detected yet in the same internode. (E,F) Stem sections of the sixth internode (I-6) showing the vascular tissue extensively colonized by hyphae of strain V− at 21 dpi. (G,H) At the same time point, hyphae of strain V+ only reached the first internode (I-1). (I,J) Stem sections of plants inoculated with the strain V− at 28 dpi showing the hyphae reaching the last stem internode (I-11) of the plant. (K,L) At the same sampling time (28 dpi), the strain V+ only reached the vascular tissue of internode 3 (I-3). (M,N) Extensive colonization of the vascular tissue up to the last plant internode (I-11) by the virus free strain at 35 dpi. (O,P) In contrast, colonization by the V+ strain did only reach internode 6 (I-6) at the same sampling time (35 dpi). Arrows indicate the presence of hyphae (green). vt, vascular tissue. Scale bar = 50 μm (A,C,E,G,I,K,M,O), 20 μm (B,D,F,H,J,L,N,P).
Analysis of stem cross sections at 21 dpi evidenced a rapid progress of the colonization by the virus-free strain, already detecting the hyphae of the fungus in the internode 6 of the carnation plants (Figures 3E,F). On the contrary, at this sampling time hyphae of the strain carrying the virus were only observed in one out of four plants sampled and reaching only the first internode (Figures 3G,H). It is worthy to mention that, at this sampling time, CLSM observations were consistent with visible symptoms of the carnation plants, which were rated quite differently among both treatments. Thus, plants inoculated with the virus-free strain V− showed visible symptoms of disease rated as 3 in a 0–5 scale, whereas plants inoculated with the virus-infected strain V+ were rated 0–1.
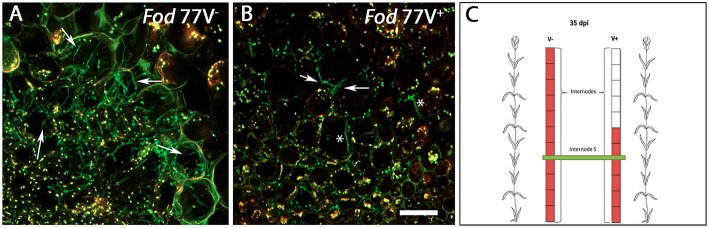
Successive observation of stem cross-sections of the sampled plants displayed a clear difference in the colonization pattern between Fod 77V− and Fod 77V+ strains. Differences were found related to the rate of progression of the plant colonization by the fungal strains. Cross sections examined at 28 and 35 dpi showed an extensive colonization of the stem including the last internode (internode 11) in plants inoculated with the virus-free strain V− (Figures 3I,J,M,N). On the contrary, plants inoculated with the virus-infected strain V+ only showed colonization of the vascular tissue up to internode 3 at 28 dpi (Figures 3K,L), and up to internode 6 at day 35 (Figures 3O,P). In addition to this delay in the colonization progress by the strain carrying the virus, the density of colonization (number of vessels colonized and hyphae inside each vessel) observed at these sampling times seemed to be lower in stem sections of plants inoculated with the virus-infected strain V+ (Figures 3I–P). As colonization progressed, both strains did also invade other plant tissues (medulla) and differences in density between both strains were detected too (Figure 4). Moreover, while hyphae of the virus-free strain V− were easily detected occupying profusely the interior of the medulla cells, hyphae of the virus-infected strain V+ were preferentially observed in the intercellular spaces of the medulla and rarely were able to invade these cells (Figure 4).
Figure 4.
Differences in the colonization pattern of the central medulla in stem sections of plants inoculated with the virus-free (V−) or the virus-infected (V+) GFP-strain of F. oxysporum f. sp. dianthi isolate 77. Images correspond to transversal sections from the same internode (fifth internode) of plants inoculated with the virus-free V− (A) and the virus-infected V+ (B) strains (green), 35 days after inoculation. (A) Hyphae of the virus free strain profusely colonizing the interior of the medulla cells. (B) Hyphae of the virus-infected strain preferentially located in the intercellular spaces of the medulla cells (arrowed) although some hyphae seem to invade the interior of the cells (asterisks). (C) Squematic representation of carnation plants indicating the level of colonization (red) by the virus-free and the virus-infected strains as well as the area shown in panels A and B corresponding to internode 5 (green). Scale bar represents 50 μm in (A) and (B).
At the end of the bioassay (40 dpi), all plants inoculated with the virus-free strain were dead (disease severity = 5) whereas plants inoculated with the virus- infected strain were still alive and maintained disease severity values ≤ 3. Analysis of the disease severity data obtained during the bioassay evidenced a significant reduction of the sAUDPC when cultivar Candela was inoculated with isolate Fod 77V+ compared to isolate Fod 77V− (Figure 5).
Figure 5.
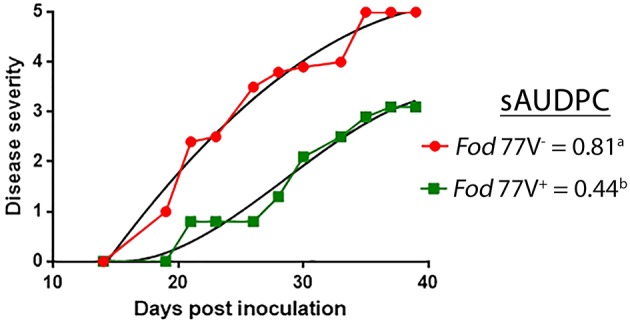
Representative graph of the progress of the disease severity symptoms. Fusarium wilt symptoms were scored for a period of 40 days, using a scale from 0 (no symptoms) to 5 (dead plant), in carnation plants inoculated with the virus-free (V−) or the virus-infected (V+) strain of F. oxysporum f. sp. dianthi isolate 77. Disease severity values were used to calculate the percentage of the standardized area under the disease progress curve (sAUDPC). Values followed by different letters are significantly different according to Fisher's least significant difference (LSD) test (P ≤ 0.05).
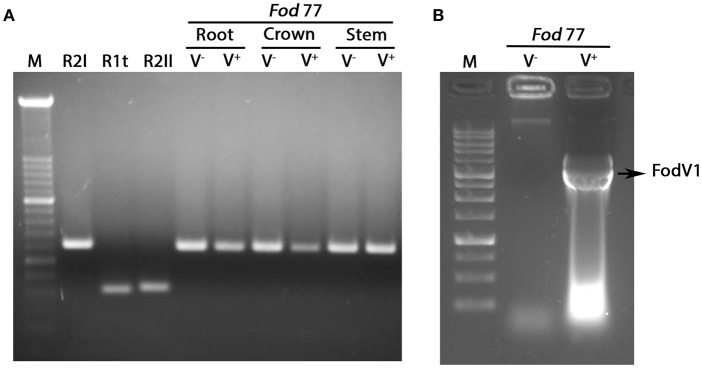
Analysis of the mycelia recovered from the inoculated plants confirmed the identity (F. oxysporum f. sp. dianthi race 2 group I) and presence or absence of viral infection in each inoculated Fod 77 strain (Figure 6).
Figure 6.
Analysis of the fungal mycelia reisolated from plants inoculated with the virus-free (V−) and the virus-infected (V+) strain of Fusarium oxysporum f. sp. dianthi isolate 77. (A) Amplification products obtained in multiplex-PCR reactions using specific primers for the different races-groups described in F. oxysporum f. sp. dianthi (Fod) and DNA from the fungal colonies recovered from root, crown, or stem of carnation plants inoculated with the virus-free strain V− or the virus-infected strain V+ of isolate Fod 77. Specific primers used and conditions of the multiplex-PCR were as described in Gómez-Lama Cabanás et al. (2012). M, molecular weight marker XIV (Roche Diagnostics). R2I, R1t, and R2II, representative of the respective race-group described in F. oxysporum f. sp. dianthi. (B) Extracts of dsRNA obtained by cellulose column chromatography using mycelia recovered from a carnation plant inoculated with the virus-free (V−) or the virus-infected (V+) strain of isolate Fod 77. M, 1Kb DNA Ladder (Nippon Genetics).
Mycovirus FodV1 Has a Low Incidence in the Collection of F. oxysporum f. sp. dianthi Isolates Analyzed
Micovirus FodV1 was detected infecting isolate Fod 116, a race 2 isolate obtained in 2008 from a carnation plant collected in Chipiona (Cádiz, Spain). To determine the incidence of FodV1-dsRNA viral infections, a collection of 221 isolates of F. oxysporum f. sp. dianthi obtained between years 2008 and 2012 from plants and soils in the same (Cádiz) or adjacent (Sevilla) geographic area was analyzed (Table 1). All the isolates had been previously characterized to race by molecular markers (Table 1), and some of them also to vegetative compatibility group (VCG) (Gómez-Lama Cabanás and Pérez-Artés, 2014).
The dsRNA-enriched extracts were obtained by cellulose column chromatography and subjected to RT-PCR amplification using specific primers for the RdRp sequence of FodV1 (Figure 7). Direct observation after agarosa gel electrophoresis of the dsRNA extracts indicated presence of FodV1 only in isolate Fod 183, an isolate obtained from a carnation plant in 2010 in Chipiona (Cádiz) (Figure 7A). Subsequent RT-PCR amplification of the dsRNA extracts showed two additional isolates infected with FodV1: isolate Fod 185, obtained from a carnation plant in 2010 in Chipiona, and isolate Fod 211, obtained from a carnation plant in 2011 in Lebrija (Sevilla) (Figure 7B). All the three new virus-infected isolates were race 2 and belonged to VCG 0021.
Figure 7.
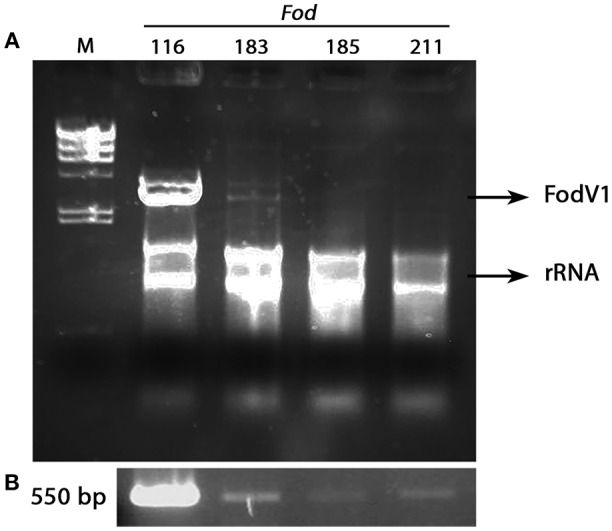
Isolates of Fusarium oxysporum f. sp. dianthi infected with mycovirus FodV1. (A) dsRNA-enriched extracts obtained by cellulose column chromatography from isolates Fod 116 (the originally infected isolate), Fod 183, Fod 185, and Fod 211. (B) Specific amplicons obtained after RT-PCR of the above dsRNA extracts using specific primers for the RdRp sequence of FodV1. M, molecular weight marker II (Roche Diagnostics) (see also Supplementary Image to Figure 7).
To confirm the identity of the mycovirus detected in isolates Fod 183, Fod 185 and Fod 211, the amplicon obtained by RT-PCR with each isolate was purified from the agarose gel and sequenced. Comparison of these sequences with that of the corresponding fragment of the RdRp sequence of FodV1 showed a 100% homology (Supplementary Figure 1).
Discussion
The majority of the studies on the induction of hypovirulence by mycoviruses of phytopathogenic fungi have been focused on the alteration of particular phenotypic traits (reviewed in Ghabrial et al., 2015) or on the modification of the gene expression pattern of the fungal host (Allen and Nuss, 2004; Cho et al., 2012; Lee et al., 2014), but the possible effect of the hypovirulence-inducing mycovirus on the fungal colonization pattern inside the plant has not yet been investigated. In this work, we have analyzed the effect of mycovirus FodV1 on the spatial and temporal dynamics of plant colonization by using confocal laser scanner microscopy. To perform this study, we have used two isogenic GFP-labeled strains of F. oxysporum f. sp. dianthi isolate 77: the original virus-free strain (V−), and a FodV1-infected strain obtained in this work (V+). Both strains were used to inoculate carnation cuttings of susceptible cultivar Candela.
Vascular pathogens are characterized by their ability to colonize the vascular system of the host. Colonization of the xylem vessels leads to a severe decrease in the transport of water and nutrients to the aerial parts of the plant causing its wilting and finally its death. A number of studies have shown that the first stage of plant colonization is common to different F. oxysporum strains and hosts, and initiate with the development of a hyphae network over the roots, followed by penetration of the root epidermis (Li et al., 2011; Jiménez-Fernández et al., 2013; Niño-Sánchez et al., 2015; Upasani et al., 2016). In this work, observation of intact roots throughout the first week after inoculation showed no appreciable differences in the root colonization pattern between the virus-free and the virus-infected strains. Conidia of both strains were observed adhered and germinating over the root surface from 1 dpi, and after 2–4 days the roots were covered by a network of hyphae, some of which appeared penetrating the root epidermis. Several works using light or confocal microscopy and other formae speciales of F. oxysporum revealed the difficulty of finding the specific structures of the fungus at the onset of root cell penetration and the impossibility to photograph them (Lagopodi et al., 2002; Jiménez-Fernández et al., 2013; Niño-Sánchez et al., 2015; Upasani et al., 2016; Pouralibaba et al., 2017). In contrast, in this work we have been able to observe the formation of specific fungal structures such as appressoria over the root surface and haustoria inside the epidermal root cell during the plant cell penetration event using CLSM and the GFP-labeled F. oxysporum f. sp. dianthi strains.
The architecture of the stem vascular system develops in the crown area, where the primordial ring of xylem vessels is formed. Independently of the strain inoculated (V− or V+), root crown sections obtained at 6, 8, 10, and 12 dpi were all free of fungal colonization. This seems to indicate that this period of approximately 1 week from the moment of penetration of the epidermal root cells was necessary for the fungus to reach the central roots and move upward to the crown area.
From 12 dpi, subsequent observations of root crown sections marked the first difference among the virus-free and the virus-infected strains. Colonization of the root crown was first detected in plants inoculated with the virus-free strain at 14 dpi, whereas root crown of plants inoculated with the strain carrying the virus remained free of fungal colonization till 21 dpi. Comparison of crown sections at different times until 28 dpi evidenced that, in addition to the delay in the colonization time, crown of plants inoculated with the virus-infected strain always showed a notably lower density of colonization. This means that, although the hyphae of both V− and V+ strains were observed inside the vascular tissue at the root crown zone, the number of vessels colonized and the number of hyphae inside them was lower in the case of V+-infected plants.
The delay observed in the time of colonization of the root crown area in plants inoculated with strain V+ compared to plants inoculated with strain V− was maintained during the colonization of the stem. Hyphae of the virus-free strain were detected colonizing the vessels in the first internode at 14 dpi, while hyphae of the virus-infected strain reached this same zone 6 days later (21 dpi). Once the hyphae of each strain were detected infecting the first internode of the plant, the progress of colonization upwards the stem was faster in plants inoculated with the virus-free strain. Cross stem sections of plants inoculated with strain V− showed fungal hyphae reaching the last internode (internode 11) at 28 dpi, whereas at the same sampling time hyphae of strain V+ had colonized only the third lower part of the stem (up to internode 3). Along with this difference in the speed of colonization of the vascular system upwards the stem, some other differences were observed related to the area colonized and the density of colonization reached. The number of vessels colonized and of hyphae inside each of them observed at the different sampling times was always notably lower in stem sections of plants inoculated with the virus-infected strain.
At advanced stages of the colonization, differences in the density and location of the hyphae of strains V− and V+ were also observed in tissues other than the vascular tissue. As colonization progressed, hyphae of both strains invaded the central area of the stem (medulla), but while hyphae of the virus-free strain V− were easily detected profusely occupying the interior of the medulla cells, hyphae of the virus-infected strain V+ were less in number and rarely were able to invade the medulla cells, being preferentially located in the intercellular spaces. This difference in the time and extent of stem colonization correlated with significant differences in the external disease symptoms so that most of the plants inoculated with strain V− were dead (disease severity = 5) at 35 dpi, whereas plants inoculated with strain V+ maintained disease severity values ≤3 at the end of the bioassay (40 dpi).
The importance of the speed in the colonization of the vascular tissue as determinant of disease severity has been described previously in a study using two strains of F. oxysporum f. sp. phaseoli differing in virulence (Niño-Sánchez et al., 2015). These authors reported differences on the spatial and temporal dynamic of colonization of a susceptible common bean cultivar infected by a highly virulent (HV) or a weakly virulent (WV) strain. Similar to our results, the main differences between both strains were found in the temporal and spatial dynamic of crown root and hypocotyl colonization, the WV strain being a much slower and less efficient colonizer of the xylem vessels (Niño-Sánchez et al., 2015). Differences in virulence between the HV and WV strains of F. oxysporum f. sp. phaseoli have been associated to the presence and expression of some transcription factors (Alves-Santos et al., 2002a,b; Ramos et al., 2007). In this work, difference in virulence between the V− and V+ strains of F. oxysporum f. sp. dianthi is associated to the presence of mycovirus FodV1. Therefore, it seems that in these two specific cases, regardless of the endogenous or exogenous cause, the hypovirulence is associated to a delay and restriction in the colonization of the vascular system of the plant.
Previous microscopic analysis related to plant colonization by F. oxysporum have been done but focused on particular host-fungus interactions (Czymmek et al., 2007; Nahalkova et al., 2008; Li et al., 2011), compatible (susceptible) or incompatible (resistant) interactions (Zvirin et al., 2010; Jiménez-Fernández et al., 2013; Lü et al., 2013; Pouralibaba et al., 2017), or related to differences in the virulence of the pathogenic strains (Niño-Sánchez et al., 2015). To the best of our knowledge, this work provides the first microscopic evidences on the effect of a hypovirulence-inducing mycovirus on the plant colonization process by its fungal host.
Another aspect analyzed in this study has been that on the incidence of FodV1-viral infections in a collection of F. oxysporum f. sp. dianthi isolates sampled between 2008 and 2012 in the geographic area where the originally infected isolate Fod 116 was obtained. In a previous work, we had demonstrated that FodV1 could be transferred by hyphal anastomosis to another vegetatively compatible isolate in the laboratory (Lemus-Minor et al., 2019). In F. oxysporum f. sp. dianthi, isolates of race 1 type and of race 2 belong to different VCGs, i.e., VCG 0022 for race 1 type isolates and VCG 0021 for race 2 isolates (Gómez-Lama Cabanás and Pérez-Artés, 2014). The majority of the isolates in the collection analyzed was race 2 and thus belonged to the same 0021 vegetative compatibility group than the virus-infected isolate Fod 116. Therefore, in theory, FodV1 should have been able to disperse among compatible hyphae in the fungal population. In spite of that, results obtained in this work demonstrate a very low incidence of FodV1 infections. Mycovirus FodV1 has been detected infecting only another three F. oxysporum f. sp. dianthi isolates. In addition to the vic genes that regulate hyphal anastomosis, the host genetic background has been shown to affect horizontal virus transmission (Cortesi et al., 2001; Choi et al., 2012; Zamora et al., 2015). Although all race 2 isolates are in VCG 0021, we have evidence that some isolates in a same VCG show a reduced compatibility, that is, they can anastomose with a few but not all the isolates in the VCG (Gómez-Lama Cabanás and Pérez-Artés, 2014). The naturally infected isolate Fod 116 could be one of those isolates with reduced compatibility, and this fact could have limited the dispersion of the mycovirus. Another factor that could be contributing to the very low dispersion rate of FodV1 is the efficiency in vertical transmission through asexual spores. Previous studies carried out with the original virus-infected isolate Fod 116V+ and the “in laboratory” virus-infected isolate Fod 77V+ showed that efficiency in vertical transmission depends on the isolate considered (Lemus-Minor et al., 2019). Thus, while isolate Fod 116V+ showed a relatively low efficiency (24%), the efficiency of isolate Fod 77V+ was 100%. This low efficiency of vertical virus-transmission showed by isolate Fod 116V+ could be contributing to restrict the presence of new virus-infected hyphae of this isolate in the population. In this way, a low density of virus-infected hyphae would decrease the probability of contact between infected and non-infected hyphae and would be limiting the dispersion of the viral infection. This evidence supports the hypothesis that selection of a specific virus-infected isolate could influence transmission and dissemination of hypovirulence (Lee et al., 2014; Lemus-Minor et al., 2019).
Finally, we must take an additional factor into consideration. From the collection analyzed, most (127) of the isolates were obtained from plants with disease symptoms, while 94 isolates were obtained from soil samples. Considering the reduction in the disease severity symptoms associated to the viral infection, this bias in the sampling could also be skewing the data of the prevalence of viral infection in the population. To avoid this inconvenient, it should be necessary to make a more exhaustive analysis with isolates collected exclusively from soil samples.
To the best of our knowledge, this work provides the first microscopic evidence about the effect of a hypovirulence-inducing mycovirus on the plant colonization process, and supplies data that reinforce the idea that the choice of the appropriate isolate is key to achieve an efficient dissemination of a viral infection in a fungal population.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
EP-A, AT-T, and PP conceived and designed the experiments. MC and MG-P obtained the GFP-tagged V+ and V− Fod strains. PP and AT-T performed the microscopic analysis. EP-A, AT-T, and PP analyzed the data. EP-A and PP wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by Grants AGL 2013-48980-R from the Spanish Ministry of Economy and Competitiveness, and AGL 2016-80048-R from the Spanish Ministry of Science and Innovation, co-funded by the European Union (FEDER funds). We acknowledge support of the publication fee by the CSIC Open Access Publication Support initiative through its Unit of Information Resources for Research (URICI). We thank Antonio Valverde for his valuable technical assistance and Emilio A. Cano (Barberet & Blanc S.A.) for providing carnation cultivars. Authors also want to acknowledge Prof. Antonio Martin for the use of the CLSM facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00051/full#supplementary-material
References
- Allen T. D., Nuss D. L. (2004). Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 78, 4145–4155. 10.1128/JVI.78.8.4145-4155.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Santos F. M., Cordeiro-Rodrigues L., Sayagués J. M., Martín-Domínguez R., García-Benavides P., Crespo M. C., et al. (2002b). Pathogenicity and race characterization of Fusarium oxysporum f. sp. phaseoli isolates from Spain and Greece. Plant Pathol. 51, 605–611. 10.1046/j.1365-3059.2002.00745.x [DOI] [Google Scholar]
- Alves-Santos F. M., Ramos B., García-Sánchez M. A., Eslava A. P., Díaz-Mínguez J. M. (2002a). A DNA-based procedure for in planta detection of Fusarium oxysporum f. sp. phaseoli. Phytopathology 92, 237–244. 10.1094/PHYTO.2002.92.3.237 [DOI] [PubMed] [Google Scholar]
- Baayen R. P. (1988). Fusarium Wilt of Carnation. Disease Development, Resistance Mechanism of the Host and Taxonomy of the Pathogen. thesis, University of Utretcht, Holland. [Google Scholar]
- Cho W. K., Yu J., Lee K. M., Son M., Min K., Lee Y. W., et al. (2012). Genome-wide expression profiling shows transcriptional reprogramming in Fusarium graminearum by Fusarium graminearum virus 1-DK21 infection. BMC Genomics 13:173. 10.1186/1471-2164-13-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G. H., Dawe A. L., Churbanov A., Smith M. L., Milgroom M. G., Nuss D. L. (2012). Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190, 113–127. 10.1534/genetics.111.133983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesi P., McCulloch C. E., Song H., Lin H., Milgroom M. G. (2001). Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 159, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czymmek K. J., Fogg M., Powell D. H., Sweigard J., Park S. Y., Kang S. (2007). In vivo time-lapse documentation using confocal and multi-photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum. Fungal Genet. Biol. 44, 1011–1023. 10.1016/j.fgb.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Darissa O., Adam G., Schafer W. (2012). A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 134, 181–189. 10.1007/s10658-012-9977-5 [DOI] [Google Scholar]
- Dobinson K. F., Grant S. J., Kang S. (2004). Cloning and targeted disruption, via Agrobacterium tumefaciens-mediated transformation, of a trypsin protease gene from the vascular wilt fungus Verticillium dahliae. Curr. Genet. 45, 104–110. 10.1007/s00294-003-0464-6 [DOI] [PubMed] [Google Scholar]
- Garibaldi A., Gullino M. L. (1987). Attempts of biocontrol of Fusarium-wilt of carnation in Italy. Phytopathology 77, 1721–1721. [Google Scholar]
- Ghabrial S. A., Caston J. R., Jiang D., Nibert M. L., Suzuki N. (2015). 50-plus years of fungal viruses. Virology 479–480, 356–368. 10.1016/j.virol.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Ghabrial S. A., Suzuki N. (2009). Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384. 10.1146/annurev-phyto-080508-081932 [DOI] [PubMed] [Google Scholar]
- Gold S. E., Paz Z., García-Pedrajas M. D., Glenn A. E. (2017). Rapid deletion production in fungi via agrobacterium mediated transformation of OSCAR deletion constructs. J. Vis. Exp. e55239. 10.3791/55239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Lama Cabanás C., Pérez-Artés E. (2014). New evidence of intra-race diversity in Fusarium oxysporum f. sp. dianthi populations based on Vegetative Compatibility Groups. Eur. J. Plant Pathol. 139, 445–451. 10.1007/s10658-014-0412-y [DOI] [Google Scholar]
- Gómez-Lama Cabanás C., Valverde-Corredor A., Pérez-Artés E. (2012). Molecular analysis of Spanish populations of Fusarium oxysporum f. sp dianthi demonstrates a high genetic diversity and identifies virulence groups in races 1 and 2 of the pathogen. Eur. J. Plant Pathol. 132, 561–576. 10.1007/s10658-011-9901-4 [DOI] [Google Scholar]
- Jiménez-Fernández D., Landa B. B., Kang S., Jiménez-Díaz R. M., Navas-Cortes J. A. (2013). Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris races. PLoS ONE 8:e61360. 10.1371/journal.pone.0061360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic O., Griffin G. J. (1998). Effect of dsRNA-containing and dsRNA-free hypovirulent isolates of Fusarium oxysporum on severity of Fusarium seedling disease of soybean in naturally infested soil. Plant Soil 201, 125–135. 10.1023/A:1004319614390 [DOI] [Google Scholar]
- Lagopodi A. L., Ram A. F., Lamers G. E., Punt P. J., Van den Hondel C. A., Lugtenberg B. J., et al. (2002). Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 15, 172–179. 10.1094/MPMI.2002.15.2.172 [DOI] [PubMed] [Google Scholar]
- Lee K. M., Cho W. K., Yu J., Son M., Choi H., Min K., et al. (2014). A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS ONE 9:e100989. 10.1371/journal.pone.0100989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemus-Minor C. G., Canizares M. C., García-Pedrajas M. D., Pérez-Artés E. (2015). Complete genome sequence of a novel dsRNA mycovirus isolated from the phytopathogenic fungus Fusarium oxysporum f. sp. dianthi. Arch. Virol. 160, 2375–2379. 10.1007/s00705-015-2509-9. [DOI] [PubMed] [Google Scholar]
- Lemus-Minor C. G., Canizares M. C., García-Pedrajas M. D., Pérez-Artés E. (2018). Fusarium oxysporum f. sp. dianthi virus 1 accumulation is correlated with changes in virulence and other phenotypic traits of its fungal host. Phytopathology 108, 957–963. 10.1094/PHYTO-06-17-0200-R [DOI] [PubMed] [Google Scholar]
- Lemus-Minor C. G., Cañizares M. C., García-Pedrajas M. D., Pérez-Artés E. (2019). Horizontal and vertical transmission of the hypovirulence-associated mycovirus Fusarium oxysporum f. sp. dianthi virus 1. Eur. J. Plant Pathol 153, 645–650. 10.1007/s10658-018-1554-0 [DOI] [Google Scholar]
- Li C., Chen S., Zuo C., Sun Q., Ye Q., Yi G., et al. (2011). The use of GFP-transformed isolates to study infection of banana with Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 131, 327–340. 10.1007/s10658-011-9811-5 [DOI] [Google Scholar]
- Liu S., Xie J., Cheng J., Li B., Chen T., Fu Y., et al. (2016). Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. U.S.A. 113, 12803–12808. 10.1073/pnas.1608013113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü G., Guo S., Zhang H., Geng L., Martyn R. D., Xu Y. (2013). Colonization of Fusarium wilt-resistant and susceptible watermelon roots by a green-fluorescent-protein-tagged isolate of Fusarium oxysporum f.sp. niveum. J. Phytopathol. 162, 228–237. 10.1111/jph.12174 [DOI] [Google Scholar]
- Marzano S. L., Nelson B. D., Ajayi-Oyetunde O., Bradley C. A., Hughes T. J., Hartman G. L., et al. (2016). Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 90, 6846–6863. 10.1128/JVI.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahalkova J., Fatehi J., Olivain C., Alabouvette C. (2008). Tomato root colonization by fluorescent-tagged pathogenic and protective strains of Fusarium oxysporum in hydroponic culture differs from root colonization in soil. FEMS Microbiol. Lett. 286, 152–157. 10.1111/j.1574-6968.2008.01241.x [DOI] [PubMed] [Google Scholar]
- Niño-Sánchez J., Tello V., Casado-Del Castillo V., Thon M. R., Benito E. P., Díaz-Mínguez J. M. (2015). Gene expression patterns and dynamics of the colonization of common bean (Phaseolus vulgaris L.) by highly virulent and weakly virulent strains of Fusarium oxysporum. Front. Microbiol. 6:234. 10.3389/fmicb.2015.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz Z., García-Pedrajas M. D., Andrews D. L., Klosterman S. J., Baeza-Montanez L., Gold S. E. (2011). One step construction of Agrobacterium-Recombination-ready-plasmids (OSCAR), an efficient and robust tool for ATMT based gene deletion construction in fungi. Fungal Genet. Biol. 48, 677–684. 10.1016/j.fgb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Pearson M. N., Beever R. E., Boine B., Arthur K. (2009). Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10, 115–128. 10.1111/j.1364-3703.2008.00503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouralibaba H. R., Pérez-de-Luque A., Rubiales D. (2017). Histopathology of the infection on resistant and susceptible lentil accessions by two contrasting pathotypes of Fusarium oxysporum f. sp. lentis. Eur. J. Plant Pathol. 148, 53–63. 10.1007/s10658-016-1068-6 [DOI] [Google Scholar]
- Prieto P., Moore G., Shaw P. (2007). Fluorescence in situ hybridization on vibratome sections of plant tissues. Nat. Protoc. 2, 1831–1838. 10.1038/nprot.2007.265 [DOI] [PubMed] [Google Scholar]
- Ramos B., Alves-Santos F. M., García-Sánchez M. A., Martin-Rodrigues N., Eslava A. P., Díaz-Mínguez J. M. (2007). The gene coding for a new transcription factor (ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet. Biol. 44, 864–876. 10.1016/j.fgb.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Sarmiento-Villamil J. L., Prieto P., Klosterman S. J., García-Pedrajas M. D. (2018). Characterization of two homeodomain transcription factors with critical but distinct roles in virulence in the vascular pathogen Verticillium dahliae. Mol. Plant Pathol. 19, 986–1004. 10.1111/mpp.12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharzei A., Banihashemi Z., Afsharifar A. (2007). Detection and characterization of a double-stranded RNA mycovirus in Fusarium oxysporum f. sp. melonis. Iran J. Plant Path. 43, 9–26. [Google Scholar]
- Upasani M. L., Gurjar G. S., Kadoo N. Y., Gupta V. S. (2016). Dynamics of colonization and expression of pathogenicity related genes in Fusarium oxysporum f. sp. ciceri during chickpea vascular wilt disease progression. PLoS ONE 11:e0156490. 10.1371/journal.pone.0156490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R. A., Nameth S. T., Jordan R. L. (1990). Analysis of double-stranded-RNA for plant-virus diagnosis. Plant Dis. 74, 255–258. [Google Scholar]
- Xie J., Jiang D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68. 10.1146/annurev-phyto-102313-050222 [DOI] [PubMed] [Google Scholar]
- Yu X., Li B., Fu Y., Jiang D., Ghabrial S. A., Li G., et al. (2010). A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 8387–8392. 10.1073/pnas.0913535107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li B., Fu Y., Xie J., Cheng J., Ghabrial S. A., et al. (2013). Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. U.S.A. 110, 1452–1457. 10.1073/pnas.1213755110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora P., Martín A. B., Dueñas M., San Martín R., Diez J. J. (2015). Cryphonectria parasitica isolates of the same vegetative compatibility type display different rates of transfer of CHV1 hypovirus. Eur. J. Plant Pathol. 143, 767–777. 10.1007/s10658-015-0727-3 [DOI] [Google Scholar]
- Zvirin T., Herman R., Brotman Y., Denisov Y., Belausov E., Freeman S., et al. (2010). Differential colonization and defence responses of resistant and susceptible melon lines infected by Fusarium oxysporum race 1·2. Plant Pathol. 59, 576–585. 10.1111/j.1365-3059.2009.02225.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the supplementary files.