Abstract
In order to elucidate the question whether resistance to nitro drugs in G. lamblia is due to common resistance markers, trophozoites of three resistant G. lamblia strains, namely C4, 1062ID10, and 713M3 were grown in the presence of the two nitro drugs metronidazole and nitazoxanide and compared to their corresponding wild-types WBC6, 106, and 713 by mass spectometry shotgun analysis of their proteomes. Depending on the strain and the nitro drug, more than 200 to 500 differentially expressed proteins were identified, but there were no common patterns across strains and drugs. All resistant strains underwent antigenic variation with distinct surface antigens like variant surface proteins or cysteine rich proteins depending on strain and nitro compound. A closer look on enzymes involved in nitroreduction and detoxification of nitro radicals, NO or O2 suggested the existence of distinct strategies for each drug and each strain. Therefore, we conclude that resistance to nitro drugs in G. lamblia is not correlated with a specific pattern of differentially expressed proteins and therefore seems not to be the result of a directed process.
Keywords: Adaptation, Drug susceptibility, Resistance, Tolerance
Graphical abstract
Highlights
-
•
Is resistance to nitro drugs in G. lamblia due to common resistance markers?
-
•
Three resistant strains were grown in the presence of two nitro drugs separately and compared to wild-types by MS shotgun analysis.
-
•
More than 200 to 500 differentially expressed proteins identified depending on strain and drug.
-
•
No common patterns across strains and drugs.
-
•
Strain specific antigenic variation and strategies linked to nitro reduction.
1. Introduction
Giardia lamblia (syn. G. duodenalis; G. intestinalis), a flagellated, amitochondrial, binucleated protozoan (Plutzer et al., 2010; Müller and Müller, 2016; Cernikova et al., 2018), is a common causative agent of persistent diarrhea in developing (Squire and Ryan, 2017) as well as in industrial (Zylberberg et al., 2017) regions. Giardiasis is commonly treated with the nitro compounds metronidazole (MET), other 5-nitroimidazole compounds, or nitazoxanide (NTZ). Albendazole (ALB) is a valuable alternative in the case of resistance to nitro drugs (Nash, 2001; Minenoa and Avery, 2003; Huang and White, 2006). Moreover, G. lamblia is susceptible to a variety of antibiotics because of its prokaryote-like transcription and translation machineries (Müller and Hemphill, 2013). According to a commonly accepted model, nitro compounds are activated by reduction yielding toxic intermediates causing nitrosative stress (Lloyd et al., 2003; Müller and Müller, 2016). The electrons are provided by pyruvate ferredoxin oxidoreductase (PFOR) as reviewed elsewhere (Brown et al., 1998; Leitsch et al., 2011). Other enzymes potentially involved in the reduction of nitro drugs are the nitroreductases NR1 (Nillius et al., 2011; Ansell et al., 2017) and NR2 (Müller et al., 2013, 2015), and thioredoxin-reductase (Leitsch et al., 2011, 2016). Moreover, the NO reducer flavohemoglobin (Mastronicola et al., 2010) may play a role as scavenger. Despite the fact that resistance formation to nitro compounds is eagerly detected both in vitro and in vivo, the molecular basis of resistance formation is far from being elucidated. Freshly obtained, resistant patient isolates would be optimal, but they are difficult to maintain in axenic culture. Therefore, most of the studies generate resistant “model” strains in vitro and compare them with isogenic wild-type strains (Upcroft, 1998; Leitsch, 2015). In accordance to the prevailing model for the mode of action of nitro drugs, one would hypothesize that resistant trophozoites have decreased amounts of enzymes involved in nitroreduction, and that this decrease is due to regulations at transcriptional and/or posttranscriptional levels. The observation that knock-down of PFOR is correlated with increased resistance to metronidazole (Dan et al., 2000) fosters this hypothesis. Studies with MET-resistant strains have revealed, however, that resistance is not always correlated with reduced PFOR activity (Müller et al., 2007, 2018). Thus mechanisms of action independent of POR activity may exist as suggested by early studies showing genome rearrangements in resistant strains (Upcroft et al., 1990; Upcroft and Upcroft, 1993). Moreover, transcriptional changes evidenced by differential analyses using microarrays followed by quantitative RT-PCR on selected transcripts (Müller et al., 2008) and strand-specific RNA sequencing (Ansell et al., 2017) reveal profound differences in gene expression between susceptible and resistant strains including different expression profiles not only of genes involved in nitroreduction, but also of genes coding for variant surface proteins (VSPs) and others. These results are backed by a recently published proteomic study (Emery et al., 2018) where – depending on the stains – more than 200 differentially regulated proteins have been identified.
Are these changes in gene expression directed or undirected? If changes in gene expression are directed, a common pattern of genes involved in resistance formation should be identified, if not, the pattern is random, and there is no common pattern of resistance markers between different strains and different nitro drugs. Our working hypothesis is that these changes are undirected.
Since gene expression patterns in G. lamblia may be influenced by biotic parameters such as strain genotypes and abiotic parameters such as the composition of the culture media, for instance, it is difficult to find an underlying common pattern by comparing studies performed by different groups on different strains. Moreover, it would be interesting to investigate whether results observed with resistant strains grown in the presence of MET can be extrapolated to other nitro compounds such as NTZ. Therefore, comparative omic approaches with various strains resistant to more than one nitro drug grown in the presence of several drugs under the same conditions are paramount. In particular, shotgun mass spectrometry is a valid tool to investigate this hypothesis as shown by previous studies (Emery-Corbin et al., 2018; Emery et al., 2018).
Here, we present a proteomic study comparing three different MET resistant strains, namely C4 derived from the wild-type strain WBC6, 1062ID10 derived from 106, to 713M3 derived from 713 to their respective wild-types. The strains belong to assemblage AI, and do not fully represent the assemblage more commonly found in humans, such as assemblage A2 and B, that have relevant differences at genomic level with assemblage AI (Emery et al., 2015). Strain C4 has been generated by successive increase of NTZ in the culture medium as described (Müller et al., 2007). The strains 1062ID10 and 713M13 have been obtained characterized in detail more than two decades ago by the Upcroft group (Upcroft et al., 1990, 1999; Townson et al., 1992; Leitsch et al., 2011). The three strains are not only resistant to both MET and NTZ, but susceptible to ALB. Therefore, we include a comparison between those strains grown on MET and on NTZ into our study.
2. Materials and methods
2.1. Biochemicals and drugs
If not otherwise stated, all biochemical reagents were from Sigma (St Louis, MO, USA). Nitazoxanide (NTZ) was synthesized at the Department of Chemistry and Biochemistry, University of Bern, Switzerland (Ch. Leumann). NTZ and metronidazole (MET) were kept as 100 mM stock solutions in DMSO at −20 °C.
2.2. Axenic culture, harvest and storage of G. lamblia trophozoites
Trophozoites from G. lamblia wild-type " (WBC6. 106, 713)" and of the NTZ/MET resistant strains " (C4, 1062ID10, 713M3)" were grown under anaerobic conditions in 10 ml culture tubes (Nunc, Roskilde, Denmark) containing modified TYI-S-33 medium as previously described (Clark and Diamond, 2002) Prior to shotgun mass spectrometry analysis, cultures from resistant strains were routinely passaged 5 times in the presence of 50 μM NTZ or MET, respectively. Subcultures were performed by inoculating 20 μl (wild-type) or 100 μl (resistant) of cells from a confluent culture detached by cooling (see below) to a new tube containing 10 ml of the appropriate medium (Müller et al., 2006). For all experiments comparing wild-type to resistant trophozoites, the medium from confluent cultures was removed one day before the harvest and replaced with fresh medium without nitro compound. Trophozoites were detached by incubation on ice for 15 min followed by centrifugation (300×g, 10 min, 4 °C). Pellets were washed three times with ice-cold PBS, counted, and stored at −80 °C for subsequent proteomic analysis or for quantification of expression of selected genes.
2.3. Proteomics
Cell pellets were lysed in 100 μL 8M urea/100 mM Tris/HCl pH 8/protease inhibitor cocktail (Roche Diagnostics, Rotkreuz, Switzerland) by incubation for 15 min at room temperature followed by 15 min in an ultrasonic water bath. Reduction, alkylation, digestion and nano-reversed-phase liquid chromatography coupled to tandem mass spectrometry (nLC-MS/MS) was performed as described elsewhere (Engel et al., 2014).
2.4. Statistical methods
The MS data were obtained from three biological replicates, with technical replicates for each biological replicate, for each strain. All MS data were processed by MaxQuant (version 1.5.4.1) with matching between runs of the same sample type, but not between different types, in order to avoid over-interpretation. The sample sets corresponding to wild-type strains WBC6, 106, and 713 and the resistant strains derived from these wild-types were interpreted separately by MaxQuant. Fragment spectra were interpreted against a recent Giardia protein sequence database in fasta format (GiardiaDB-5.0_GintestinalisAssemblageA_AnnotatedProteins_v2) using a trypsin cleavage rule with amide bond cleavage allowed after lysine and arginine if a proline follows and up to three missed cleavage sites, fixed carbamidomethylation modification of cysteines, variable oxidation of methionine and acetylation of protein N-termini. Precursor and fragment mass tolerances were set to 10 and 20 ppm, respectively. Peptide spectrum matches, peptide and protein group identifications were filtered to a 1% false discovery rate (FDR), and a minimum of two razor or unique peptides were required to accept a protein group identification. Protein identifications considered as contaminations (e.g. trypsin or BSA) as well as proteins identified only by site (considered by MaxQuant developers as very likely false positives) were removed for statistical validation. The normalized label free quantification (LFQ) protein group intensities as calculated by MaxQuant were used for relative proteome quantifications. First, we imputed missing protein LFQ values for samples in any condition group when there was at least one LFQ intensity per sample triplicate (downshift of 1.8 S.D. with a width of 0.3 S.D.). This left protein groups without values in one or the other group. For Student's T-tests, those missing protein intensities were replaced by imputed values from the very low end of intensity distributions (downshift 2.5 S.D., width of 0.3 S.D.). A permutation based FDR (=1%) procedure was then used to correct for multiple testing between sample groups. The imputed values were used for the calculation of p-values (expressed as -LOG) and q-values. The test differences in log2-fold change were then calculated on the effective median intensities and a log2-fold change of at least one was required to be considered as significant. Statistical testing and imputation were made with Perseus version 1.5.5.3 (Tyanova et al., 2016).
3. Results
3.1. Mass spectrometry analysis of proteins expressed in nitro drug-resistant and susceptible trophozoites
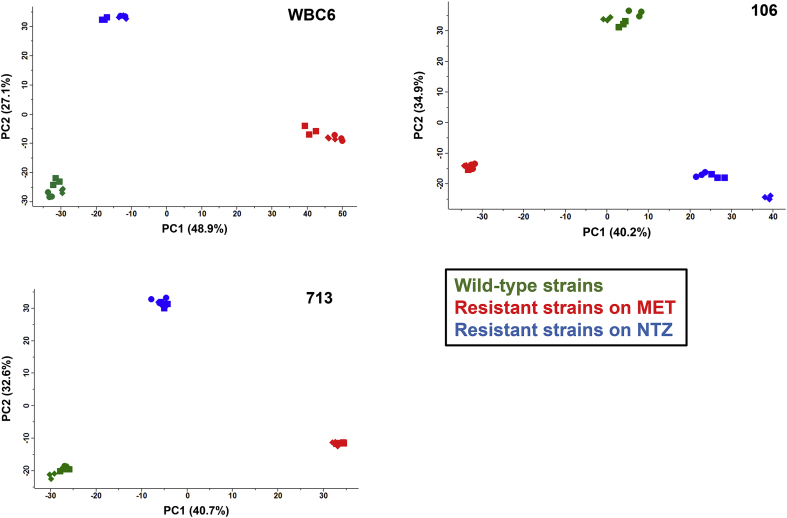
Shotgun mass spectrometry of the proteomes of trophozoites of the nitro drug-resistant strains C4, 1062ID10, and 713M3 grown in the presence of metronidazole (MET) or nitazoxanide (NTZ) and their respective wild-types WBC6, 106, and 713 allowed the identification of 1607, 1403, and 1452 proteins, respectively (Table 1). The complete datasets are accessible in Table S1. Overall analysis of the data by principal component analysis revealed three non-overlapping clusters of wild-types, resistant strains grown on MET and resistant strains grown on NTZ for all three strains tested. This suggested a very small subset of differentially expressed proteins common to both nitro compounds in each strain (Fig. 1).
Table 1.
Summary of protein quantification data. Three strains with double resistance to nitazoxanide (NTZ) and to metronidazole (MET), namely C4, 10621D10, and 713M3 and their respective wild-type strains WBC6, 106 and 713 were grown (the resistant strains in the presence of 50 μM MET or NTZ), harvested and subjected to MS shotgun analysis as described in Materials and methods. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate).
| WBC6 vs. C4 | 106 vs. 10621D10 | 713 vs. 713M3 | |
|---|---|---|---|
| Unique peptides | 21048 | 16567 | 16587 |
| Non-redundant proteins | 1607 | 1403 | 1452 |
| Differential NTZ | 225 | 248 | 304 |
| Differential MET | 510 | 287 | 216 |
Fig. 1.
Principal component analysis of proteome data set from nitro resistantG. lambliatrophozoites (C4, 10621D10, 713M3) and their corresponding wild-types (WBC6, 106, 713). Trophozoites of the resistant strains were grown in the presence of 50 μM metronidazole (MET; red symbols) or 50 μM nitazoxanide (NTZ; blue symbols) and compared to their respective wild-types (green symbols) by MS shotgun analysis as described in Materials and methods. For each strain, all technical and biological (square, circle, diamond) replicates are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Differentially expressed proteins
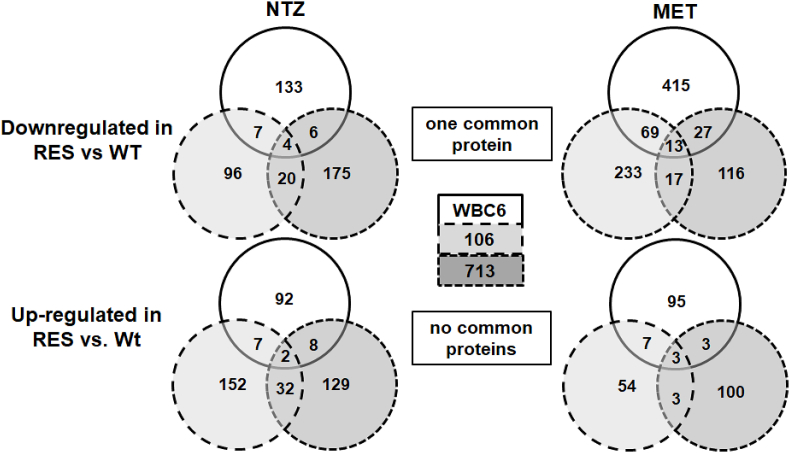
A more detailed analysis revealed between 216 and 510 proteins with different levels in wild-type and resistant strains for each drug separately (Table 1). Concerning the proteins with higher expression levels in wild-types than in resistant strains, only three proteins were commonly identified, however, with resistant strains grown on NTZ and twelve with resistant strains grown on MET. In addition, only one common protein was identified in both situations, namely the hypothetical protein p34701 with a 1985 amino acid sequence, a predicted size of ca. 220 kDa including a signal peptide with a cleavage site around amino acid 30, several transmembrane domains, and a coiled-coil domain around amino acid 1200 (Fig. S1). Concerning the opposite situation, those proteins with higher expression levels in resistant strains than in wild-types, the respective numbers were two and three with no common proteins in both subsets (Fig. 2). A closer look on these four subsets of proteins with altered expression levels in all three strains revealed that of 21 proteins in total, 8 were surface proteins (variant surface proteins, high cysteine membrane and CXC-rich proteins). Moreover, four of the seven hypothetical proteins had transmembrane domains or a putative surface localisation, six proteins had annotated functions related to gene expression, signal transduction or intracellular transport (Table 2).
Fig. 2.
Venn diagram detailing the number of differentially expressed proteins in trophozoites of wild-type (WT) and nitro drug-resistant (RES) strains. Trophozoites of the resistant strains were grown in the presence of metronidazole (MET) or nitazoxanide (NTZ; both 50 μM) and subjected to MS shotgun analysis as described in Materials and methods.
Table 2.
List of proteins with significantly different levels in trophozoites of all wild-type (WT) and all nitro drug resistant (RES) strains. The resistant strains were grown in the presence of 50 μM metronidazole (MET) or nitazoxanide (NTZ) as described in Materials and methods. Vsp, variant surface protein; nd, not detected; su, subunit. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate).
| Differential expression | Nitro compound | Annotation | Giardia DB |
|---|---|---|---|
| Down-regulated in RES vs. WT | |||
| NTZ | |||
| GTP binding ADP ribosylation factor domain-1 protein | 8140 | ||
| Intramembrane protease (minor histocompatibility antigen H13) | 8429 | ||
| Hypothetical (membrane spanning) | 114623 | ||
| MET | |||
| Hypothetical (membrane bound) | 3158 | ||
| nuclear LIM Interacting factor 1 | 4063 | ||
| High cystein membrane protein | 9620 | ||
| Dynein intermediate chain | 10254 | ||
| Hypothetical (RNA binding) | 14117 | ||
| Hypothetical | 16793 | ||
| ATP-dependent RNA helicase | 16887 | ||
| Vsp-3 | 137740 | ||
| Vsp-8 | 137618 | ||
| Vsp-77 | 137617 | ||
| Vsp-88 | 101074 | ||
| Vsp-160 | 137612 | ||
| Both | Hypothetical (transmembrane) | 34701 | |
| Up-regulated in RES vs. WT | |||
| NTZ | |||
| Phosphatase 1 regulatory subunit | 11885 | ||
| Hypothetical (serine rich adhesin) | 94542 | ||
| MET | |||
| Hypothetical (nuclear protein) | 3021 | ||
| High cystein membrane protein group 1 | 16318 | ||
| CXC-rich protein | 17476 | ||
3.3. Antigenic variation related to resistance formation
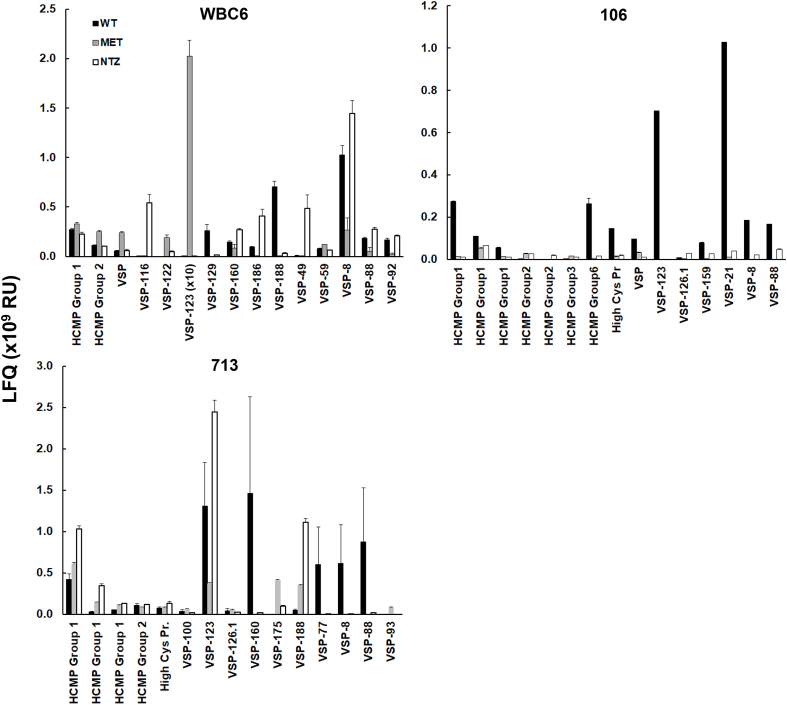
To investigate to which extent antigenic variation was affected in the resistant strains as compared to their respective wild-types, we firstly determined the complexity of surface antigens by counting the expressed high cysteine membrane proteins, CXC-rich proteins, and VSPs and classing them according to their LFQ intensities. Surprisingly, wild-types and the resistant strains expressed similar numbers of different surface antigens. In the case of WBC6, 62 surface antigens were expressed in the wild-type, 78 in C4 grown on NTZ and 62 in C4 grown on MET. For strain 106, the respective numbers were 27, 33, and 26, for strain 713, the numbers were 60, 56, and 63. Only three or less dominated this population by LFQ levels of 109 or above (Table 3). By plotting the intensities of five most predominant antigens for each strain-drug combination (thus 15 for each strain), it became clear that each wild-type strain and each resistant strain had a specific pattern of predominant surface antigens with pronounced nitro drug-dependent differences in the resistant strains (Fig. 3). In the case of WBC6 wild-type, the two most predominant antigens were VSP-8 and VSP-188. In C4 grown in the presence of NTZ, VSP-8 remained the most predominant antigen (besides VSP-49, VSP-116, and VSP-186), in C4 grown in the presence of MET, however, VSP-123 (below detection levels in wild-type and C4 on NTZ) became by far the most predominant antigen. Conversely, VSP-123 was one of the predominant antigens in the wild-type strains 106 and 713 and 713M3 on NTZ (Fig. 3).
Table 3.
Antigenic complexity in nitro drug resistant G. lamblia lines (C4, 10621D10, 713M3; grown on NTZ or MET) is not altered as compared to their corresponding wild-types (WBC6, 106, 713). The strains were grown (the resistant strains in the presence of 50 μM MET or NTZ), harvested and subjected to MS shotgun analysis as described in Materials and methods. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate). Total numbers of proteins either annotated as variant surface proteins, high cysteine surface proteins or CXC rich surface proteins are given. The proteins were classed according to their expression levels determined via the LFQ algorithm. The complete dataset concerning this group of proteins is listed in Supplementary Table S2.
| LFQ (x106) | WBC6 |
106 |
713 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | NTZ | MET | WT | NTZ | MET | WT | NTZ | MET | |
| >104 - 103 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 3 | 0 |
| 103–102 | 11 | 12 | 8 | 1 | 0 | 0 | 6 | 4 | 6 |
| 102–101 | 31 | 40 | 28 | 10 | 17 | 9 | 27 | 25 | 21 |
| 101–100 | 19 | 25 | 25 | 16 | 16 | 17 | 25 | 24 | 26 |
| total | 62 | 78 | 62 | 27 | 33 | 26 | 60 | 56 | 53 |
Fig. 3.
Quantitative assessments of the major surface antigens. Trophozoites of the resistant strains were grown in the presence of metronidazole (MET) or nitazoxanide (NTZ; both 50 μM) and subjected to MS shotgun analysis as described in Materials and methods. For all proteins, mean values ± standard errors for LFQ intensities in three biological replicates are shown.
3.4. Enzymes involved in nitroreduction or detoxification processes
Surprisingly, the dataset of commonly up- or down-regulated proteins presented in Table 2 did not contain enzymes directly or indirectly involved in reduction of nitro compounds or as detoxificators of nitro radicals or O2, as previously described by various authors (Müller et al., 2018). Therefore, it was tempting to have a closer look on the expression levels of those enzymes separately for each strain. In the resistant strain C4 derived from WBC6, the nitroreductase NR1 (annotated as Fd-NR2) was significantly down-regulated as compared to the wild-type when the strain was grown in the presence of NTZ thereby confirming previous results (Nillius et al., 2011). On MET, thioredoxin-reductase was elevated and A-type flavoprotein levels were reduced. In strain 106, these enzymes were not affected. Conversely, levels of both pyruvate-ferredoxin-oxidoreductases were significantly reduced in 1062ID10 grown on MET and PFOR1 only when the strain was grown on NTZ. Strain 713 had again a different pattern. The resistant 713M3 had higher levels of flavo-hemoglobin and on A-type flavoprotein on both nitro compounds and higher levels of thioredoxin reductase only on MET. Both NAD(P)H-oxidases remained unaffected in all strains thus serving as a control (Table 4).
Table 4.
Overview of proteins involved in reduction (and thus activation) of nitro compounds and the scavenging of radicals or other toxic intermediates as a consequence of this reduction in trophozoites of wild-type (WT) and nitro drug resistant (RES) strains, the latter grown in the presence of 50 μM metronidazole (MET) or nitazoxanide (NTZ). Cells were harvested and subjected to MS shotgun analysis as described in Materials and Methods. For each strain, three biological replicates have been tested (with three technical replicates per biological replicate). For all proteins, mean values ± standard errors for LFQ intensities (x106) in three biological replicates are given (nd, below detection limit). DB, number in GiardiaDB; Fd, ferredoxin; Hb, hemoglobin; FP, flavoprotein; LT, lateral transfer; NO, NAD(P)H oxidase; NR, nitroreductase; PFOR, pyruvate-ferredoxin oxidoreductase. *, two-sided t-test comparing the resistant strains to their respective wild-types, p < 0.001.
| Annotation | DB | WBC6 |
106 |
713 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | MET | NTZ | WT | MET | NTZ | WT | MET | NTZ | ||
| Fd-NR1 ("NR2") | 6175 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Fd-NR2 ("NR1") | 22677 | 137 ± 8 | 92 ± 5 | 58 ± 1* | 7 ± 0 | 5 ± 0 | 5 ± 0 | 42 ± 4 | 45 ± 3 | 36 ± 1 |
| NR family | 15307 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| PFOR 1 | 17063 | 4602 ± 73 | 3173 ± 100 | 4561 ± 132 | 713 ± 24 | 240 ± 23* | 292 ± 6* | 4191 ± 199 | 3735 ± 40 | 5146 ± 50 |
| PFOR 2 | 114609 | 7510 ± 193 | 4186 ± 9 | 7846 ± 110 | 987 ± 23 | 501 ± 65* | 812 ± 8 | 3623 ± 154 | 4159 ± 87 | 5071 ± 96 |
| TrxR | 9827 | 393 ± 6 | 621 ± 20* | 395 ± 25 | 122 ± 31 | 163 ± 30 | 115 ± 13 | 533 ± 62 | 1001 ± 12* | 558 ± 111 |
| Flavo-Hb | 15009 | nd | nd | nd | nd | nd | nd | nd | 2 ± 0* | 2 ± 0* |
| A-type FP | 10358 | 927 ± 35 | 606 ± 14* | 1019 ± 35 | 178 ± 6 | 178 ± 22 | 254 ± 5 | 933 ± 21 | 2076 ± 49* | 2070 ± 132* |
| NOLT | 33769 | 3631 ± 121 | 2646 ± 81 | 4463 ± 277 | 577 ± 33 | 514 ± 15 | 641 ± 27 | 2938 ± 94 | 2594 ± 113 | 2827 ± 54 |
| NO | 9719 | 451 ± 22 | 376 ± 17 | 428 ± 14 | 90 ± 4 | 85 ± 5 | 83 ± 1 | 390 ± 3 | 470 ± 6 | 425 ± 3 |
4. Discussion
In previous studies, we characterized the G. lamblia WBC6 clone C4 double-resistant to nitazoxanide (NTZ) and to metronidazole (MET) with respect to differential mRNA expression levels (Müller et al., 2007, 2008) and to physiological parameters (Müller et al., 2018) Meanwhile, other groups have published transcriptomic (Ansell et al., 2017) and proteomic (Emery-Corbin et al., 2018; Emery et al., 2018) studies with MET-susceptible and resistant strains with a different genetic background. In order to answer the question, whether resistance formation to nitro drugs in G. lamblia is directed and therefore has a common pattern of up- or down-regulated proteins on different nitro compounds, we have included two other strains with double resistance, namely 1062ID10 and 713M3, and investigated all resistant strains on both nitro compounds MET and NTZ. Overall, the responses of resistant G. lamblia strains to NTZ and to MET are clearly different from each other. This suggests that – besides a common mode of action due to the nitro groups – both compounds have additional, or different mode(s) of action, e.g. as inhibitors of various enzymes or by forming adducts on different proteins (Hemphill et al., 2013; Leitsch, 2017).
Despite the high number of proteins with different levels in wild-type vs. resistant strains in single strains on single compounds, it turns out that there are no common proteins that are up-regulated in all resistant strains on both drugs. Moreover, only one protein, the hypothetical membrane protein p34701 is down-regulated on both drugs in the resistant strains as compared to their respective wild-types. The corresponding gene is transcribed to similar extents in all strains (Table S3). Since the identification is based on two peptides at low intensities only, these results should, however, not be over-interpreted.
Regarding the drugs separately, only 15 proteins are differentially regulated in the case of MET and only 5 in the case of NTZ, the majority being surface proteins including VSPs. Common VSP patterns are found in the resistant strains upon exposure to MET, but not to NTZ. This observation suggests that double resistant strains may alter their surface protein composition depending on the nitro compound present in their culture media. Furthermore, respective data on strain 106 versus 1062ID10 imply that exposure of resistant trophozoites to both MET and NTZ resulted in an apparent overall reduction of VSP synthesis. A possible explanation is that the proteome analysis depends on G. lamblia WBC6 as genome reference strain. Therefore, the spectrum-to-peptide matches in the non-referenced isolates 106 and 713 may be limited, especially in the case of the highly variable VSPs and may have caused significant losses in VSP identifications as highlighted in a recent study depicting the limitations of differential proteomics in referenced and non-referenced isolates of G. lamblia (Emery-Corbin et al., 2018).
Concerning the reduction (and thus activation) of nitro compounds and the scavenging of radicals or other toxic intermediates as a consequence of this reduction, the three investigated strains seem to have developed three different strategies. i.) The reduction of electrons available for nitro reduction by down-regulation of pyruvate-ferredoxin-oxidoreductase prevails in the case of strain 1062ID10. ii.) The induction of the NO reducer flavohemoglobin (Mastronicola et al., 2010; Rafferty et al., 2010) and the O2-scavenger (and weak NO reducer) A-type flavor- or flavodiiron protein (Di Matteo et al., 2008; Vicente et al., 2009), thus an antioxidant stress response (Arguello-Garcia et al., 2015; Ma'ayeh et al., 2015) is the strategy of strain 713M3. iii.) In WBC6 clone C4, the previously observed down-regulation of the nitroreductase NR1 (Nillius et al., 2011; Müller et al., 2018) is confirmed on NTZ only. According to a hypothesis (Ansell et al., 2017), NR1 possibly could reduce MET by using electrons from the PFOR-ferredoxin electron transport chain. Interestingly, a recent proteomics study (Emery et al., 2018) has identified NR1 as the only enzyme potentially involved in nitroreduction downregulated in all investigated MET-resistant strains upon growth on MET although substantial downregulation was actually documented in resistant 713 strain only. Accordingly, the authors consider downregulation of NR1 as the strongest candidate for a universal passive resistance mechanism. Our present study challenges this hypothesis because here downregulation of NR1 is observed only in one nitro drug-resistant clone, namely WBC6 clone C4, and only in trophozoites grown in presence of NTZ (see Table 4). Accordingly, our data suggest that, at least as far as WBC6 clone C4 is concerned, downregulation of NR1 could be correlated to resistance formation towards NTZ but not MET. Moreover, functional studies on the recombinant enzyme revealed that NR1 is a better quinone-reductase than a nitroreductase (Müller et al., 2015). Therefore, it may be only indirectly involved in the susceptibility to nitro drugs.
On MET, levels of thioredoxin reductase are increased, a feature that this strain shares with 713M3. Thus, in these strains, thioredoxin reductase acts rather as a potential radical scavenger (Ma'ayeh et al., 2015) than as an activator of MET (Leitsch et al., 2016). This suggests that resistant strains generated by adaptation to increasing amounts of the respective drugs are different from transgenic strains expressing a specific resistance marker.
However, it should be kept in mind, that the levels of enzyme proteins may not be directly linked to the respective levels of enzyme activities. For instance, lower activities may be correlated with lower levels of essential coenzymes such as FAD, as shown for clone C4 (Müller et al., 2018) and for the resistant 106 and 713 isolates (Leitsch et al., 2011), backed by own unpublished data. Therefore, enyzmologic and metabolomics studies are complementary to genomic, transcriptomic and proteomic approaches and cannot replaced by them.
In none of the strains, peptides corresponding to the nitroreductase NR2 (annotated as Fd-NR1) have been identified. This is insofar interesting as this enzyme catalyzes the complete reduction and thus inactivation of nitro compounds in functional assays and in E. coli (Müller et al., 2013, 2015) and would therefore be a suitable candidate for up-regulation in resistant strains. Since the corresponding gene is transcribed in all strains (Müller et al., 2013), either the mRNA is subjected to post-transcriptional gene silencing, most likely by RNA interference (Prucca and Lujan, 2009; Gargantini et al., 2012) or the corresponding polypeptide is quickly degraded. We have performed immunoblots with specific antisera raised against two unique peptides of NR2 and could not detect a corresponding signal in G. lamblia trophozoite crude extracts (see Fig. S2). Similarly, a third nitroreductase homologue, the NR family protein (without N-terminal ferredoxin domain), is transcribed, but obviously not translated or quickly degraded. The recombinant protein has no nitroreductase activities, neither in functional assays, nor in E. coli (J.M and N. M., unpublished data).
This leads to the conclusion that drug resistance formation in G. lamblia is not correlated with directed changes of gene expression in the sense that targets are down- or drug scavengers are up-regulated, but rather correlated with random variation of gene expression. As reported in the context of other studies related to resistance formation (Emery et al., 2018) interaction with host cells (Emery-Corbin et al., 2018) or en- and excystation (Einarsson et al., 2016), antigenic variation, i.e. the expression of different cysteine-rich variant surface proteins (VSPs) on the surface is paramount and yields a most heterogeneous population of trophozoites. G. lamblia has several hundred genes encoding VSPs. According to a generally admitted hypothesis, there is, however, only one (major) VSP expressed on a single trophozoite (Nash, 2002). The expression of different VSPs – and thus antigenic variation - is triggered by epigenetic mechanisms involving changes of the chromatin state (Kulakova et al., 2006) and/or RNA interference (Prucca et al., 2008; Prucca and Lujan, 2009). Since there is post-transcriptional silencing of the non-expressed VSPs (Prucca et al., 2008), only proteomics can answer the question how heterogeneous a given population of trophozoites is. In our case, since the number of VSPs remains almost the same in resistant and susceptible strains (see Table 3), there is neither a broadening nor a narrowing of heterogeneity, there is only a switch to a different pattern of VSPs between those populations (as illustrated in Fig. 3). It could be that some of these VSPs have unknown enzyme activities or other functions facilitating or impairing transport of selected metabolites etc., but in general there are no such functions met in evidence - except protease activities (Cabrera-Licona et al., 2017) - to our knowledge. This direct association between nitro drug resistance formation and the immuno-evasive process of antigenic switching (Ankarklev et al., 2010; Gargantini et al., 2016) may contribute to both the establishment and persistence of resistant giardiasis in an infected host.
During resistance formation by incubation on increasing drug concentrations, these mechanisms may cause not only different VSP expression patterns, but also involve other, unrelated genes (Rivero et al., 2010) thereby generating trophozoites with gene expression patterns conferring resistance. These trophozoites are selected and enriched in subsequent cultures. The resulting resistance phenotype is multigenic and reversible. Therefore, “nitro drug resistance” is rather a “nitro drug tolerance” when compared to concepts generated from antibiotic resistance in bacteria (Brauner et al., 2016), as described in detail in a previous study (Müller et al., 2018). This observation is similar to previously reported findings where differential expression patterns of selected genes have been identified in transgenic Giardia lines in response to transfection and puromycin selection (Su et al., 2007) We hypothesize that in an untargeted transcriptomic or proteomic study on puromycin selection, antigenic variation would have been identified, as well. It should, however, be kept in mind that this study is based on resistance formation under laboratory conditions in assemblage A strains, but cannot be yet extended to naturally resistant isolates from other assemblages.
Funding
This work was supported by the Swiss National Science Foundation [grant No. 31003A_163230].
Transparency declaration
None of the authors has any competing interests in the manuscript.
Acknowledgements
We thank Dominic Rittler for his contribution in designing the scheme shown as graphical abstract.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.03.002.
Contributor Information
Joachim Müller, Email: joachim.mueller@vetsuisse.unibe.ch.
Sophie Braga, Email: sophie.braga@dbmr.unibe.ch.
Manfred Heller, Email: manfred.heller@dbmr.unibe.ch.
Norbert Müller, Email: norbert.mueller@vetsuisse.unibe.ch.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ankarklev J., Jerlström-Hultqvist J., Ringqvist E., Troell K., Svärd S.G. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 2010;8:413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- Ansell B.R., Baker L., Emery S.J., McConville M.J., Svard S.G., Gasser R.B., Jex A.R. Transcriptomics indicates active and passive metronidazole resistance mechanisms in three seminal Giardia lines. Front. Microbiol. 2017;8:398. doi: 10.3389/fmicb.2017.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello-Garcia R., Cruz-Soto M., Gonzalez-Trejo R., Paz-Maldonado L.M., Bazan-Tejeda M.L., Mendoza-Hernandez G., Ortega-Pierres G. An antioxidant response is involved in resistance of Giardia duodenalis to albendazole. Front. Microbiol. 2015;6:286. doi: 10.3389/fmicb.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- Brown D.M., Upcroft J.A., Edwards M.R., Upcroft P. Anaerobic bacterial metabolism in the ancient eukaryote Giardia duodenalis. Int. J. Parasitol. 1998;28:149–164. doi: 10.1016/s0020-7519(97)00172-0. [DOI] [PubMed] [Google Scholar]
- Cabrera-Licona A., Solano-Gonzalez E., Fonseca-Linan R., Bazan-Tejeda M.L., Raul A.-G., Bermudez-Cruz R.M., Ortega-Pierres G. Expression and secretion of the Giardia duodenalis variant surface protein 9B10A by transfected trophozoites causes damage to epithelial cell monolayers mediated by protease activity. Exp. Parasitol. 2017;179:49–64. doi: 10.1016/j.exppara.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Cernikova L., Faso C., Hehl A.B. Five facts about Giardia lamblia. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.G., Diamond L.S. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 2002;15:329–341. doi: 10.1128/CMR.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M., Wang A.L., Wang C.C. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol. Microbiol. 2000;36:447–456. doi: 10.1046/j.1365-2958.2000.01863.x. [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Scandurra F.M., Testa F., Forte E., Sarti P., Brunori M., Giuffrè A. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 2008;283:4061–4068. doi: 10.1074/jbc.M705605200. [DOI] [PubMed] [Google Scholar]
- Einarsson E., Troell K., Hoeppner M.P., Grabherr M., Ribacke U., Svard S.G. Coordinated changes in gene expression throughout Encystation of Giardia intestinalis. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery-Corbin S.J., Vuong D., Lacey E., Svard S.G., Ansell B.R.E., Jex A.R. Proteomic diversity in a prevalent human-infective Giardia duodenalis sub-species. Int. J. Parasitol. 2018;48:817–823. doi: 10.1016/j.ijpara.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Emery S.J., Baker L., Ansell B.R.E., Mirzaei M., Haynes P.A., McConville M.J., Svard S.G., Jex A.R. Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. GigaScience. 2018;7 doi: 10.1093/gigascience/giy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery S.J., Lacey E., Haynes P.A. Quantitative proteomic analysis of Giardia duodenalis assemblage A: a baseline for host, assemblage, and isolate variation. Proteomics. 2015;15:2281–2285. doi: 10.1002/pmic.201400434. [DOI] [PubMed] [Google Scholar]
- Engel H., Mika M., Denapaite D., Hakenbeck R., Muhlemann K., Heller M., Hathaway L.J., Hilty M. A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2014;58:3934–3941. doi: 10.1128/AAC.02547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargantini P.R., Serradell M.C., Torri A., Lujan H.D. Putative SF2 helicases of the early-branching eukaryote Giardia lamblia are involved in antigenic variation and parasite differentiation into cysts. BMC Microbiol. 2012;12:284. doi: 10.1186/1471-2180-12-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargantini P.R., Serradell M.D.C., Rios D.N., Tenaglia A.H., Lujan H.D. Antigenic variation in the intestinal parasite Giardia lamblia. Curr. Opin. Microbiol. 2016;32:52–58. doi: 10.1016/j.mib.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Hemphill A., Müller N., Müller J. Thiazolides, a novel class of anti-infective drugs, effective against viruses, bacteria, intracellular and extracellular protozoan parasites and proliferating mammalian cells. Antiinfective Agents. 2013;11:22–30. [Google Scholar]
- Huang D.B., White A.C. An updated review on Cryptosporidium and Giardia. Gastroenterol. Clin. N. Am. 2006;35:291–314. doi: 10.1016/j.gtc.2006.03.006. (viii) [DOI] [PubMed] [Google Scholar]
- Kulakova L., Singer S.M., Conrad J., Nash T.E. Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Mol. Microbiol. 2006;61:1533–1542. doi: 10.1111/j.1365-2958.2006.05345.x. [DOI] [PubMed] [Google Scholar]
- Leitsch D. Drug resistance in the microaerophilic parasite Giardia lamblia. Curr. Trop. Med. Rep. 2015;2:128–135. doi: 10.1007/s40475-015-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D. A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology. 2017:1–12. doi: 10.1017/S0031182017002025. [DOI] [PubMed] [Google Scholar]
- Leitsch D., Burgess A.G., Dunn L.A., Krauer K.G., Tan K., Duchêne M., Upcroft P., Eckmann L., Upcroft J.A. Pyruvate:ferredoxin oxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia. J. Antimicrob. Chemother. 2011;66:1756–1765. doi: 10.1093/jac/dkr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D., Müller J., Müller N. Evaluation of Giardia lamblia thioredoxin reductase as drug activating enzyme and as drug target. Int. J. Parasitol. Drugs Drug Resist. 2016;6:148–153. doi: 10.1016/j.ijpddr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D., Harris J.C., Maroulis S., Mitchell A., Hughes M.N., Wadley R.B., Edwards M.R. Nitrosative stress induced cytotoxicity in Giardia intestinalis. J. Appl. Microbiol. 2003;95:576–583. doi: 10.1046/j.1365-2672.2003.02008.x. [DOI] [PubMed] [Google Scholar]
- Ma'ayeh S.Y., Knorr L., Svard S.G. Transcriptional profiling of Giardia intestinalis in response to oxidative stress. Int. J. Parasitol. 2015;45:925–938. doi: 10.1016/j.ijpara.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Mastronicola D., Testa F., Forte E., Bordi E., Pucillo L.P., Sarti P., Giuffrè A. Flavohemoglobin and nitric oxide detoxification in the human protozoan parasite Giardia intestinalis. Biochem. Biophys. Res. Commun. 2010;399:654–658. doi: 10.1016/j.bbrc.2010.07.137. [DOI] [PubMed] [Google Scholar]
- Minenoa T., Avery M.A. Giardiasis: recent progress in chemotherapy and drug development. Curr. Pharmaceut. Des. 2003;9:841–855. doi: 10.2174/1381612033455260. [DOI] [PubMed] [Google Scholar]
- Müller J., Hemphill A. New approaches for the identification of drug targets in protozoan parasites. Int. Rev. Cell Mol. Biol. 2013;301:359–401. doi: 10.1016/B978-0-12-407704-1.00007-5. [DOI] [PubMed] [Google Scholar]
- Müller J., Hemphill A., Muller N. Physiological aspects of nitro drug resistance in Giardia lamblia. Int. J. Parasitol. Drugs Drug Resist. 2018;8:271–277. doi: 10.1016/j.ijpddr.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Ley S., Felger I., Hemphill A., Müller N. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J. Antimicrob. Chemother. 2008;62:72–82. doi: 10.1093/jac/dkn142. [DOI] [PubMed] [Google Scholar]
- Müller J., Rout S., Leitsch D., Vaithilingam J., Hehl A., Müller N. Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. Int J Parasitol Drugs Drug Resist. 2015;5:37–43. doi: 10.1016/j.ijpddr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Rühle G., Müller N., Rossignol J.F., Hemphill A. In vitro effects of thiazolides on Giardia lamblia WB clone C6 cultured axenically and in coculture with Caco2 cells. Antimicrob. Agents Chemother. 2006;50:162–170. doi: 10.1128/AAC.50.1.162-170.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Schildknecht P., Müller N. Metabolism of nitro drugs metronidazole and nitazoxanide in Giardia lamblia: characterization of a novel nitroreductase (GlNR2) J. Antimicrob. Chemother. 2013;68:1781–1789. doi: 10.1093/jac/dkt106. [DOI] [PubMed] [Google Scholar]
- Müller J., Sterk M., Hemphill A., Müller N. Characterization of Giardia lamblia WB C6 clones resistant to nitazoxanide and to metronidazole. J. Antimicrob. Chemother. 2007;60:280–287. doi: 10.1093/jac/dkm205. [DOI] [PubMed] [Google Scholar]
- Müller N., Müller J. Giardia. In: Walochnik J., Duchêne M., editors. Molecular Parasitology. Springer-Verlag; Vienna: 2016. pp. 93–114. [Google Scholar]
- Nash T.E. Treatment of Giardia lamblia infections. Pediatr. Infect. Dis. J. 2001;20:193–195. doi: 10.1097/00006454-200102000-00015. [DOI] [PubMed] [Google Scholar]
- Nash T.E. Surface antigenic variation in Giardia lamblia. Mol. Microbiol. 2002;45:585–590. doi: 10.1046/j.1365-2958.2002.03029.x. [DOI] [PubMed] [Google Scholar]
- Nillius D., Müller J., Müller N. Nitroreductase (GlNR1) increases susceptibility of Giardia lamblia and Escherichia coli to nitro drugs. J. Antimicrob. Chemother. 2011;66:1029–1035. doi: 10.1093/jac/dkr029. [DOI] [PubMed] [Google Scholar]
- Plutzer J., Ongerth J., Karanis P. Giardia taxonomy, phylogeny and epidemiology: facts and open questions. Int. J. Hyg Environ. Health. 2010;213:321–333. doi: 10.1016/j.ijheh.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Prucca C.G., Lujan H.D. Antigenic variation in Giardia lamblia. Cell Microbiol. 2009;11:1706–1715. doi: 10.1111/j.1462-5822.2009.01367.x. [DOI] [PubMed] [Google Scholar]
- Prucca C.G., Slavin I., Quiroga R., Elías E.V., Rivero F.D., Saura A., Carranza P.G., Luján H.D. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- Rafferty S., Luu B., March R.E., Yee J. Giardia lamblia encodes a functional flavohemoglobin. Biochem. Biophys. Res. Commun. 2010;399:347–351. doi: 10.1016/j.bbrc.2010.07.073. [DOI] [PubMed] [Google Scholar]
- Rivero M.R., Kulakova L., Touz M.C. Long double-stranded RNA produces specific gene downregulation in Giardia lamblia. J. Parasitol. 2010;96:815–819. doi: 10.1645/GE-2406.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire S.A., Ryan U. Cryptosporidium and Giardia in Africa: current and future challenges. Parasites Vectors. 2017;10:195. doi: 10.1186/s13071-017-2111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.H., Lee G.A., Huang Y.C., Chen Y.H., Sun C.H. Neomycin and puromycin affect gene expression in Giardia lamblia stable transfection. Mol. Biochem. Parasitol. 2007;156:124–135. doi: 10.1016/j.molbiopara.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Townson S.M., Laqua H., Upcroft P., Boreham P.F., Upcroft J.A. Induction of metronidazole and furazolidone resistance in Giardia. Trans. R. Soc. Trop. Med. Hyg. 1992;86:521–522. doi: 10.1016/0035-9203(92)90095-t. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Upcroft J.A., Campbell R.W., Benakli K., Upcroft P., Vanelle P. Efficacy of new 5-nitroimidazoles against metronidazole-susceptible and -resistant Giardia, Trichomonas, and Entamoeba spp. Antimicrob. Agents Chemother. 1999;43:73–76. doi: 10.1128/aac.43.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upcroft J.A., Upcroft P. Drug resistance and Giardia. Parasitol. Today. 1993;9:187–190. doi: 10.1016/0169-4758(93)90144-5. [DOI] [PubMed] [Google Scholar]
- Upcroft J.A., Upcroft P., Boreham P.F. Drug resistance in Giardia intestinalis. Int. J. Parasitol. 1990;20:489–496. doi: 10.1016/0020-7519(90)90196-t. [DOI] [PubMed] [Google Scholar]
- Upcroft P. Drug resistance in Giardia: clinical versus laboratory isolates. Drug Resist. Updates. 1998;1:166–168. doi: 10.1016/s1368-7646(98)80035-6. [DOI] [PubMed] [Google Scholar]
- Vicente J.B., Testa F., Mastronicola D., Forte E., Sarti P., Teixeira M., Giuffrè A. Redox properties of the oxygen-detoxifying flavodiiron protein from the human parasite Giardia intestinalis. Arch. Biochem. Biophys. 2009;488:9–13. doi: 10.1016/j.abb.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Zylberberg H.M., Green P.H., Turner K.O., Genta R.M., Lebwohl B. Prevalence and predictors of Giardia in the United States. Dig. Dis. Sci. 2017;62:432–440. doi: 10.1007/s10620-016-4447-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.