Abstract
The southern cattle fever tick, Rhipicephalus (Boophilus) microplus, is the most economically important ectoparasite of cattle worldwide. A limitation for sustainable control and eradication is the emergence of acaricide resistance among tick populations. Molecular diagnostic tools offer the opportunity to detect resistance rapidly, which can be complemented with confirmatory bioassays with larvae and adult ticks that are more resource and time consuming to generate. Synthetic pyrethroid resistance is one of the most prevalent and well-studied forms of resistance in arthropods, being linked with target site alterations in the sodium ion channel gene. Here, we report research on a novel molecular method to detect mutations in the para-sodium channel gene of R. microplus associated with acaricide resistance that is based on quantitative PCR high-resolution melt (HRM) analysis. Genomic DNA fragments of domains II and III of the para-sodium channel gene were amplified by real-time PCR in the presence of EVA®Green dye to test resistant and susceptible reference ticks from the U.S., Brazil, and Mexico. Larval packet tests with discriminating doses and a modified lethal time analysis were performed to confirm resistance to permethrin, cypermethrin, deltamethrin, and flumethrin in laboratory strains. Tick specimens collected from cattle that were inspected at the United States Port-of-Entry at the Texas-Mexico border were also genotyped. Previously described mutations associated with pyrethroid resistance (T170C, C190A, G184C, and T2134A) were successfully detected by qPCR-HRM in different genotypes and confirmed by sequencing. A novel non-synonymous SNP located at domain III (C2136A) and the G215T mutation in domain II, previously described only in Asian R. microplus and R. australis, were also detected with the HRM and confirmed by sequencing. This technique could be adapted for high-throughput screening, detection, and discovery of allele-specific mutations in cattle tick outbreak populations to inform eradication strategies in the USA. This knowledge could also be applied to integrated control programs in other parts of the world where R. microplus is endemic and where similar SNPs have been identified associated with pyrethroid resistance. This study highlights the existence of several mutations in the para-sodium channel gene in different combinations in field populations of R. microplus from Mexico.
Keywords: Southern cattle fever tick, Pyrethroid resistance, Molecular diagnosis, High-resolution melt analysis, Sodium channel
Graphical abstract
Highlights
-
•
Molecular detection of acaricide resistance in Rhipicephalus (Boophilus) microplus is needed.
-
•
Mutations associated with pyrethroid resistance can be detected with PCR.
-
•
A quantitative PCR with high-resolution melt (qPCR-HRM) analysis was developed.
-
•
The qPCR-HRM was successful in detecting pyrethroid resistance in R. microplus.
1. Introduction
Ticks infesting cattle and other farm animals in tropical and subtropical areas of the world are controlled primarily through the use of products containing synthetic acaricides. Resistance to one or more classes of acaricides, including pyrethroids, has been reported among populations of the southern cattle fever tick, Rhipicephalus (Boophilus) microplus (Canestrini), which is considered the most economically important ectoparasite of livestock in parts of the world where it is endemic (Grisi et al., 2014; Rodriguez Vivas et al., 2017). R. microplus is also of veterinary relevance because it is a vector of the pathogens that cause bovine babesiosis, anaplasmosis and spirochaetosis (Aubry and Geale, 2011; Pérez de León et al., 2014; Walker et al., 2003).
The characterization of pyrethroid resistance in R. microplus and other arthropod pests has been extensively explored (Rinkevich et al., 2013; Dong et al., 2014). Since the discovery of the mutation in domain III of the putative para-sodium channel gene (He et al., 1999), a number of publications documented the presence of nucleotide polymorphisms in pyrethroid resistant populations of R. microplus across different geographic areas (Guerrero et al., 2001; Morgan et al., 2009; Rosario-Cruz et al., 2009; Aguirre et al., 2010; Jonsson et al., 2010; Rodriguez-Vivas et al., 2012; Lovis et al., 2012; Nogueira Domingues et al., 2012; Kumar et al., 2013; Robbertse et al., 2016; Wyk et al., 2016; Bandara and Karunaratne, 2017; Sungirai et al., 2018). In addition to target site insensitivity, other mechanisms involved in pyrethroid resistance among R. microplus populations include detoxification enzymes such as esterases (Guerrero et al., 2002; Singh and Rath, 2014; Gupta et al., 2016; Gaur et al., 2017), and monooxygenases (Graham et al., 2016).
Bioassays like the larval packet test (Stone and Haydock (1962)), or the adult immersion test (Drummond et al., 1973) are used extensively to detect acaricide resistance phenotypes. These functional bioassays can discriminate the levels of resistance between tick populations and can be adapted to investigate possible mechanisms of resistance (Li et al., 2003). However, these bioassays require access to abundant numbers of viable engorged females that need to be collected, maintained for lengthy periods of oviposition and testing of viable larvae. Often, this can be difficult to achieve especially in areas of low infestation levels or where strict tick control measures are practiced.
Molecular methods to detect acaricide resistance can be performed with few ticks, including samples of properly conserved dead ticks (e.g. in ethanol or isopropanol). The first description of an allele-specific polymerase chain reaction (AS-PCR) assay to genotype pyrethroid resistant tick strains was reported by Guerrero et al. (2001). This AS-PCR assay detects a mutation in domain III (T2134A) and was applied to studies on the epidemiology of pyrethroid resistance in R. microplus from Mexico (Guerrero et al., 2002; Rosario-Cruz et al., 2009; Rodriguez-Vivas et al., 2012). Lovis et al. (2012) proposed a multiplex AS-PCR, aimed to detect three (C190A; G215T and T2134A) single nucleotide polymorphisms (SNPs) in the para-sodium channel gene. More refined techniques were also developed, as a Taqman® dual probe quantitative PCR diagnostic assay to detect the C190A mutation (Morgan et al., 2009), and a melt analysis of mismatch amplification mutation assay (melt-MAMA) qPCR platform, designed to detect the mutations T170C, C190A, and T2134A (Stone et al., 2014).
High-resolution melt analysis (HRM) is a closed-tube post-PCR application involving the use of a dye that fluoresces when intercalated with double-stranded DNA (dsDNA), but not when free in solution after the denaturation of DNA (Reed et al., 2007). The intercalated dye is released when dsDNA denatures, resulting in a loss of fluorescence at the melting temperature of the PCR product. HRM curves are generated by measuring the decrease of fluorescence as the temperature is slowly increased with a high level of accuracy (0.05–0.1 °C). The fluorescence decreases at each step due to the transition of the DNA from double-to single-stranded. The melting temperature of a DNA molecule is determined by its nucleic acid sequence and length (Reed et al., 2007). Differences in these nucleotide sequences between samples result in melting profiles that are unique to particular genotypes, allowing for differentiation. A PCR reaction that contains only one type of DNA sequence, such as homozygous templates from diploid organisms, produces a melt curve with one peak. Heterozygous templates result in a mixture of homopolymers and heteropolymers. Due to the imperfect binding of their strands, the melting temperature is strongly lowered in the heteropolymers, resulting in an early peak in the curve. Multi-loci SNP products will cause more complex heteropolymer formations and additional peaks in the melt curves (Mader et al., 2008).
HRM was successfully used to detect pyrethroid resistance mutations in the yellow fever mosquito Aedes aegypti (Wuliandari et al., 2015), and in the scabies mite Sarcoptes scabiei (Pasay et al., 2008). In a direct comparison with other techniques (i.e. AS-PCR and Taqman probes) the HRM had excellent performance in detecting knock down resistance (kdr) mutations in previously sequenced Anopheles gambiae standard samples. However, when the technique was used to detect those mutations in field derived samples with variable quantity and quality of DNA, its performance was not as efficient as the Taqman probes. Nevertheless, the cost of the analysis per sample was lower than the other two techniques (Bass et al., 2007).
Here, we developed an HRM assay to simultaneously detect multiple mutations in the para-sodium channel gene associated with pyrethroid-resistant Rhipicephalus microplus. Reference laboratory tick strains from Mexico, Brazil and USA, as well as field samples from Mexico were used in this study.
2. Materials and methods
2.1. Ticks
Colonies of R. microplus maintained at the USDA-ARS Cattle Fever Tick Research Laboratory (CFTRL) in Edinburg, Texas were used in this study and were reared as described by Davey et al. (1980). The Deutch strain served as the acaricide susceptible reference collected originally in Laredo, Texas and maintained as a colony at the CFTRL since 2001. Deutch strain ticks from the F58 generation were used in this study. Santa Luiza is the colony that includes ticks resistant to permethrin and amitraz, which were collected from a ranch in Rio Grande do Sul, Brazil; this colony was maintained at the Mexican National Parasitology Laboratory, Jiutepec, Morelos, Mexico, before being established at the CFTRL in 2000 (Li et al., 2004). Ticks from the F64 generation of the Santa Luiza colony were used. Ticks from the El Zamora colony are resistant to permethrin, amitraz, and fipronil, and were collected in 2010 from a ranch in the State of Tamaulipas, Mexico (Miller et al., 2013). F32 generation of El Zamora ticks were used. Ticks to establish the Yucatan colony were collected in 2014 from red deer (Cervus elaphus) at a ranch located in the State of Yucatan, Mexico (Rodríguez-Vivas et al., 2014), and are resistant to cypermethrin, coumaphos, and ivermectin. Ticks of the El Zamora F17 generation were used.
Rhipicephalus microplus field samples were obtained from cattle during inspections at the United States Ports-of-Entry located at the border of Mexico and the State of Texas. Ticks were received at the CFTRL inside 5 mL plastic assay tubes and were stored in cryovials and immediately frozen at −80 °C to be used in the molecular analysis. In total, 29 semi-engorged females were processed and those were taken from cattle with origins in different states of Mexico: Coahuila (n = 3); Nuevo León (n = 7); Tamaulipas (n = 14) and Veracruz (n = 5). No previous information about the pyrethroid resistance status was obtained from those ticks.
2.2. Preparation of ticks
Engorged female R. microplus from the susceptible (Deutch), and resistant colonies (Santa Luiza, El Zamora, and Yucatan) were collected after their natural detachment from cattle according to the protocol approved by the Institutional Animal Care and Use Committee. After collection, the ticks were washed with water and dried with paper towels. From each colony, 30 engorged females were incubated in an environmental chamber at 28 °C, and a relative humidity of 92% in a plastic petri dish (9 cm diameter) for 20 days to allow egg laying (Davey et al., 1980). Depleted females were separated from the eggs, washed with distilled water, dried with paper towels and individually frozen at −80 °C to be used in the molecular analysis. The eggs were mixed thoroughly and incubated under the same conditions in 2-dram glass vials closed with cotton plugs to allow the passage of air and moisture to permit larval hatching. The egg masses were checked daily for larval hatching. Larvae 14–21 days old were used in the bioassays.
2.3. Bioassays
The bioassays were performed with analytical standards of the following chemicals: permethrin (FMC, Philadelphia, PA, USA), cypermethrin, deltamethrin, and flumethrin (Sigma Aldrich Co., St. Louis, MO, USA). In order to characterize the phenotypic resistance to these chemicals, the larval packet test procedure was used with pre-established discriminating doses (DDs) calculated as 2 × LC99 of a susceptible reference strain. The DDs were 0.25% permethrin (Miller et al., 1999), 0.20% cypermethrin, 0.06% deltamethrin, and 0.01% flumethrin (FAO, 2004). Acaricides were diluted in a mixture containing two parts of trichloroethylene and one-part olive oil (TChE-OO) in order to prepare the impregnation solutions. A volume of 0.67 mL of each acaricide solution was used to impregnate a piece of quantitative filter paper (85 mm × 75 mm – Whatman No. 256, Whatman Inc., Maldstone, England). The material was left to dry for 2 h inside a fume hood to allow for trichloroethylene evaporation. After drying, packets of the same acaricide were wrapped in aluminum foil and maintained at 4 °C until used.
On the day of testing, filter papers were taken from the refrigerator, folded in the middle, and sealed on both sides with metal clips to form the packets. Approximately 100 tick larvae were transferred to each packet using a flat paintbrush. The packets were sealed with a third clip on top, and incubated at 27 ± 1 °C and 80–90% relative humidity. Control groups were exposed to filter papers impregnated with acaricide-free TChE-OO. After 24 h, larval mortality was determined by counting the total number of dead and viable individuals. Larvae that were paralyzed or moving only their appendices without the ability to walk were considered non-viable. Three packets impregnated with each acaricide as well as controls were prepared for each tick sample, i.e. from the Deutch, Santa Luiza, El Zamora, and Yucatan colonies. Percentage of larval mortality was determined for each of the three packets of each acaricide treatment. For each test, mean mortality and standard errors were calculated with Microsoft Excel (Microsoft Corporation, Redmond, WA).
The knockdown effect of permethrin was assessed with a lethal time bioassay. About 100–150 larvae of each strain were incubated inside the filter paper packets impregnated with 0.25% permethrin as described above. The mortality was determined at six time points: 10, 20, 30, 40, 50, and 60 min after the exposure, as described for the resistance test above. Mortality data were analyzed using the Probit model in the Polo Plus software (Version 1.0, licensed. LeOra Software, 2003). For each test, the following parameters were determined: median lethal time (LT50), with its respective 95% confidence limits (95% CL), and the slope of the regression line. The resistance ratios (RR) were obtained using the Polo-Plus software employing the formula described by Robertson et al. (2007). Comparisons were determined to be significant when the calculated 95% CL did not overlap. For visualization of data, the log time vs. probit converted mortality plots were generated using Microsoft Excel (Microsoft Corporation, Redmond, WA).
2.4. DNA extraction
The genomic DNA of ticks was extracted using a phenol-chloroform method. Frozen ticks were taken from ultracold freezer (−80 °C) and transferred to 2-mL plastic tubes containing five ceramic beads (2.8 mm diameter - Omni International, Kennesaw, GA, USA) each. The tubes also contained 600 μL of lysis buffer (10 mM Tris-HCl pH 8; 2 mM EDTA pH 8; 0.5% SDS), 3 μL proteinase K (20 mg/mL, Invitrogen, Carlsbad, CA, USA), and were processed at 4000 rpm for 15 s with a bead mill homogenizer (Omni International, Kennesaw, GA, USA). Homogenates were incubated for 12–18 h at 55 °C to allow for protein digestion. Following, the samples were incubated at 65 °C for 15 min to inactivate the proteinase K. After briefly cooling in an ice bath, 5 μL of RNase A (20 mg/mL, Invitrogen, Carlsbad, CA, USA) was added to the homogenates followed by incubation at 37 °C for 15 min to remove RNA contamination. The homogenates were submitted to a phenol-chloroform DNA extraction procedure. The DNA was precipitated in an ice-cold absolute ethanol and sodium acetate solution at −20 °C for 12–18 h. The DNA pellets were collected by centrifugation (10,000 g at 4 °C for 15 min) and washed two times with 70% ethanol. The final pellet was suspended in 50 μL Tris-EDTA, pH 8. The genomic DNA was quantified on an EON spectrophotometer (Biotek, Winooski, VT, USA), and diluted to 100 ng/μL for PCR.
2.5. PCR and sequencing
In order to identify the mutations, we used primers designed to amplify the exon region of the para-sodium channel gene domain II S4—S5 linker and the exon region of the domain III S6. The identification of the SNPs present in domains II and III was obtained by Sanger dideoxy sequencing.
Sequences of the exon encoding domain II were obtained from DNA fragments amplified with the primers designed by Morgan et al. (2009) where the forward BmNaF5 (5′TACGTGTGTTCAAGCTAGC) and reverse primer BmNaR5 (5′ACTTTCTTCGTAGTTCTTGC) yield a 167 bp product. PCRs were carried out in 50 μL volumes containing the following reagents: 100 ng of DNA template, 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 200 nM each primer and 1 U of AmpliTaq Gold II polymerase (Applied Biosystems, Carlsbad, CA, USA). The reaction was performed under the following conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s, and a final extension step of 72 °C for 7 min and final hold at 4 °C.
Sequences of the exon encoding domain III were obtained from DNA fragments amplified with the primers described by Stone et al. (2014). Initially a 135 bp fragment was amplified using the forward primer RmNaDomainIIIF1 (5′AAGAGGACCAACCGGAATACG) and reverse primer RmNaDomainIIIRS2_CON (5′TCTTCTTTTGTTCATTGAAATTGT). PCRs were carried out in 10 μL volumes containing 100 ng of DNA template, 1× PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 1 U of AmpliTaq Gold II polymerase. The reaction was performed under the following conditions: 95 °C for 10 min, followed by 40 cycles of 94 °C for 60 s, 53 °C for 30 s, 72 °C for 30 s. The PCR products from this first reaction were diluted 1:1000 and used as the template for a second PCR using tailed primers. The second PCR used forward tailed primer RmNaDomainIIIF3 (5′acccaactgaatagagagcAAGAGGACCAACCGGAATACG) and reverse tailed primer RmNaDomainIIIR3 (5′acgcacttgacttgtcttcTCTTCTTTTGTTCATTGAAATTGT) resulting in an amplicon length of 173 bp. The conditions of the second PCR were identical to the first, with exception of the annealing temperature that was 65 °C.
PCR products were visualized by electrophoresis on 2.2% agarose FlashGel DNA cassettes with a FlashGel System (Lonza Rockland, Inc., Rockland, ME, USA). PCR products were purified using Diffinity RapidTiptm (Chiral Technologies, Inc., West Chester, PA, USA), and sequenced using the primers BmNaF5 and BmNaR5 for the domain II amplicons and tail primers RmNaDomainIIIF3seq (5'acccaactgaatagagagc) and RmNaDomainIIIR3seq (5'acgcacttgacttgtcttc) for amplification through capillary electrophoresis in a 96-capillary Applied Biosystems 3730xl DNA Analyzer (Retrogen, Inc.).
The validation of the allele sequences for heterozygous individuals was performed by cloning and sequencing of PCR amplicons. PCR products were ligated into the pJET 1.2/blunt vector of the CloneJET PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's specifications. Ligations were used to transform DH5α competent cells (Invitrogen, Carlsbad, CA, USA). Up to eight colonies per individual tick were selected for PCR screening with the pJET1.2 plasmid primers. Six clones containing the correct insert size were chosen for sequencing according to the methods described above using pJET1.2 primers. Sequencing data were aligned and translated with CLC Main Workbench 7 (Qiagen Aarhus A/S, Copenhagen, Denmark). Sequences were compared to each other and analyzed for the presence of the different SNPs.
2.6. Detection of domain III mutation
Previously described AS-PCR technique was used to genotype the ticks for the T2134A mutation (Guerrero et al., 2001, 2002). PCR was performed using 20 μL reactions containing the following reagents: 100 ng of DNA template, 1× PCR buffer, 1.75 mM MgCl2, 0.2 mM dNTPs, 100 nM each primer and 0.5 U AmpliTaq Gold II polymerase (Applied Biosystems, Carlsbad, CA, USA). The reactions were carried out in a thermocycler programmed for 96 °C for 2 min followed by 37 cycles, each consisting of denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, and extension at 72 °C for 1 min. The program also included a final extension step at 72 °C for 7 min. PCR products were visualized by electrophoresis on 2.2% agarose as described above.
2.7. Quantitative PCR - HRM assays
Two high-resolution melt (HRM) assays were developed to genotype the different pyrethroid resistance related polymorphisms in the para-sodium channel gene of R. microplus. qPCR primers were designed using PRIMER3WEB v.4.0.0 (Untergasser et al., 2012). The first assay targeted the Domain II S4—S5 mutations, and the primers were designed to flank the following nucleotide substitutions: T170C, G184C, C189A (silent), C190A and G215T. Reactions with the forward primer HRM_Rm_Na-D2_Fw (5′CAAATCGTGGCCTACCCTTA) and the reverse primer HRM_Rm_Na-D2_Rv (5′GATTCCCAGGACAAAGGTCA) yielded an 88 bp amplicon. The second assay targeted the Domain III S6 mutation T2134A. Primers used on the reactions were HRM_Rm_Na-D3_Fw (5′TCTTCATTATCTTCGGCTCCTT) and HRM_Rm_Na-D3_Rv (5′TTGTCGATAATAACACCGATGAA), which produced a 61 bp amplicon.
Twenty-four ticks from each colony and the ticks from the field collections were genotyped and the reactions were performed in duplicates. Each run contained the samples, a non-template control (water), clones corresponding to wild-type and mutant genotypes and homozygous and heterozygous genomic DNA references. PCR reactions (10 μL) contained 1X Precision Melt Supermix (Biorad, Hercules, CA, USA), 200 nM each primer, and 100 ng of genomic DNA template. Reference plasmids were used at 1 ng per reaction. Samples were run on a Mic qPCR Cycler (Bio Molecular Systems Pty Ltd., Upper Coomera, Australia) using the temperature cycling conditions of 95 °C for 2 min to activate the hot-start polymerase, 44 cycles of a denaturation step of 95 °C for 10 s, and annealing/extension at 60 °C for 30 s. Fluorescence information was captured at the end of each 60 °C step. PCR products were then subjected to HRM analysis. The HRM step involved heating the PCR products to 95 °C for 30 s, cooling to 60 °C for 1 min, and then increasing the temperature to 65 °C. The temperature increased from 65 to 95 °C at a rate of 0.08 °C/s, and the change in fluorescence of EvaGreen® was recorded continuously on the green channel of the Mic qPCR Cycler.
Melt curves were generated in the “High Resolution Melt” analysis module of micPCR software (version 2.6.2, Bio Molecular Systems Pty Ltd., Upper Coomera, Australia). The parameter settings for melt curve normalization of the domain II fragment were: Pre-melt slider = 75.8–77.3 °C, and Post-melt slider = 84.5–86 °C. For domain III, the parameter settings were: Pre-melt slider = 66–67.6 °C, and Post-melt slider = 77.4–80.4 °C. The samples were assigned to a given genotype by examining normalized and difference melt plots in comparison to reference genotypes curves at a 95% confidence limit.
3. Results
3.1. Pyrethroid resistance
The larval packet discriminating dose bioassays confirmed the pyrethroid resistance phenotype of the analyzed strains. Table 1 presents the mortality data of each strain against permethrin, cypermethrin, deltamethrin and flumethrin. A high level of resistance was observed for El Zamora and Yucatan strains, across all the pyrethroids tested. The Santa Luiza strain showed to be less resistant than the other two, with approximately half of the tick larvae surviving to the discriminating dose after 24 h. All the larvae from the susceptible reference strain died after exposure to the acaricides tested.
Table 1.
Mortality of Rhipicephalus (Boophilus) microplus susceptible and pyrethroid-resistant strains submitted to the larval packet test with discriminating doses of permethrin, cypermethrin, deltamethrin and flumethrin.
| Strain | Mean percentage of mortality after 24 h (standard deviation) |
|||
|---|---|---|---|---|
| 0.25% Permethrin | 0.2% Cypermethrin | 0.06% Deltamethrin | 0.01% Flumethrin | |
| Deutch | 100 (0) | 100 (0) | 100 (0) | 100 (0) |
| Santa Luiza | 59.15 (3.53) | 51.53 (3.25) | 57.83 (3.06) | 58.35 (1.36) |
| El Zamora | 28.12 (3.77) | 6.32 (1.73) | 10.03 (1.55) | 18.03 (0.56) |
| Yucatan | 2.85 (2.35) | 0.28 (0.16) | 0.74 (0.44) | 0 (0) |
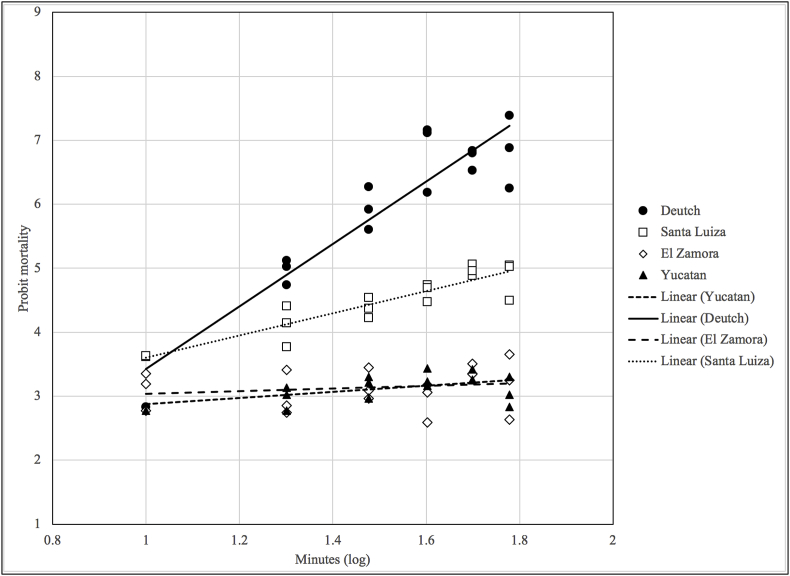
Fig. 1 and Table 2 shows the results of the lethal time bioassay with permethrin carried out with the four strains. The median lethal time (LT50) for Deutch was 21.74 min (95% CL = 18.48–24.63). After 40 min of exposure, approximately 95% of the larvae of the susceptible strain were dead. For the Santa Luiza resistant strain, the calculated LT50 was 65.02 (95% CL = 53.99–86.51), which is significantly higher than for the Deutch strain. It was not possible to calculate the LT50 for the El Zamora and Yucatan strains, as the observed mortality were found to be at very low rates (∼5%) across the observed data points (Fig. 1), supporting the “knockdown” resistance phenotype observed for both of these strains.
Fig. 1.
Mortality of R. microplus larvae of Deutch strain (susceptible) compared to that of the Santa Luiza, El Zamora and Yucatan strains (resistant) exposed to 0.25% permethrin impregnated filter papers. Mortality was evaluated at 10-min intervals for 60 min.
Table 2.
Results of lethal time bioassays conducted with susceptible (Deutch) and pyrethroid resistant (Santa Luiza, El Zamora and Yucatan) reference strains of Rhipicephalus (Boophilus) microplus against permethrin.
| Strain | N | Slope (SE) | Chi-square (DF) | H | LT50 (95%CL) | RR |
|---|---|---|---|---|---|---|
| Deutch | 1467 | 4.89 (0.23) | 112.89 (16) | 7.06 | 21.74 (18.48–24.63) | – |
| Santa Luiza | 1107 | 1.75 (0.18) | 23.56 (16) | 1.47 | 65.02 (53.99–86.51) | 2.99 |
| El Zamora | 1539 | 0.39 (0.24) | 24.31 (16) | 1.62 | >60 | a |
| Yucatan | 1531 | 0.83 (0.3) | 9.11 (16) | 0.57 | >60 | a |
N = number of individuals; SE = standard error; DF = degrees of freedom; H = heterogeneity on the Chi-square goodness of fit test; LT50 = median lethal time in minutes; 95%CL = 95% confidence limits, RR = resistance ratio. a: not calculated.
3.2. HRM genotyping of reference strains
The HRM assay was successfully used to determine the presence of previously described pyrethroid resistant SNPs in two regions of the para-sodium channel gene of R. microplus (domain II S4—S5 and domain III S6). In domain II, three SNPs were found among individuals of the tick colonies and were validated by cloning and sequencing: G184C, C189A, and C190A. The substitutions G184C and C190A resulted in amino acid changes (G62R and L64I, respectively) and C189A was a silent mutation (Fig. 2). In domain III, we detected, both by qPCR-HRM and AS-PCR, the mutation T2134A that results in a Phe to Ile change at the amino acid residue 712 of the para-sodium channel. The SNPs T170C and G215T were not detected with the HRM assay or sequencing of the fragments obtained from the ticks from the reference strains (Deutch, Santa Luiza, Yucatan, and El Zamora).
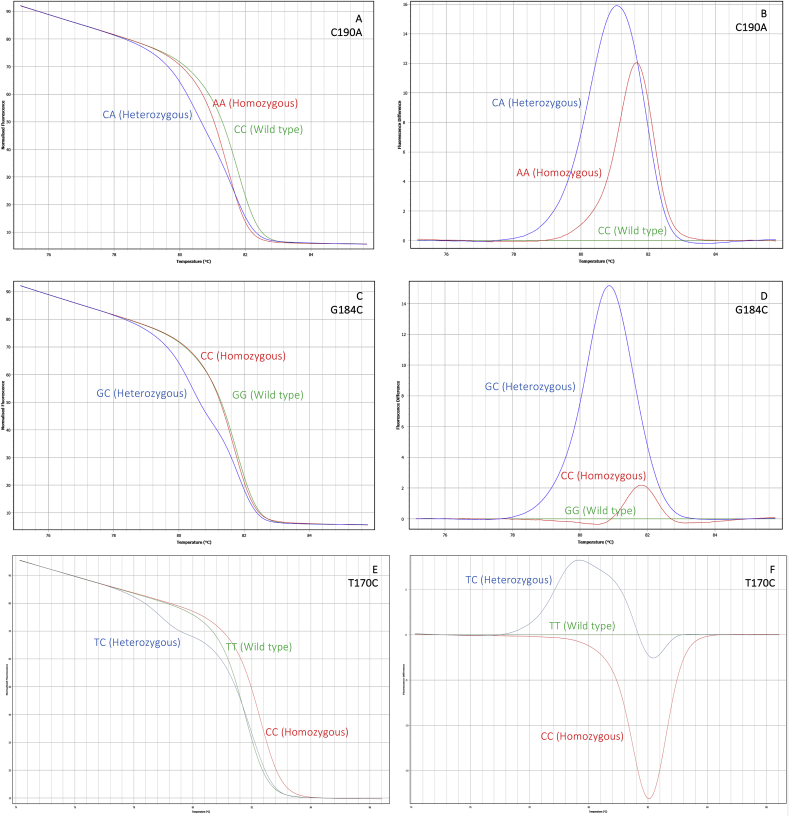
Fig. 2.
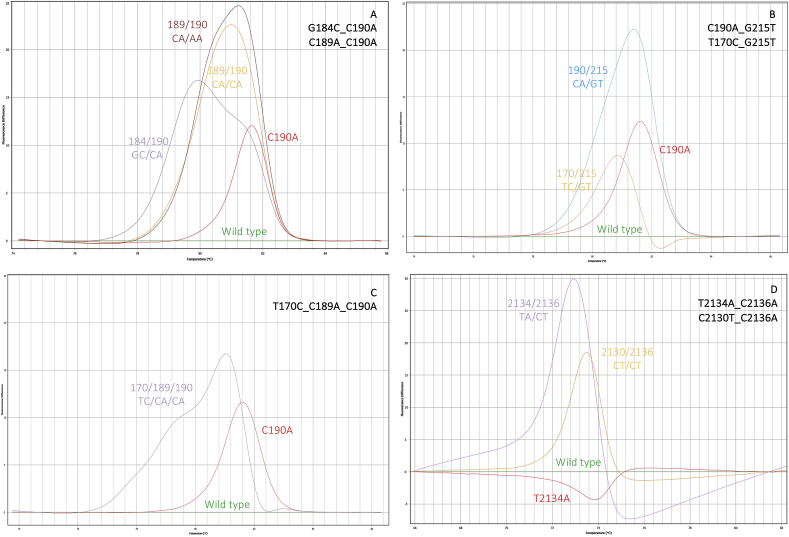
Nucleotide sequence alignment of sodium channel domains II (2A) and III (2B) DNA fragments of R. microplus reference sequence (GenBank Accession Number AF134216) with its respective translated amino acid sequences, and clone sequences obtained from pyrethroid resistant ticks: T170C, G184C, C190A, G184C_C190A, C189A_C190A, C190A_G215T, T170C_G215T, T170C_C189A_C190A, T2134A, T2134A_C2136A, C2130T_T2134A, and C2136A. Identical nucleotides are marked with dots. Numbers above sequences are based on the R. microplus para-sodium channel. Sequencing primers positions (Domain II: BmNa5F and BmNa5F; Domain III: RmNaDomainIIIF1 and RmNaDomainIIIRS2-CON) are identified as gray arrows. HRM primers (Domain II: HRM_Rm_Na-D2_F2 and HRM_Rm_Na-D2_F2; Domain III: HRM_Rm_Na_F and HRM_RmNa_F) are identified in black. Replaced amino acid residues are identified in red with one asterisk. The synonymous substitutions are identified in orange with two asterisks.
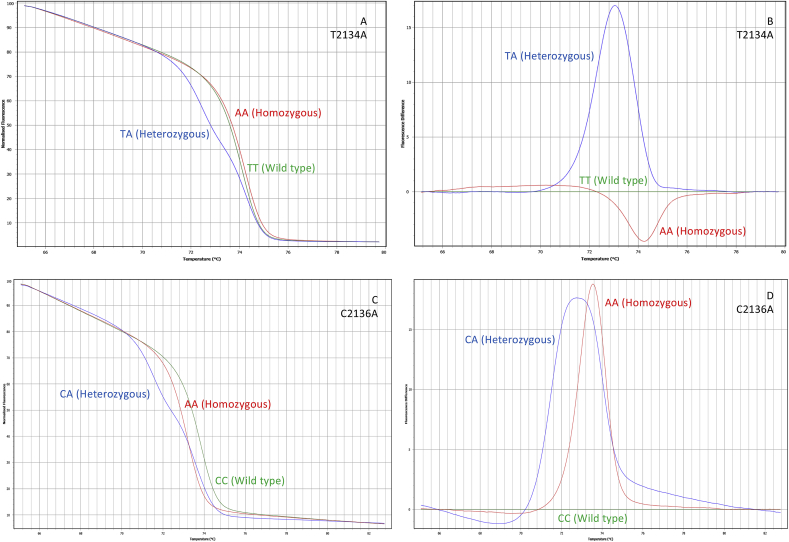
Fig. 3, Fig. 4, Fig. 5 show the normalized and difference melt plots of the fragments amplified by qPCR. On domain II, eight genotypes could be identified among the laboratory strains. All the ticks from the susceptible reference strain Deutch presented the same genotype (wt), with no substitutions of nucleotides in comparison to the reference sequence (GenBank: AF134216). The C190A and G184C substitutions were found to be associated with both homozygous and heterozygous alleles (Fig. 3A–D). The substitution in nucleotide 189 was found only in heterozygosity and always associated with C190A (Fig. 5A). The co-occurrence of G184C and C190A was also found among the tick samples, only in heterozygosity (Fig. 5A). On domain III, three genotypes were detected among the laboratory strains: wild type, heterozygous (T2134W), and homozygous (T2134A) (Fig. 4A and B), and the genotypes were confirmed with the AS-PCR technique and sequencing (Fig. 2B).
Fig. 3.
Melt and fluorescence difference curves obtained with HRM assays for pyrethroid resistance mutations detected in the domain II of para-sodium channel gene of R. microplus. 3A and 3B: mutations at position 190, (CC wild type – green line; AA homozygous – red line; and CA heterozygous – blue line). 3C and 3D: mutations at position 184 (GG wild type – green line; CC homozygous – red line; and GC heterozygous – blue line). 3E and 3F. mutations at position 170 (TT wild type – green line; CC homozygous – red line; and TC heterozygous – blue line). 4A and 4B: Melt and difference curves for the mutations at position 2134, para-sodium channel domain III (wt – green line; T2134A – red line; and T2134W – blue line).
Fig. 4.
Melt and fluorescence difference curves obtained with HRM assays for pyrethroid resistance mutations detected in the domain III of para-sodium channel gene of R. microplus. 4A and 4B: mutations at position 2134, (TT wild type – green line; AA homozygous – red line; and TA heterozygous – blue line). 4C and 4D: mutations at position 2136, (CC wild type – green line; AA homozygous – red line; and CA heterozygous – blue line).
Fig. 5.
Fluorescence difference curves obtained with HRM assays for pyrethroid resistance with multiple mutations detected in the domains II and III of para-sodium channel gene of R. microplus. 5A: G184C_C190A, and C189A_C190A; 5B: C190A_G215T, and T170C_G215T; 5C: T170C_C189A_C190A; 5D: T2134A_C2136A and C2130T_C2136A.
The designed HRM assay showed versatility in detecting a number of different genotypes. With two reactions (aimed at domains II and III), we were able to detect and assign six different genotypes (Table 3). One genotype was associated with the susceptible wild-type, and five associated with pyrethroid resistance.
Table 3.
Genotypes and their frequencies among individuals of Rhipicephalus (Boophilus) microplus susceptible and pyrethroid-resistant reference strains.
| Genotype | Amino acid sequence | SNP |
Genotype frequency (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| G184C | C190A | T2134A | Deutch | St. Luiza | El Zamora | Yucatan | ||
| Wild type | GG/LL/FF | GG | CC | TT | 100 | 0 | 0 | 0 |
| Heterozygous | GG/LI/FF | GG | CA | TT | 0 | 34.8 | 25 | 0 |
| GR/LL/FI | GC | CC | TA | 0 | 26.1 | 0 | 0 | |
| GR/LI/FI | GC | CA | TA | 0 | 21.7 | 0 | 0 | |
| Homozygous | GG/II/FF | GG | AA | TT | 0 | 8.7 | 75 | 0 |
| RR/LL/II | CC | CC | AA | 0 | 8.7 | 0 | 100 | |
G: Glycine; L: Leucine; F: Phenylalanine; R: Arginine; I: Isoleucine; SNP: single-nucleotide polymorphism.
Among the individuals of the El Zamora strain, the only detected mutation was the C190A, with 75% of the ticks being homozygous for this mutation and 25% heterozygous; no wild types were detected. Half of the sampled population (n = 12) presented the C189A silent mutation, always combined with the nucleotide substitution at position 190. All the Yucatan ticks were homozygous for the resistant mutations at domains II (G184C) and III (T2134A) simultaneously.
Ticks from the Santa Luiza strain presented the three SNPs and all the possible genotypes associated with resistance. Most of the Santa Luiza ticks (82.6%) carried at least one resistant allele with a low frequency of resistant homozygous ticks (8.7% for G184C/T2134A and C190A). Five ticks (21.7%) presented the three mutant loci simultaneously. To the best of our knowledge this is the first description of the existence of three simultaneous mutations in the para-sodium channel gene of R. microplus.
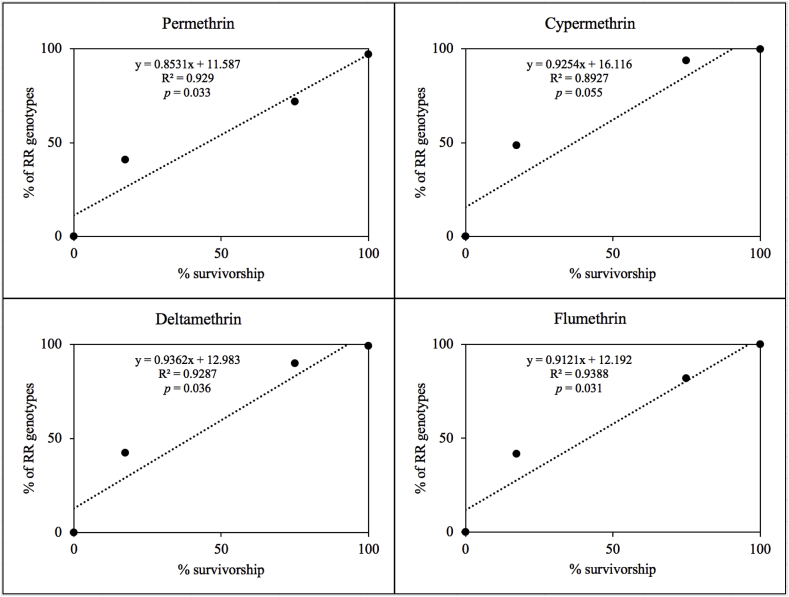
A good phenotype-genotype correlation of the assays was observed. Pearson Product Moment correlation analysis and linear regression analyses were carried out (Sigma Plot 11.0, Systat Software, 2008) by plotting the frequency of ticks carrying any of the mutations in homozygosis (GG/II/FF and RR/LL/II) against the survivorship in the bioassays with all the pyrethroids tested (Fig. 6). In a Pearson Product Moment correlation analysis, with the exception of cypermethrin (r = 0.945; p = 0.0552), there was a significant correlation between resistant-homozygous ticks (RR) and the survivorship after exposure to permethrin (r = 0.964; p = 0.0361), deltamethrin (r = 0.964; p = 0.0363), and flumethrin (r = 0.969; p = 0.0311). This observation was coherent with the results from the bioassays, showing a broad spectrum of resistance against the different pyrethroid acaricides tested (Table 1, Table 2). The high frequency of homozygous resistant ticks in the El Zamora and Yucatan strains was compatible with the “knockdown-resistance” phenotype observed in the lethal-time bioassays (Table 2, Fig. 1). This observation confirmed the association of mutations in the para-sodium channel with resistance to synthetic pyrethroids.
Fig. 6.
Linear regressions of R. microplus of susceptible (Deutch) and resistant strains (Santa Luiza, El Zamora and Yucatan), showing the relationship between the proportion of individuals carrying the mutations in homozygosis and the larval packet test survivorship using permethrin, cypermethrin, deltamethrin and flumethrin.
3.3. HRM genotyping of field samples
The developed technique was used with DNA from R. microplus field samples from northern states of Mexico (Supplementary table). The HRM was successful in detecting the previously described mutations associated with pyrethroid resistance in the tick colonies (e.g. G184C; C190A and T2134A). Three ticks were wild type for all the loci and can be considered susceptible. Two ticks presented one allele with the G215T mutation, combined with either the T170C or the C190A SNPs (Fig. 2, Fig. 5B). This is the first detection of the G215T, in the Neotropical region. This SNP results in a Gly to a Val change at the amino acid residue 72 in the domain II (Fig. 2A). A novel SNP, C2136A, was detected with the HRM in the domain III (Fig. 4C and D) resulting in a Phe to Leu change in the position 712. When this mutation simultaneously occurs with the T2134A mutation (Fig. 5D), the amino acid change was Phe to Ile (Fig. 2B). Seven ticks carry the C2136A mutation, two in homozygosis, and always associated with mutations in the domain II (T170C and C190A). One tick was heterozygous for a silent mutation, C2130T, that was detected in association with T170C, G215T and C2136A (Fig. 2). Ten ticks presented both G184C and T2134A SNPs simultaneously. Nine ticks carry at least one C190A allele, six in homozygosis. The HRM allowed the detection of the T170C (Fig. 3G and H), that results in a Met to a Thr change at the amino acid residue 57 in the domain II of the para-sodium channel (Fig. 2A). Three ticks homozygous for this mutation and one was heterozygous. Two ticks carry this mutation associated with the synonymous SNP C189A and the mutation C190A simultaneously (Fig. 5C). The more part of the ticks presented one (41%) or two mutant loci (31%) in the para-sodium channel. Two ticks presented three SNPs and three ticks presented four SNPs at the same time. The most frequent SNP was the C190A, found in 44.8% of the ticks, followed by T2134A, found in 37.9% of the samples. All the previously known and novel mutations in the para-sodium channel were detected in single ticks with two HRM reactions, aimed at the domains II and III (Fig. 3, Fig. 4, Fig. 5).
4. Discussion
Target site insensitivity is a resistance mechanism in R. microplus against pyrethroid acaricides, which is conferred by one or more mutations present in the target site, the para-sodium channel (Guerrero et al., 2014). Based on the results presented here, we successfully developed two qPCR-HRM assays to genotype the G184C, C190A and T2134A mutations, associated with resistance against four compounds of the synthetic pyrethroid class of acaricides: permethrin, cypermethrin, deltamethrin and flumethrin. We were also able to detect multiple mutations and their different combinations in two side-by-side reactions (domains II and III). Genotyping of mutations directly related to acaricide resistance could provide a useful surveillance tool to monitor resistance status promptly and accurately among R. microplus populations. This approach could also help refine integrated tick management strategies by adapting the selection of acaricides for effective and rational R. microplus control based on resistance genetic data obtained in real time.
In HRM analysis, a small region of DNA (70–100 bp) spanning the SNPs of interest is amplified by PCR in the presence of a dsDNA fluorescent dye (EVA green). This dye is used at a high concentration to achieve maximum saturation of the resulting dsDNA fragment (Bass et al., 2007). A high-resolution melt step using equipment with high thermal and optical precision is then performed in order to determine the melting temperature (Tm) of the amplicon. While the dsDNA dissociates into single strands, the dye is released and the fluorescence decreases giving a melt curve profile characteristic of the amplicon sequence (Liew et al., 2004).
The C190A, T170C and C2136A mutations are predicted to cause a relatively large change in the Tm of the amplicon (Fig. 3, Fig. 4C). In contrast, the G184C and the T2134A are predicted to cause a very small change in Tm in the melt curve making it more difficult to detect in the normalized curves (Fig. 3, Fig. 4A). However, the observation of the differences in melt plots helps in distinguishing the genotypes (3D and 4B). The combinations of different mutations (C189A/C190A; C190A/G215T; T170C/G215T; T170C/C189A/C190A; T2134A/C2136C; C2130T/C2136T) are predicted to cause even larger variations in the Tm of the melt curves (Fig. 3, Fig. 4, Fig. 5D). Using samples with known genotypes (as determined by sequencing) the developed assays were able to efficiently distinguish the different genotypes, in homozygosity and heterozygosity.
The Santa Luiza strain has been studied by synergistic bioassays and genetic studies to determine its mechanisms of resistance to permethrin (Li et al. 2007, 2008). The present study clarifies the major mechanism involved, which is the presence of one, two or three mutations in different amino acid residues of the para-sodium channel gene. Stone et al. (2014) made the first description of the presence of simultaneous mutations in the para-sodium channel gene in R. microplus populations from Mexico, where a combination of the M57T amino acid change with L64I or F712I was linked to high levels of pyrethroid resistance. Nevertheless, the mutations in the Santa Luiza ticks were mostly heterozygous (82.6%; Table 3). The cause for the lower resistance level in Santa Luiza is unknown when compared to the El Zamora and Yucatan strains.
El Zamora ticks presented mutations at the nucleotide positions 189 and 190 (kdr). Twenty-five percent of the ticks were heterozygous for the C190A mutation, and the remaining were homozygous. The C189A substitution is synonymous and was never found by itself and always in heterozygosity. The same observation was made by Stone et al. (2014) in screening populations from Mexico and the USA. These authors pointed out that the presence of this SNP could prevent PCR amplification of fragments using specific primers designed to detect the C190A mutation. The HRM domain II assay we developed was successful in detecting both polymorphisms and no amplification failures were detected. The Yucatan strain was the most resistant strain evaluated in this study (mean mortality between 0 and 2.85% depending on the acaricide tested). All the ticks genotyped were homozygous for the G62R and F712I mutations, which can explain the low mortality levels found and the knockdown resistance phenotype observed (Table 2, Fig. 1).
This assay could also be used to mitigate the risk associated with acaricide-resistant R. microplus that may infest cattle presented at ports of entry by Mexico intended for export to the U.S. (González and Hernández, 2012). Among the ticks sampled at the USA Ports-of-Entry, we found a high frequency of individuals carrying mutations in para-sodium channel gene. Most of them, with C190A or T2134A and combinations of the different mutations. The presence of multiple mutations in the para-sodium channel gene is correlated with high levels of pyrethroid resistance, and here we found ticks carrying three or four SNPs in both domains II and III (Supplementary Table). More information is required about acaricide usage or resistance status of the field samples to determine if that correlation causes high levels of pyrethroid resistance as suggested by the results of the HRM analysis.
Comparing different techniques for detection of knockdown resistance mutations in mosquitoes, Bass et al. (2007) observed that HRM analysis presented a higher failure rate than other assays, such as AS-PCR and Taqman® probes. The authors suggest that the quality and quantity of DNA could be affecting the amplification, preventing the attainment of a high signal plateau in the PCR phase, which could result in inconclusive or low resolution HRM data. In our experiments we were able to obtain satisfactory amplification and discrimination among genotypes from the tick colonies as well as from the field samples. The quality and quantity of DNA obtained with the phenol-chloroform purification was high (260/280 ratio ∼1.8, over 200 ng/μL of dsDNA per sample), which likely had a positive impact on the quality of our amplification.
The main advantage of the HRM, comparing to AS-PCR and Taqman is its capacity of screening different variants in a given gene that would be undetectable using allele specific probes and primers, as virtually any different nucleotide can be detected in a sequence using the high-resolution analysis of the melting temperatures of those fragments. This feature is particularly interesting to define samples to be analyzed by sequencing in SNP discovery studies. When investigating the field samples from Mexico, in most of the cases, we detected the previously known SNPs (C190A, 44.83%, and T2134A, 37.93%). However, we were able to detect novel melt curves (Fig. 3, Fig. 4, Fig. 5D), indicating the existence of different genotypes that were later confirmed by cloning and sequencing. Using the screening approach, we were able to detect for the first time in the Neotropical region, the mutation G215T, associated with flumethrin resistance in Australia (Jonsson et al., 2010) and a novel SNP, C2136A, located in the same codon for the Phe at the position 712. However, when this mutation is present in the absence of the T2134A mutation, the amino acid is substituted for an Ile. We hypothesize that the physiological effect of this substitution is the same as the Phe to Leu, which results in resistance to pyrethroids (He et al., 1999). While there is no information about the phenotype of the different patterns of mutations regarding the susceptibility to pyrethroids among the field samples, further research is needed to test our hypothesis.
This study provides a technique that can be used for the surveillance of pyrethroid resistance in R. microplus populations. Our findings can be used to develop a high-throughput method to genotype and detect allele-specific mutations in R. microplus populations that cause outbreaks in the U.S. The rapid turnaround of results based on a high-throughput HRM acaricide resistance assay could help Cattle Fever Tick Eradication Program personnel manage the response to R. microplus outbreaks, and also inform decisions regarding the concern with cattle presented at ports of entry by Mexico with the intention to be exported to the U.S. that may be infested with R. microplus resistant to acaricides (Pérez de León et al., 2013). This approach could also be adapted to other acaricide targets and species of ticks of medical and veterinary importance. The HRM for pyrethroid resistance SNPs could also be used in integrated control programs in other parts of the world where ticks and tick-borne diseases burden the health of humans, domestic animals, and wildlife.
5. Conclusion
A quantitative PCR-based HRM assay method was developed that detects T170C, G184C, C190A, G215T, T2134A and C2136A SNPs and their combinations in the para-sodium ion channel gene of R. microplus. Bioassays confirmed the existence of broad spectrum pyrethroid resistance correlated with the frequency of the mutations in the tick strains evaluated. This assay provides a useful methodology to screen mutations associated with susceptibility to pyrethroids in tick populations and thus facilitates surveillance for acaricide resistance.
Author contribution statement
G.M.K., R.J.M., J.P.T. and A.A.P.L., oversaw the research and planned the experiments. G.M.K. and D.S. executed the bioassays, PCR, cloning and sequencing; G.M.K. and J.P.T. analyzed the data and prepared figures. G.M.K., J.P.T., D.S., R.J.M., T.P.F., D.B.T. and A.A.P.L. prepared and reviewed the manuscript.
Declarations of interest
None.
Acknowledgements
We thank Laurence Dave Krska, Michael Moses, and Ruby Martinez for their support on the maintenance of the tick colonies, as well as Cesario Agado, James Hellums, and Homero Vazquez for the handling of animals used as tick hosts. We also thank Dr. Denise L. Bonilla, Gustavo A. Soberano, Dr. Amy Green, Dr. Camilo Potes, Dr. Albert Leslie, Dr. Amber Lassiter, Dr. Walter Howe, Dr. Kayla Wells, Dr. Lilajit Rai and Dr. Nianet Carrasquillo from USDA-APHIS for the access to the field samples. G.M.K. was funded by the USDA-ARS through the Oak Ridge Institute for Science and Education (ORISE). D.S. and T.P.F.A. were supported by the USDA AFRI National Institute of Food and Agriculture program titled “Training the next generation of agricultural scientists: coping with food security and climatic change challenges”, award number: 2016-67032-25013. Robert D. Mitchell III provided helpful comments on an earlier version of the manuscript. USDA is an equal opportunity provider and employer.
Footnotes
Note: Nucleotide sequence data reported in this paper are available in the GenBank™ database under the accession numbers: MH986340, MH986341, MH986342, MH986343.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.03.001.
Contributor Information
Guilherme M. Klafke, Email: guilherme-klafke@agricultura.rs.gov.br.
Adalberto A. Pérez de León, Email: beto.perezdeleon@ars.usda.gov.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aguirre M., Flores A.E., Alvarez G., Molina A., Rodriguez I., Ponce G. A novel amino acid substitution in the para-sodium channel gene in Rhipicephalus microplus (Acari: Ixodidae) associated with knockdown resistance. Exp. Appl. Acarol. 2010;52:377–382. doi: 10.1007/s10493-010-9371-y. [DOI] [PubMed] [Google Scholar]
- Aubry P., Geale D.W. A review of bovine anaplasmosis. Transbound. Emerg. Dis. 2011;58:1–30. doi: 10.1111/j.1865-1682.2010.01173.x. [DOI] [PubMed] [Google Scholar]
- Bandara K.M.U., Karunaratne S.H.P. Mechanisms of acaricide resistance in the cattle tick Rhipicephalus (Boophilus) microplus in Sri Lanka. Pestic. Biochem. Physiol. 2017;139:68–72. doi: 10.1016/j.pestbp.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Bass C., Nikou D., Donnelly M.J., Williamson M.S., Ranson H., Ball A., Vontas J., Field L.M. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: a comparison of two new high-throughput assays with existing methods. Malar. J. 2007;6:111. doi: 10.1186/1475-2875-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey R.B., Garza J., Jr., Thompson G.D., Drummond R.O. Ovipositional biology of the cattle tick, Boophilus annulatus (Acari: Ixodidae), in the laboratory. J. Med. Entomol. 1980;17:287–289. [Google Scholar]
- Dong K., Du Y., Rinkevich F., Nomura Y., Xu P., Wang L., Silver K., Zhorov B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014;50:1–17. doi: 10.1016/j.ibmb.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond R.O., Ernst S.E., Trevino J.L., Gladney W.J., Graham O.H. Boophilus annulatus and B. microplus: laboratory tests of Insecticides. J. Econ. Entomol. 1973;66:130–133. doi: 10.1093/jee/66.1.130. [DOI] [PubMed] [Google Scholar]
- FAO . Food and Agriculture Organization Animal Production and Health Division; Rome: 2004. Resistance Management and Integrated Parasite Control in Ruminants – Guidelines, Module 1 – Ticks: Acaricide Resistance: Diagnosis, Management and Prevention; pp. 25–77. [Google Scholar]
- Gaur R.S., Sangwan A.K., Sangwan N., Ghosh M., Kumar S. Comparative study of esterases in deltamethrin and diazinon resistant Rhipicephalus microplus and Hyalomma anatolicum ticks collected from the Trans-Gangetic plains of India. Exp. Appl. Acarol. 2017;73:115–127. doi: 10.1007/s10493-017-0175-1. [DOI] [PubMed] [Google Scholar]
- González J., Hernández R. Boophilus microplus: current status of acaricide resistance on the Mexican American border and its impact on commerce. Rev. Mex. Cienc. Pecu. 2012;3(Suppl. 1):1–8. [Google Scholar]
- Graham K.M., Sparagano O.A., Finn R.D. Isolation of the monooxygenase complex from Rhipicephalus (Boophilus) microplus - clues to understanding acaricide resistance. Ticks Tick Borne Dis. 2016;7:614–623. doi: 10.1016/j.ttbdis.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Grisi L., Cerqueira Leite R., Martins J.R.S., Medeiros de Barros A.T., Andreotti R., Duarte Cançado P.H., Pérez de León A.A., Barros Pereira J., Silva Villela H. Reassessment of economic impact by cattle parasites in Brazil. Rev. Bras. Parasitol. Vet. 2014;23:150–156. doi: 10.1590/s1984-29612014042. [DOI] [PubMed] [Google Scholar]
- Guerrero F., Pérez de León A., Rodriguez-Vivas R., Jonsson N., Miller R., Andreotti R. Acaricide research and development, resistance and resistance monitoring. In: Sonenshine D.E., Roe R.M., editors. vol. 2. Oxford University Press; New York: 2014. pp. 353–374. (Biology of Ticks 2nd Edition). [Google Scholar]
- Guerrero F.D., Davey R.B., Miller R.J. Use of an allele-specific polymerase chain reaction assay to genotype pyrethroid resistant strains of Boophilus microplus (Acari: Ixodidae) J. Med. Entomol. 2001;38:44–50. doi: 10.1603/0022-2585-38.1.44. [DOI] [PubMed] [Google Scholar]
- Guerrero F.D., Pruett J.H., Li A.Y. Molecular and biochemical diagnosis of esterase-mediated pyrethroid resistance in a Mexican strain of Boophilus microplus (Acari: Ixodidae) Exp. Appl. Acarol. 2002;28:257–264. doi: 10.1023/a:1025319104529. [DOI] [PubMed] [Google Scholar]
- Gupta S., Ajith Kumar K.G., Sharma A.K., Nagar G., Kumar S., Saravanan B.C., Ravikumar G., Ghosh S. Esterase mediated resistance in deltamethrin resistant reference tick colony of Rhipicephalus (Boophilus) microplus. Exp. Appl. Acarol. 2016;69:239–248. doi: 10.1007/s10493-016-0032-7. [DOI] [PubMed] [Google Scholar]
- He H., Chen A.C., Davey R.B., Ivie G.W., George J.E. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle tick. Biochem. Biophys. Res. Commun. 1999;261:558–561. doi: 10.1006/bbrc.1999.1076. [DOI] [PubMed] [Google Scholar]
- Jonsson N.N., Cutullè C., Corley S.W., Seddon J.M. Identification of a mutation in the para-sodium channel gene of the cattle tick Rhipicephalus microplus associated with resistance to flumethrin but not to cypermethrin. Int J Parasitol. 2010;40(14):1659–1664. doi: 10.1016/j.ijpara.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Kumar R., Nagar G., Sharma A.K., Kumar S., Ray D.D., Chaudhuri P., Ghosh S. Survey of pyrethroids resistance in Indian isolates of Rhipicephalus (Boophilus) microplus: identification of C190A mutation in the domain II of the para-sodium channel gene. Acta Trop. 2013;125:237–245. doi: 10.1016/j.actatropica.2012.10.006. [DOI] [PubMed] [Google Scholar]
- LeOra Software . In: Polo Plus Probit and Logit Analysis, User's Guide. Berkeley. Robertson J.L., Preisler H.K., Russel R.M., editors. 2003. 36 pp. [Google Scholar]
- Li A.Y., Chen A.C., Miller R.J., Davey R.B., George J.E. Acaricide resistance and synergism between permethrin and amitraz against susceptible and resistant strains of Boophilus microplus (Acari: Ixodidae) Pest Manag. Sci. 2007;63:882–889. doi: 10.1002/ps.1417. [DOI] [PubMed] [Google Scholar]
- Li A.Y., Davey R.B., Miller R.J., George J.E. Resistance to coumaphos and diazinon in Boophilus microplus (Acari: Ixodidae) and evidence for the involvement of an oxidative detoxification mechanism. J. Med. Entomol. 2003;40:482–490. doi: 10.1603/0022-2585-40.4.482. [DOI] [PubMed] [Google Scholar]
- Li A.Y., Davey R.B., Miller R.J., George J.E. Detection and characterization of amitraz resistance in the southern cattle tick, Boophilus microplus (Acari: Ixodidae) J. Med. Entomol. 2004;41:193–200. doi: 10.1603/0022-2585-41.2.193. [DOI] [PubMed] [Google Scholar]
- Li A.Y., Davey R.B., Miller R.J., Guerrero F.D., George J.E. Genetics and mechanisms of permethrin resistance in the Santa Luiza strain of Boophilus microplus (Acari: Ixodidae) J. Med. Entomol. 2008;45:427–438. doi: 10.1603/0022-2585(2008)45[427:gamopr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Liew M., Pryor R., Palais R., Meadows C., Erali M., Lyon E., Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Lovis L., Guerrero F.D., Miller R.J., Bodine D.M., Betschart B., Sager H. Distribution patterns of three sodium channel mutations associated with pyrethroid resistance in Rhipicephalus (Boophilus) microplus populations from North and South America, South Africa and Australia. Int. J. Parasitol. Drugs Drug Resist. 2012;2:216–224. doi: 10.1016/j.ijpddr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader E., Lukas B., Novak J. A strategy to setup codominant microsatellite analysis for high-resolution-melting-curve-analysis (HRM) BMC Genet. 2008;9:69. doi: 10.1186/1471-2156-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.J., Almazán C., Ortíz-Estrada M., Davey R.B., George J.E., De León A.P. First report of fipronil resistance in Rhipicephalus (Boophilus) microplus of Mexico. Vet. Parasitol. 2013;191:97–101. doi: 10.1016/j.vetpar.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Miller R.J., Davey R.B., George J.E. Characterization of pyrethroid resistance and susceptibility to coumaphos in Mexican Boophilus microplus (Acari: Ixodidae) J. Med. Entomol. 1999;36:533–538. doi: 10.1093/jmedent/36.5.533. [DOI] [PubMed] [Google Scholar]
- Morgan J.A., Corley S.W., Jackson L.A., Lew-Tabor A.E., Moolhuijzen P.M., Jonsson N.N. Identification of a mutation in the para-sodium channel gene of the cattle tick Rhipicephalus (Boophilus) microplus associated with resistance to synthetic pyrethroid acaricides. Int. J. Parasitol. 2009;39:775–779. doi: 10.1016/j.ijpara.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Nogueira Domingues L., dos Santos Alves Figueiredo Brasil B., Passos de Paiva Bello A.C., Pinto da Cunha A., Thadeu Medeiros de Barros A., Cerqueira Leite R., Silaghi C., Pfister K., Friche Passos L.M. Survey of pyrethroid and organophosphate resistance in Brazilian field populations of Rhipicephalus (Boophilus) microplus: detection of C190A mutation in domain II of the para-type sodium channel gene. Vet. Parasitol. 2012;189:327–332. doi: 10.1016/j.vetpar.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Pasay C., Arlian L., Morgan M., Vyszenski-Moher D., Rose A., Holt D., Walton S., McCarthy J. High-resolution melt analysis for the detection of a mutation associated with permethrin resistance in a population of scabies mites. Med. Vet. Entomol. 2008;22:82–88. doi: 10.1111/j.1365-2915.2008.00716.x. [DOI] [PubMed] [Google Scholar]
- Pérez de León A.A., Rodríguez-Vivas R.I., Guerrero F.D., García- Vázquez Z., Temeyer K.B., Domínguez-García D.I., Li A., Cespedes N., Miller R.J., Rosario Cruz R. Acaricide resistance in Rhipicephalus (Boophilus) microplus: impact on agro-biosecurity and cattle trade between Mexico and the United States of America. In: Domínguez-García D.I., Rosario Cruz R., Ortiz Estrada M., editors. Proceeedings, 3th International Symposium on Pesticide Resistance in Arthropods: Integrated Cattle Tick and Fly Control and Mitigation of Pesticide Resistance, 24 June 2013, Ixtapa, Zihuatanejo, Mexico. Universidad Autónoma de Guerrero Press; Chilpancingo, Mexico: 2013. pp. 18–35. [Google Scholar]
- Pérez de León A.A., Vannier E., Almazán C., Krause P.J. Tick-borne protozoa. In: Sonenshine D.E., Roe R.M., editors. vol. 2. Oxford University Press; New York: 2014. pp. 147–179. (Biology of Ticks 2nd Edition). [Google Scholar]
- Reed G.H., Kent J.O., Wittwer C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- Rinkevich F.D., Du Y., Dong K. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic. Biochem. Physiol. 2013;106:93–100. doi: 10.1016/j.pestbp.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbertse L., Baron S., van der Merwe N.A., Madder M., Stoltsz W.H., Maritz-Olivier C. Genetic diversity, acaricide resistance status and evolutionary potential of a Rhipicephalus microplus population from a disease-controlled cattle farming area in South Africa. Ticks Tick Borne Dis. 2016;7:595–603. doi: 10.1016/j.ttbdis.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Robertson J.L., Russell R.M., Preisler H.K., Savin N.E. second ed. CRC Press; Boca Raton, Florida: 2007. Bioassays with Arthropods. [Google Scholar]
- Rodriguez-Vivas R.I., Grisi L., Pérez de León A.A., Silva Villela H., Torres-Acosta J.F.J., Fragoso-Sanchez H., Romero-Salas D., Rosario-Cruz R., Saldierna F., Garcia-Carrasco D. Potential economic impact assessment for cattle parasites in Mexico. Rev. Mex. Cienc. Pec. 2017;8:61–74. [Google Scholar]
- Rodriguez-Vivas R.I., Hodgkinson J.E., Rosado-Aguilar J.A., Villegas-Perez S.L., Trees A.J. The prevalence of pyrethroid resistance phenotype and genotype in Rhipicephalus (Boophilus) microplus in Yucatan, Mexico. Vet. Parasitol. 2012;184:221–229. doi: 10.1016/j.vetpar.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Vivas R.I., Miller R.J., Ojeda-Chi M.M., Rosado-Aguilar J.A., Trinidad-Martínez I.C., Pérez de León A.A. Acaricide and ivermectin resistance in a field population of Rhipicephalus microplus (Acari: Ixodidae) collected from red deer (Cervus elaphus) in the Mexican tropics. Vet. Parasitol. 2014;200:179–188. doi: 10.1016/j.vetpar.2013.11.025. [DOI] [PubMed] [Google Scholar]
- Rosario-Cruz R., Guerrero F.D., Miller R.J., Rodriguez-Vivas R.I., Tijerina M., Dominguez-Garcia D.I., Hernandez-Ortiz R., Cornel A.J., McAbee R.D., Alonso-Diaz M.A. Molecular survey of pyrethroid resistance mechanisms in Mexican field populations of Rhipicephalus (Boophilus) microplus. Parasitol. Res. 2009;105:1145–1153. doi: 10.1007/s00436-009-1539-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Rath S.S. Esterase mediated resistance against synthetic pyrethroids in field populations of Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) in Punjab districts of India. Vet. Parasitol. 2014;204:330–338. doi: 10.1016/j.vetpar.2014.05.035. [DOI] [PubMed] [Google Scholar]
- Stone B., Haydock K. A method for measuring the acaricide-susceptibility of the cattle tick Boophilus microplus (Can.) Bull. Entomol. Res. 1962;53:563–578. [Google Scholar]
- Stone N.E., Olafson P.U., Davey R.B., Buckmeier G., Bodine D., Sidak-Loftis L.C., Giles J.R., Duhaime R., Miller R.J., Mosqueda J., Scoles G.A., Wagner D.M., Busch J.D. Multiple mutations in the para-sodium channel gene are associated with pyrethroid resistance in Rhipicephalus microplus from the United States and Mexico. Parasites Vectors. 2014;7:456. doi: 10.1186/s13071-014-0456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungirai M., Baron S., Moyo D.Z., De Clercq P., Maritz-Olivier C., Madder M. Genotyping acaricide resistance profiles of Rhipicephalus microplus tick populations from communal land areas of Zimbabwe. Ticks Tick Borne Dis. 2018;9:2–9. doi: 10.1016/j.ttbdis.2017.10.017. 2018. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer 3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A., Bouattour A., Camicas J.L., Estrada-Pena A., Horak I., Latif A., Pegram R.G., Preston P.M. second ed. Bioscience Reports; Edinburgh Scotland, U.K: 2003. Ticks of Domestic Animals in Africa: a Guide to Identification of Species. [Google Scholar]
- Wuliandari J.R., Lee S.F., White V.L., Tantowijoyo W., Hoffmann A.A., Endersby-Harshman N.M. Association between three mutations, F1565C, V1023G and S996P, in the voltage-sensitive sodium channel gene and knockdown resistance in Aedes aegypti from yogyakarta, Indonesia. Insects. 2015;6:658–685. doi: 10.3390/insects6030658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyk R.D., Baron S., Maritz-Olivier C. An integrative approach to understanding pyrethroid resistance in Rhipicephalus microplus and R. decoloratus ticks. Ticks Tick Borne Dis. 2016;7:586–594. doi: 10.1016/j.ttbdis.2016.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.