Summary
Swarming in Pseudomonas aeruginosa is a coordinated movement of bacteria over semisolid surfaces (0.5%–0.7% agar). On soft agar, P. aeruginosa exhibits a dendritic swarm pattern, with multiple levels of branching. However, the swarm patterns typically vary depending upon the experimental design. In the present study, we show that the pattern characteristics of P. aeruginosa swarm are highly environment dependent. We define several quantifiable, macroscale features of the swarm to study the plasticity of the swarm, observed across different nutrient formulations. Furthermore, through a targeted screen of 113 two-component system (TCS) loci of the P. aeruginosa strain PA14, we show that forty-four TCS genes regulate swarming in PA14 in a contextual fashion. However, only four TCS genes—fleR, fleS, gacS, and PA14_59770—were found essential for swarming. Notably, many swarming-defective TCS mutants were found highly efficient in biofilm formation, indicating opposing roles for many TCS loci.
Subject Areas: Pathogenic Organism, Biological Sciences, Microbiology
Graphical Abstract
Highlights
-
•
The swarm pattern of Pseudomonas aeruginosa is plastic and growth media dependent
-
•
P. aeruginosa swarming motility is promoted by nutrient limitation
-
•
Forty-four P. aeruginosa genes encoding two-component system modulate swarming
Pathogenic Organism; Biological Sciences; Microbiology
Introduction
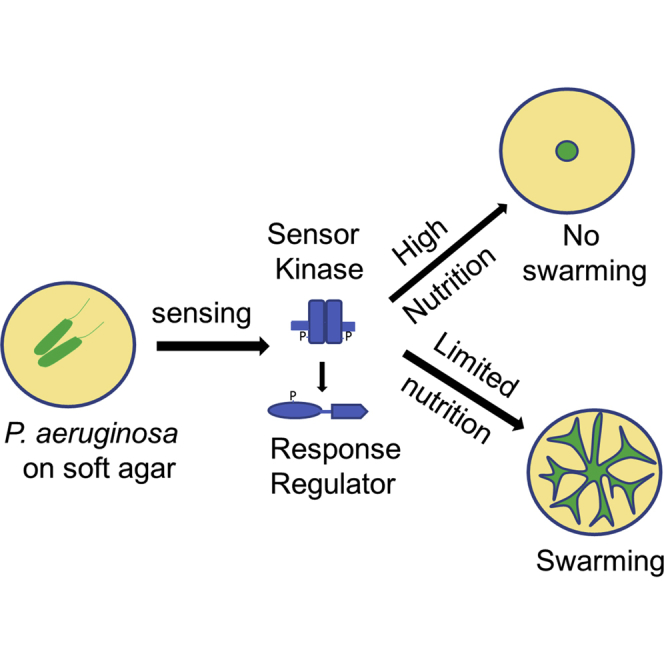
Swarming is a rapid bulk translocation behavior observed in many bacterial species, typically over semisolid agar surfaces (Harshey and Matsuyama, 1994, Henrichsen, 1972, Kearns, 2010). In many instances, bacterial swarm populations generally exhibit characteristic, macroscopic swarm patterns, which are easily recognizable (Kearns, 2010). The opportunistic human pathogen Pseudomonas aeruginosa displays a dendritic-type swarm pattern while swarming on soft agar surfaces. Flagella and quorum sensing (QS) are essential for P. aeruginosa swarming (Kohler et al., 2000, Overhage et al., 2008). Rhamnolipids, a class of glycolipid biosurfactants implicated in virulence and biofilm formation, are also critical for P. aeruginosa swarming, including tendril avoidance (Caiazza et al., 2005, Morris et al., 2011, Xavier et al., 2011). Several lines of evidence indicate that nutrient formulation, such as a change in either carbon or nitrogen sources, can have a drastic impact on rhamnolipid production (Bains et al., 2012, Kohler et al., 2000, Shrout et al., 2006). Most studies of P. aeruginosa swarming are primarily reported under minimal media conditions (M8, M9, and BM2) or in some instances, under complex media formulations such as nutrient broth, brain heart infusion (BHI), or fastidious anaerobe broth (FAB) (Baker et al., 2016, Kohler et al., 2000, Morales-Soto et al., 2015, Overhage et al., 2008, Rashid and Kornberg, 2000, Tremblay and Déziel, 2008). However, despite gross conservation in the dendritic swarm pattern on these different media formulations, the P. aeruginosa swarms often appeared distinct. Hence, these widespread, yet little described, observations strongly urge one to further examine whether the nutritional components of the growth medium can have an impact on P. aeruginosa swarm ability, particularly pattern formation.
Previously, two independent transcriptome studies showed marked dysregulation of very distinct sets of genes, virulence factors in one versus translation and energy metabolism in the other, in P. aeruginosa swarming motility (Overhage et al., 2008, Tremblay and Déziel, 2010). Such a notable divergence between these two studies might have resulted from the use of two different media (BM2 and M9), which differ in nutritional composition (Tremblay and Déziel, 2010). There also exists at least one instance where a P. aeruginosa mutant displays contrasting swarming phenotypes. A pili mutant, pilA, of P. aeruginosa is described as a non-swarmer on M8 agar (Kohler et al., 2000), reported as a swarmer on nutrient broth agar (Rashid and Kornberg, 2000), and reported as a hyper-swarmer on FAB agar (Shrout et al., 2006). These observations also suggested the possible impact of nutrient components on P. aeruginosa swarming and also the conditional requirement of several genetic regulators. An interesting question is how P. aeruginosa cells sense such changes in the nutrition that affect swarm phenotype. Environmental signals such as nutrition influence the activation of the three QS circuits—Rhl, Las, and Pqs—in P. aeruginosa (Dekimpe and Déziel, 2009, Duan and Surette, 2007, Wagner et al., 2003, Welsh and Blackwell, 2016). A few bypass signaling circuits, including a two-component system (TCS), have been implicated in such contexts (Welsh and Blackwell, 2016). For instance, the PhoB-PhoR TCS can directly activate the Rhl QS circuit under phosphate-limited condition (Jensen et al., 2006). However, a comprehensive analysis of the possible impact of various nutrients or the importance of bacterial nutrient sensors in P. aeruginosa swarming is yet to be carried out.
In the present study, we analyzed the swarming behavior of P. aeruginosa strain PA14 across six different nutrient agars and defined swarm features that can be quantified easily. We show that swarm patterns vary considerably across media with reproducible, medium-specific features. We also show that 44 genes encoding TCSs including several poorly characterized or unstudied sensor kinases (SKs) and response regulators (RRs) are required for P. aeruginosa swarming. Among these, four TCS genes are essential for swarming on all media, whereas the remaining have context-specific functions. We also find that several positive regulators of swarming have an opposite effect on biofilm formation.
Results
Phenotypic Plasticity in P. aeruginosa Swarming Is Nutrition Dependent
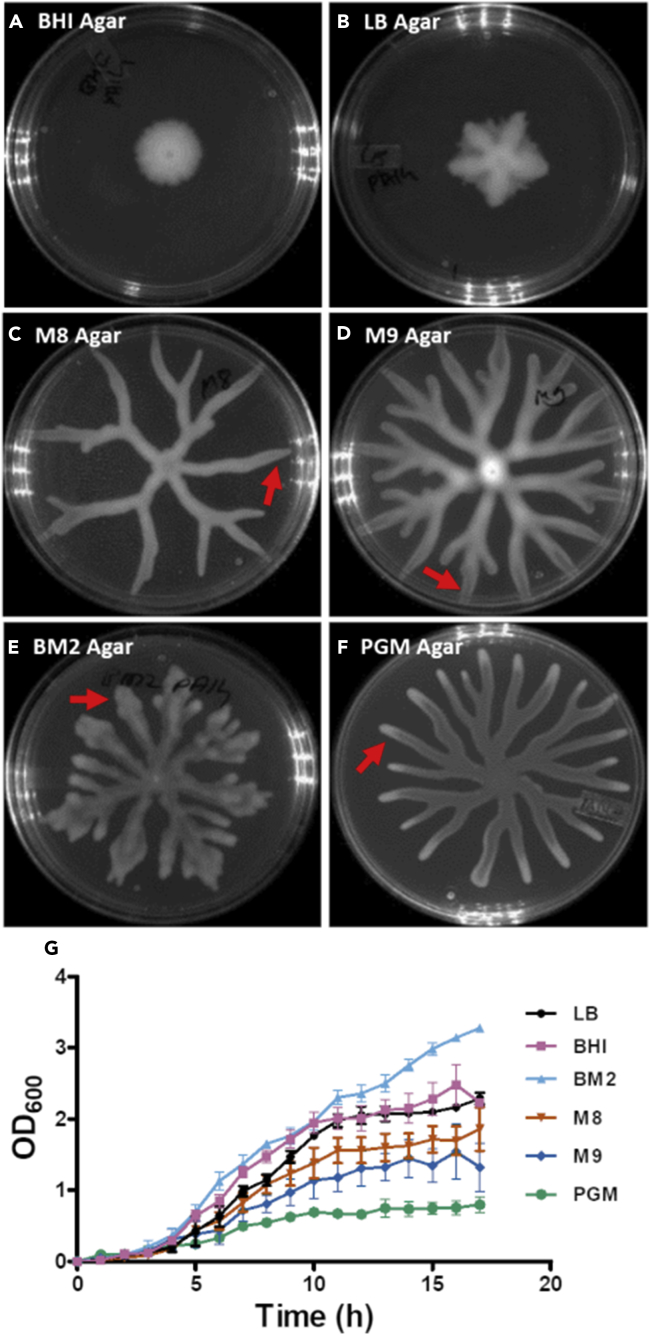
To understand whether nutrition had an impact on swarming, we analyzed P. aeruginosa swarming on six different media—Luria Bertani (LB), BHI, M8, M9, peptone growth media (PGM), and BM2—the nutrient formulations often described for P. aeruginosa growth or swarming studies (Kohler et al., 2000, Morris et al., 2011, Overhage et al., 2008, Overhage et al., 2007, Yeung et al., 2009) (Table 1). Peptone growth media (PGM), also called slow-killing medium, is used for P. aeruginosa growth in Caenorhabditis elegans infection studies (Singh and Aballay, 2006, Sun et al., 2011, Tan et al., 1999). As shown in Figures 1A–1F, we found that four media supported dendritic swarm pattern for P. aeruginosa PA14. However, BHI and LB media did not support dendrite formation, a characteristic of P. aeruginosa swarming (Figures 1A and 1B). LB and BHI agar also poorly supported isometric swarm expansion pattern exhibited by other species such as Escherichia coli, Salmonella typhimurium, or Bacillus subtilis (Harshey and Matsuyama, 1994, Kearns and Losick, 2003, Patrick and Kearns, 2009). Thus three minimal media (M8, M9, and BM2) and one undefined medium (PGM) supported swarming with distinct dendrites. All four media also supported multiple (1–3) levels of branching (Figures 1C–1F) but had medium-specific or plastic patterns.
Table 1.
Media Composition
| M8 | M9 | BM2 | PGM | |
|---|---|---|---|---|
| Magnesium sulfate (MgSO4) | 1.0 mM | 1.0 mM | 2.0 mM | 1.0 mM |
| Sodium chloride (NaCl) | 8.6 mM | 8.6 mM | – | 50 mM |
| Calcium chloride (CaCl2) | – | 1.0 mM | – | 1.0 mM |
| Ammonium chloride (NH4Cl) | – | 20 mM | – | – |
| Potassium phosphate (KPO4) | – | – | 62 mM | 10 mM |
| Potassium dihydrogen phosphate (KH2PO4) | 22 mM | 22 mM | – | – |
| Disodium hydrogen phosphate (NA2HPO4) | 12 mM | 12 mM | – | – |
| Ferrous sulfate FeSO4.7H2O | – | – | 10 μM | – |
| D-glucose | 0.2% | 0.2% | 0.4% | – |
| Casamino acids | 0.5% | 0.5% | 0.1% | – |
| Peptone | – | – | – | 0.32% |
| Cholesterol | – | – | – | 5 μg/ml |
The composition of different nutrient formulations used in this study.
Figure 1.
P. aeruginosa PA14 Swarm Pattern on Different Swarming Agar
(A–F) P aeruginosa swarm obtained at 37°C for 24 h on (A) brain heart infusion (BHI), (B) Luria Bertani (LB), (C) M8, (D) M9, (E) BM2, and (F) peptone growth media (PGM) with 0.6% agar; 6–20 plates were used for each medium.
(G) PA14 wild-type planktonic growth kinetics on various media is shown. Dendrites are indicated with arrow in (C–F).
Furthermore, we analyzed the planktonic growth kinetics of PA14 in all the six media mentioned above. Both LB and BHI broth supported better growth of PA14 (Figure 1G) compared with M8, M9, and PGM broth. The BM2 broth was also able to support good planktonic growth like LB and BHI broth. M8 and M9 broth supported moderate growth, whereas the PGM broth supported poor growth for PA14. Hence, except for the BM2 medium, our data suggest an inverse relationship between planktonic growth and propensity for swarming in P. aeruginosa PA14. Taken together, analysis of swarm agar and broth phase growth of PA14 in three undefined media and three minimal media suggests that poor media, probably lacking specific nutrients, promote swarming. These data also provide evidence for nutrition-dependent plasticity in P. aeruginosa swarm pattern.
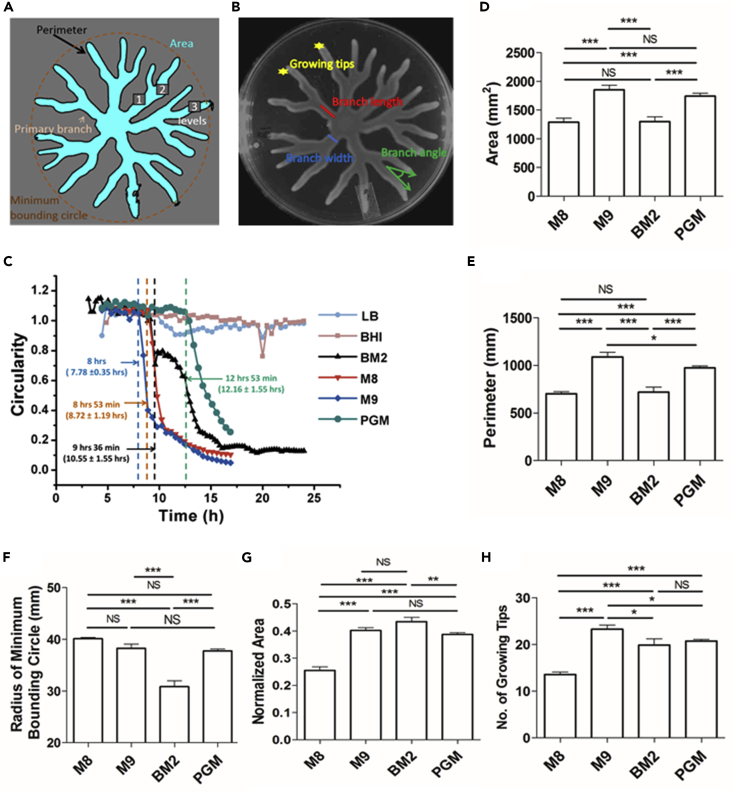
Macroscale Features Define the Plasticity of P. aeruginosa Swarm
To characterize the plasticity in PA14 swarm patterns across different media, we set out to define features of the swarm that could be quantified. We found that the conventional approach of comparing a single feature, such as the swarm diameter or bacterial cell number (Overhage et al., 2007, Xavier et al., 2011, Yeung et al., 2009), was not suitable to differentiate dendritic patterns observed on four different media in our study (Figures 1C–1F). By analyzing several swarm images for each of the media, we defined 11 measurable features of P. aeruginosa swarm illustrated in Figures 2A–2C. These included branch angle, branch width, number of growing tips, area of the swarm, swarm perimeter, normalized area, etc. (Figures 2A and 2B, see Methods). Swarm lag, the time taken to initiate branching from the time of spotting, is the shortest on M9 followed by on M8, BM2, and PGM agar (Videos S1, S2, S3, S4, S5, and S6). This is represented in Figure 2C as circularity versus time. Drop in circularity below the value of 1.0 marks the end of the swarm lag and initiation of branching. We find that the branching starts first in M9, followed by M8, and then on BM2 agar. The swarm lag is longest on PGM agar (Figure 2C). Our observations suggest that medium influences the regulatory program that dictates the set time to initiate branching.
Figure 2.
Macroscale Features of P. aeruginosa Swarm
Quantifiable features of the swarm (A) Perimeter, area, primary branch, branching levels and minimum bounding circle, and (B) Growing tips, branch length, branch width and branch angle are indicated. (see Methods for definition).
(C–H) (C) Circularity plot for swarm expansion on LB, BHI, M8, M9, BM2, and PGM swarm agar. End of swarm lag is indicated. Histogram for (D) swarm area, (E) perimeter, (F) radius of minimum bounding circle, (G) normalized area, and (H) number of growing tips. Pairwise comparison between every two media was performed by Tukey test.
p > 0.05, ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. See also Videos S1, S2, S3, S4, S5, and S6 and Figure S1.
We utilized one-way ANOVA to isolate features that account for variance across swarm patterns observed on four media that promote dendritic swarming. All 10 macroscale features—area (***), perimeter (***), radius of minimum bounding circle or RMBC (***), normalized area (***), branch length (**), branch angle (***), branch width (***), number of levels (*), number of primary branches (*), and number of growing tips (***)—could explain the variance across swarms on these media. Furthermore, we utilized Tukey's post hoc test to identify features that vary significantly between any two media (Figures 2D–2H, Figure S1). For example, perimeter and area coverage (mean ± SEM) values for swarm were significantly different (Figure 2D) for most pairwise comparisons, as well as the number of growing tips and normalized area. However, to understand the contribution of each feature to the swarm plasticity across different media, we performed a principal-component analysis (PCA). For the PCA, we used all the 10 macroscale features other than circularity of several swarms on each of the four media (Methods). Principal component 1 (branch angle, area, perimeter, normalized area, and growing tips) contributed 37% to the variance, whereas component 2 (branch width and radius of minimum bounding circle) contributed to 25% (Figure 3A) of the variance across media. Principal component 3 (branch length, number of primary branches, and number of levels) contributed only 14% to the variance (Figure 3B). Normalized area versus perimeter could also distinguish PA14 swarm patterns on M8, M9, BM2, and PGM agar into distinct centroids (Figure 3C). We have used these two features in the rest of this study. Taken together, we could define multiple quantifiable features of P. aeruginosa PA14 swarm that can be used to analyze perturbations to swarm patterns.
Figure 3.
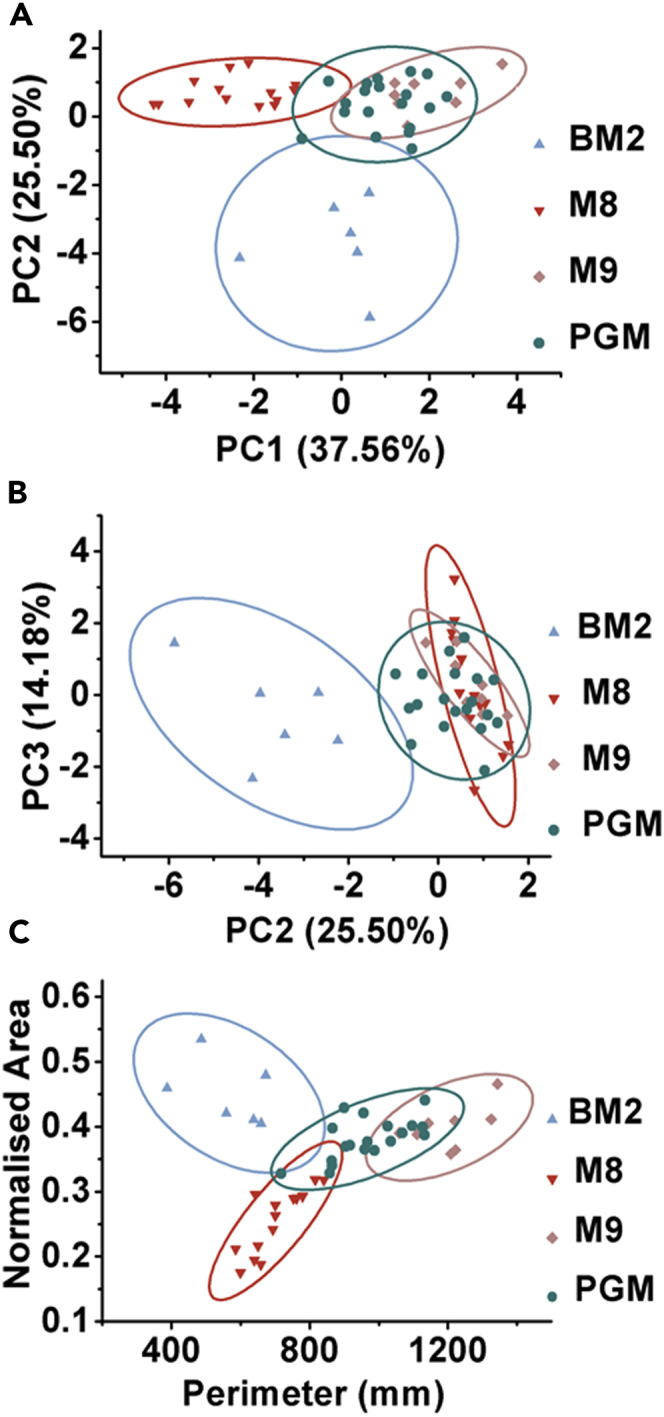
Principal-Component Analysis of Swarm Features
(A and B) (A) PC1 (branch angle, area, perimeter, normalized area, and growing tips) versus PC2 (branch width and radius of minimum bounding circle), and (B) PC2 versus PC3 (branch length, number of primary branches and number of levels) plot for dendritic swarm pattern on BM2, M8, M9, and PGM agar.
(C) Perimeter versus normalized area plot for swarm patterns on BM2, M8, M9, and PGM agar.
Centroids for each medium-specific swarm are indicated in (A–C). See also Figures S2 and S3.
Several Two-Component Genes of P. aeruginosa Are Conditional Modulators of Swarming
Media-dependent plasticity in PA14 swarming patterns strongly suggested that nutritional cues, in the growth media, might be critical for inducing swarming in P. aeruginosa. We then set out to ask how does P. aeruginosa sense such changes to execute swarming? In prokaryotes, the TCSs are predominantly involved in sensing environmental signals such as nutrition, change in pH, redox state, osmolarity, and light (Laub and Goulian, 2007, Stock et al., 2000, Zschiedrich et al., 2016). The P. aeruginosa PA14 genome encodes 160 TCS genes (Lee et al., 2006, Liberati et al., 2006) and is thought to confer exceptional adaptability of this bacterium to various environmental niches and a wide range of hosts, including humans, Drosophila, C. elegans, and plants (Barreteau et al., 2009, Clatworthy et al., 2009, D'argenio et al., 2001, Francis et al., 2017, Rodrigue et al., 2000, Tan et al., 1999).
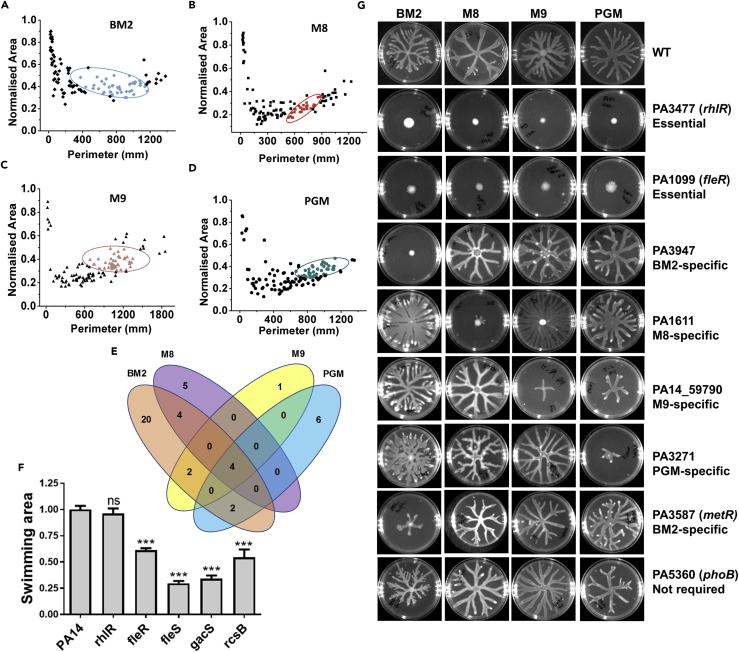
In a previous genetic screen for swarming in P. aeruginosa PA14, 12 of the candidate genes identified were TCS class regulators (Yeung et al., 2009). This study was, however, under BM2 agar condition alone. We hypothesized that several TCSs are required to sense differential nutritional signals, which promote swarming. As the media used in this study vary in both macro- and micronutrients (Table 1), we expected to find distinct TCS genes to be required for swarming on different media. To test this hypothesis, we performed a targeted genetic screen for swarming, using transposon insertion mutations affecting 113 TCS loci of PA14 (Liberati et al., 2006). The screen was performed on all six media in duplicates. To assess the effect of TCS genes on swarming, we extracted three macroscale features—area, perimeter, and normalized area—from 681 swarms using MATLAB (see Figures S2 and S3). As shown in perimeter versus normalized area plot for four media in Figure 4A–4D, many TCS genes were required for swarming (Table S1). Many of the swarm regulators were orphan, whereas there were four pairs (Table S2; images in Figure S4).
Figure 4.
Two-Component Systems of P. aeruginosa Regulate Swarming
(A–D) Perimeter versus normalized area plot for PA14 and 113 TCS mutants on (A) BM2 agar, (B) M8 agar, (C) M9 agar, and (D) PGM agar. Centroid for wild-type PA14 on respective media is shown. All mutants outside the centroids represent weak swarmers or non-swarmers.
(E) Venn diagram to show media-specific non-swarmer TCS mutants (also see Table S1).
(F) Representative images of swarm of six TCS mutants, rhlR and metR strains on BM2, M8, M9, and PGM media.
(G) Swimming phenotype of essential swarm regulators on LB-0.3% swim agar.
Mean values were compared by unpaired t test (p > 0.5, ns or not significant; ***p < 0.001). See also Figures S2–S4, and Table S1.
Irrespective of difference in nutrient formulations, we found that four TCS genes were essential for swarming on all media, whereas nine TCS genes were required for swarming on at least two media. However, the largest number of TCS genes (31 of 44) displayed medium specific role in swarming. Notably, 13 SK- and 7 RR-encoding genes were essential for swarming exclusively on BM2 agar; only 3 RR- and 2 SK-encoding genes were found essential for swarming on M8 agar alone. Swarming on PGM agar required six TCS genes, whereas a single TCS gene encoding an RR was found essential exclusively in swarming on M9 agar. Media-dependent requirement of TCS is displayed in a Venn diagram (Figure 4E). These observations suggest that swarming on the BM2 medium is dependent on signaling from many SKs, whereas swarming on other media is less reliant on them. It is also interesting to point out that in 23 cases either the RR or the SK, but not both, was enriched in the screen, indicating a possible cross talk in P. aeruginosa TCS signaling for swarming (Table S2). However, we did find four cognate pairs that displayed phenotypic correlation for swarming.
Majority of TCS mutants displayed a strong, but context-dependent, swarming phenotype across media. A few striking examples are presented in Figure 4F. PA3947/rocR had a BM2-agar-specific function (Figure 4F). Similarly, PA3271 and PA1611 had specific function in swarming on PGM agar and M8 agar, respectively. PA14_59790/pvrR, an RR found exclusively in P. aeruginosa PA14 and PA7 genomes, showed a role in swarming on M9 and weakly on PGM agar (Figure 4F). As a control, we checked the context-dependent requirement for PA3587/metR, a previously described transcriptional regulator of swarming on BM2 agar (Yeung et al., 2009). We found that metR was indeed swarming deficient on BM2 agar but swarming proficient on M8, M9, and PGM agar (Figure 4F). Interestingly, phoB required for swarming in low-phosphate (2 mM) BM2 medium (Bains et al., 2012) was swarming proficient on all the four media we tested (Figure 4F). This was expected as all media we used contain phosphate concentration of 24 mM or above.
Flagella and QS are essential for swarming in PA14 strain (Kohler et al., 2000; this study), although they have muted phenotype in PA01 strain (Gellatly et al., 2018). Indeed, quorum-defective mutant, rhlR, was a non-swarmer on all media we tested. However, rhlR had a wild-type swimming phenotype (Figure 4G). In contrast, essential swarming regulators of the TCS class were swimming defective (Figure 4G). fleS and fleR mutant showed moderate to severe swimming defect (Figure 4G), as also shown earlier (Ritchings et al., 1995). An analysis of transposon insertions in 26 flagellar genes (Liberati et al., 2006) also showed non-swarming phenotype (data not shown). Taken together, these experiments suggested that swimming ability is indeed essential for swarming under all conditions.
In all, our observations showed that 44 TCS genes are required for swarming, but in a conditional manner. The results indicated that many nutritional or environmental cues promote swarming by activating specific two-component signaling circuits.
TCS Genes Differentially Regulate Swarming and Biofilm Formation
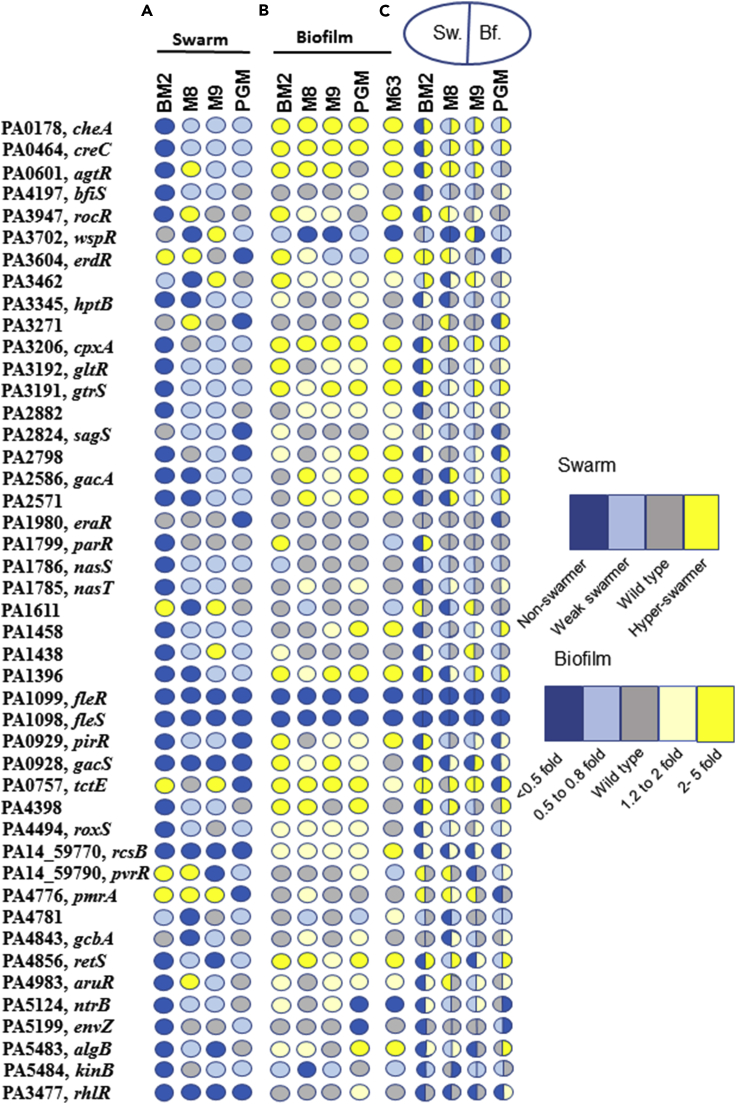
Biofilm and swarming constitute the sessile and motile population, respectively, but both rely on QS. In P. aeruginosa certain cellular components such as flagella are required for both swarming and biofilm formation (Kohler et al., 2000, O'Toole and Kolter, 1998). In contrast, some regulatory components important for biofilm formation negatively regulate swarming motility (Ueda and Wood, 2009, Bhuwan et al., 2012, Kuchma et al., 2007). For instance, higher cellular level of cyclic di- guanosine monophosphate (GMP) molecule is considered a major switch for biofilm formation in many bacteria, including P. aeruginosa (Baker et al., 2016, Romling et al., 2013, Valentini and Filloux, 2016), while suppressing swarming. Indeed, some of the genes discovered as swarm regulator in our study—gacA/gacS, retS, sagS, bfiS, wspR, and hptB—are known to be regulators of biofilm formation (reviewed in Francis et al., 2017). Yeung et al. had examined 35 swarming-defective strains and found that 19 formed better biofilm. Of these, five were TCS components—cbrA, gacS, ntrC, PA4398, and algR (Yeung et al., 2009). This raised the question whether swarm regulators discovered in this study regulate biofilm formation in PA14 and in what manner.
To understand the impact of TCS genes on biofilm formation, we assayed biofilm formation by PA14 wild-type and TCS mutants as described (O'Toole, 2011) at 24 h. We used M63 medium recommended for quantification of biofilm in addition to M8, M9, BM2, and PGM broth (Table S3). Figure 5A shows swarm phenotype of all 44 swarm mutants (see perimeter values in Table S1) and rhlR mutant on four different media in colored disk format. Figure 5B shows biofilm phenotype based on crystal violet stain of TCS mutant on five media, again in colored disk format. We found that biofilm formation was not influenced by a change in media (compare disk color in each column in Figure 5B, values in Table S3). Only three mutants were unable to form the biofilm on all media (dark blue disks, Figure 4B). These included fleS and fleR mutants defective in flagella biogenesis and wspR. sagS and bfiS are known to regulate old biofilm in PA01 strain (Petrova and Sauer, 2011), but we found no phenotype for them in young biofilm in PA14 strain. However, four TCS mutants (cheA, creC, cpxA, and tctE) showed hyper-biofilm phenotype (deep yellow disks in Figure 4B) and thus appear to be negative regulators of biofilm formation, in a media-independent manner. There were few media-specific regulators of biofilm formation as well. For instance, ntrB was essential for biofilm formation in M63 and PGM broth, whereas kinB was found essential only in M8 broth. The relationship between biofilm formation and swarm formation phenotypes is displayed in a double-faced Janus droplet map for four media in Figure 5C. The left half of the droplet represents swarm phenotype, whereas the right half represents swarming phenotype under the same condition (medium). We found that in 38 instances non-swarmer TCS mutants or a weak swarmer produced significantly better biofilm (2- to 5-fold increase in crystal violet stain) than the wild-type PA14 (16 on BM2, 8 on PGM, 9 on M9, 5 on M8, and 8 on PGM). There were 12 instances of coordinate regulation of swarming and biofilm formation, fleR, fleS, wspR, PA4781, and kinB, for both swarming and biofilm formation. In three instances, erdR and tctE were negative regulators of both biofilm and swarm (yellow Janus droplet in Figure 5C). All other events appear to indicate that TCS genes regulate one process (swarm or biofilm) but not the other. Taken together, the systematic analysis of swarm and biofilm formation on four different media (176 one-on-one comparisons) indicated that several, but not all, TCS circuits regulate switch between swarming and biofilm formation.
Figure 5.
Media-Dependent Role of TCS Genes in Swarming and Biofilm Formation
(A) Disk heatmap of swarm phenotype of 44 TCS mutants and rhlR. Heatmap is based on perimeter values in Table S1. Swarm phenotype: non-swarmer, perimeter less than 20% of PA14 swarm perimeter; weak swarmer, >20% but less than mean − SD of PA14 perimeter; hyper-swarmer, > mean + SD of PA14 swarm perimeter.
(B) Biofilm formation by 44 TCS mutants and rhlR in BM2, M8, M9, PGM, and M63 broths, measured by crystal violet stain (values in Table S3).
(C) Janus droplet representation of media-dependent biofilm and swarm phenotype. Left face shows swarming phenotype, whereas the right face reflects biofilm phenotype. Crystal violet staining for biofilm is represented as fraction of biofilm formation by PA14.
See also Figure S4, Tables S1–S3.
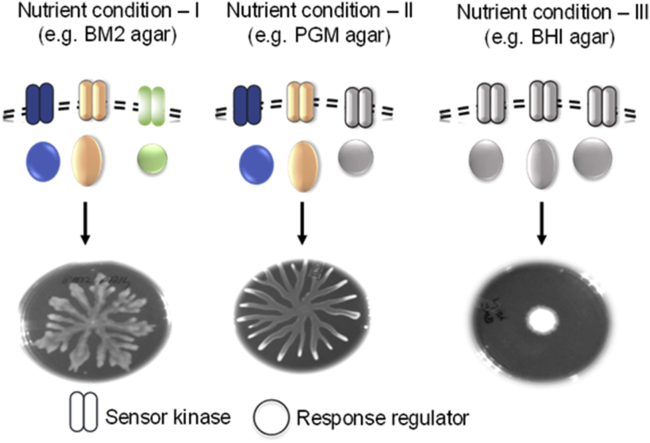
In summary, this study provides evidence that many TCS genes are critical for swarming in P. aeruginosa in a contextual manner (Figure 6). We find that PA14 swarming under one condition, such as BM2 agar, requires input from several TCS systems, whereas swarming on M8, M9, and PGM media (condition II) rely on fewer TCS circuits. Nutritionally rich media, LB and BHI, do not support dendritic swarming in P. aeruginosa PA14. Thus extrinsic nutritional cues in conjunction with bacterial SK/RR systems are critical in the modulation of P. aeruginosa swarming.
Figure 6.
Media-Dependent Plasticity in P. aeruginosa Swarming
Growth media vary in macro- and micronutrient contents. Some growth media (condition I) depend on several TCS modules to allow swarming of P. aeruginosa. Another set of media (condition II) promote swarming but rely on a smaller but specific set of TCS genes for swarming. Certain other growth media (condition III) do not allow dendritic swarming of P. aeruginosa.
Discussion
In this study, we show that P. aeruginosa exhibits a remarkable, context-dependent plasticity in its swarming behavior. This arises due to nutrient limitation in growth media and is sensed by the TCS class of SKs and their partners called response regulators. We provide a number of macroscale features of P. aeruginosa swarm to differentiate media-specific swarm patterns into distinct populations.
We were able to establish that swarm lag, the time to initiate branching or dendrite formation, is a quantifiable feature of P. aeruginosa swarming that can be represented as a change in circularity in time. Indeed, swarm lag was significantly different between M8, M9, BM2, and PGM agar. The circularity remained close to 1 for the entire duration in non-dendritic growth on LB and BHI, again making it a valuable feature. Circularity, as well as other macroscale features described in this study, can be utilized to quantify the effect of the environmental factors or genetic regulators on the swarm pattern.
One important question raised by this study is what are the nutritional cues that promote swarming? Boyle et al. (Boyle et al., 2015) have suggested that iron limitation is a requirement for swarming. Among the four media we used, only BM2 had the iron supplement. However, it does support PA14 swarming with a medium-specific pattern. The nitrogen content of the BM2 medium, however, is lower than that of M8 and M9 media (Table 1), suggesting that nitrogen limitation could also be a driver for initiating swarming under BM2 condition. Indeed, nitrogen-related TCSs—ntrB and nasS/nasT—were essential for swarming on BM2 media alone. These mutants also displayed weak swarming on other three media, suggesting that nitrogen limitation may be a contributing factor to swarming in those media as well. Phosphate limitation is a known driver for swarming (Bains M, Fernández L, 2012), but it was not relevant for the four media we tested due to the presence of phosphate in the media and dispensability of phosphate-specific TCS phoB and phoR (Table 1, Figure 4F) for swarming in this study. Involvement of low Mg2+ and cationic-peptide-inducible PmrA on swarming on PGM agar suggests that Mg2+ and cationic peptides may become relevant on certain media. Addition or removal of specific macro- and micronutrients (Table 1) to or from these four media will serve to decipher additional nutritional cues that drive TCS genes to modulate swarming. Do similar drivers exist in a P. aeruginosa infection setting in humans? A careful analysis of nutrients in body fluids of the host in different pathologies such as cystic fibrosis and diabetic foot ulcer can provide better insight into P. aeruginosa pathogenesis.
Our study raises a second question: How does nutrition impact swarming? It could be via modulation of flagellar output, modulation of rhamnolipid production, or novel pathways required for hitherto unidentified effector molecules, necessary for swarming. In planktonic growth, rhamnolipid production is induced at the end of the lag phase, which corresponds with onset of nutrient limitation (Caiazza et al., 2005). In addition, Xavier et al. have shown that rhamnolipid production is regulated by nitrogen limitation in minimal media (Xavier et al., 2011). We find that rhamnosyl transferase chain A (RhlA) transcription in much higher in PGM-broth-grown PA14 than in LB-broth-grown PA14. Transcription of rhlA is further induced on PGM-0.6% swarm agar than PGM broth (data not shown). This indicated that rhamnolipid production is dependent on media as well as surface contact. Careful analyses of rhamnolipid production in response to removal or limitation of specific nutrient one at a time would be instructive in understanding the regulation of swarming. Some of the TCS components may regulate swarming via the modulation of cyclic di-GMP levels. We found that PA4398 is a positive regulator of swarming on BM2 medium and a negative regulator of biofilm formation (Figures 5A and 5B). This is in agreement with an earlier report wherein the mutation in PA4398 led to a 50% increase in intracellular cyclic-di-GMP, which is linked to enhanced biofilm formation (Strehmel et al., 2015). Indeed, six RRs (RocR, WspR, ErdR, PvrR, PA4781, and PA4843), we identified as swarm regulator genes, do possess diguanylate cyclase (GGDEF) or phosphodiesterase (EAL and HD-GYP) domains for regulation of cyclic-di-GMP turnover in bacteria. There is also some evidence for the role of nitrogen limitation in swarming. A TCS pair (NasS/nasT) involved in nitrate assimilation is essential for swarming on BM2. In addition, SK NtrB and two ntrC domains containing RR are also essential for swarming (Table S2). A systematic study of each TCS component in a medium-specific context will help decipher the possible mechanism of modulation of swarming. Over time, it would be possible to build up the regulatory program for swarming in P. aeruginosa.
A growing body of evidence suggests that environmental signals, particularly nutritional cues, can differentially influence QS in P. aeruginosa (Duan and Surette, 2007, Jensen et al., 2006, Wagner et al., 2003, Welsh and Blackwell, 2016). The TCS class signaling was one of the few bypass activation circuits implicated in such contexts (Jensen et al., 2006, Welsh and Blackwell, 2016). We were surprised to find 32 TCS modulators of swarming on BM2 agar when compared with 12 genes uncovered in a previous screen (Yeung et al., 2009). We believe that additional TCS regulators of swarming were found due to the utilization of a large (90-mm) dish format for each strain in our study when compared with the 96-well multiplicator format deployed for the primary screen in the previous report (Yeung et al., 2009). We find that avoidance zone between two swarms occurs at about 5 mm distance, and 96-prong multiplicator does not allow enough expansion of swarm to detect all non-swarmers in our laboratory (data not shown). A recent study from our laboratory shows that P. aeruginosa can detect both bacteria and non-biological obstacles on PGM swarm agar (Kotian et al., 2018) reiterating that swarming in P. aeruginosa is sensitive to environmental cues including proximity to isogenic bacteria.
P. aeruginosa genomes exhibit expansion of the TCS-encoding genes to 160, one of the largest TCS repertoires among eubacteria. The GacS network, along with HptB and SagS branches, control biofilm, virulence, and motility. We found that six components of this extended network (gacS, gacA, retS, PA1611, hptB, sagS, and bfiS) were regulators of swarming in P. aeruginosa. Only gacS was an essential regulator of swarming. On the other hand, some of the conditional swarm modulators uncovered in this study are linked to nutrient assimilation (e.g., ntrB, nasS, and nasT). Several TCS mutants previously described as motility-, swarming-, or biofilm-related loci including PA3702/wspR (Chen et al., 2014), PA3345/hptB (Bhuwan et al., 2012, Hsu et al., 2008) PA14_59770 (rcsB) (Giraud et al., 2009), PA14_59790 (pvrR) (Giraud et al., 2009, Zheng et al., 2016), and PA4398 (Strehmel et al., 2015) enriched as swarm regulator, at least under one condition, in our study. To the best of our knowledge, the others are not known to have any swarming-related function. A very recent study analyzed TCS genes for cytotoxicity in a cell line model (Gellatly et al., 2018) and found 27 TCS genes. Only six of these were enriched as swarm regulators in our study. This indicates that TCS genes control different biological processes. Based on our study, we would like to propose that some of the P. aeruginosa TCS genes may have evolved, and would have been retained in the genome, to modulate swarming motility.
One of the surprises from this study came in the form of antagonistic regulation of swarm and biofilm formation by TCS genes and strengthens previous reports (Yeung et al., 2009). There were 38 cases of an inverse relationship between biofilm and swarm. This indicates that several TCS signaling circuits might promote swarming while suppressing biofilm formation. This leads to an interesting hypothesis that P. aeruginosa can possibly only exist in one state (either swarming or biofilm) at one time. In future, we would like to study the molecular signatures of the two states. Swarming bacteria are believed to be antibiotic resistance (Butler et al., 2010, Overhage et al., 2008). One of the directions to pursue will be to ask if swarming population of P. aeruginosa is more susceptible to antibiotics than P. aeruginosa in a biofilm. If so, could we change the state of the bacteria and make them more susceptible to antibiotics. This could be done by pharmacological intervention or simply by perturbation of host body fluids? A comprehensive analysis of transcriptional events that initiate biofilm versus initiate swarm will add to better understanding of the differences between these two quorum-dependent processes.
Limitations of the Study
Our study of the swarm pattern is limited to six different media conditions, although additional media have been described for P. aeruginosa growth. Although there are about 160 predicted two-component-related loci in P. aeruginosa PA14 genome, our study covers only 113 available in the transposon insertion library. Because of the broad-based nature of our study, we were not able to provide mechanistic details behind each of our observations. We strongly believe that our findings would be of interest to P. aeruginosa community as well as broader swarming research community.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Pseudomonas aeruginosa PA14 transposon insertion library was a gift from Prof. Frederick M. Ausubel, Massachusetts General Hospital, Boston. This work was supported by Robert Bosch Innovation Centre Grant (grant no. RBCO0014) awarded to M.V. and V.S. and DBT-IISC Partnership Program (BT/PR27952/INF/22/212/2018). S.J. was partly supported by Junior Research Fellowship from Council for Scientific and Industrial Research CSIR (CSIR-JRF 09/079(2764)/2017-EMR-1). We thank Deepak K. Saini, Sandhya S. Visveswaraiah, and Sambuddho Mukherjee for critical reading of the manuscript.
Author Contributions
A.M.K, S.J., D.P., H.S.K., D.P., A.M., M.V., and V.S. conceptualized the study. S.J., A.M.K., D.P., and A.M. performed all the experiments. S.J., A.M.K., D.P., and A.M. analyzed the data. H.K. and D.B. performed MATLAB-based processing of images and videos. H.K. and D.B. derived swarm features from swarm images and performed PCA analysis. S.J., A.M.K., H.K., D.P., M.V., and V.S. wrote the manuscript.
Declaration of Interests
Authors declare no conflict of interest.
Published: March 29, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.02.028.
Supplemental Information
References
- Bains M., Fernández L., Hancock R.E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012;78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A.E., Diepold A., Kuchma S.L., Scott J.E., Ha D.G., Orazi G., Armitage J.P., O’Toole G.A. A PilZ domain protein FlgZ mediates c-di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J. Bacteriol. 2016;198:1837–1846. doi: 10.1128/JB.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H., Bouhss A., Fourgeaud M., Mainardi J.L., Touzé T., Gérard F., Blanot D., Arthur M., Mengin-Lecreulx D. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J. Bacteriol. 2009;191:3657–3664. doi: 10.1128/JB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuwan M., Lee H., Peng H., Chang H. Histidine-containing phosphotransfer protein-B ( HptB ) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J. Biol. Chem. 2012;287:1903–2114. doi: 10.1074/jbc.M111.256586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle K.E., Monaco H., van Ditmarsch D., Deforet M., Xavier J.B. Integration of metabolic and quorum sensing signals governing the decision to cooperate in a bacterial social trait. PLoS Comput. Biol. 2015;11:1–26. doi: 10.1371/journal.pcbi.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.T., Wang Q., Harshey R.M. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U S A. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza N.C., Shanks R.M.Q., O’Toole G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.I., Dolben E.F., Okegbe C., Harty C.E., Golub Y., Thao S., Ha D.G., Willger S.D., Toole G.A.O., Harwood C.S. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014;10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A.E., Lee J.S.W., Leibman M., Kostun Z., Davidson A.J., Hung D.T. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V., Déziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- Duan K., Surette M.G. Environmental regulation of Pseudomonas aeruginosa PAO1 las and Rhl quorum-sensing systems. J. Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'argenio D.A., Gallagher L.A., Berg C.A., Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 2001;183:1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis V.I., Stevenson E.C., Porter S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017;364:1–22. doi: 10.1093/femsle/fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly S.L., Bains M., Breidenstein E.B.M., Strehmel J., Reffuveille F., Taylor P.K., Yeung A.T.Y., Overhage J., Hancock R.E.W. Novel roles for two-component regulatory systems in cytotoxicity and virulence-related properties in Pseudomonas aeruginosa. AIMS Microbiology. 2018;4:173–191. doi: 10.3934/microbiol.2018.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud C., Filloux A., Mikkelsen H. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R.M., Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U S A. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., Chen H., Peng H., Chang H. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen V., Löns D., Zaoui C., Bredenbruch F., Meissner A., Dieterich G., Münch R., Häussler S. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 2006;188:8601–8606. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D.B., Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kohler T., Curty L.K., Barja F., Van Delden C., Pechere J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotian H.S., Harkar S., Joge S., Mishra A., Zafal A., Singh V., Varma M.M. Spatial awareness of a bacterial swarm. bioRxiv. 2018 doi: 10.1101/341529. [DOI] [Google Scholar]
- Kuchma S.L., Brothers K.M., Merritt J.H., Liberati N.T., Ausubel F.M., O’Toole G.A. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Lee D.G., Urbach J.M., Wu G., Liberati N.T., Feinbaum R.L., Miyata S., Diggins L.T., He J., Saucier M., Déziel E. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Soto N., Anyan M.E., Mattingly A.E., Madukoma C.S., Harvey C.W., Mark A., Déziel E., Kearns D.B., Shrout J.D. Preparation, imaging, and quantification of bacterial surface motility assays. J. Vis. Exp. 2015 doi: 10.3791/52338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.D., Hewitt J.L., Wolfe L.G., Kamatkar N.G., Chapman S.M., Diener J.M., Courtney A.J., Leevy W.M., Shrout J.D. Imaging and analysis of Pseudomonas aeruginosa swarming and rhamnolipid production. Appl. Environ. Microbiol. 2011;77:8310–8317. doi: 10.1128/AEM.06644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J., Lewenza S., Marr A.K., Hancock R.E.W. Identification of genes involved in swarming motility using a mutant library. J. Bacteriol. 2007;189:2164–2169. doi: 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J., Bains M., Brazas M.D., Hancock R.E.W. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 2008;190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Patrick J.E., Kearns D.B. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J. Bacteriol. 2009;191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova O.E., Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M.H., Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchings B.W., Almira E.C., Lory S., Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A., Quentin Y., Lazdunski A., Méjean V., Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- Romling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout J.D., Chopp D.L., Just C.L., Hentzer M., Givskov M., Parsek M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Singh V., Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U S A. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Strehmel J., Neidig A., Nusser M., Geffers R., Brenner-Weiss G., Overhage J. Sensor kinase PA4398 modulates swarming motility and biofilm formation in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 2015;81:1274–1285. doi: 10.1128/AEM.02832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R., Aballay A. Neuronal GPCR controls innate immunity by regulating non- canonical unfolded protein response genes. Science. 2011;332:729–732. doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.-W., Mahajan-Miklos S., Ausubel F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J., Déziel E. Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 2008;48:509–515. doi: 10.1002/jobm.200800030. [DOI] [PubMed] [Google Scholar]
- Tremblay J., Déziel E. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics. 2010;11:587. doi: 10.1186/1471-2164-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A., Wood T.K. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA. PLoS Pathog. 2009;5:1–15. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M., Filloux A. Biofilms and Cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V.E., Bushnell D., Passador L., Brooks A.I., Iglewski B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M.A., Blackwell H.E. Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 2016;23:361–369. doi: 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier J.B., Kim W., Foster K.R. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A.T.Y., Torfs E.C.W., Jamshidi F., Bains M., Wiegand I., Hancock R.E.W., Overhage J. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 2009;191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Tsuji G., Opoku-temeng C., Sintim H.O. Inhibition of P. aeruginosa c-di-GMP phosphodiesterase RocR and swarming motility by a benzoisothiazolinone derivative. Chem. Sci. 2016;9:6238–6244. doi: 10.1039/c6sc02103d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich C.P., Keidel V., Szurmant H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.