Abstract
Quantitative radiomics features, extracted from medical images, characterize tumour-phenotypes and have been shown to provide prognostic value in predicting clinical outcomes. Stability of radiomics features extracted from apparent diffusion coefficient (ADC)-maps is essential for reliable correlation with the underlying pathology and its clinical applications. Within a multicentre, multi-vendor trial we established a method to analyse radiomics features from ADC-maps of ovarian (n = 12), lung (n = 19), and colorectal liver metastasis (n = 30) cancer patients who underwent repeated (<7 days) diffusion-weighted imaging at 1.5 T and 3 T. From these ADC-maps, 1322 features describing tumour shape, texture and intensity were retrospectively extracted and stable features were selected using the concordance correlation coefficient (CCC > 0.85). Although some features were tissue- and/or respiratory motion-specific, 122 features were stable for all tumour-entities. A large proportion of features were stable across different vendors and field strengths. By extracting stable phenotypic features, fitting-dimensionality is reduced and reliable prognostic models can be created, paving the way for clinical implementation of ADC-based radiomics.
Introduction
Diffusion-weighted magnetic resonance imaging (DWI) is widely used in oncology for identification and characterization of tumours1, as well as localization2. Signal attenuation in DWI arises from Brownian motion of water molecules and reflects their interaction with cellular barriers and tissue macromolecules that restrict their mobility. DWI is used for tumour characterization and as an indirect biomarker of tissue cellularity3. By incorporating a number of diffusion-sensitizing gradients with varying strength, duration and time interval (i.e., b-values) into the MR pulse sequence, parametric apparent diffusion coefficients (ADC) maps can be derived. In oncology, ADC maps are used to determine tumour malignancy and assess early treatment response by quantifying the diffusion-related attenuation of MR signal intensity3,4. There is no consensus regarding the threshold value below which ADC is indicative of tumour, however ADC values around 1000 × 10−6 mm2/s are considered normal, while lower values generally reflect restricted diffusion that could relate to hyper-cellularity or hyper-viscosity characteristic of tumour tissue, and higher ADC values represent fluid filled regions where water diffusion is unrestricted (e.g., cystic lesions, necrotic tissue). Unfortunately, an overlap between ADC values characteristic of active and treated tumours often reduces the utility of ADC in clinical decision-making and variations in estimates of ADC resulting from lack of standardized DWI protocols do not allow the integration of absolute values of ADC as an objective, quantifiable biomarker for personalised healthcare in the clinic5–7.
Radiomics may provide complementary information from ADC maps by high-throughput extraction of quantitative tumour phenotypic features (i.e., shape, texture, signal intensity, and wavelet features) that have previously been correlated with tissue pathology and treatment response prediction8,9. Please view https://youtu.be/Tq980GEVP0Y for more information. This methodology has shown promising results for CT imaging in oncology and in the development of prediction modelling for treatment response and outcome10,11. For DWI, the variability of ADC across different MR systems, vendors and magnetic field-strengths potentially compromises the stability of radiomics features, limiting the clinical implementation of MR radiomics. In addition, there is a realistic risk of overfitting when building a model where the numbers of features extracted greatly outnumber the size of the study cohort. The majority of published radiomics analyses remove features due to redundancy, either because features correlate highly with each other, or because they do not correlate with the endpoint being analysed. Reducing the number of unstable features substantially increases the reliability of radiomics analyses and may not only improve correlation with underlying pathology and tumour biology, but may also allow construction of improved prognostic or predictive models12–14. Elimination of features with poor clinical reproducibility is critical for achieving a high radiomics quality score15.
The purpose of this analysis was to evaluate the stability of radiomics features extracted from ADC maps, derived from standardized test-retest DWI acquisitions embedded in prospective multicentre trials. DWI for each subject were acquired twice within 7 days, under similar conditions. We hypothesized that selected ADC-based features are generalizable and unaffected by different sources of data variability (i.e., tumour type, MR system, magnetic field strength) when applying standardized protocols on quality-assured MR scanners across multiple clinical centres. Establishing this methodology and presenting the results derived from the underpinning study would not only stimulate the clinical implementation of MR radiomics using ADC as a biomarker for tumour phenotyping16 but would also outline a generalizable method for other quantifiable MRI parameters.
Results
Feature stability
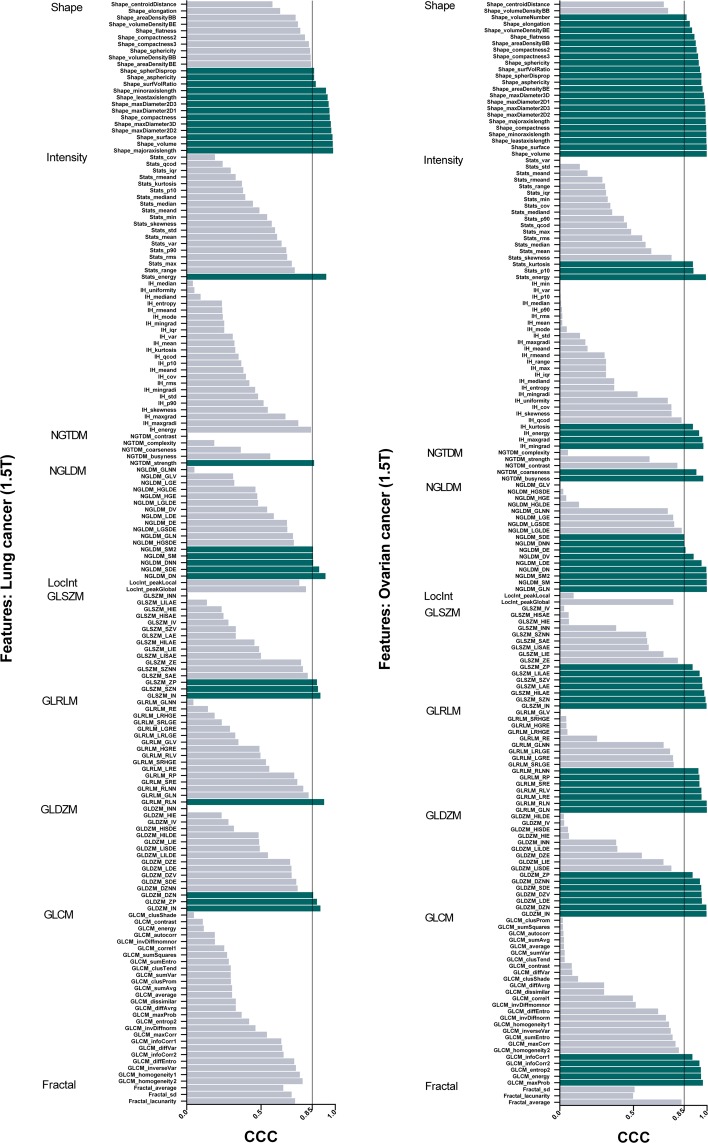
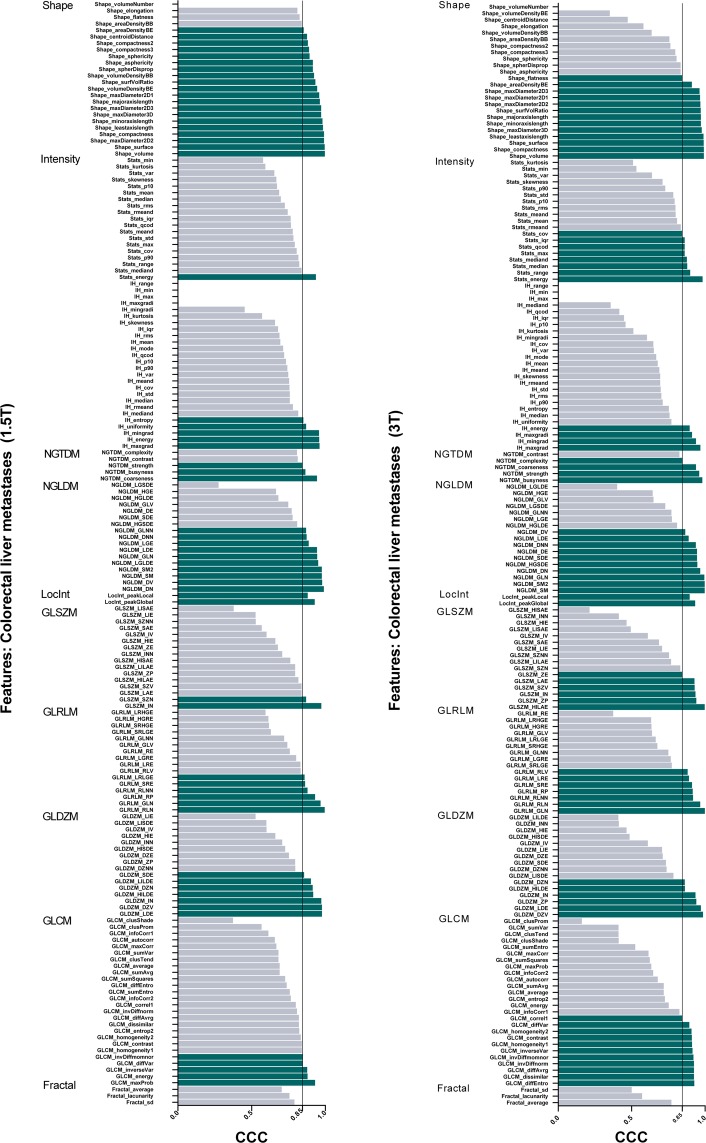
Based on previous work, we selected a threshold of 0.85 for the concordance correlation coefficient (CCC), whereby features above this threshold were considered stable between test and retest scans17. Figures 1 and 2 show an overview of the stability of test-retest radiomics features for all patient cohorts.
Figure 1.
Stability of test-retest radiomics features for all lung and ovarian. cancers The threshold for stability was set at concordance correlation coefficients (CCC) greater than 0.85.
Figure 2.
Stability of test-retest radiomics features for all collorectal liver metastases acquired at 1.5 T and 3 T. The threshold for stability was set at concordance correlation coefficients (CCC) greater than 0.85.
Tumour-type differences at 1.5 T
In 20 ovarian cancer lesions from 12 patients, 29% of all features (378/1322) were stable in test-retest ADC maps (Table 1). Of the unfiltered features, most stable features were related to geometric shape (22/24, 92%) and texture (37/99, 37%). A comprehensive list of all features is presented as supplementary information. After wavelet decomposition, 144 additional radiomics features were calculated for all 8 wavelet-filters, resulting in 27% (312/1152) of filtered features being stable. In 17 colorectal liver metastases from 17 patients, 25% of all extracted radiomics features (330/1322) demonstrated CCC-values greater than 0.85 in 1.5 T ADC-maps. Of these stable features, 36% (61/170) of unfiltered and 23% (269/1152) of wavelet-filtered features were stable in colorectal liver metastases. For 22 lung cancer lesions from 19 patients, 25% of all features (330/1322) showed stability matching our specified threshold. In contrast to colorectal liver metastases, only 16% (27/170) of unfiltered radiomics signatures were stable, while the percentage of stable wavelet-filtered features were 26% (303/1152).
Table 1.
Stable features in ADC maps acquired at 1.5 T over different tumour-entities (i.e., 20 ovarian cancer lesions, 17 colorectal liver metastases, 22 lung cancer lesions).
| Tumour type (1.5 T) | ADC Mean ± SD (10−6 mm2/s) | Stable features (CCC > 0.85) | ||||
|---|---|---|---|---|---|---|
| Unfiltered | Wavelet filtered | ALL (unfiltered + wavelet) | ||||
| Intensity | Shape | Texture | ||||
| Ovarian | 1086.2 ± 191.9 | 7/47 | 22/24 | 37/99 | 312/1152 | 378/1322 (29%) |
| Colorectal Liver | 979.2 ± 420.9 | 8/47 | 20/24 | 33/99 | 269/1152 | 330/1322 (25%) |
| Lung | 1340.2 ± 412.5 | 1/47 | 13/24 | 13/99 | 303/1152 | 330/1322 (25%) |
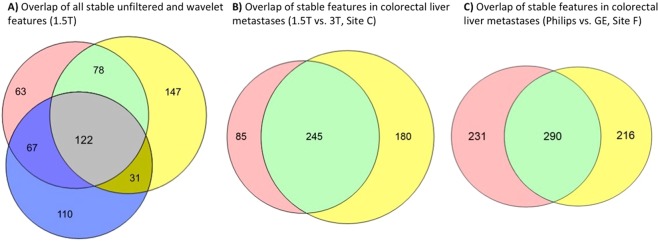
122 features (23 unfiltered and 99 wavelet-filtered) were regarded as stable in all three tumour entities and 298 features (49 unfiltered and 249 wavelet-filtered) were stable for at least 2 tumour-types on 1.5 T ADC maps (Fig. 3A, Supplementary Fig. S1). Statistically significant differences in CCC from all features (unfiltered and wavelet-filtered) were found between ovarian tumours and colorectal liver metastases (P < 0.0001), and between colorectal liver metastases and lung cancer (P = 0.0051) but not between ovarian and lung cancer (P = 0.56).
Figure 3.
Overlapping results in feature stablility extracted from 1.5 T-MR images of (A) all tumour-entities (i.e., colorectal liver metastases (red), ovarian cancer (yellow), and lung cancer (blue)); derived from MR images of colorectal liver metastases (B) acquired at 1.5 T (red) and 3 T (yellow); and obtained from 3 T-MR images of colorectal liver metastases (C) acquired on a Philips Ingenia (red) and GE Discovery (yellow).
Magnetic field strength differences
The effects of magnetic field strength differences were analysed in ADC-maps acquired at 1.5 T and at 3 T in 17 and 13 patients with colorectal liver metastases, respectively. On ADC maps acquired at 3 T, 32% (425/1322) of radiomics features were stable over 13 segmented lesions from test-retest scans (Table 2). These consisted of 71/170 unfiltered features (i.e., 13/24 (54%) geometric shape features, 44/99 (44%) texture features, and 14/47 (30%) intensity features) and 355/1152 (29%) of wavelet-filtered features.
Table 2.
Test-retest feature stability of colorectal liver metastases measured on 1.5 T (n = 17) and 3 T ADC maps (n = 13).
| Magnetic field (Colorectal Liver) | ADC Mean ± SD (10−6 mm2/s) | ADC Median (10−6 mm2/s) | Stable features (CCC > 0.85) | ||
|---|---|---|---|---|---|
| Unfiltered | Wavelet filtered | ALL (unfiltered + wavelet) | |||
| 1.5 T | 979.2 ± 420.9 | 953.4 | 61/170 | 269/1152 | 330/1322 (25%) |
| 3 T | 1353.3 ± 409.8 | 1202.6 | 71/170 | 355/1152 | 425/1322 (32%) |
No statistically significant differences were found in stability between features extracted from ADC maps acquired at 1.5 T and 3 T (P = 0.51). Correspondingly, 245 extracted features (42 unfiltered and 204 filtered) were shown to be stable in both populations, regardless of magnetic field strength (Fig. 3B, Supplementary Fig. S2). Furthermore, comparable mean and median ADC values were derived from both 1.5 T- and 3 T-ADC maps of the entire cohort (Table 2).
Cross-vendor differences
Vendor-specific subgroups were analysed within the dataset of colorectal liver metastases acquired at 3 T (site F). As shown in Table 3, feature stability of ADC maps acquired on a Philips Ingenia and a GE Discovery MR-system presented CCC-values > 0.85 for 521/1322 (39%) and 506/1322 (38%) features, respectively. No statistically significant differences were found in the number of features that exceeded the CCC-threshold (P = 0.49). However, 290 features (79 unfiltered and 211 filtered) presented high stability across both vendors’ test-retest data (Fig. 3C, Supplementary Fig. S3). Furthermore, 154 features (34 unfiltered, 120 filtered) were stable in both cross-vendor and in both 1.5 T and 3 T datasets.
Table 3.
Test-retest feature stability of colorectal liver metastases measured on 3 T ADC maps acquired on a Philips Ingenia (n = 10) and GE Discovery (n = 8) MR systems at the same clinical centre.
| Site F (Colorectal Liver, 3 T) | ADC Mean ± SD (10−6 mm2/s) | ADC Median (10−6 mm2/s) | Stable features (CCC > 0.85) | ||
|---|---|---|---|---|---|
| Unfiltered | Wavelet filtered | ALL (unfiltered + wavelet) | |||
| Philips | 1237.9 ± 324.4 | 1129.8 | 106/170 | 415/1152 | 521/1322 (39%) |
| GE | 1752.3 ± 395.6 | 1882.9 | 100/170 | 406/1152 | 506/1322 (38%) |
A detailed description of stable features is described in Supplementary Tables S1-S4 for each subgroup and listed as supplementary information (online-only).
Correlations between features and tumour volume
Across all tissue types and centres, the mean absolute of Spearman’s r (|r|) was 0.34 ± 0.23. The distribution of Spearman’s r values for all features can be seen in Supplementary Fig. 4. A total of 73 features were found to correlate highly with tumour volume (|r| > 0.8), 10 of which were shape features such as maximum diameter and surface, and the remaining 63 were made up of 7 texture features and their 56 wavelet filtered equivalents. A further 137 features correlate strongly (|r| > 0.6) with volume, of which 11 are texture features, two intensity histogram features, and one is a statistical feature, their associated 112 wavelet features, an additional shape feature, and 10 further wavelets of texture features. For all unfiltered features, Supplementary Fig. 5 shows the correlation between |r| and CCC for all tumour sites while Supplementary Fig. 6 details the strength of the correlations with tumour volume.
Discussion
Within a multicentre trial, we present a method of data analysis to evaluate the stability of radiomics features derived from parametric MRI. This approach has shown that a substantial fraction of ADC-based radiomics features (25–29%) presented test-retest stability over a variety of tissues, MR-systems, and vendors. In addition, 122 features were stable over all tissues and could be regarded to be independent of tumour origin. These results regarding radiomics feature stability are in line with studies that analysed repeatability of absolute ADC values, a correlation that could be attributed to the low coefficient of variance (CoV) presented in these studies5,18,19. The methodology for stable feature selection and volume correlation presented in this analysis, together with the list of repeatable ADC radiomics features, facilitate the development of reliable MR-based radiomics signatures, and future clinical implementation across multiple centres. We therefore postulate that this method of analysis can be generalized to a larger field of quantifiable MR imaging features.
Shape features describe the volume contained within the segmentation, rendering it volume-dependent. Mismatch in shape features between test and retest scans could thus be attributed to differences in tumour segmentation and inter-observer variability although the same observer outlined the test-retest data in this study20. Especially in lung cancer, accurate tumour segmentation is complicated by respiratory motion and motion-related MR artefacts which vary between test-retest studies. Since DWI protocols in the lung were acquired during free-breathing, low stability of shape features is expected. However, these results were also present in radiomics analyses of CT-images where the acquisition is done in breath-hold17. Tumorous lesions in the liver and ovaries are less subjected to respiratory motion and as expected produced shape features with higher stability.
Texture features describe the pattern distribution of the voxels and quantify intra-tumour heterogeneity in all three dimensions21. As it takes into consideration the spatial relation to nearby voxels, stability across test-retest were similar to those of the shape features. In lung cancer, MR-related susceptibility artefacts are more common in DWI with distortions at the boundaries of tumour and air-filled lung tissue11. Hard transitions between tumours and normal lung tissue further complicate tumour segmentation as small delineation differences could have large impact on ADC measurements20. Nevertheless, radiomics features derived from lung cancer ADC maps achieved comparable reproducibility after wavelet-filtering, which can alleviate boundary inhomogeneity, justifying the use of ADC-based radiomics in multi-centre trials in lung cancer.
Intensity features in lung cancer also showed more variance compared to colorectal liver metastases and ovarian cancer. As the DWI protocol in lung cancer was acquired in free-breathing, variation in partial volume effects during test-retest are very likely to have been responsible for the high variation of the intensity features. There are several other reasons for variability in intensity feature stability in MRI. Unlike CT, variations in signal-intensity result from differences in RF coil sensitivity and coil placement during patient repositioning. Despite attempting to mitigate this by using ADC rather than absolute values of signal-intensity on high b-value images, intensity-features generally showed low stability. Although no treatment was administered to our patients between test-retest scans, eliminating the possibility of treatment-related physiological effects3,22, inflammatory processes associated with the tumour (e.g., lung-cancer atelectasis) and small molecular fluctuations of thermal diffusion, may also have affected the stability of the ADC intensity features23.
Stability of radiomics features was unaffected by differences in magnetic field strength, matching the field-independent nature of ADC3. In the literature, no significant difference has been reported between 1.5 T and 3 T ADC values measured in multiple organs24,25. However, higher mean CoVs have been reported in 3 T abdominal ADC24. Potentially, this variability is associated with increased difficulty in maintaining homogenous excitation pulses and gradient linearity at higher magnetic fields, and to the presence of artefacts related to magnetic susceptibility and eddy currents25.
Surprisingly, no statistically significant differences in feature stability were found between vendors, despite potential technical differences (e.g., imaging algorithms, shimming, fat suppression, and ADC reconstruction) (P = 0.49)23,26. Previously, studies have shown low longitudinal inter-vendor ADC variability when evaluating an ice/water phantom (CoV = 1–4%) and cancer patients (CoV ≤ 10%)16,27,28. High repeatability and low inter-scanner variation of ADC measurements could have a direct positive effect on radiomics feature stability.
Radiomics features derived from CT-images have been shown to have a prognostic value9. For CT, these included tumour intensity (‘energy’), texture (‘Grey-level non-uniformity’), wavelet (Grey-level non-uniformity HLH’) and shape (‘compactness’). These features were also highly stable in all tumour types in our data with CCC-values ranging from 0.89–0.99 (Fig. 1). This opens the possibility that similar radiomics features derived from ADC data might be useful in model-building, or in complementing the currently used method of detecting treatment-related changes by analysing absolute ADC metrics (i.e., histogram analysis of centiles, kurtosis and skewness)18.
Radiomics studies in CT and positron emission tomography have suggested that some reproducible features could be a surrogate of tumour volume29,30. This appears to hold true for some but certainly not the majority of the stable features presented in this research, and knowledge of which features correlate highly with volume is important for any radiomics study moving forward. Since radiomics should be purely quantitative imaging, no prognostic or diagnostic features should be excluded a priori, including simple ones such as volume or those that correlate with it.
Our analysis methods and underpinning study set-up had some limitations. Although efforts were made to minimise sources of variability by using a quality-assured standardized protocol and excluding b-values below 100 mm/s2 from ADC reconstruction to reduce perfusion-related MR-signal, the use of a standardised protocol within a multi-site study does not permit optimization of data from individual MR-systems26. Furthermore, DWI protocols did not include respiratory triggering or motion correction. Motion artefacts, predominantly seen in lung cancer patients and patients with colorectal liver metastases (site C, 3 T), had an adverse effect on ADC feature stability. However, for the colorectal liver metastases data acquired at 3 T, the protocol was adjusted from the one specified at 1.5 T to avoid specific image-artefacts24. For example, a larger bandwidth of 1500–2650 Hz/pixel was set to minimise geometric image distortion. In addition to standardised DWI acquisition, we did not specifically reduce ADC variability through post-processing to further improve the stability of radiomics features31. In a recent multi-centre study by Pathak et al., the percentage change in test-retest ADC measurements decreased from 21.1% to 2.7% in colorectal liver metastases using a standardization-strategy to account for measurement uncertainty (i.e., error modelling). This type of approach has the potential to further improve the stability of radiomics features. In addition, radiomics stability could benefit from improvements in tumour segmentation (i.e., reducing inter- and intra-observer variability) and image quality (i.e., increased signal-to-noise ratio and reduced image artefacts)5,6,20,31. Although we controlled for observer segmentation (same observer segmented each test-retest), image reconstruction and processing algorithms varied between centres and scanner vendors. Neither the assessment of differences between medical centres nor the number of reproducible features as a function of CCC cut-off (as performed in17) were included in this study owing to the small number of patients from each site and should be addressed in future work. However, good agreement in ADC-measurements between centres previously has been reported27. Also, image pre-processing can be regarded as another variable in the feature extraction workflow, and as such is also able to influence feature variability, so minimal pre-processing was performed, using common values from radiomics literature. Performing test-retest experiments are crucial in order to ensure that only stable features are selected for meaningful analysis and inclusion of parametric MR-radiomics as a clinical tool15.
In conclusion, we have presented the assessment of stability of radiomics features from parametric ADC based on standardized test-retest measurements. This methodology enables selection of stable features that quantitatively represent phenotypic features and enables the exciting use of high-quality radiomics analysis to attain reliable biomarkers complementary to other clinical/imaging data. As extracted ADC-based radiomics features are stable across multiple centres, tumour types, 1.5 T-3 T systems, and MR-vendors, this analysis can be widely included in multicentre trials. The implementation of such quantitative analysis of tumour phenotype will facilitate the development of diagnostic and theragnostic models that could help detect cancers earlier than the current standard, predict early treatment response and improve treatment decision-making towards personalized healthcare.
Methods
Patient population
As part of prospective clinical trials to qualify ADC imaging biomarkers and stability performed by the Quantitative Imaging in Cancer: Connecting Cellular Processes with Therapy (QuICConCePT) consortium (lung and liver) and the Cancer Research UK (ovary), sixty-one patients were included from 6 university hospitals across the UK, Italy, France, and the Netherlands (site A-C, and E-G). Included patients were diagnosed with either lung cancer stage III18, ovarian cancer (Winfield et al., personal communication), or colorectal liver metastases32, and had a minimum of two imaging sessions maximally 7 days apart before start of treatment. Patient cohorts are summarized in Table 4. This study was approved by the institutional medical ethics committee of each centre (Medical Ethical Committee VU University Medical Centre, Ethics Committee Humanitas Milan, INSERM Ethics Committee, University Research Ethics Committee at the University of Manchester, Commissie Mensgebonden Onderzoek regio Arnhem – Nijmegen (WMO) at the Radboud University Medical Centre Nijmegen, and the Research Ethics Committee for The Royal Marsden Hospital Sutton). Formal written informed consent was recorded for each participant and all data analyses were compliant to the Medical Research Involving Human Subjects Act (WMO).
Table 4.
Main patient cohort characteristics.
| Tumour site | Tumour type | Number | Age range | Treatment received |
|---|---|---|---|---|
| Lung | NSCLC/metastases | 19 | 41–86 | 5 naïve, 14 previously treated |
| Liver | Colorectal metastases | 30 | 44–77 | No treatment within 6 months |
| Ovary | High grade serous | 12 | 31–77 | Naïve |
Image acquisition
Patients were scanned twice within 7 days before the start of treatment. In total, DWIs were acquired on 4 different MRI systems of 1.5 Tesla (T) and 3 different MRI systems of 3 T using a common scan protocol per tumour region (Table 5): on 1.5 T - GE Signa HDxt (site A), Philips Achieva DS (site B,G) Siemens Magnetom Avanto (site C,E), and on 3 T Siemens Magnetom Trio Tim (site C), Philips Ingenia (site F), and GE Discovery 750 w (site F). The applied MR-protocol was comprised of T1/T2-weighted images for anatomical imaging and diffusion-weighted sequences with three b-values. For DWI of each tumour-type, a common and quality-assured protocol was applied by all centres27 (Table 5). ADC maps were constructed by mono-exponential linear fitting of diffusion data. Images with b-values smaller than 100 s/mm2 were excluded to minimize components of blood perfusion in parametric ADC maps.
Table 5.
Diffusion-weighted MR scan protocol. (*) Philips Ingenia and GE Discovery.
| Lung Cancer (site A, B, E, G) | Colorectal liver metastases (1.5 T) (site A, B, C, E) | Colorectal liver metastases (3 T) (site C, F*) | Ovarian cancer (site E) | |
|---|---|---|---|---|
| Sequence | ss-EPI | ss-EPI | ss-EPI | ss-EPI |
| TR (ms) | ≥8000 | ≥8000 | 5000 | ≥8000 |
| TE (ms) | minimum | minimum | minimum | minimum |
| NSA | 4 | 4 | 2–4 | 4 |
| FOV (mm2) | 380 × 273 | 380 × 380 | 380 × 273 | 332 × 380 |
| Matrix | 128 × 112 | 128 × 128 | 128 × 128 | 128 × 112 |
| Bandwidth (Hz/px) | 1400–1800 | 1400–1800 | 1500–2650 | 1400–1800 |
| Slice thickness (mm) | 5 | 5 | 5–6 | 6 |
| Slice gap (mm) | 0 | 0 | 0 | 0 |
| Pixel size (mm2) | 3 × 3 | 1.5 × 1.5 | 1.5 × 1.5 | 1.5 × 1.5 |
| b-values | 100, 500, 800 | 100, 500, 900 | 150, 400, 800 | 100, 500, 900 |
| Fat saturation | yes | yes | yes | yes |
| Parallel imaging | yes | yes | yes | yes |
Segmentation
Volumes-of-interest (VOI) were manually delineated over all primary tumours and metastases on DWI images with high b-value. The gross tumour volume (GTV) was determined at central review by experienced radiologists (with a minimum of 2 years experience) using all diagnostic information available, and saved as binary masks. For each patient, the same observer segmented the same lesion in both test-retest images, while at the same time making sure that no large anatomical variations occurred. After voxel-wise rigid registration, tumour delineations on DWI were transferred to corresponding ADC maps. If ≥2 tumours were present, the 2 largest lesions were delineated while excluding cystic or necrotic regions from segmentations. The same lesion was delineated separately on retest-data while blinded from test-data. In total, 72 lesions were included for analysis. All segmentations were performed using OsiriX (Pixmeo SARL, Bernex, SUI), Mirada RX (Mirada Medical, Oxford, UK), or Adept (in-house software, Institute of Cancer Research, London, UK).
Feature extraction
Radiomics features were retrospectively extracted from each VOI in the test-retest ADC dataset. ADC maps were pre-processed in two steps: (1) in order to reduce image noise and grey-level matrix (GLM) size, images were rescaled using a bin-size of 25 grey levels; (2) in order to standardise voxel size across all datasets, images were rescaled using a linearly resampled into isotropic voxel-sizes of 3 × 3 × 3 mm3,33. A total of 1322 radiomics features were obtained using an in-house developed software-toolbox in MATLAB 2014a (Mathworks, Natick, USA)21. These features included signal intensity features (n = 47), geometric features (n = 24), and texture features (n = 99), which respectively described the histogram-distribution of voxel intensity-values (i.e, first-order grey-level statistics, local intensity (Locint), and intensity histogram (IH) features), the 3D shape of delineated volumes, and the spatial distribution of fractal dimensions and voxel intensities using 6 texture matrices (i.e., grey-level co-occurrence (GLCM, 26 features)34, grey-level distance-zone (GLDZM, 16 features)35, grey-level run-length (GLRLM, 16 features)5,36, grey-level size-zone (GLSZM, 16 features)37, neighbouring grey-level dependence (NGLDM, 17 features)38, and neighbourhood grey-tone difference matrix (NGTDM, 5 features)39. Furthermore, 3D wavelet decompositions of the original image resulted in additional 1152 features focusing on different spatial frequency ranges within tumour values9.
A mathematical description of all features was previously published in9,21,40 and were presented as supplemental material with permission of the corresponding authors. Most features used in this study are in compliance with feature definitions as described by the Imaging Biomarker Standardization Initiative (IBSI). Where features differ, a note has been added specifying the difference.
Statistical analysis
To select stable radiomics features, the pairwise concordance correlation coefficient (CCC) was calculated between data derived from the test and retest ADC images41. CCC-values range from −1 to +1 and describe the negative or positive agreement between 2 datasets. Based on previous work, features with a minimum CCC of 0.85 were regarded as statistically stable and robust17,40,42. Stability is defined as the closeness of agreement between measured quantity values obtained by replicate measurements performed under the same conditions (e.g., patient, scanner, imaging protocol)18. Statistical differences in stable features between tumour types, and between MR-systems with different magnetic field strengths were tested using Kruskal-Wallis 1-way ANOVA with Dunn’s correction for multiple testing. Differences between MR-systems from different vendors were tested for statistical significance using a Mann-Whitney test. All statistical analyses were performed using GraphPad Prism version 6.01 (GraphPad, USA). P-values < 0.05 were considered statistically significant.
Feature correlation with tumour volume
Features with a constant value (or near-zero variance) across all images in the test dataset were excluded, and the remainder were examined for correlations with the tumour volume using Spearman’s rho statistic to estimate a rank-based measure of association. The Spearman coefficients of all unfiltered features were plotted against the feature stability as measured by the CCC for all tumour types and field strengths.
Supplementary information
Acknowledgements
Authors acknowledge the contribution of data from the QuiC-ConCePT partners at VUMC, Amsterdam, Humanitas Milan, INSERM Paris, University of Manchester, Nijmegen Medical Center and The Royal Marsden Hospital, Sutton UK. Particular thanks to Prof S. Stroobants, Prof A. Chiti for their support and to P.G. Arteiser, J. Wakefield, E. Pace, A. Weller for their help with patient recruitment and region-of interest selection. Special thanks to J.E. van Timmeren, R.T.H. Leijenaar, R.T.H.M. Larue, A. Jochems for their insight in radiomics and statistical analysis. This research received financial support from ERC advanced grant (ERC-ADG-2015, n° 694812 - Hypoximmuno) and the QuIC-ConCePT project, which is partly funded by EFPI A companies and the Innovative Medicine Initiative Joint Undertaking (IMI JU) under Grant Agreement No. 115151. This research is also supported by the Dutch technology Foundation STW (grant n° 10696 DuCAT & n° P14–19 Radiomics STRaTegy), which is the applied science division of NWO, and the Technology Programme of the Ministry of Economic Affairs. Authors also acknowledge financial support from the EU 7th framework program (ARTFORCE - n° 257144, REQUITE - n° 601826), SME Phase 2 (RAIL - n°673780), EUROSTARS (SeDI, CloudAtlas, DART, DECIDE, COMPACT), the European Program H2020-2015-17 (BD2Decide - PHC30-689715, ImmunoSABR - n° 733008 and PREDICT - ITN - n° 766276), Interreg V-A Euregio Meuse-Rhine (“Euradiomics”). We acknowledge funding from Cancer Research UK BIDD grant C1353/A12762 to the Cancer Imaging Centre at the Institute of Cancer Research.We acknowledge funding from Cancer Research UK BIDD grant C8742/A18097 to the Cancer Imaging Centre at Cambridge and Manchester.
Author Contributions
We are submitting original research and declare that all named authors have read the manuscript and have agreed to submit it in the present form. All named authors have made a sufficient contribution to the work. J.P. and H.C.W. wrote the manuscript, which was iterated intensely by all authors. J.P. prepared all figures and tables, except for supplementary figures 4 and 5, which were prepared by H.C.W. Table 5 was made with the help of J.M.W. Image acquisition was performed by J.M.W., B.E.V.B., A.H. and A.J. Image analyses were performed by J.P., H.C.W. and A.I. Statistical analyses were performed by J.P. and H.C.W. The multi-center collaboration was facilitated by the efforts of J.E.W., F.M.M., N.M.d.S. and P.L. Both senior authors, N.M.d.S. and P.L., contributed equally and provided overall supervision and invaluable guidance.
Competing Interests
Dr. Philippe Lambin reports, within and outside the submitted work, grants/sponsored research agreements from Varian medical, Oncoradiomics, ptTheragnostic, Health Innovation Ventures and DualTpharma. He received an advisor/presenter fee and/or reimbursement of travel costs/external grant writing fee and/or in kind manpower contribution from Oncoradiomics, BHV, Merck and Convert pharmaceuticals. Dr. Lambin has shares in the company Oncoradiomics SA and Convert pharmaceuticals SA and is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248, PCT/NL2014/050728) licensed to Oncoradiomics and one issue patent on mtDNA (PCT/EP2014/059089) licensed to ptTheragnostic/DNAmito, three non-patentable invention (softwares) licensed to ptTheragnostic/DNAmito, Oncoradiomics and Health Innovation Ventures. Dr. Woodruff has (minority) shares in the company Oncoradiomics. Dr. Wildberger reports institutional grants from Agfa, Bard, Bayer, GE, Optimed, Philips, Siemens and personal fees (Speaker’s Bureau) from Bayer and Siemens outside the submitted work.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nandita M. DeSouza and Philippe Lambin contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41344-5.
References
- 1.Peerlings J, et al. The Diagnostic Value of MR Imaging in Determining the Lymph Node Status of Patients with Non-Small Cell Lung Cancer: A Meta-Analysis. Radiology. 2016;281:86–98. doi: 10.1148/radiol.2016151631. [DOI] [PubMed] [Google Scholar]
- 2.Pollard JM, Wen Z, Sadagopan R, Wang J, Ibbott GS. The future of image-guided radiotherapy will be MR guided. The British journal of radiology. 2017;90:20160667. doi: 10.1259/bjr.20160667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koh DM, Collins DJ. Diffusion-weighted MRI in the body: applications and challenges in oncology. AJR. American journal of roentgenology. 2007;188:1622–1635. doi: 10.2214/ajr.06.1403. [DOI] [PubMed] [Google Scholar]
- 4.Padhani AR, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia (New York, N.Y.) 2009;11:102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pathak R, et al. A data-driven statistical model that estimates measurement uncertainty improves interpretation of ADC reproducibility: a multi-site study of liver metastases. Scientific reports. 2017;7:14084. doi: 10.1038/s41598-017-14625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deSouza NM, et al. Implementing diffusion-weighted MRI for body imaging in prospective multicentre trials: current considerations and future perspectives. European radiology. 2017 doi: 10.1007/s00330-017-4972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinkus R, Van Beers BE, Vilgrain V, DeSouza N, Waterton JC. Apparent diffusion coefficient from magnetic resonance imaging as a biomarker in oncology drug development. Eur J Cancer. 2012;48:425–431. doi: 10.1016/j.ejca.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 8.van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P. Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncol. 2017;56:1537–1543. doi: 10.1080/0284186x.2017.1350285. [DOI] [PubMed] [Google Scholar]
- 9.Aerts HJ, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Timmeren JE, et al. Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother Oncol. 2017;123:363–369. doi: 10.1016/j.radonc.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Bowen, S. R. et al. Tumor radiomic heterogeneity: Multiparametric functional imaging to characterize variability and predict response following cervical cancer radiation therapy. Journal of magnetic resonance imaging: JMRI, 10.1002/jmri.25874 (2017). [DOI] [PMC free article] [PubMed]
- 12.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150–166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fave X, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys. 2015;42:6784–6797. doi: 10.1118/1.4934826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambin P, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 16.Winfield JM, et al. A framework for optimization of diffusion-weighted MRI protocols for large field-of-view abdominal-pelvic imaging in multicenter studies. Medical physics. 2016;43:95. doi: 10.1118/1.4937789. [DOI] [PubMed] [Google Scholar]
- 17.Larue R, et al. 4DCT imaging to assess radiomics feature stability: An investigation for thoracic cancers. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2017;125:147–153. doi: 10.1016/j.radonc.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Weller A, et al. Diffusion-weighted (DW) MRI in lung cancers: ADC test-retest repeatability. European radiology. 2017;27:4552–4562. doi: 10.1007/s00330-017-4828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winfield JM, et al. Modelling DW-MRI data from primary and metastatic ovarian tumours. European radiology. 2015;25:2033–2040. doi: 10.1007/s00330-014-3573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leijenaar RT, et al. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52:1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambin P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padhani AR, Koh DM, Collins DJ. Whole-body diffusion-weighted MR imaging in cancer: current status and research directions. Radiology. 2011;261:700–718. doi: 10.1148/radiol.11110474. [DOI] [PubMed] [Google Scholar]
- 23.Koh DM, et al. Whole-body diffusion-weighted MRI: tips, tricks, and pitfalls. AJR. American journal of roentgenology. 2012;199:252–262. doi: 10.2214/ajr.11.7866. [DOI] [PubMed] [Google Scholar]
- 24.Rosenkrantz AB, Oei M, Babb JS, Niver BE, Taouli B. Diffusion-weighted imaging of the abdomen at 3.0 Tesla: image quality and apparent diffusion coefficient reproducibility compared with 1.5 Tesla. Journal of magnetic resonance imaging: JMRI. 2011;33:128–135. doi: 10.1002/jmri.22395. [DOI] [PubMed] [Google Scholar]
- 25.Donati OF, et al. Diffusion-weighted MR imaging of upper abdominal organs: field strength and intervendor variability of apparent diffusion coefficients. Radiology. 2014;270:454–463. doi: 10.1148/radiol.13130819. [DOI] [PubMed] [Google Scholar]
- 26.Taouli B, et al. Diffusion-weighted imaging outside the brain: Consensus statement from an ISMRM-sponsored workshop. Journal of magnetic resonance imaging: JMRI. 2016;44:521–540. doi: 10.1002/jmri.25196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winfield JM, et al. Development of a diffusion-weighted MRI protocol for multicentre abdominal imaging and evaluation of the effects of fasting on measurement of apparent diffusion coefficients (ADCs) in healthy liver. The British journal of radiology. 2015;88:20140717. doi: 10.1259/bjr.20140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winfield JM, et al. Extracranial Soft-Tissue Tumors: Repeatability of Apparent Diffusion Coefficient Estimates from Diffusion-weighted MR Imaging. Radiology. 2017;284:88–99. doi: 10.1148/radiol.2017161965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch ML, et al. Vulnerabilities of radiomic signature development: The need for safeguards. Radiotherapy and Oncology. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 30.Orlhac F, et al. Tumor Texture Analysis in 18F-FDG PET: Relationships Between Texture Parameters, Histogram Indices, Standardized Uptake Values, Metabolic Volumes, and Total Lesion Glycolysis. Journal of Nuclear Medicine. 2014;55:414–422. doi: 10.2967/jnumed.113.129858. [DOI] [PubMed] [Google Scholar]
- 31.Guyader JM, et al. Influence of image registration on apparent diffusion coefficient images computed from free-breathing diffusion MR images of the abdomen. Journal of magnetic resonance imaging: JMRI. 2015;42:315–330. doi: 10.1002/jmri.24792. [DOI] [PubMed] [Google Scholar]
- 32.Kumar V, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafiq-Ul-Hassan M, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics SMC- 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 35.Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng. 2014;61:630–637. doi: 10.1109/TBME.2013.2284600. [DOI] [PubMed] [Google Scholar]
- 36.Galloway MM. Texture analysis using gray level run lengths. Computer Graphics and Image Processing. 1975;4:172–179. doi: 10.1016/s0146-664x(75)80008-6. [DOI] [Google Scholar]
- 37.Thibault, G. et al. Texture indexes and gray level size zone matrix application to cell nuclei classification. Pattern Recognit Inf Process, 140–145 (2009).
- 38.Sun C, Wee WG. Neighboring gray level dependence matrix for texture classification. Computer Vision, Graphics, and Image Processing. 1983;23:341–352. doi: 10.1016/0734-189X(83)90032-4. [DOI] [Google Scholar]
- 39.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics. 1989;19:1264–1274. doi: 10.1109/21.44046. [DOI] [Google Scholar]
- 40.van Timmeren JE, et al. Test-Retest Data for Radiomics Feature Stability. Analysis: Generalizable or Study-Specific? Tomography. 2016;2:361–365. doi: 10.18383/j.tom.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 42.Zhao B, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.