Abstract
PURPOSE: Accumulation of PIK3CA, ESR1, and GATA3 mutations results in resistance to endocrine therapy in breast cancer patients; however, the response of these genes to chemotherapy is unclear. Therefore, we sought to evaluate the genetic response of circulating tumor DNA (ctDNA) to chemotherapy in metastatic breast cancer patients. METHODS: The mutation frequency of 1021 genes was examined prior to chemotherapy in ctDNA of 44 estrogen receptor–positive metastatic breast cancer patients. These genes were evaluated again in a subset of patients (n=24) following chemotherapy. Mutation frequency was defined as the percentage of mutations found in ctDNA compared to total cell-free DNA. RESULTS: Prior to chemotherapy, PIK3CA was the most commonly mutated gene, with mutation found in 22 of the metastatic breast cancer patients. Following chemotherapy, 16 patients exhibited progressive disease (PD), and 8 patients experienced no progression (non-PD). PIK3CA mutation frequency increased in 56.25% (9/16) of the PD patients but decreased in 62.5% (5/8) of the non-PD patients. As a result, more PD patients exhibited increased PIK3CA mutation frequency than non-PD patients (56.25% vs 0%, P=.002). Further, ESR1 and GATA3 mutations correlated with PIK3CA mutation. Interestingly, patients receiving the mTOR inhibitor everolimus exhibited a lower progression rate (0% vs 62.5%, P=.001), and the combination of everolimus and chemotherapy effectively suppressed PIK3CA, ESR1, and GATA3 gene mutations. CONCLUSION: Together, these results suggest that mTOR inhibition may be a useful chemotherapy adjuvant to suppress chemotherapy-induced gene mutations that render tumors resistant to endocrine therapy in metastatic breast cancer patients with PD.
Introduction
Although the 5-year mortality rate of breast cancer has dropped by 34% since 1990, it remains the leading cause of tumor-related death among women [1]. The majority of breast cancer patients benefit from the initial therapy; however, they may eventually develop more aggressive tumor forms, such as metastasized recurrence, that are generally resistant to the treatment [2], [3]. In breast cancers, metastasis and drug resistance are often accompanied by genomic instability, alteration of tumor gene subclone changings, and microenvironmental selective pressure [4]. The constantly varied variation in gene mutations leads to tumor heterogeneity, limiting the effectiveness of targeted therapy.
In breast cancer patients, about 70% of the tumors express estrogen receptor (ER) and are treated with endocrine therapy [5]. Endocrine therapies include nonsteroidal aromatase inhibitors (anastrozole and letrozole), steroidal aromatase inhibitors (exemestane), serum ER modulators (tamoxifen, or toremifene), ER downregulators (fulvestrant), etc. However, after 1-5 years of treatment, almost all advanced breast cancer patients eventually become resistant to endocrine therapy [6]. For example, ESR1 mutation plays a key role in the resistance to aromatase inhibitors. Prior to endocrine therapy, ESR1 mutations are rare (<1%) [7], but in advanced patients with previous aromatase inhibitors (AIs) treatment, ESR1 mutations occur more frequently (22%) [8]. Moreover, some studies have reported that ESR1 mutation is an independent predictor of poor prognosis for progression-free survival (PFS) and overall survival [9], [10], [11].
GATA3 is another gene which is expressed differentially in ESR1-positive and -negative breast cancers [12]. GATA3 is essential for hormone-driven cancers, and low GATA3 expression is a prognostic indicator of aggressive disease and poor survival [13]. GATA3 mutation occurs approximately 10% for the patients with breast cancer [7]. GATA3 mutation and consequential abnormal expression result in ESR1 ligand activation, leading to endocrine therapy resistance [14].
In addition to the mutation of ESR1 and GATA3, activation of the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR signaling pathway also facilitates endocrine therapy resistance in breast cancers [15]. PI3K-AKT-mTOR signaling is one of the most active pathways in breast cancer, and this pathway plays an important role in cell growth, proliferation, survival, and metabolism [16]. Mutations in genes associated with the PI3K-mTOR pathway are common in ER-positive breast cancers. In particular, PIK3CA mutation occurs in approximately 20%-30% of breast cancers [17], [18]. Mutations in the PI3K-mTOR pathway can lead to tumor resistance to multiple antitumor agents, including paclitaxel, tamoxifen, trastuzumab, etc. [19]. In addition, the PI3K and ER pathways often play a synergistic role in the tumor progression [20], [21].
Derived from cell-free DNA (cfDNA) testing, circulating tumor DNA (ctDNA) analysis is a powerful surveillance tool for effective and continuous detection of potential drug-resistant gene mutations [22], [23], [24], [25]. Compared with imaging and serum biomarkers, ctDNA testing provides valuable and sensitive information about gene mutations in tumors after the drug-based therapies. For example, in ER-positive breast cancer patients, mutations in PI3K/AKT pathway genes and ESR1 were detected in 15.1% and 2.7% of patients, respectively, and these mutations predicted treatment failure [26].
In this study, 44 ER-positive metastatic breast cancer (MBC) patients were recruited, and their genetic response to chemotherapy was detected using ctDNA testing. The accumulation of PIK3CA, PIK3R2, TP53, NOTCH2, ERBB2/3, ESR1, and GATA3 gene mutations existed after chemotherapy in resistant patients. Among these genes, accumulation of PIK3/AKT, ESR1 and GATA3 mutations may significantly increase the risk of endocrine therapy resistance. Therefore, our findings suggest that drug resistance to endocrine therapy might emerge after chemotherapy in the progressed ER-positive MBC patients via accumulation of the mutations in the specific genes.
Materials and Methods
Patient Cohort and Clinical Data Collection
This study was approved by the Ethics Committee in Hunan Cancer Hospital. A total of 44 ER-positive MBC patients, who were treated from January 2016 to March 2018, were enrolled in this study. Informed consent was obtained from each patient prior to the study onset. All the recruited patients were diagnosed with ER-positive stage IV primary breast malignant tumor or MBC. Patients were aged between 18 and 70 years old, and the heart, liver, and renal functions of the patients were determined to be adequate enough to tolerate chemotherapy. Basic demographic and clinical information, including age, pathology, laterality, stage, metastatic sites, HR/HER2 status, imaging records, and treatment history, were collected from the patients at the beginning of the study.
Immunohistochemistry (IHC) Classification
According to the American Society of Clinical Oncology/College of American Pathologists guidelines, ER- and progesterone receptor (PR)–positive tumors were defined as having a minimum of 1% of invasive tumor cells that stained positive for ER and PR. HER2-positive status was defined as “HER2 IHC 3+” or with HER2 copy number or HER2:CEP17 amplification by fluorescent in situ hybridization. ER-positive breast cancer patients were divided into ER-positive/HER2-negative and ER-positive / HER2-positive subtypes.
Blood Sample Collection and DNA Extraction
Peripheral blood samples were collected 7 days before the treatment and at the time of the chemotherapy completion (6 months after the initiation of the treatment). Peripheral blood samples were collected in Streck tubes (Streck, Omaha, NE) and centrifuged within 72 hours to separate the plasma from peripheral blood cells. The cfDNA was extracted from plasma based on a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Genomic DNA (gDNA) was extracted from peripheral blood cells based on a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Both DNA extractions were performed according to the manufacturer’s instructions. The gDNA was sequenced as the control sample.
Target Capture and Next-Generation Sequencing
Both cfDNA and gDNA libraries were constructed with the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA) according to the manufacturer’s protocol. Capture probes were designed to cover the coding sequences and the hot exons of 1021 genes that are frequently mutated in the solid tumors. A detailed description of the capture experiments has been reported previously [27]. Libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Integrated DNA Technologies, Coralville, IA). DNA sequencing was performed using the HiSeq 3000 Sequencing System (Illumina, San Diego, CA) with 2×101-bp paired-end reads. Clonal hematopoietic mutations, including those in DNMT3A, IDH1, and IDH2, and specific alterations within ATM, GNAS, and JAK2, were filtered as previously described [28]. Passenger mutations were not filtered, as these alterations are somatic.
Sequencing Data Analysis
Terminal adaptor sequences and low-quality reads were removed from the raw data. BWA (version 0.7.12-r1039) was used to align clean reads to the reference human genome (hg19), and Picard (version 1.98) was used to mark PCR duplicates. Realignment and recalibration were performed using GATK (version 3.4-46-gbc02625). Single nucleotide variants were identified using MuTect (version 1.1.4) and NChot, a software developed in-house to review hotspot variants [27]. Small insertions and deletions (indels) were also identified using GATK. Somatic copy number alterations were identified with CONTRA (v2.0.8). Significant copy number variation was expressed as the ratio of adjusted depths between ctDNA and control gDNA. The final candidate variants were all manually verified using the Integrative Genomics Viewer. This sequencing method was found to be credible with simulated cfDNA in a previous report [27]. Therefore, we did not validate the mutations found in ctDNA by sequencing tumor biopsies.
ctDNA Gene Mutation Frequency
Total cfDNA included ctDNA and other normal cfDNA. Mutations in ctDNA were identified by comparison to the reference genome (hg18) and gDNA. The ctDNA mutation frequency was defined as the proportion of ctDNA gene mutations in the total cfDNA. For example, PIK3CA mutation frequency was 46.6%, indicating 46.6% cfDNA clones contained PIK3CA ctDNA mutation.
Image Evaluation and Definition of Drug Resistance
MRI/CT image evaluation was performed every two to three treatment cycles according to RECIST 1.1 standards. In targeted therapy-based treatment trials of MBC patients, PFS closely correlates with overall survival [29], [30]. Therefore, in this study, PFS was used to evaluate the drug treatment response. Drug resistance was defined as disease progression within 6 months of treatment.
Statistical Data Analyses
Continuous variables were summarized as the mean (standard deviation) and median (interquartile range). Categorical variables were reported as counts (percentage). An analysis of variance (ANOVA) was used to compare the continuous variables with symmetrical distributions across subgroups. Chi-square tests or Fisher’s exact tests (n<5) were used to compare differences among subgroups. Kaplan-Meier curves were used to estimate survival distributions against progression, and the log-rank test was used to assess differences in PFS among subgroups. Fisher’s exact tests (n<5) were used to compare the gene mutation trends between progressive disease (PD) and non-PD groups after treatment. Due to the small sample size, Fisher’s exact tests (n<5) were performed to evaluate the effect of the mTOR inhibitor. All statistical tests were two-tailed and conducted at a significance level of .05. Statistical analyses were conducted using SAS 9.4 (Cary, NC).
Results
Demographic and Clinical Features of Patients
A total of 44 ER-positive MBC patients were included in this study. The ER/HER2 subtypes, demographic characteristics, and clinical features of the patients are summarized in Table S1. All patients were female. The average age at the first diagnosis of breast cancer was 44.1 years old. Most patients (95.45%) underwent primary tumor surgery, and half of the total patients received radiotherapy. According to the biopsy IHC results, 31 patients had ER-positive/HER2-negative breast cancer, and 13 patients had ER-positive /HER2-positive breast cancer. Patients progressed to MBC in an average of 4 years and provided samples for the follow-up ctDNA test analysis (Table S1).
At the time of the recruitment, there were no significant differences in lymph nodes, distant metastases, or treatment history observed among patients within each ER/HER2 subtype. Anti-HER2 target therapy was only performed in ER-positive/HER2-positive patients. After MBC was diagnosed, the majority of patients had received one to three rounds (lines) of chemotherapy, endocrine therapy, or targeted therapy. Unfortunately, the response of these patients to treatment was not satisfactory, resulting in continued tumor growth continuation and an unclear treatment plan.
Baseline Mutation Profiling of ER-Positive MBC Patients
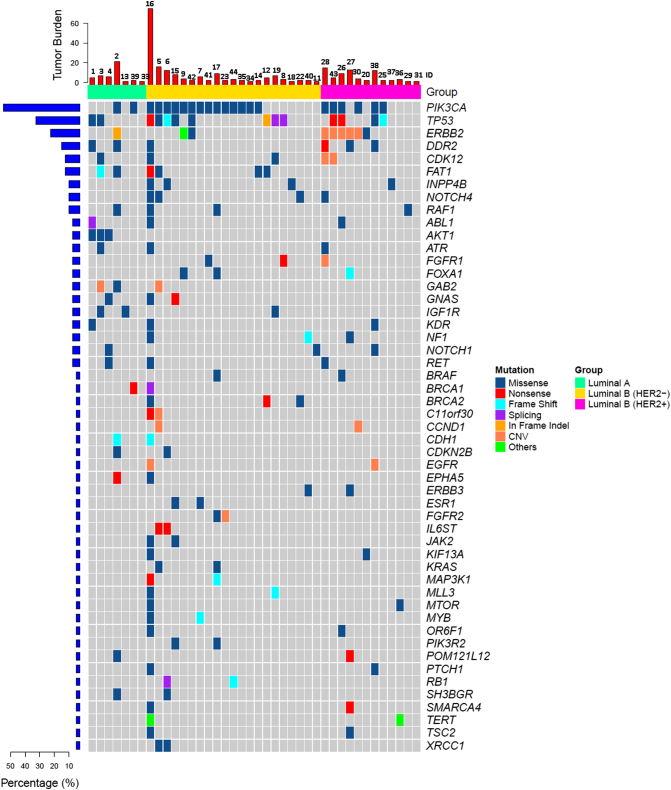
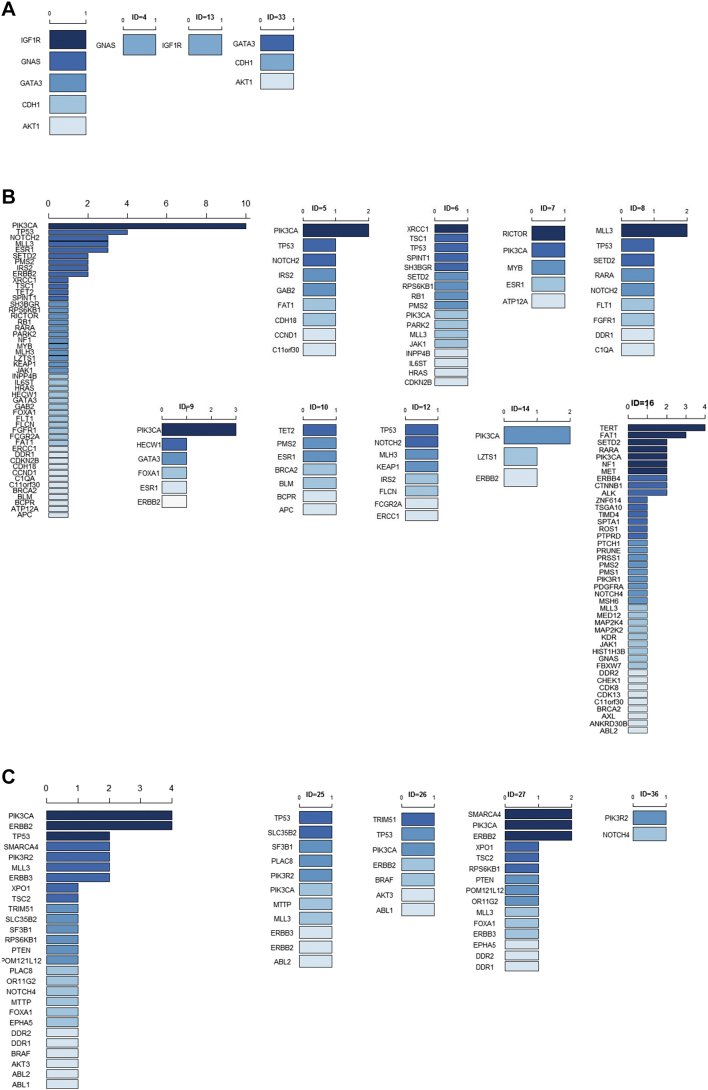
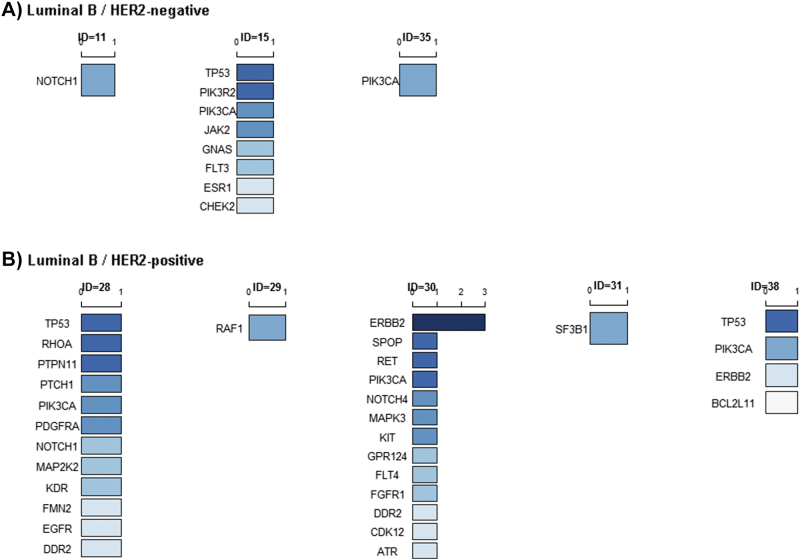
ctDNA testing was performed prior to beginning a new treatment regimen. Among the 44 recruited patients, 28/31 (90.32%) ER-positive/HER2-negative patients and 12/13 (92.31%) ER-positive /HER2-positive patients had tumor gene mutations (Figure 1). PIK3CA was the most frequently mutated gene in both HER2-negative and HER2-positive MBC patients (Table S2). TP53 mutation was also common in ER-positive/HER2-negative and ER-positive/HER2-positive patients. More specifically, 3/31 ER-positive/HER2-negative patient had an ERBB2 mutation (Figure 2A), with one frame shift (p.Y772_A775dup, ID=2), one missense mutation (p.S310F, ID=42), and one in-frame indel (p.G776delins, ID=9). ERBB2 mutations were present in 6/13 (46.15%) ER-positive/HER2-positive patients, with 5 ERBB2 amplifications and 1 missense mutation (p.S280F, ID=20, Figure 2B). In the ER-positive /HER2-positive subtype, one patient had both an ERBB2 amplification and a missense mutation (p.E844K, ID=27), and one patient had an ERBB2 amplification and two other missense mutations (p.F279I and p.V670G, ID=3, Figure 2B). All AKT1 mutations were concentrated in the ER-positive/HER-positive subtype (4/31, 12.90%, Figure 2A).
Figure 1.
Circulating tumor DNA (ctDNA) gene mutation profiles in 44 ER-positive MBC patients. Each column (labeled with ID number) represents an individual patient.
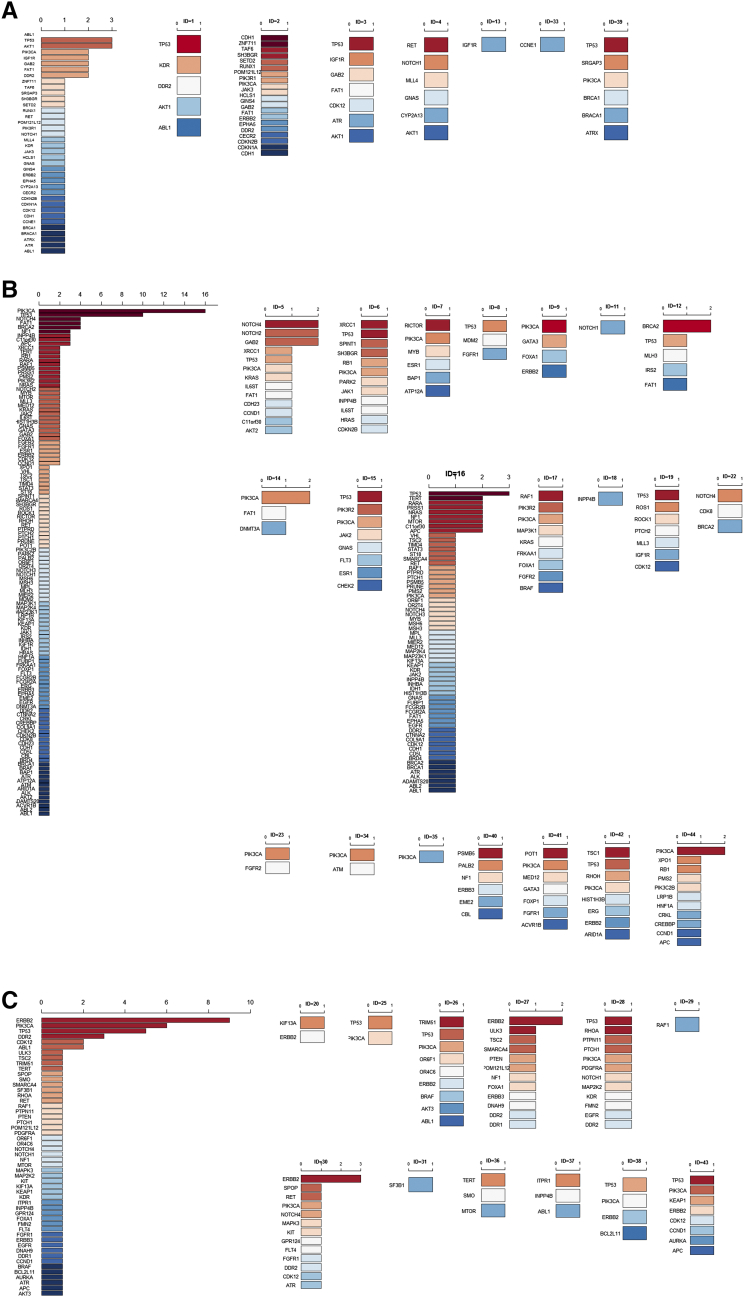
Figure 2.
Baseline circulating tumor DNA (ctDNA) gene mutations in ER-positive/HER2-negative and ER-positive/HER2-positive MBC patients. Dark red represents the most common mutated genes, and dark blue represents the rarest mutations. If the mutated genes appeared at the same frequency, they are ranked in alphabetic order.
(A) Rank of the baseline ctDNA gene mutations in all ER-positive/HER2-negative patients (left) and in seven individual patients (ID=1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, 22, 23, 33, 34, 35, 39,40, 41, 42, 44).
(B) Rank of the baseline ctDNA gene mutations in all ER-positive/HER2-positive patients (left) and in 12 individual patients (ID=20, 25, 26, 27, 28, 29, 30, 31, 36, 37, 38, 43).
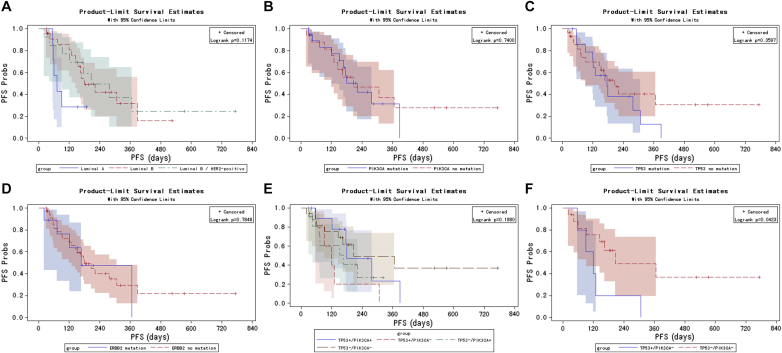
In this study, PFS was used to evaluate the response to drug treatment. For both ER-positive/HER2-negative and ER-positive/HER2-positive subtypes, TP53, PIK3CA, or ERBB2 mutation was not significantly associated with PFS (Figure S1). However, the small sample size may have contribution to this finding. The TP53+/PIK3CA− subgroup (TP53 mutation and PIK3CA wild-type) showed a significantly poorer PFS compared to the TP53−/PIK3CA− subgroup (wild-type TP53 and PIK3CA). These findings suggest that gene mutations may be potential risk factors for poor prognosis.
Figure S1.
Kaplan-Meier curves for PFS probabilities stratified by ER/HER2 subtypes (A) and by baseline ctDNA mutations in TP53 (B), PIK3CA (C), ERBB2 (D), and TP53/PIK3CA (E). Kaplan-Meier curves for PFS stratified based on the presence and absence of TP53 mutations in patients without PIK3CA mutations (TP53+/PIK3CA− and TP53−/PIK3CA−, respectively; F).
PIK3CA Mutation Frequency Increases in Patients with PD and Decreases in Drug-Sensitive Patients
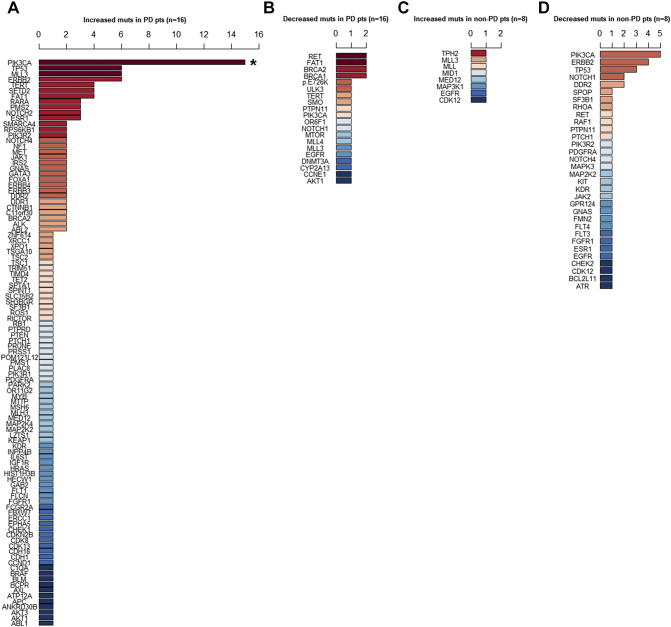
To further investigate the relationship between ctDNA mutation and drug response, we compared the overall trend of ctDNA mutation frequency with disease progression in the 24 patients with ctDNA surveillance results. Within 6 months of chemotherapy completion, 16 had PDs, and 8 patients had non-PD. In the PD group (n=16), the top three genes with increased mutation frequencies were PIK3CA, TP53, and ERBB2 (Figure 3A). In contrast, the mutation frequencies of RET, FAT1, and BRCA1/2 decreased (Figure 3B). Significantly more PD patients had increased PIK3CA mutation frequency than non-PD patients (56.25% vs 0%, P=.002, Figure 3A). On the other hand, although rare mutations, such as TPH2, MLL3, MED12, EGFR, etc., increased in the non-PD (drug-sensitive) group (n=8) (Figure 3C), the frequencies of PIK3CA, ERBB2, and TP53 mutations primarily decreased (Figure 3D). These findings suggest that alteration in PIK3CA mutation frequency is most strongly associated with disease progression and drug response; the mutation frequency of PIK3CA increased as disease progressed and decreased when treatment was effective.
Figure 3.
The ctDNA gene mutation frequencies changed in patients with PD and in those sensitive to treatment (non-PD). PD was defined in patients who had disease progression within 6 months. Non-PD was defined in patients who were sensitive to treatment and had no disease progression within 6 months. Dark red represents the most common mutated genes and dark blue represents the rarest mutations. If the mutated genes appeared at the same frequency, they are ranked in alphabetic order.
(A) In 16 PD patients, the frequencies of mutations in 102 genes were increased. The most common gene with increased mutations was PIK3CA [asterisk (*) indicates that significantly more PD patients had increased PIK3CA mutation than non-PD patients (56.25% vs 0%, P=.002)].
(B) In 16 PD patients, the frequencies of mutations in 20 genes were decreased. The most common genes with decreased mutations were RET, FAT1, and BRCA1/2.
(C0 In eight therapy-sensitive patients, only nine genes had increased frequencies of mutation. No mutation was more common.
(D) In 8 therapy-sensitive patients, 32 genes had decreased frequencies of mutations. The most common gene with decreased mutations was PIK3CA.
Gene Mutations Render Tumors Resistant to Endocrine Therapy
In breast cancer, an activating mutation of the PIK3CA gene is an upstream event in oncogenic activation of the PI3K/AKT/mTOR pathway [31], and PI3K-mTOR pathway activation further promotes PD and endocrine therapy resistance [15], [19]. Therefore, we next examined the mutation frequency of endocrine therapy-related genes, such as ESR1, GATA3, and PI3K-mTOR–related genes, in the 16 PD patients. In this study, 15/16 PD patients received chemotherapy (Table S3). In the ER-positive/HER2-negative patients who received chemotherapy, 6/12 (50%) (ID=5, 6, 7, 9, 14, 16; Figure 4A) exhibited increased PIK3CA gene mutation frequency, including a few instances of multiple mutations in the PIK3CA gene. In addition to the PIK3CA, chemotherapy also increased the frequency of ESR1 mutation in 3/9 (33.33%) patients (ID=7, 9, 10; Figure 4A). Furthermore, chemotherapy increased the frequency of GATA3 mutation in two patient (ID=9, 33, Figure 4A) and AKT1 mutation in one patient (ID=33, Figure 4A). Interestingly, all of these patients also had PI3K-mTOR–related gene mutations (Figure 4A).
Figure 4.
Mutated genes with increased frequencies were ranked in ER-positive/HER2-negative and ER-positive / HER2-positive PD patients. Dark blue represents the most common increased mutations in PD patients.
(A) Rank of mutated ctDNA genes with increased frequencies in ER-positive/HER2-negative PD patients (n=3, left) and in individual PD patients (ID=4, 5, 6, 7, 8, 9, 10, 12, 13, 14, 16, 33).
(B) Rank of increased mutation ctDNA genes in ER-positive/HER2-positive PD patients (n=4, left) and in individual PD patients (ID=25, 26, 27, 36).
In the four ER-positive/HER2-positive PD patients, 3/4 (75%) exhibited increased PIK3CA mutation frequency, and the remaining patient had increased PIK3R2 mutation frequency (Figure 4B). In all, these results indicate that endocrine therapy-related gene mutations (ESR1, GATA3, and PI3K-mTOR–related genes) emerged or mutation frequencies increased in 12/15 (80%) PD ER-positive MBC patients after chemotherapy. The remaining three PD patients had increased mutation frequencies of IGF1R (ID=13, Figure 4A) or TP53/NOTCH2 (ID=8, 12, Figure 4A). These gene mutations were most often found in patients with PD, suggesting that they are likely to affect the drug resistance of MBC.
Everolimus Decreases PIK3CA Mutation Frequency
The mTOR inhibitor everolimus is a clinically approved anticancer drug used to treat ER-positive patients [32]. In this study, approximately 36% of the ER-positive/HER2-negative and ER-positive/HER2-positive patients had PIK3CA gene mutations prior to chemotherapy (Table S2). A few of these patients received everolimus and chemotherapy (ID=15, 28, 30, 35, 38). The patients who received everolimus in addition to chemotherapy responded to treatment, and their PIK3CA mutation frequency decreased compared to baseline mutations (Figure 5). In addition, the mutation frequency of ESR1 decreased in patient ID15 (Figure 5A). As shown in the Table 1, patients with everolimus treatment exhibited a lower rate of PD within 6 months (0% vs 62.5%, Fisher’s exact test, P=.001). Table S4 listed the treatment regimen for non-PD patients with decreased ctDNA mutation frequencies. No significant differences in rates of PD were observed between PIK3CA mutations and wild-type PIK3CA at baseline; however, significantly more PD patients exhibited increased PIK3CA mutation frequency compared to non-PD patients following chemotherapy (56.25% vs 0%, Fisher’s exact test, P=.002). Together, these results suggest that everolimus decreased PI3KCA mutation frequencies.
Figure 5.
Decreased gene mutations in sensitive MBC patients (n=8). Dark blue represents the most common decreased mutations in sensitive patients.
(A) In ER-positive /HER2-negative therapy-sensitive patients (n=3), the frequencies of mutations in ESR1 (ID=15) and PIK3CA (ID=15, 35) decreased.
(B) In ER-positive /HER2-positive non-PD patients (n=5), the frequencies of mutations in PIK3CA decreased in three patients (ID=28, 30, 38). Two patients had decreases in the frequencies of RAF1 and SF3B1 gene mutation.
Table 1.
Effect of Everolimus Treatment on PFS
| Subgroups | ||||
|---|---|---|---|---|
| Variable | PD (n=16) | Non-PD (n=8) | P value⁎ | |
| Treatment | Everolimus+chemotherapy | 0 (0%) | 5 (62.5%) | .001 |
| Chemotherapy | 16 (100%) | 3 (37.5%) | ||
| Baseline PIK3CA mutation | Yes | 8 (50%) | 5 (62.5%) | .68 |
| PIK3CA mutation frequency | Increased | 9 (56.25%) | 0 (0%) | .002 |
| Decreased | 1 (6.25%) | 5 (62.5%) | ||
P value was calculated using Fisher’s exact test (n<5) for subgroup comparison between the indicated subgroups.
Discussion
In MBC patients with massive tumors, clinicians always recommend chemotherapy at first in order to suppress the tumor burden or relieve symptoms as fast as possible. However, unexpected problems may emerge due to the chemotherapy itself. For example, clinicians have found that patients who were resistant to an initial chemotherapy may also be resistant to the endocrine therapy during the subsequent treatment. Investigation of the underlying mechanism of this resistance suggested that mutation of PI3K-AKT pathway–related genes may be a factor contributing [33], [34].
We analyzed the baseline ctDNA mutations in 44 ER-positive MBC patients prior to chemotherapy. We found that PI3K-AKT pathway–related genes were frequently mutated in these patients. Therefore, the mTOR inhibitor everolimus was recommended in these patients with PI3K-AKT pathway–related gene mutations. A phase III clinical trial (BOLERO-3) has demonstrated that the addition of everolimus to trastuzumab plus vinorelbine significantly improves PFS for patients with trastuzumab-resistant and taxane-pretreated HER2-positive, advanced breast cancer [35]. The underlying mechanism for this effect was reportedly due to the sensitization of tumor cells to chemotherapy by suppression of the functional activation of mutated PIK3CA [20], [36], [37], [38]. Although some patients complied with the suggested addition of everolimus to their treatment strategy, others declined due to economic reasons. In patients who received everolimus, mTOR pathway–related gene (PIK3R2 and PIK3CA) mutation frequencies decreased (Figure 5), whereas patients who did not receive everolimus had increased PIK3CA, PIK3R2, and AKT1 mutation frequencies (Figure 4). These results support that ER-positive MBC patients might benefit from everolimus in conjunction with chemotherapy.
Apart from the PI3K-AKT pathway genes, ESR1 and GATA3 are endocrine therapy resistance genes in breast cancer [6], [14], [19]. In ER-positive patients, the presence of ESR1 mutations following endocrine therapy indicates treatment resistance [39], [40], [41], [42], [43]. In this study, 31/44 (70.45%) patients had a history of endocrine treatment (Table S1); however, ESR1 mutation was not common, with only two ER-positive/HER2-negative (ID=7, 15) patients presenting with baseline ESR1 mutations. Interestingly, both of these patients also had baseline PIK3CA mutations. One of these patients (ID=7, Table S3) received chemotherapy alone and progressed within 6 months with increased mutation frequencies for both ESR1 and PIK3CA (Figure 4). In contrast, the other non-PD patient (ID=15, Table S4) received everolimus plus chemotherapy and was controlled without progression and with decreased mutation frequencies for both ESR1 and PIK3CA (Figure 5). Moreover, in one patient (ID=9) who had no ESR1 mutation prior to treatment (Figure 2B), ESR1 mutation emerged as PIK3CA and GATA3 mutation frequencies increased and the disease progressed (Figure 4B). These findings suggest that the mutation of PIK3CA and ESR1 varies with progression in MBC patients.
Furthermore, the frequencies of GATA3 gene mutation also increased in two PD patients (ID=33, 9). In one of these patient (ID=33), the GATA3 mutation frequency increased as AKT1 mutation frequency increased. In the other patient (ID=9), the GATA3 mutation frequency increased as both PIK3CA and ESR1 mutation frequencies increased (Figure 4, A and B). Therefore, both GATA3 and ESR1 mutations seem to be coupled with mutation of the PI3K-AKT pathway genes.
Interestingly, in this study we observed chemotherapy-induced selection of preexisting mutations as well as new mutations that arose after chemotherapy. More specifically, some patients had preexisting mutations, and these ctDNA mutation frequencies increased after treatment, indicating the resistance of mutation bearing clones to chemotherapy in patients with PD. In addition, some patients did not have preexisting mutations prior to chemotherapy, and tumor gene mutations emerged after treatment. For example, patient ID33 (Figure 4A) did not have a GATA3 mutation before chemotherapy, but after treatment, the GATA3 mutation frequency was 1.6%. Occasionally, these two phenomena coexisted. For example, patient ID5 did not have a PIK3CA amplification mutation before chemotherapy, but after capecitabine treatment, the PIK3CA amplification mutation frequency was 3.2%. Moreover, this patient also had a PIK3CA H1047R mutation frequency of 42.4% before treatment that increased to 59.6% after chemotherapy.
ctDNA mutation is complicated, with both time and space heterogeneity. It is difficult to divide ctDNA mutation into just two or three types. Each mutation may be significant for an individual patient. In this study, we summarized the overall trend of ctDNA mutations following chemotherapy in ER-positive patients. We aimed to provide important and valuable clues for clinicians.
At baseline, PIK3CA gene mutations were common in both HER2-negative and HER2-positive patients. After treatment, the mutation frequency of PIK3CA increased in the majority of ER-positive PD patients. Chemotherapy is a common choice for treatment of MBC; however, baseline TP53 mutation predicts a poor response to chemotherapy in breast cancer patients [44]. Indeed, we found that PIK3CA wild-type patients with baseline TP53 mutation had a significantly poorer PFS than patients without baseline TP53 mutation (P=.04, Figure S1F). PIK3CA mutation has been found to be positive prognostic indicator for overall survival and breast cancer–specific survival in 590 patients (at a single center) and 2587 patients (from 12 independent studies) [45], [46]. However, due to the small sample size of this study, this benefit was not significant, and patients with baseline PIK3CA mutation failed to show improved PFS (Figure S1B).
Tumors acquire resistance to systemic treatment as a result of clonal selection [47]. PIK3CA p.E545K mutation is associated with chemoresistance in breast epithelial cells [48], and its mutation frequency increases significantly after paclitaxel treatment [47]. Our study further confirmed this finding, supporting an alternative treatment regimen involving everlolimus in ER-positive patients by inhibiting the mTOR pathway.
Conclusions
In ER-positive MBC patients with tumor progression, the baseline ctDNA mutation patterns varied across ER/HER2 subgroups. After chemotherapy, ESR1 and GATA3 mutations were coupled with PI3K-AKT1 pathway–related gene mutations. Everolimus treatment in conjunction with chemotherapy suppressed PIK3CA, ESR1, and GATA3 gene mutation. In conclusion, gene mutations that render tumors resistant to endocrine therapy may be suppressed by concomitant mTOR inhibitor treatment. Since our study was limited by a relatively small sample size, future studies should include a larger cohort to compare ctDNA mutation within treatment subgroups and focus on investigation of the underlying effects of everolimus on ctDNA mutations in chemotherapy-resistant MBC patients.
The following are the supplementary data related to this article.
Demographics and Clinical Features of Patients
Gene Mutation Comparison in ER-Positive/HER2-Negative and ER-Positive/HER2-Positive Subgroups
Treatment Regimen for Patients with Increased ctDNA Mutations
Treatment Regimen for Sensitive Patients with Decreased ctDNA Mutations
Acknowledgments
Acknowledgements
We would like to thank the researchers from Geneplus Beijing Institute as these individuals sequenced all patients’ ctDNA and gDNA samples for this study.
Author Contributions
Dr. Quchang Ouyang had full access to all data in the study and takes responsibility for the integrity and accuracy of the data analysis.
Study concept and design: Quchang Ouyang and Zheyu Hu
Data acquisition: Yu Tang
Data analysis and interpretation: Zheyu Hu
Drafting of the manuscript: Zheyu Hu
Critical revision of the manuscript for important intellectual content: All authors
Compliance with Ethical Standards
Disclosure of potential conflicts of interest
All authors declared none potential conflicts of interest.
Research Involving Human Participants and/or Animals
This study involved human participants and was approved by the Ethics Committee at Hunan Cancer Hospital. Informed consent was obtained from each patient prior to study onset.
Contributor Information
Quchang Ouyang, Email: oyqc1969@126.com.
Zhe-Yu Hu, Email: huzheyu@hnszlyy.com.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014 Jan-Feb;64(1):52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Hu W, Tan C, He Y, Zhang G, Xu Y, Tang J. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. 2018;11:1529–1541. doi: 10.2147/OTT.S152462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Driscoll L, Clynes M. Biomarkers and multiple drug resistance in breast cancer. Curr Cancer Drug Targets. 2006 Aug;6(5):365–384. doi: 10.2174/156800906777723958. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013 Feb 28;368(9):842–851. doi: 10.1056/NEJMra1204892. [DOI] [PubMed] [Google Scholar]
- 5.Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6(3):181–202. [PubMed] [Google Scholar]
- 6.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013 Dec;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comprehensive molecular portraits of human breast tumours Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu D, Paoletti C, Gersch C, VanDenBerg DA, Zabransky DJ, Cochran RL. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res. 2016 Feb 15;22(4):993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016 Sep 1;34(25):2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 10.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016 Oct 1;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clatot F, Perdrix A, Augusto L, Beaussire L, Delacour J, Calbrix C. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016 Nov 15;7(46):74448–74459. doi: 10.18632/oncotarget.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X. Mutation of GATA3 in human breast tumors. Oncogene. 2004 Oct 7;23(46):7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 13.Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005 Dec 15;65(24):11259–11264. doi: 10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 14.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013 Jan;23(1):12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, Chen H. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010 Jul;120(7):2406–2413. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015 Apr 15;7(283):283ra51. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004 Aug;3(8):772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 18.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005 Apr 1;65(7):2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 19.Brown KK, Toker A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep. 2015;7:13. doi: 10.12703/P7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012 Feb 9;366(6):520-9. [DOI] [PMC free article] [PubMed]
- 21.Zardavas D, Fumagalli D, Loi S. Phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway inhibition: a breakthrough in the management of luminal (ER+/HER2-) breast cancers? Curr Opin Oncol. 2012 Nov;24(6):623–634. doi: 10.1097/CCO.0b013e328358a2b5. [DOI] [PubMed] [Google Scholar]
- 22.Cheng H, Liu C, Jiang J, Luo G, Lu Y, Jin K. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int J Cancer. 2017 May 15;140(10):2344–2350. doi: 10.1002/ijc.30650. [DOI] [PubMed] [Google Scholar]
- 23.Guibert N, Mazieres J, Delaunay M, Casanova A, Farella M, Keller L. Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget. 2017 Jun 06;8(23):38056–38060. doi: 10.18632/oncotarget.16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015 Aug;26(8):1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan K, Rata M, Cunningham D, Koh DM, Tunariu N, Hahne JC. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut. 2017 Aug:08. doi: 10.1136/gutjnl-2017-314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Mol Cancer. 2018 Feb 26;17(1):67. doi: 10.1186/s12943-018-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Chu Y, Zhang R, Han Y, Zhang L, Fu Y. Technical validation of a next-generation sequencing assay for detecting clinically relevant levels of breast cancer–related single-nucleotide variants and copy number variants using simulated cell-free DNA. J Mol Diagn. 2017 Jul;19(4):525–536. doi: 10.1016/j.jmoldx.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017 Aug;9(403):16. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saad ED, Katz A. Progression-free survival and time to progression as primary end points in advanced breast cancer: often used, sometimes loosely defined. Ann Oncol. 2009 Mar;20(3):460–464. doi: 10.1093/annonc/mdn670. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Pan Z. Progression-free survival and time to progression as real surrogate end points for overall survival in advanced breast cancer: a meta-analysis of 37 trials. Clin Breast Cancer. 2017 Jul;25 doi: 10.1016/j.clbc.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Raphael J, Desautels D, Pritchard KI, Petkova E, Shah PS. Phosphoinositide 3-kinase inhibitors in advanced breast cancer: a systematic review and meta-analysis. Eur J Cancer. 2018 Mar;91:38–46. doi: 10.1016/j.ejca.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Moynahan ME, Chen D, He W, Sung P, Samoila A, You D. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(−) advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017 Mar 14;116(6):726–730. doi: 10.1038/bjc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen IC, Hsiao LP, Huang IW, Yu HC, Yeh LC, Lin CH. Phosphatidylinositol-3 kinase inhibitors, buparlisib and alpelisib, sensitize estrogen receptor-positive breast cancer cells to tamoxifen. Sci Rep. 2017 Aug 29;7(1):9842. doi: 10.1038/s41598-017-10555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabine VS, Crozier C, Brookes CL, Drake C, Piper T, van de Velde CJ. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J Clin Oncol. 2014 Sep 20;32(27):2951–2958. doi: 10.1200/JCO.2013.53.8272. [DOI] [PubMed] [Google Scholar]
- 35.Andre F, O'Regan R, Ozguroglu M, Toi M, Xu B, Jerusalem G. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014 May;15(6):580–591. doi: 10.1016/S1470-2045(14)70138-X. [DOI] [PubMed] [Google Scholar]
- 36.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010 Aug;120(8):2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng T, Dou QP. Everolimus inhibits growth of gemcitabine-resistant pancreatic cancer cells via induction of caspase-dependent apoptosis and G2 /M arrest. J Cell Biochem. 2017 Sep;118(9):2722–2730. doi: 10.1002/jcb.25921. [DOI] [PubMed] [Google Scholar]
- 38.Martin LA, Pancholi S, Farmer I, Guest S, Ribas R, Weigel MT. Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast Cancer Res. 2012 Oct 17;14(5):R132. doi: 10.1186/bcr3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnedos M, Vicier C, Loi S, Lefebvre C, Michiels S, Bonnefoi H. Precision medicine for metastatic breast cancer—limitations and solutions. Nat Rev Clin Oncol. 2015 Dec;12(12):693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 40.Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016 May 13;7 doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fribbens C, Garcia Murillas I, Beaney M, Hrebien S, O'Leary B, Kilburn L, et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol. 2018 Jan 1;29(1):145-53. [DOI] [PMC free article] [PubMed]
- 42.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015 Nov 11;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fribbens C, Garcia Murillas I, Beaney M, Hrebien S, O'Leary B, Kilburn L. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol. 2017 Oct:4. doi: 10.1093/annonc/mdx483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson J, Larsson L, Klaar S, Holmberg L, Nilsson J, Inganas M. Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann Oncol. 2005 May;16(5):743–748. doi: 10.1093/annonc/mdi150. [DOI] [PubMed] [Google Scholar]
- 45.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009 Aug 15;15(16):5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 46.Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: an analysis of 2587 patients with breast cancer. Chin J Cancer. 2012 Jul;31(7):327–334. doi: 10.5732/cjc.012.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013 May 2;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 48.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005 Dec 1;65(23):10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographics and Clinical Features of Patients
Gene Mutation Comparison in ER-Positive/HER2-Negative and ER-Positive/HER2-Positive Subgroups
Treatment Regimen for Patients with Increased ctDNA Mutations
Treatment Regimen for Sensitive Patients with Decreased ctDNA Mutations