Abstract
Nicotinamide adenine dinucleotide (reduced form: NADH) serves as a vital redox-energy currency for reduction-oxidation homeostasis and fulfilling energetic demands. While NADH exists as free and bound forms, only free NADH is utilized for complex I to power oxidative phosphorylation, especially important in neurons. Here, we studied how much free NADH remains available for energy production in mitochondria of old living neurons. We hypothesize that free NADH in neurons from old mice is lower than the levels in young mice and even lower in neurons from the 3xTg-AD Alzheimer’s disease (AD) mouse model. To assess free NADH, we used lifetime imaging of NADH autofluorescence with 2-photon excitation to be able to resolve the pool of NADH in mitochondria, cytoplasm, and nuclei. Primary neurons from old mice were characterized by a lower free/bound NADH ratio than young neurons from both non-transgenic (NTg) and more so in 3xTg-AD mice. Mitochondrial compartments maintained 26 to 41% more reducing NADH redox state than cytoplasm for each age, genotype, and sex. Aging diminished the mitochondrial free NADH concentration in NTg neurons by 43% and in 3xTg-AD by 50%. The lower free NADH with age suggests a decline in capacity to regenerate free NADH for energetic supply to power oxidative phosphorylation which further worsens in AD. Applying this non-invasive approach, we showed the most explicit measures yet of bioenergetic deficits in free NADH with aging at the subcellular level in live neurons from in-bred mice and an AD model.
Keywords: NADH, Aging brain, Alzheimer’s disease, FLIM, Mitochondria, Redox states
Introduction
Aging is generally accompanied by progressive loss in cognitive function (Squier 2001). The Epigenetic Oxidative Redox Shift (EORS) theory of aging proposes that a sedentary low-energy state with aging triggers an oxidative shift and mitochondrial dysfunction and further causes metabolic disturbances (Brewer 2010). Indeed, aging increases the risk for neurodegenerative diseases, including Alzheimer’s disease (AD). A 2016 report projects that by 2050, the number of people above 65 years of age with AD will grow from 5.4 million to 13.8 million (Alzheimer’s Association 2016). Aging and AD share some common characteristics such as oxidative stress (Zhu et al. 2015), mitochondrial impairment (Lin and Beal 2006), and bioenergetic deficits (Parihar et al. 2008), which are associated with cognitive deficits or dementia. In the 3xTg-AD mouse model that we used, Fattoretti et al. (2010) reported decreased mitochondrial numeric density (number of mitochondria/μm3 of cytoplasm) in CA1 in 10-month-old mice, but the volume and average length increased. Martins et al. (2017) also found decreased mitochondrial number with age in the hippocampus along with abnormal mitochondrial morphology with elongation, swelling and disorganized cristae (Martins et al. 2017). Manczak and Reddy (2012) found decrements in the motility of mitochondria in the 3xTg-AD model due to changes in Drp1 levels for fission. We propose that these mitochondrial structural changes reflect functional deficits of free NADH as a bioenergetic substrate for oxidative phosphorylation (OXPHOS) that initiates a vicious cycle involving redox imbalance, energetic shortage, and mitochondrial dysfunction.
The brain consumes large amounts of energy to fire action potentials (Attwell and Laughlin 2001). Neurons primarily depend on the energy-transducing capacity of mitochondria to meet redox and phosphorylation demands. Since the reduced form of nicotinamide adenine dinucleotide (NADH) is the major intracellular redox currency and the substrate for generation of ATP as the energy currency via oxidative phosphorylation (OXPHOS) in mitochondria, mitochondrial NADH availability to the electron transport chain (ETC) becomes a critical issue (Brewer 2010). NADH functions as a pivotal metabolite that bridges redox states, metabolic pathways, and energy (ATP). Neurons respond to higher energy demands by increasing free NADH production in the TCA cycle as electron donors to feed the electron transport chain (ETC) for ATP generation. We hypothesized that aging brain neurons exhibit an oxidized NADH redox state with decreased free NADH and these free NADH levels are further decremented by the genetic burden of AD-like mutations in 3xTg-AD mouse neurons. Antioxidant defenses also depend on NADH and its reductive conversion to NADPH and GSH. For instance, the forward reaction of nicotinamide nucleotide transhydrogenase (NNT) oxidizes NADH and reduces NADP+ to generate NADPH (Ronchi et al. 2016). However, during oxidative shifts, the backward reaction of NNT drives NADH production at the expense of NADPH (Ronchi et al. 2016). NADPH also donates electrons to GSSG for regeneration of GSH. Evidence for which of these is upstream comes from titrated inhibition of enzymes for their synthesis and supplementation of precursors (Ghosh et al. 2014). From these studies in neurons, NAD(P) H appears upstream of GSH, since decreasing NAD(P) H lowers GSH levels more than the reverse and neuron survival was more dependent on NAD(P) H, especially in neurons from old mice and those from an Alzheimer model mouse brain (Ghosh et al. 2014). Therefore, determination of the availability of free NAD(P) H in mitochondria was evaluated to provide a possible mechanism of age-related and Alzheimer’s associated decrements in energy supply and oxidative redox shifts.
Although NADH and NAD+ are reduced and oxidized forms of nicotinamide adenine dinucleotide, only NADH has intrinsic fluorescence (Chance and Thorell 1959). Different excited state lifetimes of free and bound NADH enable determination of the free NADH available from that bound to enzymes (Lakowicz et al. 1992). The long intrinsic fluorescent lifetime of NADH is due to the protein-bound state (Yu and Heikal 2009; Stringari et al. 2012). The ratio of free to protein-bound NADH correlates with the NADH/NAD+ ratio (Bird et al. 2005). Our previous work measured steady state levels of NAD(P) H without discriminating between free and bound forms (Ghosh et al. 2012, 2014). These studies found age-related and AD-associated declines in NAD(P) H concentrations, NADH regenerating capacity, NAMPT and NNT gene expression for regeneration in neurons from non-transgenic (NTg), and an age-matched triple transgenic Alzheimer’s mouse (3xTg-AD). We can do this because of techniques for isolating neurons from any age animal and culturing them in a common serum-free environment (Brewer 1998; Brewer and Torricelli 2007). These neurons are thus removed from an aging immune system, age-related changes in hormones, and the vasculature. In vitro, we demonstrated the age-related increased susceptibility to biochemical stressors as a critical mechanism in aging (Brewer 1998, 2010). Based on measures in aged rat neurons challenged with glutamate, steady state NADH and glutathione decline with age before declines in ATP (Parihar et al. 2008). Flux control experiments indicated that the often-reported declines in mitochondrial function with age are caused by lower supplies of NADH substrate, rather than problems with the complex I enzyme that uses the NADH to generate a proton gradient across the mitochondrial membrane to power generation of ATP (Jones and Brewer 2010). In mitochondria isolated from 3xTg-AD brains, respiratory capacity was shown to be impaired (Yao et al. 2009). Conventional biochemical approaches performed on the cell lysates as a snapshot of redox states to measure NADH concentrations in cells introduces errors as the NADH is oxidized when exposed to air (Uppal and Gupta 2003; Klaidman et al. 1995) and rapidly consumed by mitochondrial demand for ATP. Further, these studies could not discriminate neuronal from glial mitochondria and did not examine the age-dependence or how the proportion of free to bound NADH dynamically changes in living neurons during aging and AD. Since the distribution of NADH and NAD+ are highly compartmentalized within cells (Xiao et al. 2018), here we determine how aging and AD affect the re-distribution of free to bound NADH ratios. Though intrinsic fluorescence does not discriminate between NADH and NADPH due to identical photophysical properties (Yu and Heikal 2009), the autofluorescence of intracellular NADH levels contributes the majority of the NAD(P) H intensity (Eng et al. 1989). Here, we applied a non-invasive, sensitive method that utilizes the intrinsic NADH fluorescence lifetimes as a natural probe to identify the shifts in free NADH levels, free to bound NADH ratios (Van Munster and Gadella 2005) and re-distribution of NADH among subcellular compartments in NTg and age-matched 3xTg-AD mouse neurons from young to old ages.
Materials and methods
Mouse model
We used LaFerla’s triple transgenic mouse model of AD (3xTg-AD) with human transgenes βAPP (SWE), PS1 (M146V), and Tau (P301L) to mimic the neuropathological features of AD (Oddo et al. 2003). Non-transgenic (NTg) C57BL/6 mice were used as controls. All mice underwent genotyping before using in experiments.
Primary neuron culture
Adult hippocampal neurons were isolated from NTg and 3xTg-AD age-matched mouse brains at young (3–4 months old), middle ages (9–10 months old), and old ages (18, 21, 22, 23 months old) (Brewer and Torricelli 2007). Briefly, hippocampus of each hemisphere were sliced at 0.5 mm and combined in Hibernate A (BrainBits LLC, Springfield, IL), 2% B27 supplement (Invitrogen), and 0.5 mM Glutamax (Invitrogen) for 8 min at 30 °C. The slices were transferred and digested with 2 mg/mL papain (Worthington) in Hibernate without B27 for 25 min at 30 °C. After trituration, neurons were separated from debris and microglia on an Optiprep (Sigma-Aldrich) density gradient. The neuron-enriched fraction was collected and viable neurons counted by exclusion of trypan blue. Neurons were plated at 50,000 cells/cm2 on 15 mm glass coverslips (Assistent; Brand, Carolina Biologicals). Slip were precoated overnight with poly-D-lysine, 100 μg/mL in 18 MΩ deionized water. Neurons were cultured in NbActiv1 (BrainBits) with 5 ng/mL mouse FGF2 and 5 ng/mL mouse PDGFbb (Invitrogen) for trophic support for 9–12 days at 37 °C in 5% CO2, 9% O2 at saturated humidity. Viability in our neuronal cultures was similar to previous studies of neurons from young, middle, and old ages of both genotypes (Ghosh et al. 2012). The neuronal densities of all ages and genotypes in culture were similar without fragmented axons or dendrites. Further, the strong TMRE intensity of mitochondria indicated negative membrane potentials, without perinuclear fragmentation for no obvious age- or AD-related differences or qualitative changes in number of stained mitochondria per neuron. Furthermore, in follow-up experiments, the intracellular NADH levels of old neurons of both genotypes were remarkably responsive to an external imposed reductive and oxidative stress in cultures (Dong and Brewer in preparation).
TMRE staining of mitochondria
To label mitochondria with minimal interference in their membrane potential (Ward et al. 2000), tetramethylrhodamine ethyl ester (TMRE; Molecular Probes) stock solution was made in DMSO at 5 mM. After series dilutions with culture medium (NbActiv1), neurons were incubated with 10 nM TMRE in NbActiv1 for 20 min under 5% CO2 at 37 °C. After incubation, the coverslip was rinsed twice gently with warm NbActiv1. The cell slip was mounted in a slip-holder (Warner Instruments). Experiments were conducted on at least three coverslips from independent cultures with 10–20 neurons for young, middle, and old ages.
FLIM imaging
Fluorescence lifetime images were acquired on a Zeiss LSM 710 microscope (Carl Zeiss, Jena, Germany) using a 63× oil immersion objective, 1.2 N.A. (Carl Zeiss, Oberkochen, German). The environment was controlled at 5% CO2 and 37 °C at saturated humidity while collecting FLIM images. The 2-Photon excitation Titanium:Sapphire MaiTai laser (Spectra-Physics, Mountain View, CA) was modelocked at 740 nm generating ~ 120 fs pulses with a repetition rate of 80 MHz. Image scan speed was 25.21 μs/pixel and images were collected at 256 × 256 pixels. The emission from the excited native NADH in the cultured neurons was filtered with a bandpass at 460/80 nm. FLIM data was acquired using SimFCS32/64 FLIMBox (ISS, Champaign, IL). FLIM lifetime calibration of the system used a Coumarin 6 solution (at 100 μM) with a known lifetime as 2.5 ns. One hundred counts per pixel were collected for one FLIM image of the same field of view.
FLIM phasor data analysis
As described previously (Stringari et al. 2012, 2015), every pixel of the FLIM image is transformed in one pixel in a phasor plot by performing a Fast Fourier Transform (FFT) of the intensity decay I(t). The coordinates g and s in the phasor plot are the real and imaginary part of the FFT as follows (Stringari et al. 2015):
where the intensity signal (I) at indices i and j identify a pixel of the image and ω frequency (ω = 2πƒ), with ƒ the laser repetition rate (80 MHz) and t is the period of the laser, 12.5 ns. Based on the linearity of the phasor coordinates, the g and s position of each pixel represents the fraction of free to bound NADH in the image (Stringari et al. 2015).
We examined regions of interest (ROI) for mitochondrial, cytoplasmic, and nuclear subcellular compartments segmentation using circles. We conducted measurements of g, s, and NADH bound fraction from five ROIs on each compartment of each neuron. We then calculated the average of five ROIs in each compartment per cell and further averaged the values of g, s, and bound fraction of 10 neurons from each mouse (n = 20 neurons/age/genotype for female and n = 10 neurons/age/genotype for male). Free NADH fraction was given by one minus the bound NADH fraction.
Free NADH calibration
β-Nicotinamide adenine dinucleotide (NADH, Sigma-Aldrich, Inc) was prepared fresh at 450 μM in a 0.2 mM Tris-HCl buffer, pH 7.5, and stored at 4 °C for use. As the free NADH solution gradually oxidizes in air, the absolute concentration of the prepared free NADH solution for calibration was determined in a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific Inc). After blanking with 100 mM Tris-HCl pH 7.4, the absorbance of the free NADH was measured at 340 nm wavelength. Calculated the actual free NADH concentration for calibration based on the Beer-Lambert law: A=εbc, where A is the readout absorbance value from NanoDrop; ε is the extinction coefficient of NADH (6.22 at 340 nm); b is the length of light path (1 mm for NanoDrop) and c refers to concentration of NADH in the solution. Each time after imaging neurons, with the laser power and exposure parameters unchanged, we loaded the free NADH solution in the microscope system for absolute free NADH calibration measurements.
Measurements of absolute NADH concentrations in mitochondria (Ma et al. 2016)
Before calibration with free NADH for absolute NADH quantification, we calibrated the instrument response function with a known standard, 100 μM Coumarin 6 with lifetime at 2.5 ns. In the phasor plot, the pure free NADH lifetime is short of 0.4 ns (green cursor in Fig. 1). Whereas, the NADH bound to LDH (lactate dehydrogenase) has a longer lifetime of 3.4 ns (red cursor in Fig. 1). The lifetime distribution of the NADH signal from the cells represents the fractional combination of free and bound NADH along the line between the pure free NADH and bound NADH (green line in Fig. 1). To measure the total NADH in the neurons, we first acquired a FLIM image of a known concentration of free NADH calibrated using the absorbance spectrophotometer. We then corrected for the difference between a lower quantum yield of the free and higher quantum yield of the bound form of NADH as described by Ma et al. (2016). To determine the mitochondrial free NADH concentration, we multiplied the measured total NADH by the corresponding free NADH fraction in mitochondria. Masks of mitochondrial, nuclear, and cytoplasmic regions were made individually.
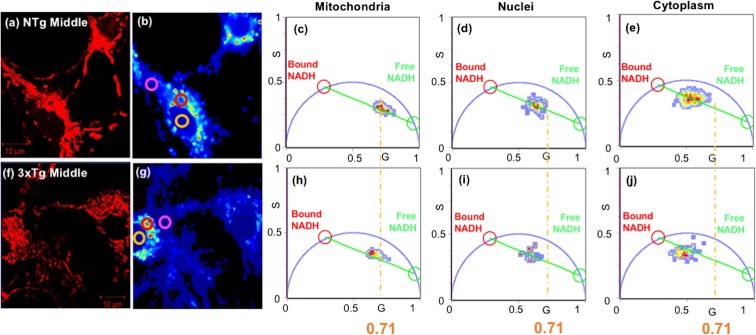
Fig. 1.
Highest free NADH fraction in mitochondria compared to nuclei and cytoplasm in each genotype. AD genotype shows lower free NADH fraction than the corresponding compartment levels of NTg neurons. (a, f) TMRE stain for mitochondrial localization of neurons from (a) NTg and (f) 3xTg-AD 9-month (middle-age) female mouse. (b, g) NADH-FLIM intensity images of the corresponding neurons. To determine the proportion of free and bound NADH in each compartment, ROIs were chosen relative to the TMRE image for analysis on mitochondrial (red circle), nuclear (yellow), and cytoplasmic (pink) compartments. (c–e, h–j) Phasor plots present compartment-specific differences in the ratios of free to bound NADH redox states from the corresponding NTg and 3xTg middle age neurons. Pure, free NADH with lifetime of 0.4 ns is shown by green circle and bound NADH with lifetime of 3.4 ns with a red circle on the semicircle. All the NADH compartmental clusters fall along the lines between pure free NADH (green cursor) and protein-bound NADH (red cursor). The vertical orange lines are centered on the NTg young mitochondrial distribution for reference at G = 0.71
Statistics
Data are presented as means and S.E. One-way or two-way ANOVA was used to assess the difference of means and variances in Excel. The number of replicates is indicated in the legends. The level of significance was set at p < 0.05 to reject the null hypothesis. Multiple comparison ANOVAs were analyzed by ProStat software (Poly Software, Pearl River, NY) using Fisher’s LSD method.
Results
Distribution of free and bound NADH in neuronal mitochondria, cytoplasm, and nucleus
We isolated and cultured neurons in a uniform medium from mice at young, middle, and old ages, removed from a complicated aging context in vivo including age-related inflammation, hormones, and vasculature. Prior studies showed no significant age-related differences in neuronal viability (Patel and Brewer 2003; Ghosh et al. 2012), basal respiration, complex I activity with excess substrate (Jones and Brewer 2010), resting ATP levels (Parihar et al. 2008), or mitochondrial number per cell (Ghosh et al. 2012), but significant age-associated oxidative shifts of steady state NADH/FAD redox ratio after middle age (Ghosh et al. 2012). Better than steady state fluorescence measures of NADH, two-photon lifetime imaging of NADH discriminates between free and bound NADH portions of the total NADH as well as neuronal subcellular distributions in live cells. Mitochondria were localized within neurons by fluorescent staining with tetramethyl rhodamine ethyl ester (TMRE), which partitions into mitochondria by their negative membrane potential (Fig. 1a, f; Ward et al. 2000). In the same field of view, we investigated the distribution of intrinsic NADH of individual neurons by two-photon fluorescence lifetime imaging microscopy (FLIM) (Fig. 1b, g), which show the distribution of two-photon intensities within neuronal sub-regions segregated into mitochondria (red circle ROIs), nuclei with their central circular appearance (yellow circles), and elsewhere in regions lacking mitochondria as cytoplasmic compartments (pink circles). Each pixel in the FLIM intensity image was transformed into phasor space by a Fast Fourier Transform (FFT) to determine the distribution of bound and free forms of NADH within each compartment of middle age neurons from NTg (Fig. 1c–e) and 3xTg-AD mice (Fig. 1h–j) (Stringari et al. 2015). The real and the imaginary parts of the FFT transformation are represented as G and S coordinates in the phasor plots. Each compartmental NADH cluster presented in the phasor plot (Fig. 1c–e, h–j) is composed of a combination of free and bound NADH with lifetimes of 0.4 ns (green circle in the phasor plot) and 3.4 ns (red circle) respectively (Datta et al. 2015). Cellular pixels are distributed along the line joining the pure free and protein-bound NADH (green line). The free/bound NADH ratio serves as an indicator of NADH redox state. A lower free/bound NADH fraction indicates a more oxidized NADH state and a higher free/bound NADH ratio implies a more reduced NADH redox state. Mitochondria distributed closest to the free NADH lifetimes (green cursor) compared to nuclei and cytoplasm, indicating the highest free/bound NADH ratio, highest free NADH fraction and more reductive NADH redox state in the mitochondria. Conversely, the cytoplasm showed the highest fraction of bound NADH with less free NADH, consistent with a more oxidized redox state. A smaller genotype effect was suggested by a small shift toward more bound NADH (red circle) in 3xTg-AD age-matched neurons for each compartment as indicated by the vertical yellow lines, centered on the NTg mitochondria at G = 0.71 (Fig. 1).
Age- and AD-related oxidative shifts from free to more bound NADH
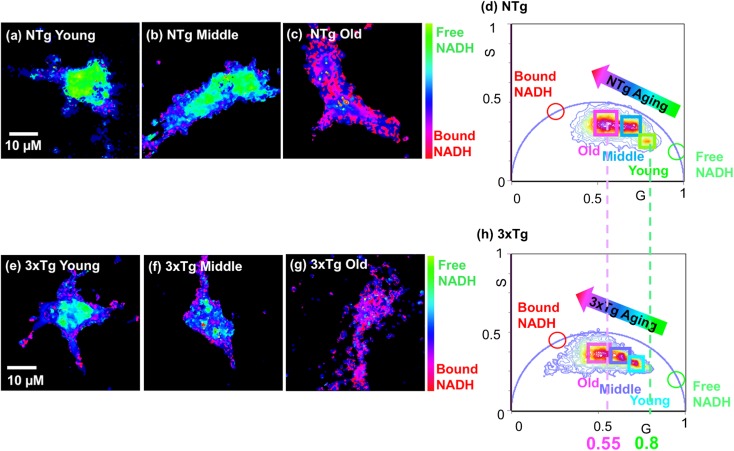
With aging, NADH regenerating capacity declines (Ghosh et al. 2012) which could promote oxidative and energetic shifts. In vivo, NADH exists as either bound to enzymes or free in solution. The free/bound NADH ratio is altered by the cellular redox, metabolic and energetic states. Normally, the free NADH levels maintain balanced between the activity of glycolysis for producing more free NADH and more reliance on oxidative phosphorylation (OXPHOS) for consuming free NADH (Yu and Heikal 2009). To determine whether aging or the 3xTg-AD genotype affects the NADH redox states, we compared free/bound NADH ratios from different age neurons of both genotypes (Fig. 2). To illustrate the distribution of free/bound NADH in FLIM images of neurons, we present NADH color maps of neurons with different ages for NTg (Fig. 2a–c) and 3xTg-AD mouse neurons (Fig. 2e–g) respectively. In these color maps, cyan-green color reflects more free NADH and pink-purple color indicates more bound NADH. With aging, in both NTg (Fig. 2a–c) and 3xTg-AD (Fig. 2e–g) neurons, the higher cyan-green free NADH shifted toward more pink-purple bound NADH, indicating depletion of free NADH and shift toward a more oxidized NADH redox state with age. Furthermore, 3xTg-AD neurons presented comparatively a more pink-purple bound NADH distribution than the age-matched NTg neurons, meaning an even lower free NADH in 3xTg-AD neurons. The free/bound NADH distribution of whole neurons at ages of young, middle, and old were quantitatively transformed into one phasor plot for NTg (Fig. 2d) and 3xTg-AD neurons (Fig. 2h). In genotypes, age drives substantial shifts from free to more bound NADH along the free-bound trajectory. The cluster of young age neuron presents much closer to the free NADH form (green cursor) whereas the cluster of old age neuron displays a shift toward bound NADH (red cursor). This indicates that young age neurons contained the highest free NADH fractions and old-aged neurons contained the most bound NADH with depletion in free NADH. Compared to the clusters of NTg neurons (Fig. 2d), those of the age-matched 3xTg-AD neurons were shifted toward more bound NADH, implying the AD genotype accelerated the age-related shifts toward more bound NADH and depletion of free NADH.
Fig. 2.
Aging promotes shifts toward more bound and less free NADH in NTg and 3xTg-AD neurons. FLIM color maps of young, middle, and old age neurons from NTg (a–c) and 3xTg-AD (e–g) mice illustrate a dramatically age-dependent depletion in free NADH level of both genotypes shown as the color shifts from more green-cyan (free NADH) to more pink-purple (bound NADH) with aging. Comparatively, the 3xTg-AD neurons present more bound NADH than the age-matched NTg neurons seen as a more pink-purple color in the age-matched 3xTg-AD neuron. (d, h) Phasor plots show the shift in distribution from free to bound NADH in neurons from young, middle, and old age neurons of NTg and 3xTg-AD brains, respectively. Each cluster that falls along the free-bound NADH trajectory (line between free and bound NADH) represents the combination of free and bound NADH from the corresponding single neuron in the colormap
Mitochondria show the most reduced state with highest free NADH fraction among subcellular compartments
To statistically validate these results, we selected five regions of interest for each compartment of 10–20 neurons in separate cultures of each age and genotype and sex. We expected compartmental differences in neurons because compartments of HeLa cells measured by thiol/disulfide redox states showed that mitochondria were the most reduced followed by nuclei and cytoplasm, which was comparatively the most oxidized (Hansen et al. 2006). In addition, as the distribution of NADH is highly compartmentalized (Xiao et al. 2018), we further investigated how the free/bound NADH ratios change in mitochondria, cytoplasm, and nuclei with aging of both NTg and 3xTg-AD neurons.
Age effect
Aging strongly drove a decline in the free NADH fractions in all subcellular compartments, p < 0.001, in both genotypes and both genders. In male NTg mouse neurons (Fig. 3a), from young to old age, mitochondrial free NADH fractions declined 23%, nuclear free NADH fractions decreased 12%, and cytoplasmic free NADH levels dropped 23%. Similarly, in female neurons (Fig. 3b), aging drove least decline in nuclear free NADH fractions of NTg female neurons with only 18%, but a 26% drop in mitochondrial free NADH fraction (Fig. 3b). In male and female 3xTg-AD neurons, the slopes of age-associated declines in free NADH fractions present similar changes in percentages to that of gender-matched NTg neurons in each compartment, but further drops in the AD genotype neurons in response to aging. The age-related depletion of free NADH levels in all compartments suggests an impaired capacity for free NADH regeneration and lower capacity for energetic supplement in old-aged neurons of both NTg and 3xTg-AD mouse brains. With aging, the nuclear compartment declined the least compared to that of mitochondria and cytoplasm in both NTg and 3xTg-AD of both genders, indicating a better buffering capacity and protective mechanism in nuclei against age-related oxidative shifts and depletion in free NADH levels.
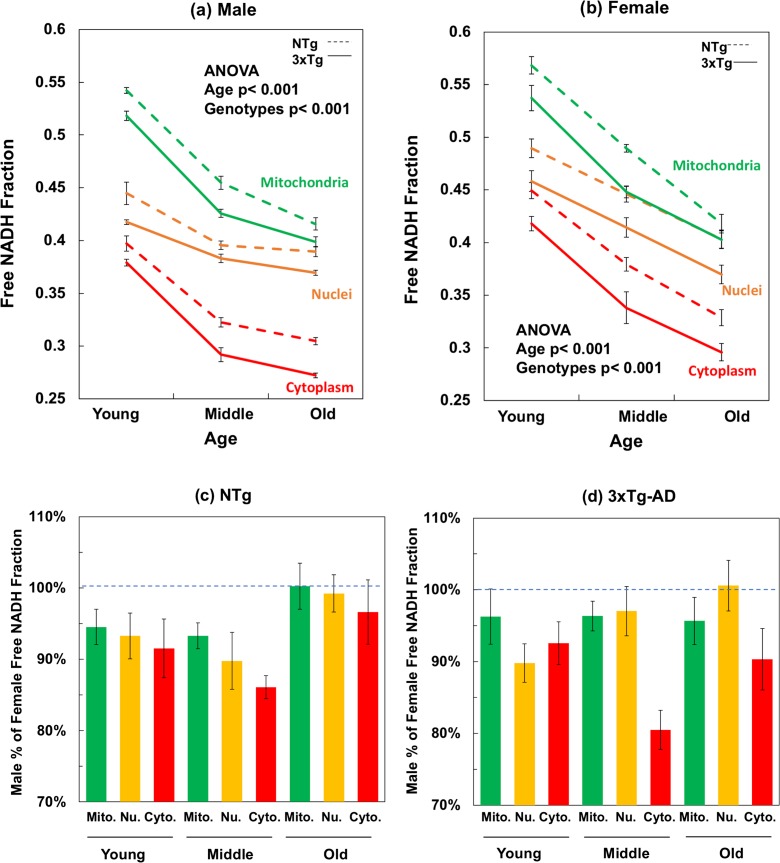
Fig. 3.
Mitochondrial, nuclear, and cytoplasmic free NADH fractions decline with age are further depleted with AD genotype of both a male and b female mouse neurons. For the compartment effect, mitochondria were the most reduced with the highest free NADH fractions and cytoplasm presents the most oxidized NADH state with the lowest free NADH levels (ANOVA for the compartment-differences, in male, NTg F(2,89) = 397, p < 0.001, 3xTg-AD F(2, 89) = 927, p < 0.001; in female, NTg F(2,179) = 154, p < 0.001, 3xTg-AD F(2,179) = 106, p < 0.001). Aging depleted free NADH levels in all compartments of both genotypes. ANOVA for male, mitochondria F(2,59) = 338, p < 0.001, nuclei F(2,59) = 56, p < 0.001, cytoplasm F(2,59) = 262, p < 0.001; for female, mitochondria F(2,119) = 161, p < 0.001, nuclei F(2,119) = 51, p < 0.001, and cytoplasm F(2,119) = 96, p < 0.001). The 3xTg-AD genotype demonstrated significantly more bound NADH and more oxidized NADH redox state than the age-matched NTg neurons in each compartment. By gender (c, d), male neurons were lower in free NADH fraction than the female neurons at young and middle ages, but the multiple comparison results indicated no significant differences in old ages between female and male neurons. Two-way ANOVA for gender effect on compartments, mitochondria F(1,59) = 13, p < 0.001; nuclei F(1,59) = 13, p < 0.001; cytoplasm F(1,59) = 25, p < 0.001). In 3xTg-AD neurons, male NADH fraction was lower than that in female neuron of each compartment (mitochondria F(1,59) = 7, p < 0.01; nuclei F(1,59) = 8, p < 0.01; cytoplasm F(1,59) = 51, p < 0.001) and each age (young F(1,59) = 18, p < 0.001; middle F(1,59) = 26, p < 0.001; old F(1,59) = 8, p < 0.01)
Compartment effects
Figure 3 a and b show that mitochondria have the highest free NADH fraction while cytoplasm displayed the lowest free NADH among the three subcellular compartments (ANOVA for compartment effect, in male, both NTg and 3xTg-AD p < 0.001; for female, NTg and 3xTg-AD p < 0.001). In NTg male young neurons (Fig. 3a), free NADH fraction in mitochondria was 37% higher than the levels in cytoplasm. This difference reached a maximum of 41% higher in the middle age followed by a 36% higher in mitochondrial free NADH in old aged neurons. 3xTg-AD male young neurons presented the same 37% higher in mitochondrial free NADH fraction compared to that of cytoplasm, with further increases in the mitochondria-cytoplasm differences to 46% in middle and old ages. Similar magnitude changes in free NADH with age and genotype were seen in female neurons (Fig. 3b). The relative order of compartmental free to bound NADH redox states was independent of age and genotype. Overall, the largest changes in free NADH were associated with age, then compartment, genotype, and gender. The highest free NADH fraction in mitochondria suggests that mitochondria have the highest capacity for free NADH production compared to nuclei and cytoplasm.
Genotype effect
As illustrated in Fig. 3a, b, the age-matched 3xTg-AD neurons present consistently lower free NADH levels compared to the free NADH levels of NTg neurons in each compartment of each gender. The cytoplasm responded most to the genetic loads of 3xTg-AD, ranging from 6% at young age to 12% at old age lower in free NADH levels of both male and female (young, p < 0.05; old p < 0.01). Under the genetic loads of 3xTg-AD, free NADH levels in mitochondria and nuclei were 5–10% lower than the age-matched NTg neurons from young to old ages of each gender (p < 0.001). The comparatively lower free NADH levels in 3xTg-AD neurons than that of the age- and gender-matched NTg neurons suggests an AD genotype-driven decline in capacity of free NADH regeneration of 3xTg-AD mouse brains. We infer that the genetic load of 3xTg-AD promoted age-related oxidative shifts to more bound NADH redox states by either consuming more free NADH or impairing the capacity for free NADH production.
Male free/bound NADH fraction slightly less than that of female neurons
To gain insights into gender differences in free NADH depletion during aging in the NTg and 3xTg-AD mouse model, we compared the free NADH fractions in male and female neurons of mitochondria, nucleus, and cytoplasm as a function of age and genotype. To more easily compare gender effects, the same data was expressed as male percent of female free NADH (Fig. 3c, d). Male neurons displayed less reducing free NADH states compared to the age-matched female neurons in these compartments (Fig. 3c, d), but these gender differences in free NADH diminished in old age. The average free NADH levels of male neurons was significantly 7% lower than that in female, across all ages and compartments of both genotypes (p < 0.01). Multiple comparison analysis demonstrated gender differences in the young and middle age of all compartments, but no significant gender differences at old ages. This suggests that higher reductive capacity in females than males is gradually lost in old age.
Mitochondrial free NADH concentrations decline with age and AD genotype in hippocampal neurons
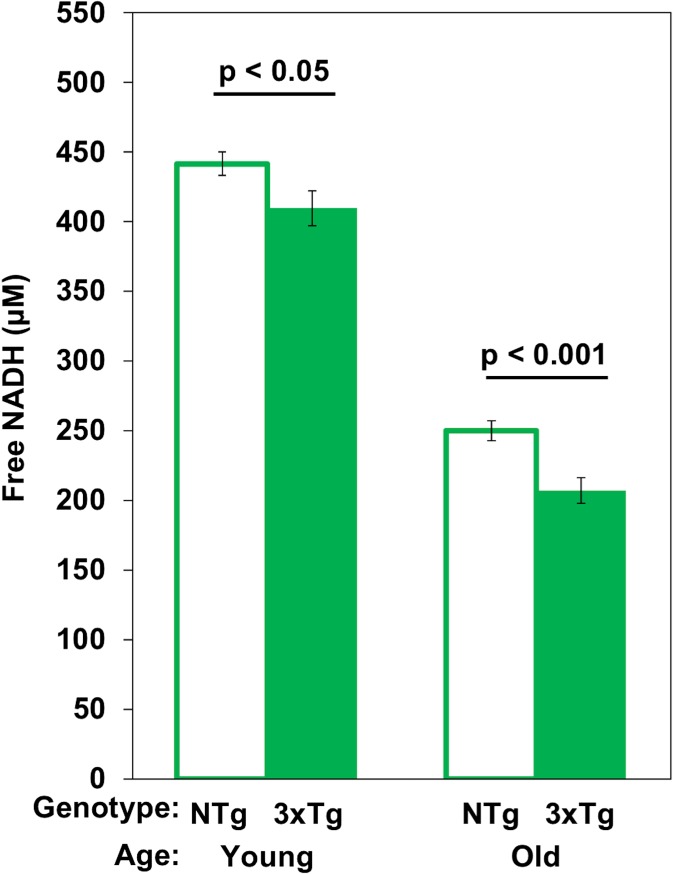
A variety of approaches have been reported for measurement of NAD(P) H concentrations (Sporty et al. 2008; Zhu et al. 2015; Coremans et al. 1997; Yu and Heikal 2009; Ghosh et al. 2012). Neither HPLC or steady state fluorescence measures distinguish the bound form with a higher quantum yield from the free form (Ma et al. 2016; Ghosh et al. 2012). However, absolute measurement of concentration of both the free and bound NADH in situ is possible by a FLIM phasor method (Ma et al. 2016). We determined the absolute free NADH concentrations in live neurons from young and old age mouse hippocampal neurons specifically in mitochondria (Fig. 4). In NTg female neurons from young to old ages, mitochondrial free NADH concentrations declined 43% from 442 to 250 μM (Fig. 4). In 3xTg-AD neurons, age drove a roughly 50% loss in mitochondrial free NADH levels from 410 down to 207 μM. In addition, comparison of genotype effects indicated 8% less free NADH in the young (p < 0.05) and 21% in the old (p < 0.001). With a lower NAD total pool size in aging and AD, the declines in free/bound NADH fraction with age result from larger declines in free NADH concentration. In our results, we observed 43–50% age-related declines in concentrations of mitochondrial free NADH, which were much larger than the 23–26% declines in free NADH fractions. Thus, the concentration of mitochondrial free NADH was depleted twofold more with aging in the 3xTg-AD than the NTg neurons. These results suggest both an age- and AD-related decrease in capacity for NADH regeneration or increase in consumption of NADH.
Fig. 4.
Free NADH concentrations decline from young to old mitochondria in situ exacerbated by the 3xTg-AD genotype. Two-way ANOVA shows age effects F(1,63) = 466, p < 0.001 and for genotype effects F(1,63) = 16, p < 0.001 (n = 16 neurons/age/genotype)
Discussion
Age- and AD-dependent decline in free NADH
Free NADH serves as a vital redox-energy intermediate, produced from NAD+ in glycolysis, the TCA cycle and transhydrogenase. Energy generation at complex I of oxidative phosphorylation in neurons requires at least three elements: sufficient levels of complex I protein, favorable regulation of this complex, and sufficient free NADH substrate. The 3-min lability of NADH (Chance and Thorell 1959) likely precludes its measurement in human surgical samples or postmortem. Our previous work in cultured rat neurons reported here suggested that NADH substrate was at least part of the age-related limitation (Jones and Brewer 2010). Here, we examined the free NADH levels as a function of age and AD-like genotype. With aging, we found a 43% loss of free NADH concentration at rest, which was diminished further to 50% under the load of the transgenes in the 3xTg-AD neurons. These numbers are larger than the steady state fluorescence declines with aging (Ghosh et al. 2012) in the same neuron preparations because free NADH gets oxidized when exposed in air during tissue extraction. The lower NADH concentrations with age are consistent with an age-dependent lower capacity of NADH regeneration in the same live neuron preparations (Ghosh et al. 2012) or increased NADH consumption. In 3xTg-AD mouse brain mitochondria with age, the activity of cytochrome c oxidase (COX) decreased by 51% and PDH by 55%, which exceeded the 31% age-driven declines in NTg mitochondria (Yao et al. 2009). These assays are performed with excess substrate. Therefore, a lower NAD pool could further slow these activities. Judged by steady state fluorescence, neuronal NAD(P) H concentration peaks in middle age compared to young and old age neurons (Ghosh et al. 2012). Similar to mouse brains, human postmortem AD brains exhibited 27–57% declines in PDHC, ICDH, and KGDHC activities relative to age-matched controls (Bubber et al. 2005). As these enzymes produce NADH in the TCA cycle, AD-related declines in these dehydrogenases would lead to lower free NADH production in mitochondria, in line with our findings of lower free NADH with age and further decreased with AD genetic load.
There can be concerns for how well primary cultures from aged brains represent the aging brain with age-related changes in hormones, inflammatory mediators and the vasculature. Many fundamental properties of brain neurons are maintained in culture such as dendrite and axon regeneration (Brewer 1997), synapse formation (Brewer et al. 2009), action potential firing (Evans et al. 1998), NMDA receptor modulation (Cady et al. 2001), and response to redox shifts (Ghosh et al. 2014). Although a lower NAD pool can be measured in old brain homogenates (Gomes et al. 2013), the lability of NADH makes in vivo measurements difficult. In vivo circadian oscillation of NADH have been detected in stem cells within the epidermal basal layer of mice by two-photon excitation (Stringari et al. 2015). A controlling influence of NAD+ levels on behavior was demonstrated in mice without the NAD-hydrolase CD38 (Sahar et al. 2011). As NADH needs to be measured non-invasively, studies on neuron cultures will be useful in attempts to remedy age and AD-related deficits in free NADH.
The age-related declines in NADH could be caused by age-associated sedentary behavior (Figueiredo et al. 2009; Lee et al. 2015; Stolle et al. 2018) that sets epigenetic controls for metabolic changes (Walker et al. 2013; Intlekofer et al. 2013). Metabolic expression of enzymes of the TCA cycle and ETC that could impede NADH regeneration and ATP production need to be measured in activity-controlled studies of aging and AD. This type of study in aging mouse skeletal muscle showed a decline in respiratory activity with aged sedentary behavior (Figueiredo et al. 2009). A review of rat cardiac energy function highlighted decreased mitochondrial energetics with age (Yaniv et al. 2013). As compensatory effects, pathways may re-direct fluxes to replenish the decreasing NADH levels. The NNT reverses the direction of reaction to generate more NADH at the cost of NADPH. This begins a vicious cycle because NADPH is needed to regenerate GSH as the major redox buffer, when GSH levels are decreased by age-associated oxidative stress (Ghosh et al. 2014). Age- and AD-related oxidative shifts stimulate up-regulation of glycolytic pathway to feed the NADH and energetic demand. Alternatively, to maintain redox balance within a viable range, NADH needs to be recycled via either lactate dehydrogenase (with associated acidosis) or the electron transport chain (with minimal oxyradicals) or plasma membrane NAD(P) H oxidoreductase (NOX, with one oxyradical per NADH) (Brewer 2010; Aon et al. 2010). Moreover, rat neurons are more susceptible to stressors with aging (Brewer 1998) and old mouse neurons are more vulnerable than young neurons to limits on NADH resynthesis and GSH depletion (Ghosh et al. 2014). An upstream possible cause is the age-related 50% decline in the total NAD(P) H pool of whole 3xTg-AD neurons compared to 27% decrease in age-matched NTg neurons (Ghosh et al. 2012). The mitochondrial free NADH concentrations of NTg old neurons surpassed those of 3xTg-AD old neurons by 21%, further indicating an even lower capacity to maintain free NADH levels with genetic load in 3xTg-AD old mouse brains.
NAD pool size diminished with aging and AD
Under normal physiology, the concentrations of NADH and NAD+ are balanced by the NAD-synthesizing and NAD-consuming enzymes. In aging and AD, evidence suggests that consumption exceeds recycling and synthetic capabilities. NAD+ levels are maintained via three biosynthetic pathways: the Preiss-Handler, de novo biosynthesis and salvage pathways (Verdin 2015). Age- and AD-related changes in consumption and resynthesis of NAD+ diminish the levels of free NADH (Prolla and Denu 2014), which could further lead to a decrease in capacity for ATP synthesis. Either the age- and AD-related oxidative shift or the energy crisis with demand is insufficient to cause many downstream dysfunctions including amyloid and tau processing, LTP, calcium handling (Green and LaFerla 2008), mitochondrial function and motility (Gibson and Blass 1976; Calkins et al. 2011), DNA repair (Imam et al. 2006), and autophagy (Nixon 2013).
With aging, the activity of NAD biosynthesis decreases while the activities of NAD+-consuming enzymes increase such as CD38 (Camacho-Pereira et al. 2016) and PARP (Bai and Canto 2012) which together contribute to the age-associated decline in NAD pool size. PARP inhibitors together with NAD+ precursors boost ATP levels and increased gene expression of enzymes in TCA cycle and glycolysis, such as citrate synthase and hexokinase (Mouchiroud et al. 2013). Supplementation of the NAD+ precursor, nicotinamide riboside improves learning and memory in AD/Polβ mouse model of mitochondrial DNA damage (Hou et al. 2018). Mounting evidence supports the beneficial effects of nicotinamide (NAM) to ameliorate cognitive decline in AD mouse models associated with declines in Sirt3 activity (Green et al. 2008; Liu et al. 2013). Gomes et al. (2013) reported the reversibility of mitochondrial dysfunction in skeletal muscle of old mice by raising NAD+ levels via the SIRT-1-HIF-1α-c-Myc pathway. Ghosh et al. (2012) showed that neurons pretreated with nicotinamide reverse NAD(P) H levels and NADH regenerating capacity of both NTg and 3xTg-AD neurons (Ghosh et al. 2012). Nicotinamide pre-treatment improved recovery of hippocampal neuronal function, enhanced NADH reduction and ATP content in an acute model of hypoxia in rat (Shetty et al. 2014).
Tau protein is critical to stabilize microtubules of axons. Overexpression of tau in P301L tau mice alters the distribution of organelles including mitochondria that are dependent on microtubule motor proteins for transport and decrease the distribution of mitochondria in synapses of neurons (Rhein and Eckert 2007). Furthermore, in P301L tau mice together with reduced NADH-ubiquinone oxidoreductase activity with age were found to impair mitochondrial respiration and ATP synthesis (Rhein and Eckert 2007; David et al. 2005). Our results of decreased free NADH levels with age could impair energetic capacity to remove amyloid and tau through autophagy (Brewer 2010; Barnett 2011; Brewer submitted 2019).
Age-related oxidized shifts in NADH/NAD+ in subcellular compartments
The subcellular distribution of NADH and their signaling pathways are highly compartmentalized and function somewhat independently (Koch-Nolte et al. 2011). In our study with aging, free NADH shifted toward more bound and less free NADH in both NTg and 3xTg-AD mouse neurons. Of note, all ages of 3xTg-AD neurons display comparatively more oxidized redox state with less free NADH than the age-matched NTg neurons, indicating a further impaired in capacity of free NADH regeneration. The genetic load of 3xTg-AD neurons promoted age-related depletion in free NADH levels and oxidative shifts in NADH redox states starting as early as in young age. The NADH/NAD+ redox states also mediates crosstalk of signaling between neurons and astrocytes in brain (Winkler and Hirrlinger 2015). Both NADH and NAD+ levels drop remarkably with aging and AD. However, growing evidence demonstrates the age- and AD-induced oxidative shifts in NADH/NAD+ redox states (Zhu et al. 2015; Parihar et al. 2008; Ghosh et al. 2012). An important issue is whether redox state controls ROS production or ROS levels influence redox state. Based on more neurodegeneration from titrated inhibition of NADH production than GSH synthesis, we concluded that NADH redox state is upstream of ROS generation (Ghosh et al. 2014).
We found nuclear free NADH to be less affected by age than cytoplasmic or mitochondrial NADH. Cytoplasmic and nuclear NAD+ pools can communicate by diffusion via the nuclear pore (Verdin 2015). Given the important role of NADH shuttles in the communication between compartments, Easlon et al. (2008) reported beneficial effects on extending lifespan in yeast by overexpression of the functional component of the NADH malate-aspartate shuttle and glycerldehyde-3-phosphate shuttle in the calorie restriction (CR) pathway. In addition to comparatively lower consumption of NADH in the nucleus, the nuclear-localization of NMNAT1, with its lower Km and higher Vmax than mitochondrial NMNAT3 (Berger et al. 2005), could be sufficient to regenerate NAD+ consumed by Sirt1 and PARPs. Control of many redox-sensitive transcription factors (Lavrovsky et al. 2000) could buffer or exacerbate age-related changes in free NADH and the NAD pool. We found mitochondrial free NADH to be the most affected by age. The mitochondrial membrane is impermeable to both NAD+ and NADH. Normal ratios of NADH/NAD+ are maintained by the malate-aspartate and glutamate-aspartate shuttles and transhydrogenase. NAD+ precursors such as nicotinamide and nicotinamide riboside can traverse the mitochondrial membrane (Pittelli et al. 2011) to be used for NADH/NAD+ regeneration by nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenyl transferase (NMNAT) (Revollo et al. 2004) via the salvage pathway and transhydrogenase.
Redox states of NADH/NAD+ reflect metabolic states of neurons
The alterations of free/bound NADH provide insight on kinetic switches for modulation of metabolism based on NADH/NAD+ redox states. We find in mouse hippocampal neurons across the age span, both genotypes and both genders, that the NADH redox state is more reduced in mitochondria, than nuclei and that cytoplasm was the most oxidized. This order replicates the relative thiol redox states from most reducing to most oxidizing in Hela cells: mitochondria > nuclei > cytoplasm (Hansen et al. 2006). As the cytoplasmic and mitochondrial NADH/NAD+ ratios are metabolic readouts (Christensen et al. 2014), a shift from free to more bound NADH suggests a metabolic shift from glycolysis to more oxidative phosphorylation with more consumption of free NADH (Stringari et al. 2015). As we observed an age- and AD-related depletion of free NADH in mouse neurons, an oxidative shift in NADH/NAD+ redox states predicts a downstream switch and re-direction in metabolic fluxes from higher TCA-dependent to higher reliance on glycolysis for energy supply and lactate generation for redox balance.
Relationship to caloric restriction (CR)
Caloric restriction (CR) and fasting can extend lifespan and increase NAD levels in the Alzheimer’s mouse brain (Qin et al. 2006), rat or mouse liver (Yang et al. 2007; Nakagawa et al. 2009; Hayashida et al. 2010), and skeletal muscles (Canto et al. 2010) of animal models by inducing the expression of NAMPT in the salvage pathway. Protein restriction (PR), especially, methionine restriction was also found to extend lifespan (Schiff et al. 2011; Lopez-Torres and Barja 2008) and decrease mitochondrial reactive oxygen species (ROS) production in both brain and kidney mitochondria of rats (Caro et al. 2009) as well as free radical leak in rat mitochondria (Lopez-Torres and Barja 2008). Paradoxically, the enzymatic activities needed by the brain for energy production, complex I and III are decreased in rat brain with MetR (Naudi et al. 2007). Martin et al. (2016) also found that CR decreased cytochrome c oxidase activity together with 40% increases in the levels of NAD(P) H in the molecular and polymorphic layers of the mouse dentate gyrus of the hippocampus. This CR paradox of lower activities of the electron transport chain so critical to brain energetics may be explained if CR forces higher turnover of leaky mitochondria so that higher efficiency is maintained (Yang et al. 2016). CR mitochondria of higher efficiency could produce more energy with less consumption of NADH. Thus, CR could cause free NADH levels to rise above the age-related depletion that we observed. The overall effects can be described by antagonistic pleiotropy in which restriction results in higher autophagic quality control to maintain efficient function (Barnett and Brewer 2011; Yang et al. 2016).
Gender differences in age-related declines of free NADH levels
We found that levels of free NADH in male neurons were significantly lower than those of age- and genotype-matched female mouse brains, particularly in the cytoplasmic compartment. This protective effect of higher free NADH levels in female neurons appears to lessen with age, ending in similar free NADH levels in old neurons of female and male. This direction of lower male NADH levels follows the results of (Guevara et al. 2009) who found lower mitochondrial capacity of male brain mitochondria with lower respiratory capacity and lower antioxidant enzyme activities compared to that in aged female brains. Consistent with these studies, Grimm and Eckert (2017) summarized a number of sex-specific changes with age in mitochondria, including mitochondrial dynamics and mitophagy (Guebel and Torres 2016), peroxide production (Borras et al. 2003), respiratory control rate, pyruvate dehydrogenase, and cytochrome c oxidase (COX) activity (Yao et al. 2012). Many of these changes could be downstream of changes in the bioenergetics of NADH. However, in our studies with lower free NADH in males, sex differences were smaller than the age and genotype effects. This protection seems to be lost after menopause in females or just older age in males (Borras et al. 2003; Brewer et al. 2006).
Conclusions
To our knowledge, this is the first time mitochondrial free NADH concentrations were measured in sub-compartments of live primary neurons using FLIM. The ability to distinguish free NADH from total NADH in live neurons across the age span from NTg and 3xTg-AD mice identified critical age-related deficits in energetic states of the neurons. Furthermore, imaging the intrinsic fluorescence of NADH in live neurons by two-photon excitation avoids modifying or introducing any extra fluorophore probe or perturbation. Combined with subcellular analysis, we found an oxidative shift in free NADH levels in mitochondria, cytoplasm, and nuclei of neurons as a function of age and AD-like genotype. The observed depletion of free NADH levels with age and AD genotype suggests that less free NADH will be available for demand-critical energy supply to oxidative phosphorylation. The drop in free NADH could be caused by any combination of increased consumption by resting demand for ATP, decreased regeneration of NAD+ by de novo and nicotinamide salvage pathways or relative metabolic changes, in glycolysis, lactate generation. Together with this work, our earlier work in rat neurons that found ATP levels unchanged with age but significantly lower NAD(P) H levels with age (Parihar et al. 2008) suggests that an age-associated oxidative shift in NADH redox state is upstream of the age-related metabolic shifts in rat and mouse brains. However, the consumption of NAD+ by glycolysis or the TCA cycle for NADH demand at lower NAD pool sizes together with increased lactate production may contribute to a lower redox ratio of NADH/NAD+ (Brewer 2010). Discrimination between excess consumption and regeneration in free NADH with age and genotype will require interventional studies (Dong and Brewer in preparation). Since free NADH is the substrate for numerous redox reactions as well as energy generation, NADH target engagement and reversal of low free NADH levels could counter AD and extend lifespan via a series of NADH-sensor mechanisms involving oxidative stress, DNA repair, and mitochondrial function.
Acknowledgements
We appreciate Prof. Enrico Gratton for inspirational discussions and the help of Rachel Cinco-Hedde, Ning Ma, and Sara Sameni at Laboratory for Fluorescence Dynamics, UC Irvine.
Funding information
This work was supported by the UC Irvine Foundation, NIH P41-GM103540 and a grant from the NIH RF1 AG058218.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alzheimer’s A 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 2016;12:459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Aon MA, Cortassa S, O’Rourke B. Redox-optimized ROS balance: a unifying hypothesis. Biochim Biophys Acta. 2010;1797:865–877. doi: 10.1016/j.bbabio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Barnett A, Brewer GJ. Autophagy in aging and Alzheimer’s disease: pathologic or protective? J Alzheimers Dis. 2011;25:385–394. doi: 10.3233/JAD-2011-101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, White JG, Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme. NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- Borras C, Sastre J, Garcia-Sala D, Lloret A, Pallardo FV, Vina J. Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic Biol Med. 2003;34:546–552. doi: 10.1016/S0891-5849(02)01356-4. [DOI] [PubMed] [Google Scholar]
- Brewer GJ. Isolation and culture of adult rat hippocampal neurons. J Neurosci Methods. 1997;71:143–155. doi: 10.1016/S0165-0270(96)00136-7. [DOI] [PubMed] [Google Scholar]
- Brewer GJ. Age-related toxicity to lactate, glutamate, and beta-amyloid in cultured adult neurons. Neurobiol Aging. 1998;19:561–568. doi: 10.1016/S0197-4580(98)00091-8. [DOI] [PubMed] [Google Scholar]
- Brewer GJ. Epigenetic oxidative redox shift (EORS) theory of aging unifies the free radical and insulin signaling theories. Exp Gerontol. 2010;45:173–179. doi: 10.1016/j.exger.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR. Isolation and culture of adult neurons and neurospheres. Nat Protoc. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Reichensperger JD, Brinton RD. Prevention of age-related dysregulation of calcium dynamics by estrogen in neurons. Neurobiol Aging. 2006;27:306–317. doi: 10.1016/j.neurobiolaging.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Boehler MD, Pearson RA, DeMaris AA, Ide AN, Wheeler BC. Neuron network activity scales exponentially with synapse density. J Neural Eng. 2009;6:014001. doi: 10.1088/1741-2560/6/1/014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- Cady C, Evans MS, Brewer GJ. Age-related differences in NMDA responses in cultured rat hippocampal neurons. Brain Res. 2001;921:1–11. doi: 10.1016/S0006-8993(01)03063-3. [DOI] [PubMed] [Google Scholar]
- Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum Mol Genet. 2011;20:4515–4529. doi: 10.1093/hmg/ddr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Pereira J, Tarragó MG, Chini CCS, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A, Chini EN. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro P et al. (2009) Effect of 40% restriction of dietary amino acids (except methionine) on mitochondrial oxidative stress and biogenesis, AIF and SIRT1 in rat liver Biogerontology 10:579–592. 10.1007/s10522-008-9200-4 [DOI] [PubMed]
- Chance B, Thorell B. Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry. J Biol Chem. 1959;234:3044–3050. [PubMed] [Google Scholar]
- Christensen CE, Karlsson M, Winther JR, Jensen PR, Lerche MH. Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. J Biol Chem. 2014;289:2344–2352. doi: 10.1074/jbc.M113.498626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coremans JM, Ince C, Bruining HA, Puppels GJ. (Semi-)quantitative analysis of reduced nicotinamide adenine dinucleotide fluorescence images of blood-perfused rat heart. Biophys J. 1997;72:1849–1860. doi: 10.1016/S0006-3495(97)78831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R, Alfonso-Garcia A, Cinco R, Gratton E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Sci Rep. 2015;5:9848. doi: 10.1038/srep09848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Hauptmann S, Scherping I, Schuessel K, Keil U, Rizzu P, Ravid R, Dröse S, Brandt U, Müller WE, Eckert A, Götz J. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J Biol Chem. 2005;280:23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J, Lynch RM, Balaban RS. Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophys J. 1989;55:621–630. doi: 10.1016/S0006-3495(89)82859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MS, Collings MA, Brewer GJ (1998) Electrophysiology of embryonic, adult and aged rat hippocampal neurons in serum-free culture J Neurosci Meth 79:37–46 [DOI] [PubMed]
- Fattoretti P, Balietti M, Casoli T, Giorgetti B, di Stefano G, Bertoni-Freddari C, Lattanzio F, Sensi SL. Decreased numeric density of succinic dehydrogenase-positive mitochondria in CA1 pyramidal neurons of 3xTg-AD mice. Rejuvenation Res. 2010;13:144–147. doi: 10.1089/rej.2009.0937. [DOI] [PubMed] [Google Scholar]
- Figueiredo PA, Powers SK, Ferreira RM, Amado F, Appell HJ, Duarte JA. Impact of lifelong sedentary behavior on mitochondrial function of mice skeletal muscle. J Gerontol A Biol Sci Med Sci. 2009;64:927–939. doi: 10.1093/gerona/glp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, LeVault KR, Barnett AJ, Brewer GJ. A reversible early oxidized redox state that precedes macromolecular ROS damage in aging nontransgenic and 3xTg-AD mouse neurons. J Neurosci. 2012;32:5821–5832. doi: 10.1523/JNEUROSCI.6192-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Levault KR, Brewer GJ. Relative importance of redox buffers GSH and NAD(P) H in age-related neurodegeneration and Alzheimer disease-like mouse neurons. Aging Cell. 2014;13:631–640. doi: 10.1111/acel.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GE, Blass JP. Impaired synthesis of acetylcholine in brain accompanying mild hypoxia and hypoglycemia. J Neurochem. 1976;27:37–42. doi: 10.1111/j.1471-4159.1976.tb01540.x. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KN, LaFerla FM. Linking calcium to Abeta and Alzheimer’s disease. Neuron. 2008;59:190–194. doi: 10.1016/j.neuron.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm A, Eckert A. Brain aging and neurodegeneration: from a mitochondrial point of view. J Neurochem. 2017;143:418–431. doi: 10.1111/jnc.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guebel DV, Torres NV. Sexual dimorphism and aging in the human hyppocampus: identification, validation, and impact of differentially expressed genes by factorial microarray and network analysis. Front Aging Neurosci. 2016;8:229. doi: 10.3389/fnagi.2016.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara R, Santandreu FM, Valle A, Gianotti M, Oliver J, Roca P. Sex-dependent differences in aged rat brain mitochondrial function and oxidative stress. Free Radic Biol Med. 2009;46:169–175. doi: 10.1016/j.freeradbiomed.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- Hayashida S, Arimoto A, Kuramoto Y, Kozako T, Honda S, Shimeno H, Soeda S. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol Cell Biochem. 2010;339:285–292. doi: 10.1007/s11010-010-0391-z. [DOI] [PubMed] [Google Scholar]
- Hou Y, Lautrup S, Cordonnier S, Wang Y, Croteau DL, Zavala E, Zhang Y, Moritoh K, O’Connell JF, Baptiste BA, Stevnsner TV, Mattson MP, Bohr VA. NAD(+) supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc Natl Acad Sci U S A. 2018;115:E1876–E1885. doi: 10.1073/pnas.1718819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging. 2006;27:1129–1136. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Intlekofer KA, Berchtold NC, Malvaez M, Carlos AJ, McQuown SC, Cunningham MJ, Wood MA, Cotman CW. Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor-dependent mechanism. Neuropsychopharmacology. 2013;38:2027–2034. doi: 10.1038/npp.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TT, Brewer GJ. Age-related deficiencies in complex I endogenous substrate availability and reserve capacity of complex IV in cortical neuron electron transport. Biochim Biophys Acta. 2010;1797:167–176. doi: 10.1016/j.bbabio.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaidman LK, Leung AC, Adams JD., Jr High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem. 1995;228:312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Fischer S, Haag F, Ziegler M. Compartmentation of NAD+-dependent signalling. FEBS Lett. 2011;585:1651–1656. doi: 10.1016/j.febslet.2011.03.045. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound. NADH Proc Natl Acad Sci U S A. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrovsky Y, Chatterjee B, Clark RA, Roy AK. Role of redox-regulated transcription factors in inflammation, aging and age-related diseases. Exp Gerontol. 2000;35:521–532. doi: 10.1016/S0531-5565(00)00118-2. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim J, Han ES, Chae S, Ryu M, Ahn KH, Park EJ. Changes in physical activity and cognitive decline in older adults living in the community. Age (Dordr) 2015;37:20. doi: 10.1007/s11357-015-9759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liu D, Pitta M, Jiang H, Lee JH, Zhang G, Chen X, Kawamoto EM, Mattson MP. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. Neurobiol Aging. 2013;34:1564–1580. doi: 10.1016/j.neurobiolaging.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Torres M, Barja G. Lowered methionine ingestion as responsible for the decrease in rodent mitochondrial oxidative stress in protein and dietary restriction possible implications for humans. Biochim Biophys Acta. 2008;1780:1337–1347. doi: 10.1016/j.bbagen.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Ma N, Digman MA, Malacrida L, Gratton E. Measurements of absolute concentrations of NADH in cells using the phasor. FLIM method. Biomed Opt Express. 2016;7:2441–2452. doi: 10.1364/BOE.7.002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Reddy PH. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum Mol Genet. 2012;21:2538–2547. doi: 10.1093/hmg/dds072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, et al. Regional metabolic heterogeneity of the hippocampus is nonuniformly impacted by age and caloric restriction. Aging Cell. 2016;15:100–110. doi: 10.1111/acel.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins IV, Rivers-Auty J, Allan SM, Lawrence CB. Mitochondrial abnormalities and synaptic loss underlie memory deficits seen in mouse models of obesity and Alzheimer’s disease. J Alzheimers Dis. 2017;55:915–932. doi: 10.3233/JAD-160640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO. Signal Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naudi A, et al. Methionine restriction decreases endogenous oxidative molecular damage and increases mitochondrial biogenesis and uncoupling protein 4 in rat brain. Rejuvenation Res. 2007;10:473–484. doi: 10.1089/rej.2007.0538. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/S0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- Parihar MS, Kunz EA, Brewer GJ. Age-related decreases in NAD(P) H and glutathione cause redox declines before ATP loss during glutamate treatment of hippocampal neurons. J Neurosci Res. 2008;86:2339–2352. doi: 10.1002/jnr.21679. [DOI] [PubMed] [Google Scholar]
- Patel JR, Brewer GJ. Age-related changes in neuronal glucose uptake in response to glutamate and beta-amyloid. J Neurosci Res. 2003;72:527–536. doi: 10.1002/jnr.10602. [DOI] [PubMed] [Google Scholar]
- Pittelli M, Felici R, Pitozzi V, Giovannelli L, Bigagli E, Cialdai F, Romano G, Moroni F, Chiarugi A. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol. 2011;80:1136–1146. doi: 10.1124/mol.111.073916. [DOI] [PubMed] [Google Scholar]
- Prolla TA, Denu JM. NAD+ deficiency in age-related mitochondrial dysfunction. Cell Metab. 2014;19:178–180. doi: 10.1016/j.cmet.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rhein V, Eckert A. Effects of Alzheimer’s amyloid-beta and tau protein on mitochondrial function -- role of glucose metabolism and insulin signalling. Arch Physiol Biochem. 2007;113:131–141. doi: 10.1080/13813450701572288. [DOI] [PubMed] [Google Scholar]
- Ronchi JA, Francisco A, Passos LA, Figueira TR, Castilho RF. The contribution of nicotinamide nucleotide transhydrogenase to peroxide detoxification is dependent on the respiratory state and counterbalanced by other sources of NADPH in liver mitochondria. J Biol Chem. 2016;291:20173–20187. doi: 10.1074/jbc.M116.730473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 2011;3:794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff M, Benit P, Coulibaly A, Loublier S, El-Khoury R, Rustin P. Mitochondrial response to controlled nutrition in health and disease. Nutr Rev. 2011;69:65–75. doi: 10.1111/j.1753-4887.2010.00363.x. [DOI] [PubMed] [Google Scholar]
- Shetty PK, Galeffi F, Turner DA. Nicotinamide pre-treatment ameliorates NAD(H) hyperoxidation and improves neuronal function after severe hypoxia. Neurobiol Dis. 2014;62:469–478. doi: 10.1016/j.nbd.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin SJ, Bench G. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J Sep Sci. 2008;31:3202–3211. doi: 10.1002/jssc.200800238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squier TC. Oxidative stress and protein aggregation during biological aging. Exp Gerontol. 2001;36:1539–1550. doi: 10.1016/S0531-5565(01)00139-5. [DOI] [PubMed] [Google Scholar]
- Stolle S, Ciapaite J, Reijne AC, Talarovicova A, Wolters JC, Aguirre-Gamboa R, van der Vlies P, de Lange K, Neerincx PB, van der Vries G, Deelen P, Swertz MA, Li Y, Bischoff R, Permentier HP, Horvatovitch PL, Groen AK, van Dijk G, Reijngoud DJ, Bakker BM (2018) Running-wheel activity delays mitochondrial respiratory flux decline in aging mouse muscle via a post-transcriptional mechanism. Aging Cell 17. 10.1111/acel.12700 [DOI] [PMC free article] [PubMed]
- Stringari C, Nourse JL, Flanagan LA, Gratton E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS One. 2012;7:e48014. doi: 10.1371/journal.pone.0048014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen B, Gratton E. In vivo single-cell detection of metabolic oscillations in stem cells. Cell Rep. 2015;10:1–7. doi: 10.1016/j.celrep.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal A, Gupta PK. Measurement of NADH concentration in normal and malignant human tissues from breast and oral cavity. Biotechnol Appl Biochem. 2003;37:45–50. doi: 10.1042/BA20020052. [DOI] [PubMed] [Google Scholar]
- van Munster EB, Gadella TW. Fluorescence lifetime imaging microscopy (FLIM) Adv Biochem Eng Biotechnol. 2005;95:143–175. doi: 10.1007/b102213. [DOI] [PubMed] [Google Scholar]
- Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- Walker MP, LaFerla FM, Oddo SS, Brewer GJ. Reversible epigenetic histone modifications and Bdnf expression in neurons with aging and from a mouse model of Alzheimer’s disease. Age (Dordr) 2013;35:519–531. doi: 10.1007/s11357-011-9375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MW, Rego AC, Frenguelli BG, Nicholls DG. Mitochondrial membrane potential and glutamate excitotoxicity in cultured cerebellar granule cells. J Neurosci. 2000;20:7208–7219. doi: 10.1523/JNEUROSCI.20-19-07208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler U, Hirrlinger J. Crosstalk of signaling and metabolism mediated by the NAD(+)/NADH redox state in brain cells. Neurochem Res. 2015;40:2394–2401. doi: 10.1007/s11064-015-1526-0. [DOI] [PubMed] [Google Scholar]
- Xiao W, Wang RS, Handy DE, Loscalzo J. NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid Redox Signal. 2018;28:251–272. doi: 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Licastro D, Cava E, Veronese N, Spelta F, Rizza W, Bertozzi B, Villareal DT, Hotamisligil GS, Holloszy JO, Fontana L. Long-term calorie restriction enhances cellular quality-control processes in human skeletal muscle. Cell Rep. 2016;14:422–428. doi: 10.1016/j.celrep.2015.12.042. [DOI] [PubMed] [Google Scholar]
- Yaniv Y, Juhaszova M, Sollott SJ. Age-related changes of myocardial ATP supply and demand mechanisms. Trends Endocrinol Metab. 2013;24:495–505. doi: 10.1016/j.tem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Irwin R, Chen S, Hamilton R, Cadenas E, Brinton RD. Ovarian hormone loss induces bioenergetic deficits and mitochondrial beta-amyloid. Neurobiol Aging. 2012;33:1507–1521. doi: 10.1016/j.neurobiolaging.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Heikal AA. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B. 2009;95:46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci U S A. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]