Abstract
The development of genetic engineering in the 1970s marked a new frontier in genome-editing technology. Gene-editing technologies have provided a plethora of benefits to the life sciences. The clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/ Cas9) system is a versatile technology that provides the ability to add or remove DNA in the genome in a sequence-specific manner. Serious efforts are underway to improve the efficiency of CRISPR/Cas9 targeting and thus reduce off-target effects. Currently, various applications of CRISPR/Cas9 are used in cancer biology and oncology to perform robust site-specific gene editing, thereby becoming more useful for biological and clinical applications. Many variants and applications of CRISPR/Cas9 are being rapidly developed. Experimental approaches that are based on CRISPR technology have created a very promising tool that is inexpensive and simple for developing effective cancer therapeutics. This review discusses diverse applications of CRISPR-based gene-editing tools in oncology and potential future cancer therapies.
Introduction
The CRISPR/Cas9 system
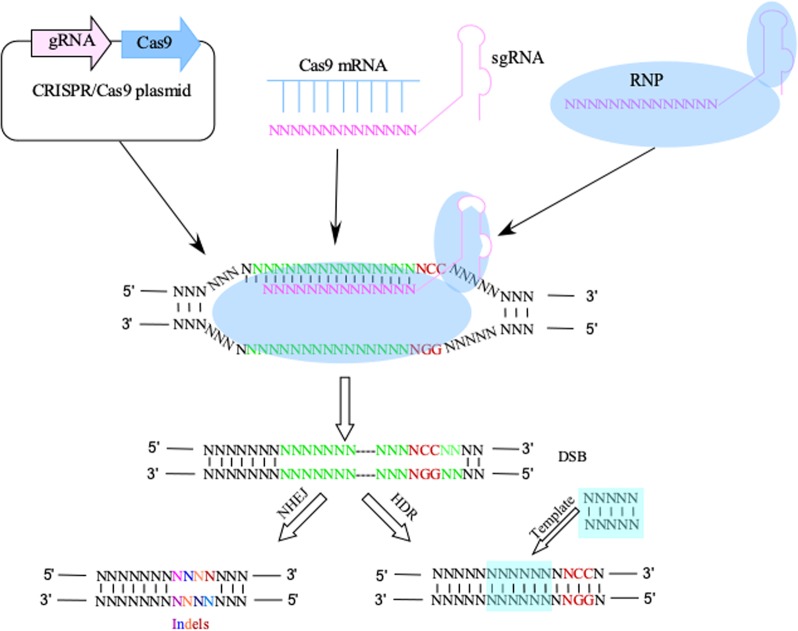
CRISPR/Cas9 is a prokaryotic, adaptive immune system that consists of a programmable RNA molecule that helps guide an associated Cas9 endonuclease to specific exogenous genetic invaders based on recognized sequences.1 The CRISPR-Cas9 system consists of two components, a Cas9 endonuclease and a single-stranded guide RNA (sgRNA).2,3 The sgRNA directs the Cas9 endonuclease to cleave both DNA strands in a sequence-specific manner (Fig. 1). DNA cleavage occurs at a sequence 3 base pairs upstream of an “NGG” protospacer adjacent motif (PAM).4 Following the double-strand break (DSB), the genome is repaired by DNA-DSB repair mechanisms. Using the CRISPR/Cas9 system, targeted genome modifications can be made, such as the introduction of small insertions and deletions (indels) mediated through the relatively error-prone non-homologous end-joining (NHEJ) pathway or the high fidelity homology-directed repair (HDR) pathway.5 Genes of interest can be easily targeted using a 17–21 nucleotide-targeting sequence. To identify genes that are important for a particular phenotype, a pooled population of sgRNAs can be introduced into Cas9-expressing cells by phenotype-based screening of genomic changes.6 In this review, we provide examples of current applications of this technology and speculate on future applications in cancer biology and oncology.
Fig. 1.
CRISPR/Cas9-based gene modification. Common methods of delivering the CRISPR system include a plasmid-based method and Cas9 protein complex with sgRNA or RNP. After the sgRNA binds to the target site of genomic DNA, the Cas9 protein creates a DSB around the PAM site. Random indels or precise modifications introduced into the genomic DNA by the NHEJ or HDR pathway
CRISPR/Cas9 variations
Many variations of the CRISPR/Cas9 system have been developed (Table 1). The Cas9 protein consists of a bi-lobed architecture and the sgRNA is captured between the alpha-helical and nuclease lobes. In the nuclease lobe are two functional domains, HNH and RuvC. The RuvC domain belongs to the retroviral integrase superfamily of proteins and it cleaves the non-target DNA strand whereas the HNH domain cuts the targeted strand of the specific DNA. Normally, the HNH and RuvC domains generate a DSB.7 The inactivation of both domains by a mutation at H840A and D10A in the HNH and RuvC domains, respectively, results in a catalytically inactive Cas9 (dCas9). However, a single mutation of HNH or RuvC results in the generation of a single-strand break rather than a DSB. The Cas9 H840A and D10A mutants also have nickase activity wherein the RuV mutant D10A nicks the targeting strand and the HNH mutant H840A nicks the non-targeting strand. Because dCas9 is enzymatically inactive, it cannot cleave DNA. However, it retains its RNA-guided DNA binding ability, which has led to several innovative applications.8 dCas9, when fused to a transcriptional repressor peptide such as KRAB (Kruppel associated box), can be used to knockdown gene expression by guiding RNA. This fusion system can block the initiation of transcription and elongation and is referred as CRISPRi. The dCas9-KRAB fusion protein, when co-expressed with a target-specific sgRNA, binds the sgRNA, and the entire complex binds to the DNA strand, blocking the initiation of transcription and elongation resulting in depletion of transcripts of interest.9 In a similar approach, dCas9 can also be used to activate gene expression if it is fused with an activator peptide such as the VP64 and VPR activation domains. This complex is called CRISPRa and can increase transcription of target gene transcripts. CRISPRi and CRISPRa provide new tools for investigating human genome functions, transcriptome research, and regulation of functional factors in cancer biology and oncology. This differs from the canonical CRISPR system that often causes meaningless mutations or leads to a chaotic phenotype.10 Compared with other CRISPR approaches, dCas9-based CRISPRi and CRISPRa are inducible, reversible, have fewer off-target effects, and low toxicity. These approaches have advantages in long non-coding (lnc) RNA knockdown and overexpression.11 In cancer research, precise regulation of gene expression is a very useful approach and scientists have developed and expanded different systems, such as RNAi (RNA interference) and ORF (open reading frame) expression for loss or gain-of-function studies.11 RNAi has played a critical role in biological studies mainly because it has deterministic outcomes and is easy to deliver into mammalian cells. On the other hand, CRISPR system-based tools are often difficult to deliver into mammalian cells. Two components are required compared with a single component RNAi. However, RNAi can also lead to unpredicted non-specific toxicity and strong siRNA/shRNA can cause extensive off-target effects.12 Thus, even though CRISPR has disadvantages, CRISPR-based loss-of-function approaches are widely used because CRISPRi shows less endogenous off-target effects compared to RNAi, and also provides a high specificity of gene knockdown by blocking distinct promoters. It can also be useful for targeting lncRNA, whereas RNAi may be inefficient.13
Table 1.
Variations of the CRISPR system
| Variations | Features | Effects | Advantages | Disadvantages | Applications in cancer research |
|---|---|---|---|---|---|
| CRISPR/Cas9 | WT Cas9; sgRNA | Double-strand break at the target site | Versatile; effective; stable; easy accessibility | Off-target; PAM limited; different modified alleles | Set up research model; functional gene study; drug target identification |
| CRISPR/Cas9 Nickase | Mutant Cas9 H840A or D10A; sgRNA | Single-strand break | Convenient; efficient; flexible; precise, scalable; robust | PAM limited; 2 sgRNA for KO |
Manipulate epigenetic modifications; Simultaneous activation and repression |
| CRISPRi | dCas9; repressor peptide; sgRNA | Block transcription elongation or knockdown transcripts | Inducible; reversible; low off-target effects; low toxicity |
PAM limited; off-target effects at bidirectional promoters | Genome and transcriptome research; LncRNA knockdown and overexpression |
| CRISPRa | dCas9; activator peptide; sgRNA | Increase transcription | PAM limited; complicated to deliver the multiple components |
CRISPR/Cas9 and basic research
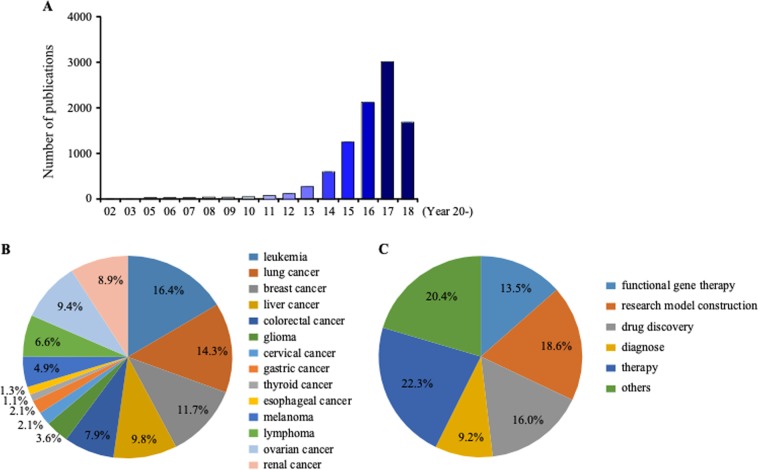
CRISPR/Cas9 is a rapidly developing gene-editing tool that has revolutionized many areas of research. An online search ranging from 2002 until 2018 (5/26/2018) was conducted on PubMed using the term “CRISPR/Cas9” (Fig. 2). The search results revealed an increasing number of research articles (9332 publications) on PubMed over those years (Fig. 2a). In 2017, publications regarding CRISPR/Cas9 numbered 2889. Among these publications, 8.6% are associated with cancer and most of those focus on leukemia (16.4%), lung cancer (14.3%), breast cancer (11.7%), or liver cancer (9.8%, Fig. 2b). A search was also conducted using key words “CRISPR/Cas9” and “cancer” along with “model”, “functional gene”, “drug target”, “diagnosis”, or “therapy”. Most papers were focused on cancer therapy (22.3%), model construction (18.6%), drug validation (16%), or functional gene studies (13.5%; Fig. 2c). Clearly, since its initial discovery, the CRISPR/Cas9 system has been integrated into cancer research as a useful tool to identify oncogenes and mediators of cancer.14
Fig. 2.
Publications focusing on CRISPR identified from 2002 to 2017 on PubMed reflect great interest and cancer applications. a The number of publications on PubMed with the keyword “CRISPR” from 2002 to 2018. b CRISPR/Cas9 applications in different cancers as reported on PubMed. c CRISPR/Cas9 applications with different approaches for cancer. Both b and c results are shown as a percentage
The occurrence and development of cancer is a highly complex process with multi-gene and multi-path interactions.15 Many scientists have been attempting to decipher the mechanisms of cancer occurrence, development, and metastasis and clearly, CRISPR has accelerated research efforts.16 Presently, CRISPR technology is used to investigate the genetic mechanisms in almost all areas of cancer, from prevention to prognosis and treatment,14,17 which greatly promotes transition to the clinic. For example, CRISPR has been applied to breast cancer diagnosis, treatment, and even drug resistance research.18 Based on the CRISPR-dCas9 system, researchers have fused a DNA methyltransferase effector to dCas9 and infected it by lentivirus into healthy breast cells.19 The results showed that the cyclin dependent kinase inhibitor 2A (CDKN2A) gene is a key driver in causing abnormally rapid cell division, which might be an early diagnostic marker in breast cancer. Another group attempted to apply the CRISPR system in breast cancer therapy by targeting the HER2 gene.20 Results showed that targeting HER2-expressing cells inhibited growth and attenuated tumorigenicity, compared to non-targeted cells, suggesting a new therapeutic choice against breast cancer. Additionally, CRISPR was also used to confirm the BRCA1-delta11q alternative splice isoform as a primary factor in breast cancer resistance to treatment.21 The CRISPR/Cas9 system is becoming a widespread, practical, and useful tool against many types of tumors and it promises to accelerate cancer research.14
Applications of CRISPR/Cas9 in cancer research
Generation of cancer models
Cancers are driven by processes influenced by underlying genes.22 Being able to decipher the molecular genetics of disease is crucial to elucidate the underlying mechanisms.23 Cell lines and animal models are invaluable for dissecting the relationship of the genotype, chemotherapeutic effects, and immune microenvironment. CRISPR genetically engineered cancer models now can be produced rapidly, efficiently, and inexpensively24 (Table 2). Leukemia models were generated by targeting several inactivated genes through a lentivirus-delivered Cas9–sgRNA system in primary hematopoietic stem and progenitor cells (HSPCs).25 The pooled lentiviruses target several genes, including Tet2, Runx1, Dnmt3a, Nf1, Ezh2, and Smc3. With a fluorescent marker, multiple targeted HSPCs were selected that are involved in the development of myeloid malignancy. The CRISPR/Cas9 technology has been used to generate several other cancer models.26 By introducing mutations of APC, SMAD4, TP53, KRAS, and/or PIK3CA, an organoid model of colon cancer was built with CRISPR technology.27 In the future, the generation of precision cancer models would greatly stimulate the study of functional cancer genomics and enhance the development of precision cancer medicine.28
Table 2.
CRISPR applications in cancer research
| Application | Targets | sgRNA design | Vehicle for delivery | Features | Advantages | Disadvantages |
|---|---|---|---|---|---|---|
| Generate cancer model | HSPCs; healthy human organoids | Targeting the model type-related suppressors oncogenes | Pooled lentivirus | Disrupt suppressors or edit oncogenes | Rapid, efficient, and inexpensive | Special delivery techniques; tissue limited |
| Synergistic gene study | Cells | Targeting optional drug target from database | Lenti-double sgRNA library | Together with deep sequencing | Effective, low cost, innovative approach | Double sgRNA construction; need highly efficient sgRNA; special analysis |
| Target validation | Drug or anticancer reagent resistant cells | Lentiviral library from Addgene; or optional targets | Plasmid | Identify the target from resistant cells by sequencing | Effective | False-positives |
| Gene diagnose | Genome | Target sensitive genes | Lentivirus | Together with Cas13a or Cas12a to induce collateral effects | Sensitive, rapid, low cost | Certain template concentration |
Synergistic gene studies using CRISPR/Cas9
The CRISPR/Cas9 system also provides an effective strategy for the identification of synergistic gene interactions, which could be used to block drug resistance. A CRISPR-based double knockout (CDKO) system in K562 leukemia cells has been developed using a double sgRNA library system to screen for combinatorial genes and identify pairs of synthetic lethal drug targets. Together with deep sequencing, phenotype measurement and gene analysis have identified interactions between synergistic drug targets, like BCL2L1 and MCL1.29 Another simple and efficient strategy called CombiGEM (combinatorial genetics en masse)-CRISPR was also developed to analyze combinatorial gene function.30 It is similar to the CDKO system in which two pooled sgRNA libraries were combined in one vector. With this approach, some genetic hits (e.g., KDM6B + BRD4) were discovered. Depleting these genes using the CombiGEM system has shown stronger synergistic efficacy against ovarian cancer cell proliferation compared to a reported small-molecule inhibitor.30 Compared to drug inhibition, the CRISPR system costs less.9 The power of the CRISPR library to screen functional variants could thus play an important role in precision cancer medicine.31 Indeed, this innovative approach could allow for the development of personalized genotype-based therapies built upon genotype-specific targets.32
Functional gene screening using CRISPR/Cas9
Precision cancer medicine has resulted in the development of many targeted drugs to treat different cancers. Targeted therapy already has shown enormous potential but several challenges still exist. Only patients who exhibit a certain mutation or altered gene expression respond to the targeted drug treatment and drug resistance to the therapy still occurs.33 Functional genome-screening approaches using the CRISPR system could reveal gene expression changes after treatment and pinpoint genes associated with resistance to the targeted drugs, thereby identifying new biomarkers for precision therapy and providing new insights into cancer development.34 One successful example involved screening for a cancer metastasis-related gene with a CRISPR-Cas9-mediated loss-of-function screen.35 This group infected a non-metastatic lung cancer cell line with a mouse genome-scale CRISPR knockout (mGeCKO) sgRNA library and the transduced cells were subcutaneously transplanted into immunocompromised mice. Six weeks later, the mice exhibiting lung cancer metastasis were selected for sequencing the enriched sgRNA. Finally, several candidate genes associated with lung metastasis were identified and validated, including the already reported genes Pten,36 miR-152,37 and miR-345,38 and several new genes like Nf2, Trim72, and Fga. Other loss-of-function screens were also applied to examine suppressor genes in liver tumors.39 In this study, p53−/−/Myc mouse embryonic liver progenitor cells were infected with an mGeCKO library and transplanted into nude mice. sgRNAs that were increased 8-fold were chosen as candidates and Nf1, Plxnb1, Flrt2, and B9d1 were identified as new tumor suppressors involved in liver cancer formation. Another group applied CRISPR interference (CRISPRi) to screen for functional lncRNA loci, which could modify cell growth.40 They designed a comprehensive sgRNA library to target the lncRNA transcription start site (TSS). The library was transduced into several different cell lines and together with sequence analysis, 499 lncRNAs were identified as associated with cell growth.
CRISPRi can repress the transcription of targeted genes by recruiting the complex of dCas9 and a repressor to the TSS, which is more suitable for lncRNA gene research.10 In summary, combining CRISPR-based functional genetic screening is a powerful approach to validate alternative genes associated with a specific phenotype.41
Target validation by CRISPR/Cas9
Revealing the mechanism of action for small-molecule drugs is a time-consuming and laborious process.42 Efficiently identifying new drug targets with a CRISPR-Cas-based genetic screening system (CRISPRres) containing large sgRNA libraries is now possible.43 If the molecular binding site is depleted or mutated, cancer cells commonly acquire resistance, but the molecular target could be clearly identified by sgRNA sequencing. CRISPR was used to successfully identify nicotinamide phosphoribosyl transferase as the primary target of KPT-9274, an anticancer agent. Based on CRISPR/Cas9 technology, another team set up a system called DrugTargetseqR to identify direct physiologic targets by mutating potential targets for drug resistance.44 In this study, kinesin-5 was confirmed as the direct target of ispinesib by mutating kinesin-5 D130V or A133P in HeLa cells. Targeting the exons that encode functional protein domains rather than the 5′ exons reportedly is a better method using the CRISPR system for identifying drug targets.45 In this case, a negative selection system was constructed with a GFP reporter and used to screen hundreds of chromatin regulatory domains in leukemia cells. The sgRNAs targeting of functional domains, like the ATPase domain or DNA-binding domain, led to a stronger negative selection phenotype compared to targeting the 5′ exon. Obviously, combining CRISPR genome-editing with deep sequencing or cellular biophysical assays could produce new insights into target validation studies.46,47
Gene diagnosis
Genetic diagnostics to determine sensitive genes is critical for cancer prevention.48 Although a low frequency mutation is not easily determined by sequencing, a CRISPR-based diagnostic system called SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing), has been established.49 A key factor in this system is Cas13a, an RNA-guided RNase, which induces robust non-specific single-stranded DNA (ssDNA) trans-cleavage as a collateral effect.50 Another essential element is the reporter signal, which is released after RNA cleavage. This method has been used to detect two cancer mutants, BRAF V600E and EGFR L858R, and appears to be a highly sensitive detection approach. A similar system referred to DETECTR (DNA endonuclease-targeted CRISPR trans reporter) has been developed.51 In this system, another Cas family member, Cas12a, is used, and acts similar to Cas13a. An additional enzyme, RPA (recombinase polymerase amplification), is used to amplify micro-samples. RPA can be used as a detection tool for screening for infections in cancers. The system was used to detect HPV types 16 and 18 in lung carcinomas and appears to be a rapid and inexpensive approach.52
Anticancer applications in clinical trials
Based on promising results of pre-clinical studies, the CRISPR/Cas9 system could also potentially be used clinically to target cancer-causing genes. At this time, eleven clinical trials are underway testing the effectiveness of CRISPR for cancer therapies (Table 3). Seven of the eleven trials are immunotherapies that target program cell death-1 (PD-1) protein expression. The PD-1 protein and programmed cell death ligands (PD-Ls) are important for the negative regulation of the immune system, specifically on T-cells. Their attenuation of the immune response helps tumor cells survive by evading the immune system.53 Pembrolizumab, a monoclonal antibody against PD-1, confirmed that blocking PD-1 and PD-L1 in the immune system could significantly increase the overall survival rate in cancer patients.54 PD-1 is thus an attractive target for immunotherapy and PD-1 inhibitors have been recently approved by the U.S. Food and Drug Administration (FDA) for cancer immunotherapy. Coincidentally, a team in China has gone one step further by using CRISPR/Cas9 to directly target PD-1 in patients (NCT02793856). Using CRISPR/Cas9, they disabled PD-1 expression in cells harvested from a metastatic non-small-cell lung cancer patient. They expanded the cells in a large culture system and then injected the modified cells back into the patient.55 Based on the results of a dose-escalation study, the safety of PD-1 knockout-engineered T-cells in treating metastatic non-small cell lung cancer will be evaluated. Similar trials targeting PD-1 expression in T-cells are being conducted in prostate (NCT02867345), bladder (NCT02863913), and renal cell cancers (NCT02867332). Another phase II clinical study has applied the same PD-1 knockout on T-cells for esophageal cancer (NCT03081715).
Table 3.
Anticancer applications in clinical trials
| Applications | Target site | Study phase | Editing strategy | Clinical trials identification |
|---|---|---|---|---|
| Advanced esophageal cancer | PD-1 | Phase II | PD-1 knockout | NCT03081715 |
| Castration resistant prostate cancer | PD-1 | Phase I | PD-1 knockout | NCT02867345 |
| Muscle-invasive bladder cancer | PD-1 | Phase I | PD-1 knockout | NCT02863913 |
| Metastatic non-small cell lung cancer | PD-1 | Phase I | PD-1 knockout | NCT02793856 |
| EBV associated malignancies | PD-1 | Phase I Phase II |
PD-1 knockout | NCT03044743 |
| Metastatic renal cell carcinoma | PD-1 | Phase I | PD-1 knockout | NCT02867332 |
| Relapsed or refractory leukemia and lymphoma | CD19 and CD20 or CD22 | Phase I Phase II |
Edit CD19 and CD20 or CD22 | NCT03398967 |
| Human papillomavirus-related malignant neoplasm | HPV16-E6/E7 HPV18 E6/E7 |
Phase I | HPV16-E6/E7 or HPV18 E6/E7 knockout |
NCT03057912 |
| CD19 + leukemia and lymphoma | TCR B2M |
Phase I Phase II |
TCR and B2M knockout |
NCT03166878 |
| Tumor of the central nervous system | NF1 | — | Fix NF1 mutation allele | NCT03332030 |
| Multiple myeloma Melanoma Synovial sarcoma Myxoid/round cell liposarcoma |
TCR PD-1 |
Phase I | TCR and PD-1 knockout | NCT03399448 |
Generating chimeric antigen receptor (CAR) T-cells by CRISPR/Cas9 is another ex vivo approach in clinical trials. Researchers from the University of Pennsylvania organized the first-in-human trial to test the effect of HLA-A*0201 restricted NY-ESO-1 redirected engineered T-cells in a wide range of cancer types, including relapsed refractory multiple myeloma (MM), melanoma, synovial sarcoma, and myxoid/round cell liposarcoma (NCT03399448). Tumor rejection activity might be enhanced by eliminating endogenous TCR and PD-1 with the use of CRISPR. Another clinical trial (NCT03166878) focused on CD19+ leukemia and lymphoma. Allogeneic CD19-directed CAR T-cells were generated using the lentiviral delivery of the CAR receptor and CRISPR RNA by electroporation to disrupt the endogenous TCR and B2M genes. This approach might assist in evading host-mediated immunity and thus deliver anti-leukemic effects to patients without having to worry about graft-versus-host-disease (GVHD). Unfortunately, a subset of patients relapse due to the loss of CD19 in tumor cells. Thus another clinical trial is focusing on CRISPR-edited dual specificity CD19 and CD20 or CD22 CAR T-cells, which could recognize and kill the CD19-negative malignant cells through recognition of CD20 or CD22 (NCT03398967). This may be a complementary approach for a wide range of patients. In another application, CRISPR/Cas9 was used to disrupt the human papillomavirus-16 (HPV16) E7 protein, an oncogenic protein that is important for the maintenance of the malignant phenotype in cervical cancer. When E7 was disrupted in HPV-positive cervical cancer cells, inhibition of tumor growth inhibition, and induction of apoptosis occurred.56 These promising results have led to a phase I clinical trial (NCT03057912) to evaluate the safety and efficacy of TALEN-HPV E6/E7 and CRISPR/Cas9-HPV E6/E7 in treating HPV persistent and HPV-related cervical intraepithelial neoplasia. In this study, a CRISPR/Cas9 plasmid targeting HPV16-E6/E7T1 or HPV18 E6/E7T2 was administered twice a week for four weeks to disrupt the expression.
Another clinical trial using CRISPR technology has been designed to screen and identify drugs (NCT03332030). In this trial, an induced pluripotent stem cell (iPSC) bank was established from phenotypically well-characterized patients with Neurofibromatosis type 1 (NF1). NF1 is a frequent neurocutaneous syndrome which easily causes various benign or malignant tumors.57 To identify a NF1 specific target drug, CRISPR/Cas9 was used to develop different cell lines, NF1 wild type (NF1+/+), NF1 heterozygous (NF1+/−), and NF1 homozygous (NF1−/−). By examining the reverse or alleviated phenotypes, NF1-specific drugs might be identified. Although results from CRISPR/Cas9 clinical studies might be promising, more work is needed to assure that CRISPR/Cas9 is a safe and effective tool for treating human cancers.
Future applications
Cancer is characterized by numerous genetic alterations and multiple mutations. The CRISPR system provides unparalleled precise control to correct cancer-associated mutations in the genome compared with the original ZFNs or TALENs technologies.58,59 One could easily imagine that directly correcting abnormal genes through the HDR pathway will be an effective therapeutic strategy against cancer.60 Recent work has provided strong evidence for this vision. TMEM135-CCDC67 and MAN2A1-FER fusion genes have been identified as cancer-derived genes in human prostate cancer and hepatocellular carcinoma.61,62 These genes were replaced by the HSV1-TK death-promoting gene using CRISPR-Cas9 technology. HSV1-TK is a phosphotransferase that can block DNA synthesis as a suicide gene.63 In this study, an adenoviruses delivery system was used to disrupt the dysfunctional gene by dCas9 and two sgRNAs. One sgRNA was designed to target the gene locus and the other was targeted to the breakpoint region of the fusion gene. In this way, CRISPR-Cas9 only induces DSBs in cancer cells (not normal cells) that contain the TMEM135-CCDC67 and MAN2A1-FER fusion genes, which greatly reduced the off-target effects. The DSBs were repaired by an HSV1-TK suicide gene template, which contained the same flanking sequence as the HDR donor.63 With this method, the TMEM135-CCDC67 and MAN2A1-FER fusion genes will be inactivated or replaced by the HSV1-TK suicide gene with no side effects on normal cells.
Another promising application of the CRISPR/Cas9 system is personalized therapy, which requires rapid and systematic screening to identify the genotype-specific changes in the patient’s genome.64 Combining guide RNA libraries with Cas9 nuclease could assist in screening a broad range of potential gene targets and identify genes responsible for the altered phenotypes, including genetic mutations and dysfunctional signaling pathways, which might initiate cancer or promote progression.65 Therapeutic strategies can subsequently be developed based on the results of the gene screening. The treatment of EGFR-mutant lung cancer is a typical example for personalized therapy.66 One method is to target the tumor-driving mutations by tyrosine kinase inhibitors (TKIs). Because of the inevitable drug resistance problems, other “molecular surgeries” using CRISPR technology to directly correct or disrupt the mutated site to inactivate the oncogenic activity, might be a more permanent strategy for EGFR-mutant lung cancer treatment.67 The CRISPR technology is poised to open the door to effective personalized cancer treatment from specific applications of genome screening to therapeutic strategies.68
The CRISPR-Cas9 system is a new rapidly developing technology with potential to completely revolutionize scientific research including transformation from basic research to actual clinical application providing more opportunities for a greater understanding of cancer biology and treatment.
Conclusions and perspectives
The advent of CRISPR technology has revolutionized the biological sciences and provided cancer biologists with a powerful gene-editing method that can alter the genetic make-up of cells in unprecedented ways. Indeed, promising results have been achieved in diverse disciplines, from basic research to the development of potential therapies against cancer, congenital defects, and other chronic diseases. Many challenges associated with CRISPR technology still exist, mainly in clinical use associated with delivery and safety.69 Improved strategies will be required to increase the targeting efficiency and to minimize off-target effects. Ensuring that CRISPR/Cas9 has precise genome-editing ability is also important. Following CRISPR/Cas9 mediated DSBs, DNA repair can be achieved by either the “error-prone NHEJ” or “precise repair through HDR” in the genome. In an effort to promote HDR over NHEJ, a study by Maruyama et al. used the NHEJ inhibitor Scr7 and observed that Scr7 treatment did indeed increase the levels of HDR-mediated genome editing over NHEJ.70 In addition to concerns regarding off-target effects, considerations regarding the immune response to CRISPR-mediated gene-editing must also be considered. The Cas9 protein is bacterial in origin and thus might elicit an immune response, which could in turn affect its gene-editing efficiency. Also, the exogenous sgRNAs may be cleared by immune cells like monocytes and macrophages.71 The delivery approach for CRISPR/Cas9 thus depends upon the objective as well as the target. Transient expression of the sgRNA and the Cas9 protein through microinjection might be safer than other non-viral or viral delivery methods. Viral delivery has the highest efficiency for the expression of the Cas9 protein and sgRNA, but comes with risks. During one in vivo study, the lentivirus particles elicited a strong immune response in treated animals, which affected the efficacy of the lentiviral vector.72 Another issue with lentiviral delivery is also the possibility that the lentiviral vector could randomly integrate into the cellular genome. To overcome this issue, integrase-deficient lentiviral vectors, nanoparticle carriers, or an inducible system could be used for delivery of CRISPR/Cas9.
The prohibitively high costs for CAR T-cell therapy have made this treatment unavailable to a large section of society. CAR T-cell therapy offered by Novartis, the first approved by the FDA, costs $475,000 per treatment; however, further advances in CRISPR technology might help reduce costs and make CAR T-cell therapy available to more patients. So far, the majority of CRISPR clinical trials are being conducted in China. A recent article in the Wall Street Journal suggested that this may be due to the fact that fewer rules and restrictions exist in China. Indeed, Dr. Shixiu Wu from the Hangzhou Cancer Hospital, who was featured in the article,73 has been able to use CRISPR technology-based treatments on his cancer patients, without the need for national regulatory approval and has few reporting requirements. Many scientists worry that the technology has the potential to harm patients and that unintended consequences from using CRISPR on patients without sufficient oversight could hinder progress in the whole field. Dr. Wu recognized that he was undertaking risks in using CRISPR-based treatments for his cancer patients, but was considering their limited available survival time. The thought is that being able to live for additional time is better than imminent death. Persistent post-treatment monitoring of patients could help to eliminate or treat any unwanted consequences. Indeed, treatment-related observations could potentially save many lives of those who are on the brink of death.
We need consider the limitation as seriously as the potential benefits of this technology. A new human trial, in which the team took a crash course in bioethics and created CRISPR babies, brought the ethical issues and controversies into the public. One hundred and twenty-two Chinese scientists and many other scientists worldwide have already condemned the trial. How to use this powerful without overstepping the ethical bottom line is a serious question that needs careful consideration. We might expect more positive reports from clinical trials as well as the development of improved approaches that will bring new hope for personalized therapy.
Acknowledgements
This work was supported by grant funding from National Institutes of Health, U.S.A., CA187027, CA 166011, and CA 196639 and the Key program of Henan Province, China, Grant NO. 161100510300 (Z.G.D.) and the National Natural Science Foundation of China NSFC81672767 (M.H.L.), and the Henan Provincial Government, China.
Author contributions
X.T. and M.H.L. contributed to the literature search and collection of articles, assisted with designing the figures and writing; T.G. and S.P. assisted in the literature search and advised which articles were most appropriate; A.M.B. edited the manuscript; Z.D. supervised the studies and allocated the funding.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xueli Tian and Tingxuan Gu
Contributor Information
Mee-Hyun Lee, Phone: +86-371-65587008, Email: mhyun_lee@hanmail.net.
Zigang Dong, Phone: +1-507-437-9600, Email: zgdong@hi.umn.edu.
References
- 1.Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 6.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson MH, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2018;13:406–416. doi: 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boettcher M, McManus MT. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol. Cell. 2015;58:575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biagioni A, et al. Type II CRISPR/Cas9 approach in the oncological therapy. J. Exp. Clin. Cancer Res. 2017;36:80. doi: 10.1186/s13046-017-0550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan, T., Rindtorff, N., Betge, J., Ebert, M. P. & Boutros, M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol (2018). [DOI] [PubMed]
- 15.Maemura K, Natsugoe S, Takao S. Molecular mechanism of cholangiocarcinoma carcinogenesis. J. Hepatobiliary Pancreat. Sci. 2014;21:754–760. doi: 10.1002/jhbp.126. [DOI] [PubMed] [Google Scholar]
- 16.Gulei D, Berindan-Neagoe I. CRISPR/Cas9: a potential life-saving tool. What’s next? Mol. Ther. Nucleic Acids. 2017;9:333–336. doi: 10.1016/j.omtn.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget. 2016;7:12305–12317. doi: 10.18632/oncotarget.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, et al. Break breast cancer addiction by CRISPR/Cas9 genome editing. J. Cancer. 2018;9:219–231. doi: 10.7150/jca.22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunderson EA, et al. Hit-and-run epigenetic editing prevents senescence entry in primary breast cells from healthy donors. Nat. Commun. 2017;8:1450. doi: 10.1038/s41467-017-01078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017;385:137–143. doi: 10.1016/j.canlet.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, et al. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76:2778–2790. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen D, Xu T. The expanding role of mouse genetics for understanding human biology and disease. Dis. Model Mech. 2008;1:56–66. doi: 10.1242/dmm.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayin VI, Papagiannakopoulos T. Application of CRISPR-mediated genome engineering in cancer research. Cancer Lett. 2017;387:10–17. doi: 10.1016/j.canlet.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Heckl D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 28.Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015;7:53. doi: 10.1186/s13073-015-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han K, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong AS, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl Acad. Sci. USA. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baliou S, et al. CRISPR therapeutic tools for complex genetic disorders and cancer (Review) Int J. Oncol. 2018;53:443–468. doi: 10.3892/ijo.2018.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn R, et al. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp. Hematol. 2015;43:838–848 e833. doi: 10.1016/j.exphem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franco-Tormo MJ, et al. CRISPR/Cas9, the powerful new genome-editing tool for putative therapeutics in obesity. J. Mol. Neurosci. 2018;65:10–16. doi: 10.1007/s12031-018-1076-4. [DOI] [PubMed] [Google Scholar]
- 34.Bester AC, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–664 e620. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Z, Ma R, Tan W, Zhang L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp. Mol. Med. 2014;46:e112. doi: 10.1038/emm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang JT, et al. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis. 2011;32:1207–1215. doi: 10.1093/carcin/bgr114. [DOI] [PubMed] [Google Scholar]
- 39.Song CQ, et al. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology. 2017;152:1161–1173 e1161. doi: 10.1053/j.gastro.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu SJ, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:pii: aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geerts S, Gryseels B. Drug resistance in human helminths: current situation and lessons from livestock. Clin. Microbiol Rev. 2000;13:207–222. doi: 10.1128/CMR.13.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neggers JE, et al. Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes. Nat. Commun. 2018;9:502. doi: 10.1038/s41467-017-02349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasap C, Elemento O, Kapoor TM. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat. Chem. Biol. 2014;10:626–628. doi: 10.1038/nchembio.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arrowsmith CH, et al. The promise and peril of chemical probes. Nat. Chem. Biol. 2015;11:536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munoz L. Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 2017;16:424–440. doi: 10.1038/nrd.2016.266. [DOI] [PubMed] [Google Scholar]
- 48.Rahman N. Mainstreaming genetic testing of cancer predisposition genes. Clin. Med (Lond.) 2014;14:436–439. doi: 10.7861/clinmedicine.14-4-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gootenberg JS, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abudayyeh OO, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JS, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 53.Fife BT, Pauken KE. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann. N. Y Acad. Sci. 2011;1217:45–59. doi: 10.1111/j.1749-6632.2010.05919.x. [DOI] [PubMed] [Google Scholar]
- 54.Reck M, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 55.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 56.Hu Z, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed. Res Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbaum T, Wimmer K. Neurofibromatosis type 1 (NF1) and associated tumors. Klin. Padiatr. 2014;226:309–315. doi: 10.1055/s-0034-1382021. [DOI] [PubMed] [Google Scholar]
- 58.Niu J, Zhang B, Chen H. Applications of TALENs and CRISPR/Cas9 in human cells and their potentials for gene therapy. Mol. Biotechnol. 2014;56:681–688. doi: 10.1007/s12033-014-9771-z. [DOI] [PubMed] [Google Scholar]
- 59.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen ZH, et al. MAN2A1-FER fusion gene is expressed by human liver and other tumor types and has oncogenic activity in mice. Gastroenterology. 2017;153:1120–1132 e1115. doi: 10.1053/j.gastro.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu YP, et al. Novel fusion transcripts associate with progressive prostate cancer. Am. J. Pathol. 2014;184:2840–2849. doi: 10.1016/j.ajpath.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen ZH, et al. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat. Biotechnol. 2017;35:543–550. doi: 10.1038/nbt.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esplin ED, Oei L, Snyder MP. Personalized sequencing and the future of medicine: discovery, diagnosis and defeat of disease. Pharmacogenomics. 2014;15:1771–1790. doi: 10.2217/pgs.14.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evers B, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 66.Wu K, House L, Liu W, Cho WC. Personalized targeted therapy for lung cancer. Int J. Mol. Sci. 2012;13:11471–11496. doi: 10.3390/ijms130911471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang H, Shrager JB. CRISPR/Cas-mediated genome editing to treat EGFR-mutant lung cancer: a personalized molecular surgical therapy. EMBO Mol. Med. 2016;8:83–85. doi: 10.15252/emmm.201506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ratan ZA, et al. CRISPR-Cas9: a promising genetic engineering approach in cancer research. Ther. Adv. Med Oncol. 2018;10:1758834018755089. doi: 10.1177/1758834018755089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maruyama T, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waller MC, Bober JR, Nair NU, Beisel CL. Toward a genetic tool development pipeline for host-associated bacteria. Curr. Opin. Microbiol. 2017;38:156–164. doi: 10.1016/j.mib.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loisel-Meyer S, et al. Potent induction of B- and T-cell immunity against human carcinoembryonic antigen-expressing tumors in human carcinoembryonic antigen transgenic mice mediated by direct lentivector injection. Mol. Cancer Ther. 2009;8:692–702. doi: 10.1158/1535-7163.MCT-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rana, P., A. D. M. a. W. F. (The Wall Street Journal, 1211 Avenue of the Americas, 2018).