Abstract
IKZF1 deletion (ΔIKZF1) is an important predictor of relapse in both childhood and adult B-cell precursor acute lymphoblastic leukemia (B-ALL). Previously, we revealed that COBL is a hotspot for breakpoints in leukemia and could promote IKZF1 deletions. Through an international collaboration, we provide a detailed genetic and clinical picture of B-ALL with COBL rearrangements (COBL-r). Patients with B-ALL and IKZF1 deletion (n = 133) were included. IKZF1 ∆1-8 were associated with large alterations within chromosome 7: monosomy 7 (18%), isochromosome 7q (10%), 7p loss (19%), and interstitial deletions (53%). The latter included COBL-r, which were found in 12% of the IKZF1 ∆1-8 cohort. Patients with COBL-r are mostly classified as intermediate cytogenetic risk and frequently harbor ETV6, PAX5, CDKN2A/B deletions. Overall, 56% of breakpoints were located within COBL intron 5. Cryptic recombination signal sequence motifs were broadly distributed within the sequence of COBL, and no enrichment for the breakpoint cluster region was found. In summary, a diverse spectrum of alterations characterizes ΔIKZF1 and they also include deletion breakpoints within COBL. We confirmed that COBL is a hotspot associated with ΔIKZF1, but these rearrangements are not driven by RAG-mediated recombination.
Introduction
B-cell precursor acute lymphoblastic leukemia (B-ALL) comprises multiple subtypes defined by structural and numerical chromosomal alterations. These initiating lesions include aneuploidy and chromosomal rearrangements, leading to expression of ectopic fusion proteins and/or to deregulation of gene expression. During the evolution of B-ALL, a series of secondary genomic alterations, such as DNA copy number alterations (CNAs) and sequence mutations, usually emerges [1]. These secondary events commonly involve deletions of genes that encode regulators of cell-cycle (CDKN2A and CDKN2B) and B-cell development (PAX5 and IKZF1) [2].
IKZF1 deletions occur in ~15% patients with B-ALL and are more recurrent in the BCR-ABL1 and Philadelphia-like subgroups, affecting more than 70% and 40% of the patients, respectively [3]. IKZF1 is located within chromosome band 7p12 and codifies the transcription factor IKAROS. It has two main domains: a DNA-binding zinc finger domain (encoded by exons 4-6) and a zinc finger dimerization domain (encoded by exon 8). Some intragenic IKZF1 deletions impair its DNA-binding domain resulting in a dominant-negative effect (e.g. Δ4-7), while deletions of the whole gene (Δ1-8), as well as deletions of exons 1-2 (e.g. Δ2-3) lead to the gene haploinsufficiency. IKZF1 deletions have been associated with dismal prognosis in both pediatric and adult B-ALL [4], [5], despite the type of deletion [6]. Interestingly, sequencing of IKZF1 breakpoints suggested that aberrant recombination activating gene (RAG)–mediated recombination is responsible for these deletions [7]. Due to their clinical relevance, the identification of IKZF1 deletions has become an essential prognostic biomarker over the last decade. Hence, aiming to detect these deletions, we developed a rapid and efficient polymerase chain reaction (PCR)–based method [8]. Recently, by delineating the genetic landscape of IKZF1 Δ1-8, we identified COBL (COBL cordon-bleu WH2 repeat protein) gene as an originating hotspot for IKZF1 deletions [9]. Here we established an international collaboration to enable a detailed genetic and clinical characterization of B-ALL with COBL rearrangements (COBL-r).
Material and Methods
Patients
This study included 146 patients diagnosed with B-ALL and IKZF1 deletions. This patient cohort comprised two independent groups. The first one included 133 patients with 1) IKZF1 complete deletion, Δ1-8 (n = 104); or 2) intragenic deletion of IKZF1 including exon 8, Δn-8 (n = 29). The second group referred to 13 patients already diagnosed with IKZF1 deletions and COBL-r at collaborating centers [8], [9], [10]. Patient samples and information were obtained through the cooperative efforts of nine centers worldwide, and the project was conducted in accordance with the declaration of Helsinki.
Detection of IKZF1 Deletions and CNAs
IKZF1 deletions and CNAs in EBF1, JAK2, CDKN2A, CDKN2B, PAX5, ETV6, BTG1, RB1, and the pseudoautosomal region 1-PAR1 (SHOX, CRLF2, C2F2RA, IL3RA, and P2RY8) were determined by either multiplex ligation-dependent probe amplification (MLPA), or single nucleotide polymorphism (SNP) array analyses. The SALSA MLPA P335-(A3-B2) and/or SALSA MLPA P202-B1 (MRC Holland) were used for MLPA experiments, and the data analyses were performed using Coffalyser software. The CytoScan HD Array (Affymetrix) assessed the occurrence of CNAs in the Brazilian (partially), British, and French cohorts. Data were analyzed with Chromosome Analysis Suite software, version 3.2 (Applied Biosystems), and the GRCh38/hg38 build of the Human Genome Assembly.
Characterization of COBL Breakpoints
We developed customized MLPA assays for the screening of CNAs within chromosome 7. The probes’ design and assay conditions have been previously described [9]. To validate our SNP array and MLPA findings, we performed either multiplexed long-distance PCR (M-PCR) or long-distance inverse (LDI)-PCR. These PCR approaches allowed us to confirm COBL-r and to determinate the breakpoints at nucleotide level.
Patient Data and Survival Analyses
For the comparison of laboratory and clinical data between patients with or without COBL-r, we used the Pearson χ2 test. Overall survival (OS) was defined as the time in months from the date of diagnosis to death or to the last follow-up assessment for patients alive. Kaplan-Meier method was used to estimate OS rates of patients according to COBL status with differences compared by the log-rank test. Statistical analyses were performed using R software, version 3.5.2, and P values < .05 were considered statistically significant.
Analysis of Activation-Induced Deaminase (AID) or RAG Recognition Signal Sequences in Recombined COBL Alleles
An agnostic search for motifs located within fragments spanning 50 bp from the breakpoint junctions of COBL-r was performed using MEME. The limit of output motifs was set to 5, and width ranged from 2 to 15 bp. The FIMO (Find Individual Motif Occurrences) tool was used for the analysis of WGCW and CG sequences, and the recombination signal sequence (RSS) consensus sequence was used to search for cryptic RSS sequences [11], [12].
Results
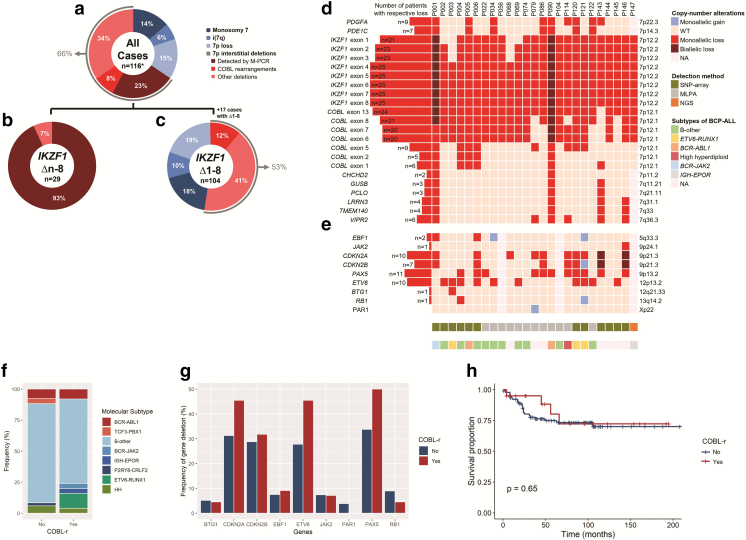
Most of the deletions encompassing IKZF1 exon 8 were classified as whole-gene deletions (Δ1-8: 78% vs. Δn-8: 22%). The CNAs found in the cohort were associated with monosomy 7 (14%; n = 16); isochromosome 7q (6%; n = 7): 7p loss (15%; n = 17), and interstitial deletions within 7p (66%; n = 76) (Figure 1A). Among the Δn-8 group, most of the alterations (93%) were IKZF1 deletions detectable by the M-PCR, i.e., Δ2-8 or Δ4-8 (Figure 1B). Alternatively, 47% of the Δ1-8 group presented with aberrations involving loss of the whole 7p, such as monosomy 7 (18%; n = 19), isochromosome 7q (10%; n = 10), and 7p loss (19%; n = 20). The remaining 55 patients (53%) harbored interstitial deletions; COBL-r were found in 12% of the Δ1-8 cohort (Figure 1C).
Figure 1.
Genetic alterations associated with IKZF1 deletions in B-ALL. (A) Patients with IKZF1 deletions spanning its exon 8 presented four types of aberrations on chromosome 7: monosomy 7; isochromosome 7q, i(7q); 7p loss; and interstitial deletions within 7p arm. *One cohort of 17 cases comprising only complete deletions of IKZF1 was excluded from this analysis because they would lead to a biased estimative of CNAs' frequencies among patients with IKZF1 deletions overall. Then, patients were divided into two groups: (B) intragenic deletions of IKZF1 spanning its exon 8 (n = 29) and (C) complete deletions of IKZF1 (n = 104). (D) The COBL rearrangements were identified in 25 patients. Most of the deletions spanned between IKZF1 exon 1 and COBL intron 5, although they presented variable size. (E) These patients had additional alterations, such as ETV6, PAX5, CDKN2A, and CDKN2B deletions. NA, not available. (F) Distribution of molecular subtypes of B-ALL among patients with or without COBL-r. Legend colors illustrate alterations associated with high risk (red), intermediate risk (blue), and good risk (green). HH, high hyperdiploidy. (G) The frequency of deletions in genes associated with B-ALL was compared according to COBL status. PAX5, CDKN2A, CDKN2B, and ETV6 deletions were recurrently observed; however, the frequency of these CNAs was similar for all these genes, as indicated by P values above each bar. PAR1, pseudoautosomal region 1. (H) No significant difference was observed when comparing the overall survival of B-ALL patients with COBL-r vs. COBL wild type (P = .65).
We identified COBL-r in 25 B-ALL cases with IKZF1 deletions (Figure 1D). COBL-r were detected by MLPA screening in 133 patients with B-ALL and IKZF1 deletions (n = 12) or SNP array/NGS investigation performed at collaborating centers (n = 13), as described in the methodology. The loss of genes located within 9p locus — PAX5 (n = 11), CDKN2A (n = 10), and CDKN2B (n = 7) — and ETV6 deletions (n = 10) were the most recurrent additional alterations among these patients (Figure 1E). Demographic and laboratory data for the 25 patients with COBL-r showed they included 17 males and 8 females who were mainly children and adolescents (n = 21), with a median age at diagnosis of 5.5 years (range 1-59 years) and a median white blood cell count of 7.5 × 109/l (range 1.5-459.6 × 109/l) (Table 1). The patients were treated on diverse therapy protocols (Table 2). Seven patients relapsed within a median of 5.5 years (range 1.4-16.3 years), and six of them experienced isolated disease recurrence in the bone marrow (n = 5) or testes (n = 1). One patient relapsed at both sites: bone marrow and central nervous system. The median follow-up was 5.1 years (range 0.3-16.3 years), and the five deaths (mainly in patients with early relapse) occurred at a median 4.2 years (range 0.3-5.5 years) following diagnosis. In addition, the comparison between patients with COBL-r vs. COBL wild-type revealed that both groups presented similar laboratory and clinical characteristics (Table 3). Among the B-ALL cytogenetic abnormalities, TCF3-PBX1 and ETV6-RUNX1 were exclusively found in patients without or with COBL-r, respectively (Figure 1F). Although we did not observe any significant difference in the frequency of additional gene deletions when comparing patients with vs. without COBL-r, it is worthy of note that CDKN2A, ETV6, and PAX5 deletions were more frequent in patients with COBL-r (Figure 1G). Follow-up data were available for 111 B-ALL patients with (n = 20) or without COBL-r (n = 91). The OS of patients with COBL-r was similar to those with IKZF1 deletion only (hazard ratio, 1.278; 95% CI, 0.35-4.68; P = .646) (Figure 1H).
Table 1.
Demographic and Laboratory Characteristics of B-ALL Cases with COBL Rearrangements.*
| Patient | Age (Years) | Gender | WBC (×109/l) | Blasts at BM | 5′ Breakpoint | 3′ Breakpoint | Detection Method | Karyotype |
|---|---|---|---|---|---|---|---|---|
| P004 | 1 | F | 96.000 | 95% | ELMO1 intron 14 | Upstream COBL | SNP array | 46,XX,+2,t(3;16)(p21;p13),del(4)(q22),−7,der(12)t(7;12)(q11;p13),−13,+16,−18,−19,+mar1,+mar2 |
| P034 | 1 | M | 3.400 | 98% | RAD50 intron 5 | COBL intron 5 | LDI-PCR | NA |
| P086 | 1 | M | 5.000 | 95% | 7pter | COBL intron 7 | MLPA/LDI-PCR | NA |
| P120 | 1 | F | 459.600 | 80% | 7p14 | COBL intron 5 | SNP array/M-PCR | NA |
| P056 | 2 | F | 1.600 | 98% | SPATA intron 1 | COBL intron 5 | LDI-PCR | 46,XX |
| P122 | 2 | M | 13.400 | 40% | 7pter | COBL intron 7 | MLPA | NA |
| P145 | 2 | F | 92.000 | 98% | TCRGC2 intron 1 | COBL intron 13 | SNP array | 47,XX,del(7)(p11),+10,add(12)(p13),del(12)(p11.2)[4] |
| P022 | 4 | F | 7.500 | 85% | 7p12 | COBL intron 2-5 | MLPA | 46,XX |
| P003 | 5 | M | 10.000 | 95% | IKZF1 intron 3 | COBL intron 5 | SNP array/M-PCR | 46,XY |
| P074 | 5 | M | 16.400 | 78% | 7p12.1 | COBL intron 5‡ | LDI-PCR | NA |
| P079 | 5 | F | 5.700 | 88% | 7p | COBL intron 8-13 | MLPA | NA |
| P121 | 5 | F | 7.470 | 50% | 7p12.2 | COBL intron 5 | SNP array/M-PCR | NA |
| P144 | 5 | M | 31.000 | NA | 7p14.2 | COBL intron 5 | SNP array | 47,XY,+X,del(16)(q13),i(17)(q10),ider(21)(q10)dup(21)(q?)[3]/ 47,idem,add(7)(p1)[3] |
| P002 | 6 | M | 115.000 | 97% | 7p12.2 | COBL intron 5 | SNP array/M-PCR | 46,XY,del(6)(q21q25),der(12)del(12)(p11p13)t(12;17)(p13;q11),der(17)t(12;17) |
| P069 | 10 | F | 204.000 | 89%⁎ | IKZF1 intron 1 | COBL intron 5 | LDI-PCR | NA |
| P146 | 11 | M | 1.500 | 90% | SPATA intron 2 | COBL intron 5 | SNP array | 48,XY,+X,?t(6;20)(p1;q1),?t(7;9)(p1;p2),i(9)(q10),+12,der(21)dup(21)(q?)[9] |
| P068 | 12 | M | NA | NA | IKZF1 intron 3 | COBL intron 5 | RNA-seq/LDI-PCR | 48,XY,+X,del(4)(q25),−7,del(9)del(9)t(4;9)(q25;p13),+21,+mar[cp8]/ |
| P104 | 13 | M | 1.680 | 84% | 7p14 | COBL intron 5 | MLPA | 46,XY,del(7)(p14),der(9;12)(q10;q10),+mar[6] |
| P090 | 15 | M | 4.500 | 83% | 7p22 | COBL intron 5 | MLPA | NA |
| P114 | 16 | M | 55.000 | 45% | 7p | COBL intron 2-5 | MLPA | NA |
| P143 | 16 | M | 6.200 | 96% | COBL intron 1† | 7p12.1 | SNP array | 46,XY,idic(9)(p13)[1]/ 45,idem,−7[3]/ 45,idem,add(2)(p25),−7[9] |
| P147 | Adolescent | M | NA | NA | SUN3 intron 5 | COBL intron 5 | NGS | NA |
| P005 | 21 | M | 2.820 | 90% | 7pter | Upstream COBL | SNP array | 46,XY,t(9;22)(q34;q11) |
| P006 | 38 | M | 3.040 | 67% | VWC2 intron 3 | Upstream COBL | SNP array | 46,XY |
| P001 | 59 | M | 58.000 | 75% | 7p12 | COBL intron 7§ | SNP array | 45,XY,−7,t(9;22;15)(p24;q11;q21),add(9)(p13) |
BM, bone marrow; LDI-PCR, long-distance inverse PCR; M-PCR, multiplex PCR; MLPA, multiplex ligation-dependent probe amplification; NA, not available; WBC, white blood cell count.
In this case, blast percent was estimated from peripheral blood.
Patient had COBL rearrangement and 7p12.3::GRB10 intron 5 deletion.
Patient had an inversion within COBL intron 5.
Patient had COBL rearrangement and monosomy 7.
Table 2.
Clinical Characteristics and Outcome of B-ALL Cases with COBL Rearrangements
| Patient | Clinical Trial | CNS Disease | MRD Day 33 | MRD Day 78 | PR | CMR Day 33 | Relapse | Outcome | Follow-Up (Months) |
|---|---|---|---|---|---|---|---|---|---|
| P147 | AALL0232_1 | NA | NA | NA | NA | NA | NA | NA | NA |
| P068 | AALL0331/UKALLR3 | No | Negative | Negative | NA | Yes | BM | Dead | 45 |
| P034 | AIEOP-BFM ALL2000 | No | Negative | Negative | Good | Yes | No | 1st CR | 96 |
| P022 | AIEOP-BFM ALL2000 | No | Negative | Negative | Good | Yes | No | 1st CR | 79 |
| P114 | AIEOP-BFM ALL2000 | No | NA | NA | Good | Yes | NA | Dead | NA |
| P120 | AIEOP-BFM ALL2000 | No | Positive | Positive | Poor | No | No | Dead | 4 |
| P104 | ALL IC 2009 | Yes | Negative | Negative | Good | Yes | No | 1st CR | 38 |
| P145 | UKALL2003 | No | Negative | Negative | NA | Yes | No | 1st CR | 105 |
| P146 | UKALL2003 | No | Negative | NA | NA | Yes | BM, CNS | Dead | 66 |
| P144 | UKALL97 | No | NA | NA | NA | Yes | No | 1st CR | 26 |
| P143 | UKALL97 | No | NA | NA | NA | Yes | No | 1st CR | 167 |
| P003 | CAALLF01 | No | Negative | NA | Good | Yes | NA | 1st CR | 4 |
| P079 | COALL 05-92 | No | NA | NA | NA | Yes | No | 1st CR | 161 |
| P086 | COALL 06-97 | No | Negative | NA | NA | Yes | Testes | 2nd CR | 195 |
| P074 | COALL 06-97 | No | Negative | NA | NA | Yes | BM | 2nd CR | 154 |
| P069 | COALL 07-03 | No | Positive | NA | NA | No | No | 1st CR | 123 |
| P004 | EORTC 58081 | No | Negative | Negative | Good | Yes | No | 1st CR | 49 |
| P090 | EsPhALL | No | Positive | NA | Good | Yes | Isolated BM | Dead | 56 |
| P006 | FRALLE 93 | No | Positive | Positive | Poor | Yes | Yes | Relapse | 192 |
| P002 | FRALLE BT | No | Positive | Negative | Poor | Yes | No | 1st CR | 15 |
| P122 | GBTLIALL99 | No | NA | NA | Good | Yes | NA | Alive | NA |
| P121 | GBTLIALL99 | No | Negative | Negative | Good | Yes | No | Alive | NA |
| P005 | GRAAPH | No | Positive | Positive | Not evaluable | Yes | No | 1st CR | 27 |
| P056 | NA | No | Positive | NA | Good | Yes | BM | Lost follow-up | 17 |
| P001 | NA | No | Positive | NA | Good | Yes | No | SCT; alive | 9 |
BM, bone marrow; CNS, central nervous system; CMR, complete morphological remission; MRD, minimal residual disease; NA, not available; PR, prednisone response; SCT, stem-cell transplantation; WBC, white blood cell count. MRD-negative status was defined as <0.01% leukemic cells in bone marrow and peripheral blood.
Table 3.
Demographic and Laboratory Data of Patients with B-ALL*
|
COBL Rearrangement |
P Value⁎ |
||
|---|---|---|---|
| No n = 121 |
Yes n = 25 |
||
| n (%) | n (%) | ||
| Gender | 0.473 | ||
| Male | 48 (39.7) | 8 (32.0) | |
| Female | 73 (60.3) | 17 (68.0) | |
| Age at diagnosis | 0.701 | ||
| <1 year | 2 (1.7) | 0 (0.0) | |
| 1-9 years | 49 (41.5) | 12 (48.0) | |
| ≥10 years | 67 (56.8) | 13 (52.0) | |
| WBC (×109/L) | 0.948 | ||
| <50 | 85 (70.2) | 16 (69.6) | |
| ≥50 | 36 (29.8) | 7 (30.4) | |
| NCI risk | 0.797 | ||
| Standard | 69 (58.0) | 14 (60.9) | |
| High | 50 (42.0) | 9 (39.1) | |
| CNS disease | 0.741 | ||
| No | 96 (94.1) | 23 (95.8) | |
| Yes | 6 (5.9) | 1 (4.2) | |
| Prednisone response | 0.143 | ||
| Good | 66 (91.7) | 11 (78.6) | |
| Poor | 6 (8.3) | 3 (21.4) | |
| Relapse | 0.607 | ||
| No | 75 (70.8) | 13 (65.0) | |
| Yes | 31 (29.2) | 7 (35.0) | |
| Outcome | 0.604 | ||
| Alive | 94 (77.7) | 20 (83.3) | |
| Dead | 27 (22.3) | 4 (16.7) | |
WBC, white blood cell count; NCI, National Cancer Institute of US; CNS, central nervous system.
Pearson χ2 calculation.
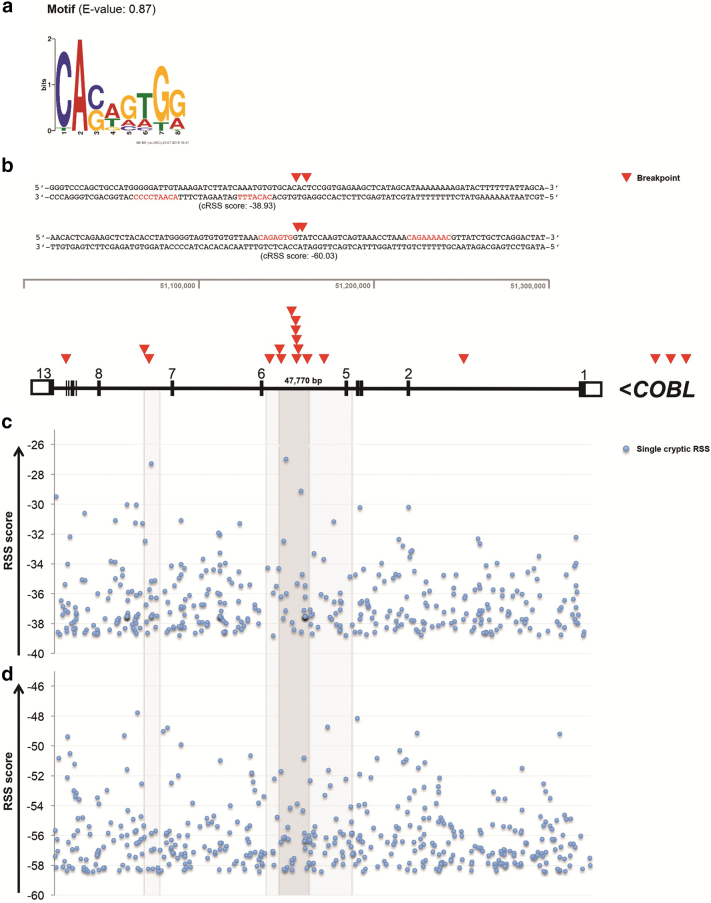
The breakpoints of the COBL-r were determined at nucleotide level in 11 of the 25 cases, and 56% of breakpoints were located within COBL intron 5 (Figure 2B). To address the possible causes of these breakpoints, we first performed an agnostic motif search. This analysis identified the motif CASWGTGG (E-value = 0.87) within all 22 breakpoint sequences of COBL-r (Figure 2A; Supplementary Table S1). CASWGTG is similar to the heptamer of the RAG RSS, which is composed of a heptamer (5′-CACAGTG-3′) and a nonamer (5′-ACAAAAACC-3′) sequence, interspaced by 12 or 23 random nucleotides associated to RAG-type rearrangements. Since the nonamer was absent in our first analysis, we then investigated the presence of complete motifs associated with the occurrence of rearrangements in leukemia: cryptic RSS sequence (RAG-type fusions), WGCW (AID-type fusions), and CG sequences. WGCW and CG sequences were not found; however, we identified cryptic RSS in 5 out of 22 breakpoint regions, although none of them were spanning breakpoints within COBL (Supplementary Table S2). Additionally, we performed a robust analysis for the identification of cryptic RSSs within the whole sequence of COBL (Figure 2, C-D). The results revealed a broad distribution of this motif throughout the gene and no enrichment for the breakpoint cluster located in intron 5 of COBL.
Figure 2.
Identification of motifs within the breakpoint sequences. (A) An agnostic motif search using MEME identified the sequence CASWGTGG (E-value = 0.87) among 22 breakpoint sequences. (B) The map of 19 deletion breakpoints (red triangles) within COBL revealed a hotspot located at intron 5. Three breakpoints were detected within a downstream region of 7p12.1. The sequences highlight two breakpoint clusters located at COBL intron 5. The mapped cryptic recombination signal sequences were not statistically significant. The cryptic recombination signal sequences (cRSS) with a spacer of 12-bp (c) and 23-bp (d) were mapped along COBL gene. The highest RIC scores represent cRSS (blue dots) associated with RSS functionality. The gray area highlights the COBL intron 5 and breakpoint cluster regions.
Discussion
In this study, among the Δn-8 group, the majority of the alterations were IKZF1 deletions which had already been detected by the M-PCR method [8]. Since the remaining Δn-8 samples harbored deletions restricted to IKZF1, either they had DNA fusions outside the breakpoint cluster region of IKZF1 or M-PCR failed to detect them. Among the Δ1-8 group, COBL-r were found in 12% of the patients. This result revealed that COBL-r are more frequent among Δ1-8 but rarely related to Δn-8. Although COBL-r were not detected within the Δn-8 group for the cases included in the current proposal, we found these deletions in patients with COBL-r from a previous study [9]. These patients had IKZF1-COBL fusions, which involved IKZF1 intron 1 (n = 1) or intron 3 (n = 2) and COBL intron 5.
Confirming the idea that COBL represents a genomic hotspot for IKZF1 deletions in B-ALL [9], we identified COBL-r in 25 B-ALL cases with IKZF1 deletions. Although either good (ETV6-RUNX1, high hyperdiploid) or high-risk (BCR-ABL1) cytogenetic groups have been observed in some of these patients, most of them had the so-called “B-other” subclassification with either normal or other abnormal (BCR-JAK2, IGH-EPOR) cytogenetic profile. In the current genetic risk stratification, many cases with COBL-r would then be classified as intermediate cytogenetic risk. We found that these patients also presented other secondary abnormalities commonly identified in B-ALL, such as PAX5 and CDKN2A/B and ETV6 deletions. This combination is of special interest because these patients could benefit from the newly proposed combined risk stratification strategies, such as IKZF1plus or the UKALL-CNA classifiers [13], [14], [15]. Considering that patients with B-ALL share similar laboratory and clinical characteristics regardless of COBL involvement, IKZF1 deletions may play a major role on risk stratification for these patients.
Regarding the breakpoints of the COBL-r, it is remarkable that 56% of them were located within COBL intron 5, although this region represents only 16% (47,770 out of 300,587 bp) of the entire gene. Based on this observation, we formulated two hypotheses to explain the presence of a hotspot for breakpoints within COBL: 1) the production of a truncated COBL protein, encoded by exons 1-5 only, could have a role on leukemogenesis, or 2) there is a breakage mechanism involving COBL intron 5, thus enriching this area for gene rearrangements.
COBL protein has three Wiskott-Aldrich syndrome protein homology 2 domains for actin binding. It shows substantial expression in neurons and muscle cells, although levels are low in blood [16]. COBL functions as an actin nucleator, controlling neuronal morphology and development [17]. Considering that COBL does not play a direct role in lymphoid development, the enrichment of COBL-r in B-ALL is more likely to be related to mechanisms controlling DNA breakage and promotion of genetic fusions.
Usually, genetic rearrangements in lymphoid malignancies are caused by either AID or the RAG complex. Therefore, we searched for motifs located within the breakpoint junctions of COBL-r, which could potentially provide a rational explanation for the observed chromosomal rearrangements. RSSs are recognized by RAG enzymes during V(D)J recombination, and previous studies have located cryptic RSS immediately internal to the breakpoints of intragenic deletions of IKZF17. Although the mutual motif within sequences spanning the breakpoints was similar to the heptamer sequence, our results do not support the idea that aberrant RAG-mediated recombination is the mechanism responsible for IKZF1 and COBL codeletions.
In summary, our results highlight COBL as a hotspot for interstitial deletions within chromosome 7, especially for deletions including the IKZF1 gene. Most of the COBL-r arose within COBL intron 5, leading to complete deletion of IKZF1; nevertheless, we also observed fusions between both genes. The analysis of the breakpoint sequences revealed a common motif resembling the heptamer of recombination signal sequence, but the analysis of the whole consensus sequence did not provide evidence for RAG-mediated recombination. Lastly, our study demonstrates that patients with IKZF1 deletions are associated with worse outcome regardless of COBL-r.
The following are the supplementary data related to this article.
Map of the Agnostically Identified Motif Within Breakpoint Sequences
Identification of Cryptic Recombination Signal Sequences Spanning Breakpoint Sequences
Funding
This research was funded by CNPq (PQ-2017#305529/2017-0) and FAPERJ-JCNE (E_26/201.539/2014 and E_26/203.214/2017) awarded to M. E. B. A. L. was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and the Alexander von Humboldt Foundation. G. C. was supported by the Italian Association for Cancer Research (AIRC).
Conflict of Interest
The authors declare no conflict of interest.
Author Contribution
Conception and design: B. A. L., C. M., R. M., M. E. Experiments, data analysis, and interpretation: B. A. L., C. M., T. C. B., C. P. P., N. D., M. B., U. S., C. P., N. C. V., R. M., M. E. Provision of samples, data collection, and assembly: B. A. L., M. B. M., N. D., C. J.H., U. S., M. H., M. S. P. O., G. C., R. S., C. N. A., G. T., S. G., S. B. Writing and/or revision of the manuscript: B. A. L., R. M., M. B. M., C. J. H., G. C., R. S., C. N. A., M. E. Final manuscript approval: all authors.
References
- 1.Greaves M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer. 2018;18:471–484. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa TC, Terra-Granado E, Quezado Magalhaes IM, Neves GR, Gadelha A, Guedes Filho GE, Souza MS, Melaragno R, Emerenciano M, Pombo-de-Oliveira MS. Frequency of copy number abnormalities in common genes associated with B-cell precursor acute lymphoblastic leukemia cytogenetic subtypes in Brazilian children. Cancer Genet. 2015;208:492–501. doi: 10.1016/j.cancergen.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 4.Dorge P, Meissner B, Zimmermann M, Moricke A, Schrauder A, Bouquin JP, Schewe D, Harbott J, Teigler-Schlegel A, Ratei R. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98:428–432. doi: 10.3324/haematol.2011.056135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boer JM, van der Veer A, Rizopoulos D, Fiocco M, Sonneveld E, de Groot-Kruseman HA, Kuiper RP, Hoogerbrugge P, Horstmann M, Zaliova M. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia. 2016;30:32–38. doi: 10.1038/leu.2015.199. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 8.Meyer C, Zur Stadt U, Escherich G, Hofmann J, Binato R, Barbosa TC, Emerenciano M, Pombo-de-Oliveira MS, Horstmann M, Marschalek R. Refinement of IKZF1 recombination hotspots in pediatric BCP-ALL patients. Am J Blood Res. 2013;3:165–173. [PMC free article] [PubMed] [Google Scholar]
- 9.Lopes BA, Meyer C, Barbosa TC, Zur Stadt U, Horstmann M, Venn NC, Heatley S, White DL, Sutton R, Pombo-de-Oliveira MS. COBL is a novel hotspot for IKZF1 deletions in childhood acute lymphoblastic leukemia. Oncotarget. 2016;7:53064–53073. doi: 10.18632/oncotarget.10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duployez N, Nibourel O, Ducourneau B, Grardel N, Boyer T, Bories C, Darre S, Coiteux V, Berthon C, Preudhomme C. Acquisition of genomic events leading to lymphoblastic transformation in a rare case of myeloproliferative neoplasm with BCR-JAK2 fusion transcript. Eur J Haematol. 2016;97:399–402. doi: 10.1111/ejh.12752. [DOI] [PubMed] [Google Scholar]
- 11.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, den Boer ML, Boer JM, Braun M, Dalla Pozza L. Validation of the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv. 2019;3:148–157. doi: 10.1182/bloodadvances.2018025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorman AV, Enshaei A, Schwab C, Wade R, Chilton L, Elliott A, Richardson S, Hancock J, Kinsey SE, Mitchell CD. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444. doi: 10.1182/blood-2014-03-562918. [DOI] [PubMed] [Google Scholar]
- 15.Stanulla M, Dagdan E, Zaliova M, Moricke A, Palmi C, Cazzaniga G, Eckert C, Te Kronnie G, Bourquin JP, Bornhauser B. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol. 2018;36:1240–1249. doi: 10.1200/JCO.2017.74.3617. [DOI] [PubMed] [Google Scholar]
- 16.G.T. Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of the Agnostically Identified Motif Within Breakpoint Sequences
Identification of Cryptic Recombination Signal Sequences Spanning Breakpoint Sequences