Abstract
Cancer arises from the acquisition of multiple genetic and epigenetic changes in host cells over the span of many years, promoting oncogenic traits and carcinogenesis. Most cancers develop following random somatic alterations of key oncogenic genes, which are favoured by a number of risk factors, including lifestyle, diet and inflammation. Importantly, the environment where tumours evolve provides a unique source of signalling cues that affects cancer cell growth, survival, movement and metastasis. Recently, there has been increased interest in how the microbiota, the collection of microorganisms inhabiting the host body surface and cavities, shapes a micro-environment for host cells that can either promote or prevent cancer formation. The microbiota, particularly the intestinal biota, plays a central role in host physiology, and the composition and activity of this consortium of microorganisms is directly influenced by known cancer risk factors such as lifestyle, diet and inflammation. In this Review, we discuss the pro- and anticarcinogenic role of the microbiota, as well as highlighting the therapeutic potential of microorganisms in tumourigenesis. The broad impacts, and, at times, opposing roles of the microbiota in carcinogenesis serve to illustrate the complex and sometimes conflicted relationship between microorganisms and the host—a relationship that could potentially be harnessed for therapeutic benefits.
Microorganisms are present in all terrestrial and aquatic habitats and this ubiquitous environmental presence has produced symbiotic, mutualistic and parasitic coevolution between hosts and microorganisms across kingdoms. In humans, this evolutionary process has led to the acquisition of a rich and diverse set of microorganisms, comprising Bacteria, viruses, fungi, protozoa and Archaea inhabiting every surface and cavity of the human body.
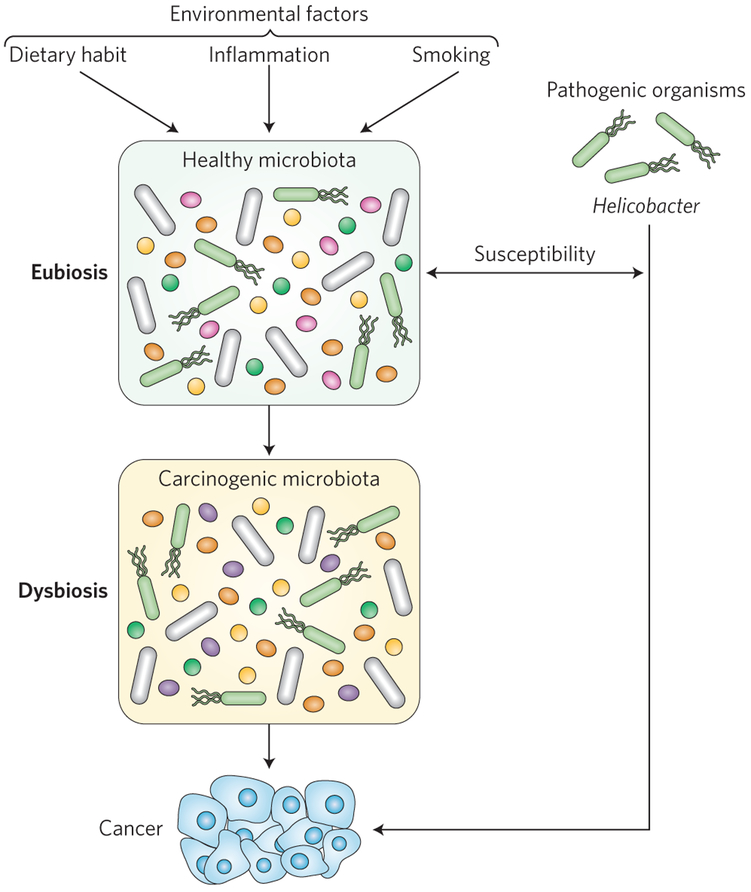
Although essential for life, a narrow segment of the microbial world is categorized as pathogenic and exposure to these microorganisms causes various infectious diseases, some leading to the development of cancer1,2. For example, subjects infected with microorganisms that are classified as class 1 carcinogens, such as Helicobacter pylori, hepatitis B or C viruses, Epstein-Barr virus or Kaposi-sarcoma-associated herpesvirus infection, were found to develop various cancers, including lymphoma, leukaemia, gastric cancer and hepatocellular carcinoma (HCC)3. In addition to the presence of specific pathogens, host environmental conditions such as smoking4, inflammation5-7, antibiotics8 and diet9 can promote changes in microbial community composition or metabolic activity, which may lead to conditions favouring neoplastic changes (Fig. 1). To add another layer of complexity, these host–microbiota interactions also modulate host susceptibility to infectious bacteria (Helicobacter)10 and viruses (such as norovirus, rotavirus, poliovirus)11-13, suggesting that cancer susceptibility depends on the interactions between the normal microbiota and infectious entities.
Figure 1 ∣. Environmental changes can promote dysbiosis and pathogen-derived susceptibility to cancer.
Disruption of the healthy microbiota by environmental factors, such as inflammation, can both increase host susceptibility to carcinogenic pathogens (such as Helicobacter) and expose the host to the cancer risks associated with dysbiosis.
Efforts to better understand the microbial communities of humans have become a major focus of research over the last decade. Initial characterization studies were mostly directed at compositional changes within bacterial communities living at different body sites using high-throughput nucleic acid sequencing techniques, but more recent studies have included more comprehensive sampling and analysis strategies, such as the generation of RNA sequences from metagenomes (such as metatranscriptomes)14,15. Among the various body locations subjected to these studies, the digestive tract, mouth to anus, was found to harbour the most abundant and rich repertoire of microorganisms, with the lower intestine (colon) showing the highest density of microorganisms16—although the ratio of bacteria to human cells is probably lower than the 10:1 ratio often cited17. These microorganisms have been studied in the context of health and disease and compared to a state of normalcy or eubiosis, and intestinal bacterial communities have been characterized as dysbiotic in many diseases, including allergy, asthma, rheumatoid arthritis, cardiovascular diseases, metabolic syndrome, obesity, inflammatory bowel diseases (IBDs) and colorectal cancer (CRC)18–23. Therefore, the intestinal bacterial community appears to influence local as well as extraintestinal function. This review will focus on recent discoveries on how the intestinal microbiota exerts its modulatory role on carcinogenesis, particularly CRC, and whether and how microorganisms could be integrated into the therapeutic landscape.
Bacteria as a modulating agent of carcinogenesis
The role of bacteria in carcinogenesis is complex, as both pro- and anticarcinogenic functions have been attributed to microorganisms24. For example, the pro-neoplastic activity of bacteria in cancer has been known for decades as exemplified in the promotion of gastritis and cancer by pathogenic Helicobacter—the first bacteria classified as a group 1 carcinogen25. Studies using Helicobacter have demonstrated that toxin cytotoxin-associated gene A (CagA)-induced DNA damage and the promotion of host-derived inflammatory mediators and growth factors are direct risk factors for carcinogenesis26. Helicobacter toxin CagA is an oncoprotein that enhances DNA damage through host-mediated overproduction of reactive oxygen species (ROS)27–29. Vaculolating cytotoxin A (VacA) alters membrane permeability and can lead to increased rates of apoptosis, and a recent meta-analysis has confirmed its association with increased gastric cancer risk30,31. Remarkably, the link between gastric cancer and Helicobacter appears specific, as a recent study suggests that no other single bacteria has a major influence on the development of some gastric cancers32. Helicobacter colonization has been shown to increase cancer rates by an estimated six times, possibly through induction of high levels of nitric oxide release from immune cells33. However, recent literature suggests that Helicobacter can also play protective roles against cancers34–37. For example, a meta-analysis of H. pylori and oesophageal cancer risk demonstrated a statistically significant decrease in risk associated with infection, although the mechanism for such a protective effect remains unknown38. This duality of cancer promotion and protection at the single-species level indicates that host health outcomes that are associated with microorganisms are highly context dependent. Salmonella typhi has also been associated with increasing gallbladder cancer risk39, and recent evidence has shown that effector proteins delivered through the type III secretion system of this bacterium induce MAPK (mitogen-activated protein kinase) and Akt signalling—critical host responses that lead to cellular transformation40.
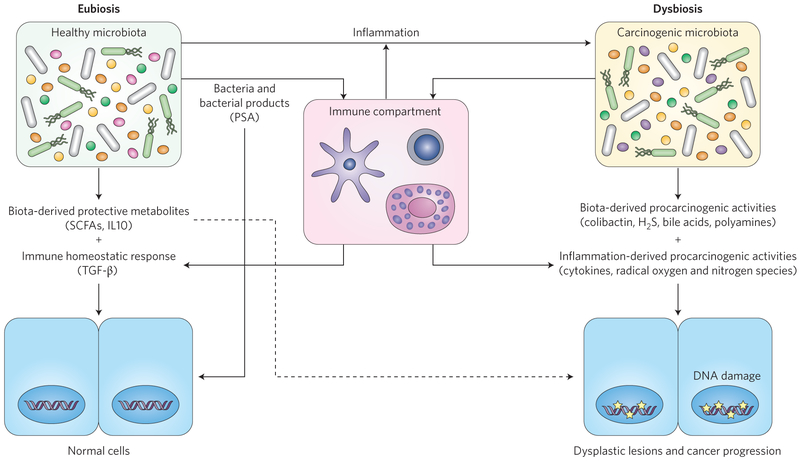
While pathogens such as Helicobacter can directly promote cancer risk, an indirect mechanistic link between the microbiome and cancer is inflammation41. This major environmental stress, derived from either chronic or para-inflammation, is strongly implicated with increased cancer risk41–45. At steady state (eubiosis), the microbiota participates in homeostasis by generating metabolites such as short-chain fatty acids (SCFAs) and by engaging protective innate and adaptive immune responses16,46 (Fig. 2). However, inflammation is associated with the expansion of disease-promoting bacteria and depletion of protective bacteria, leading to a state of microbial dysbiosis47 with the potential for positive-feedback loops promoting further inflammation. The mechanisms by which inflammation contributes to change in microbial composition and activities are still unclear, but it is likely that they depend on the ability of bacteria to adapt to an inflammatory environment and utilize unique resources present in this environment (see next paragraph). For example, patients with chronic intestinal inflammation displayed reduced overall microbial diversity with increased representation of specific families such as Enterobacteriaceae and Fusobacteriaceae, whose species, including adherent invasive Escherichia coli and Fusobacterium nucleatum, are well-documented pro-inflammatory bacteria48. Patients with IBDs also showed reduced abundance of Lachnospiraceae, a family containing SCFA-producing members (for example, group IV and XIVa Clostridia). These microbial metabolites modulate immune responses and play a critical function in epithelial cell energy balance, a key element in host homeostasis49,50. This depletion of SCFA-producing bacteria may not only alter host-dependent immune signalling but may also create favourable conditions for the emergence of potential carcinogenic strains. For example, antibiotic-mediated depletion of butyrate-producing Clostridia leads to expansion of Salmonella enterica serovar Typhimurium due to decreased hypoxic conditions at the epithelium level51. SCFAs, however, do not always promote beneficial effects on the host. For example, microbiota-derived acetate production leads to metabolic syndrome in TLR5 (toll-like receptor 5) gene-deficient mice, a mechanism that is associated with increased liver glucogenesis52. More specific to cancer, a recent study showed that microbial-derived butyrate promotes carcinogenesis by enhancing intestinal epithelial cell proliferation in a MutS homologue 2 gene-deficient mouse crossed to a multiple intestinal neoplasia (Min or Apcmin/+) model (ref. 53). It is possible that the impact of SCFAs on the host is beneficial at steady state but promotes deleterious effects under specific genetic or inflammatory conditions.
Figure 2 ∣. Microbial interactions with the immune system modulate cancer risk.
Microbiota promote homeostasis directly through metabolites and bacterial products, which influence both the epithelial and immune cell response. In addition, dysregulated immune-host interaction favours the development of dysbiosis, which contributes to carcinogenesis through metabolic activities and activation of immune responses. Some protective microbiota (SCFAs) may promote cellular proliferation of cancer-initiated cells (dashed arrow). Brackets contain example compounds. PSA, polysaccharide A; TGF-β, transforming growth factor-β. Stars indicate DNA damage.
Facultative anaerobic pathogen and pathobiont strains thrive in an inflammatory environment due, in part, to their ability to utilize inflammation-derived molecules such as nitrites and oxides as electron acceptors—a feature not shared by many symbionts54. In addition, microbial metabolism is altered under dysbiotic conditions, conferring new microbial phenotypes such as enhanced cellular adherence and invasion, mucus utilization, and production of metabolites and toxins (such as H2S, bile acids and genotoxins)1,2,55. Furthermore, it has recently been recognized that the induction of inflammation allows bacteria and tumour cells to communicate via peptides associated with quorum sensing, which could contribute to metastasis56. Taken together, these studies demonstrate that inflammation creates host-derived (cytokines, growth factors, radical oxygen and nitrogen species) and bacteria-derived (genotoxins, H2S, bile acids, radical species, and so on) pro-carcinogenic conditions that provide an ideal landscape for cancer development (Fig. 2).
Establishing an ‘ecosystem’ profile of cancer dysbiosis appears to be an important first step toward the identification of the microbial community implicated in the promotion or protection of carcinogenesis. Microbial dysbiosis has been observed in oral, lung, breast and liver cancers (Box 1)57. However, functional experiments causally linking the state of dysbiosis to carcinogenesis have not been extensively performed in these organs. In contrast, microbial dysbiosis has been extensively studied in patients with CRC, and preclinical models have shown clear functional consequences. For example, an altered microbial community is observed between tumour and normal flanked tissue of CRC patients58,59, distal versus proximal tumours, and across the neoplastic progression from adenoma to adenocarcinoma60,61. Across this spectrum, specific changes within the intestinal microbial community are observed in CRC patients, such as increased abundance of Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia and Methanobacteriales, while representation of Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium, Roseburia and Treponema decreased23,62,63. Bifidobacterium and Lactobacillus spp. have been shown to possess antitumourigenic effects in preclinical models64. Changes in the microbial community in CRC patients may be robust enough to serve as potential non-invasive biomarkers to predict carcinogenic stages65–67. Reproducibility across studies remains a serious challenge, however, although the field is beginning to address these concerns with new tools that evaluate the generalizability of models across studies68.
Box 1 ∣. Microbiota and extra-intestinal cancer.
The microbiome has attracted attention as an environmental factor that is implicated in carcinogenesis144. The first microbiome encountered in the digestive tract, that of the oral cavity has long been indicated as an intermediary between diet and cancer risk145,146. Oral squamous cell carcinomas have shown changes in microbiota diversity between tumour and healthy tissue, and taxa that are associated with disease status have been identified, though cohort sizes in such studies have been small147–149. The relative diagnostic ease of sampling the oral microbiota has the potential to yield biomarkers for colorectal, pancreatic, GI and other cancers150–153. Similarly, the lung is a nexus between environmental factors154, microbial dysbiosis155 and cancer with its own potential biomarkers156,157. In contrast to these ‘open environments in human physiology Hieken et al. recently confirmed differences in microbiota abundance profiles between aseptically collected cancerous and healthy breast tissue that reduce the concerns of contamination driving profile differences in similar studies158–160. The far-reaching influence of gut bacteria such as Helicobacter can also be seen through the promotion of cancers distant from the gut, such as breast cancer, through Helicobacter’s interactions with neutrophils161. Helicobacter has also been indicated, with some contention, in the development of HCC, warranting further investigations of microbiota–HCC associations162–164. The relationship between the microbiome and HCC is also one of inflammation and dysbiosis, and there has been some evidence of probiotics restoring eubiosis and reducing tumour growth165,166.
Numerous factors can influence intestinal microbiota composition, including diet, medication, disease and health parameters69. Host genetics has also been shown to influence human intestinal microbiota composition70. It is still unclear whether microbial dysbiosis observed in human CRC patients is a consequence of the pathology or a leading factor promoting the disease. Animal models suggest microbial change occurs before development of neoplasis. For example, at pre-neoplastic stage, the colonic mucosa of Apcmin/+ mice showed increased relative abundance of Bacteroidetes spp. compared to wild-type mice. In addition, microbial composition changed during inflammation onset in IL10−/− (interleukin 10 knockout) mice, which preceded development of invasive CRC71. Moreover, deletion of the antimicrobial peptide lipocalin-2 (Lcn2) gene in Il10−/− mice (Il10−/− Lcn2−/−) leads to intestinal inflammation, microbial dysbiosis and increased tumourigenesis compared to Il10−/− mice72. Cross-fostering experiments between Il10−/− Lcn2−/− mother and Il10−/− newborn mice showed that tumourigenesis is communicable72. In a separate study, faecal transfer of a dysbiotic community from tumourbearing mice into germ-free mice lead to higher tumour numbers compared to healthy community transfer73. Whether human cancer-associated dysbiotic microbiota is functionally implicated in carcinogenesis remains unclear. Recent studies showed that colitis developed in germ-free Il10−/− mice transplanted with stools from dysbiotic IBD patients but not from eubiotic healthy controls74. Surprisingly, germ-free mice colonized with stools from CRC patients displayed lower tumour burdens than mice colonized with stools from healthy subjects after exposure to azoxymethane (AOM) and the inflammatory agent dextran sodium sulfate (DSS)75. As the AOM–DSS chronic wound-healing model was used for these experiments, it is possible that dysbiotic bacteria from CRC patients enhanced tissue repair, thereby attenuating DSS-induced wound healing—a critical component of carcinogenesis in this model76.
To add further complexity to the relationship between the microbial community and development of CRC, bacterial biofilms were identified in 50% of tumour and paired adjacent normal tissue samples from human CRC patients77. In addition, biofilm microbial organization is a feature of patients with both ulcerative colitis and Crohn’s disease. Bacteria with the biofilm feature are detected in 95% of IBD patients, compared to 35% for healthy subjects78. Microorganisms growing in biofilms frequently express phenotypes that are different from their non-adherent planktonic counterparts79. Interestingly, higher levels of acetylated polyamines, essential metabolites for cellular growth and proliferation80, were reported in biofilm-positive cancer tissue when compared to biofilm-negative cancer tissue81. Moreover, functional experiments using germ-free Il10−/− Apcmin/+ mice showed that mice colonized with biofilm-positive but not biofilm-negative bacteria obtained from CRC patients developed tumours82. These findings highlight the key roles played by the microbiota in influencing neoplastic changes in the intestine, which may also play a role in different forms of cancer (Fig. 2; Box 1).
Bacteria as therapeutic tools for cancer
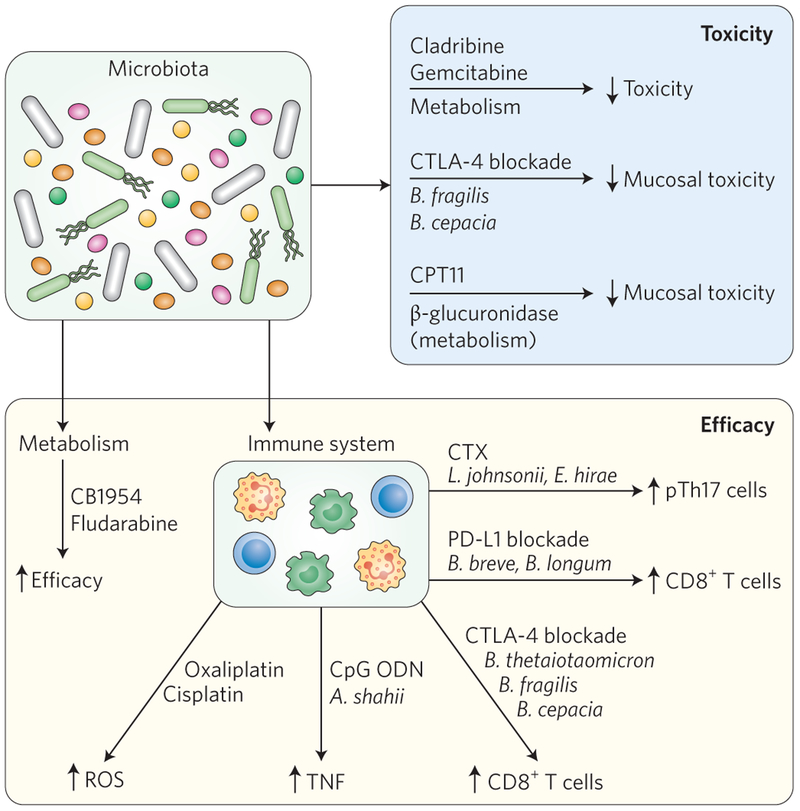
Despite substantial recent progress in understanding the molecular mechanisms of tumourigenesis, cancer remains stubbornly difficult to treat. Even with some encouraging successes in rationally derived drugs, rates of clinical trial failures for new cancer drugs remain high83. There is an urgent need to develop new anticancer therapies or to improve current ones, and the potential roles of bacteria in this mission has been the focus of considerable research. An emerging frontier in cancer therapeutics focuses on how the interaction between bacteria, individual metabolism and immune response and established antitumour drugs could shape cancer management. This integration of bacteria to the therapeutic landscape forms the essence of ‘pharmacomicrobiomics’84, where microbial bioactivities and direct interaction with the host become important variables influencing drug toxicity and efficacy (Fig. 3).
Figure 3 ∣. The microbiota influences drug toxicity and efficacy.
Efficacy and toxicity of anticancer drugs can be modulated by the microbiota through numerous mechanisms, including microbial metabolism and host-mediated immune responses. Small arrows indicate the direction of the response (increased or decreased).
Interaction between immunotherapy and bacteria
Targeting co-inhibitory or co-stimulatory receptor–ligand systems, such as programmed death 1 (PD-1)-programmed death-ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), on T lymphocytes has emerged as a powerful means to recruit the host immune system in the fight against tumour cells85,86. Antibodies against CTLA-4 and PD-1 have been approved for treatment of advanced melanomas and have shown strong therapeutic response, and various clinical trials are ongoing to test their efficacy in other forms of cancer, including ovarian, prostate, bladder, renal and lung cancer87. Unsurprisingly new antibody-based immune targets have also been developed and are currently being tested for their antitumour efficacy87,88. With the recognized interaction between bacteria and host immune responses, a logical step was to investigate the impact of microorganisms on immunotherapy As with many cancer treatments, utilization of immunotherapy agents requires balancing their effectiveness in eliminating cancer with the burden the drugs place on the patient89. In the following section, we discuss in detail how consideration of microbial function may impact the toxicity and effectiveness of this class of drugs.
Bacteria and toxicity.
The inhibitory antibody ipilimumab binds the immune checkpoint protein CTLA-4, impeding regulatory T (Treg)-cell function and consequently promoting T-cell activation. This antibody is highly effective in the treatment of metastatic melanoma but its dampening effect on Treg function causes numerous adverse effects, including intestinal inflammation90. In the case of colitis, the deleterious effect of inhibiting CTLA-4 signalling was attenuated by reconstituting microbiota-depleted mice with Bacteroides fragilis and Burkholderia cepacia91, which is consistent with the observation that patients who are resistant to ipilimumab-induced colitis show a high abundance of bacteria belonging to the Bacteroidetes phylum92. The mechanism by which these microorganisms alleviate the deleterious effect of ipilimumab treatment is unclear, but increased Treg function is probably not implicated as Bacteroides fragilis is critical for increased drug efficacy (as discussed in the following section).
Bacteria and efficacy.
Recent literature suggests that bacteria may not only alleviate drug toxicity but could also enhance the efficacy of immune checkpoint inhibitors. The potential role of bacteria in drug efficacy has been highlighted in experiments where microbial content was manipulated through antibiotics, faecal transfer and gnotobiotics. For example, the presence of bacteria appears to be essential for ipilimumab (anti-CTLA-4) therapy as this treatment was less effective in preventing subcutaneous growth of MCA205 sarcoma, MC38 colon carcinoma and Ret melanoma in mice maintained in germ-free conditions or exposed to broad-spectrum antibiotics to eliminate the intestinal biota91. When administered by oral feeding, specific strains of bacteria (Bacteroides thetaiotaomicron, B. fragilis and B. cepacia) were found to stimulate CTLA-4-induced antitumour immune response, thereby increasing therapeutic efficacy91. In humans, the intestinal microbiota of metastatic melanoma patients was sampled before and after treatment with ipilimumab. The microbiome of these patients was found to form three major clusters: one dominated by Alloprevotella or Prevotella, while the other two were characterized by distinct Bacteroides spp. Interestingly, there was some evidence that ipilimumab treatment caused patients to shift between the two Bacteroides-dominated clusters. Faecal transfer of these different clusters into germ-free animals appeared to have different consequences for tumour formation in the recipient animals, with increased drug efficacy correlating with the cluster that also produced increased colonization of the immunogenic bacteria B. fragilis and B. thetaiotaomicron in the animal host. Data from these experiments are consistent with the hypothesis that ipilimumab treatment increases the abundance of immunogenic Bacteroides spp., which in turn improves the efficacy of the drug. Although less efficient than live B. fragilis, oral administration of polysaccharides isolated from B. fragilis caused an immunostimulatory effect and antitumour activity suggesting that the enhanced antitumour efficacy is not dependent on a microbial metabolite. The immunostimulatory effect of B. fragilis on dendritic cells and the resulting CD8+ cell response in this system is somewhat surprising as previous observations showed an immunosuppressive effect of a B. fragilis bacterial strain and associated polysaccharides93,94. These results again highlight the complex nature of host immune system interactions with bacteria, where the final outcomes, immunostimulation or immunosuppression, are dictated by complex combinations of cellular signal processing.
The efficacy of another immune checkpoint blocker, PD-L1, was also shown to depend on bacteria. Microbial genomic analysis and faecal transplantation in mice identified Bifidobacterium spp. (Bifidobacterium breve and Bifidobacterium longum) as important contributors of a PD-L1 blockade-mediated antitumour effect on subcutaneous B16.SIY melanoma growth95. Bifidobacterium enhanced the antigen-presentation function of dendritic cells, which resulted in augmented activation of CD8+ T cells’ antitumour activity. Remarkably, even in the absence of PD-L1 treatment, melanoma tumour growth was impaired in mice colonized with B. breve and B. longum, suggesting a direct engagement of the host immune antitumour response. Although both immune checkpoint inhibitors (CTLA-4 and PD-L1) synergize with bacteria to achieve antitumour effects, their mechanisms of action appear somewhat different as CTLA-4 may rely on bacteria–tumour cross-reactivity, whereas bacteria-induced antigen presentation is probably a key component of PD-L1 efficacy96.
Over a century ago, William Coley administered heat-killed bacterial extracts to patients with different forms of inoperable cancer, as part of what is now considered the first immunotherapy experiment97. It is not clear which components of this original microbial cocktail activated the immune system, but cell wall components and nucleic acids, such as DNA, all have immunostimulatory properties. For example, unmethylated CpG oligodeoxynucleotides (CpG ODN) have potent immunostimulatory effects that could be utilized for the treatment of various pathologies including cancer98. Interestingly, the gut microbiota modulates the antitumour effect of a combined anti-IL10 receptor (anti-IL10R)-antibody–CpG-ODN immunotherapy99. Similar to CTLA-4 and PD-L1, the antitumour efficacy of an anti-IL10R-antibody–CpG-ODN immunotherapy against subcutaneous tumour growth (EL4 lymphoma, MC38 colon carcinoma or B16 melanoma) was diminished in germ-free mice or broad-spectrum antibiotic-treated mice, an effect that correlated with the host’s ability to generate tumour necrosis factor-α (TNFα). Specific bacteria were positively (Alistipes, Ruminococcus) or negatively (Lactobacillus) correlated with TNF production and activation of cytotoxic CD8+ T-cell response in the tumour environment. Microbiota-depleted mice colonized with Alistipes shahii increased TNF production by tumour-associated myeloid cells following anti-IL10R-antibody–CpG-ODN exposure. It is remarkable that although microorganisms work in concert with immunotherapeutic drugs (CTLA-4, PD-L1, CpG ODN) to enhance their efficacy, a ‘pairing’ system between each drug and bacteria seems to take place. Therefore, the presence of specific microbial species may determine patient response to a given drug treatment (Fig. 3).
Interaction between chemotherapeutics and bacteria
Chemotherapeutic drugs essentially induce cytotoxicity of rapidly dividing cells, which is a common feature of cancer cells but also of specific healthy cells present in bone marrow and the gastrointestinal (GI) tract. Consequently, these drugs cause significant collateral damage to the patient, generally leading to a state of immunosuppression and severe diarrhoea. Therefore, it has been a challenge to reach maximum antitumour effects with such side effects.
Bacteria and toxicity.
Camptothecin-11 (CPT-11) is an analogue of the natural alkaloid CPT, an inhibitor of topoisomerase I, which is required for DNA replication100. CPT-11 (irinotecan) is mainly used in patients with CRC and is biotransformed into the active topoisomerase I inhibitor SN-38 to exert its antitumour effect101. The compound is subjected to further metabolism in the liver, where a glucuronide group is added to generate the inactive derivative SN-38G (ref. 101). This derivative is excreted via biliary ducts into the GI tract, where the bacterially derived β-glucuronidase enzyme reactivates the compound to its cytotoxic SN-38 form, thereby causing damage to intestinal epithelial cells and leading to severe diarrhoea. In order to prevent microbially derived reactivation of SN-38 in the intestine, E. coli-derived β-glucuronidase inhibitors were screened and successfully tested in mice to alleviate CPT-11-induced GI toxicity102,103. Targeting microbially derived β-glucuronidase did not alter irinotecan pharmacokinetics, suggesting that drug toxicity and efficacy could be uncoupled by targeting bacterial enzymes104.
Similar to CPT-11, methotrexate (MTX) is another chemotherapeutic compound that causes significant GI toxicity in patients105. Antibiotic-mediated microbiota depletion increased MTX-induced mucosal injury in mice, an effect linked to TLR2 signalling106. The microbiota has also been shown to modulate radiation-induced GI toxicity107,108 and one could envision the use of selective bacteria to alleviate toxicity due to cancer treatment109.
Bacteria and efficacy.
As mentioned previously, bacteria modulate antitumour drug efficacy through their influence on host immune responses. Cyclophosphamide (CTX) is a prodrug that, once activated, acts as an alkylating cytotoxic compound, effective in numerous forms of solid tumours110. The CTX-mediated antitumour effect in a subcutaneous tumour growth model (P815 mastocytoma and MCA205 sarcoma) was reduced in germ-free, antibiotic-treated or vancomycin (specific for Gram-positive bacteria)-treated mice111. Importantly, CTX failed to induce an antitumour response in a vancomycin-treated transgenic mouse model of lung adenocarcinoma, suggesting that the interplay between bacteria and host immune response is not restricted to xenograft models. Interestingly, administration of CTX led to the disruption of small intestinal barrier function, enabling the translocation of intestinal bacteria, particularly Gram-positive and vancomycin-sensitive Lactobacillus johnsonii, Lactobacillus murinus and Enterococcus hirae, into mesenteric lymph nodes and spleen111. This extra-intestinal interaction between bacteria and immune cells was critical for the induction of pathogenic T helper 17 (pTh17) cells expressing interferon-γ in the spleen, which were responsible for the drug’s therapeutic effect111. Oral administration of L. johnsonii and E. hirae restored the pTh17 response in the spleen of antibiotic-treated mice. Hence, although CTX causes damage to the intestinal barrier as part of its toxic effect, it allows for translocation of specific Gram-positive intestinal bacteria, a necessary process for the activation of peripheral immune cells and anticancer efficacy.
Oxaliplatin and cisplatin are both alkylating agents that are widely used in cancer therapy112. As observed with CTX, microbiota depletion by broad-spectrum antibiotic exposure reduced the antitumour efficacy of these compounds in subcutaneous tumour growth (EL4 lymphoma and MC38 colon carcinoma)99. The reduced antitumour efficacy was linked to reduced production of ROS by myeloid cells, a critical step for the platinum-induced antitumour effect.
The microbiota possesses a large collection of genes with an impressive metabolic potential that not only participates in energy harvest, but also metabolizes numerous xenobiotics113. For example, after the preincubation of 30 chemotherapeutic drugs with non-pathogenic E. coli Nissle 1917 or Listeria welshimeri serovar 6B SLCC533410, 10 out of 30 chemotherapeutic drugs (for example, gemcitabine, cladribine, daunorubicin) showed reduced cytotoxic activity against the Lewis lung carcinoma cell line, while 6 compounds showed increased efficacy (for example, fludarabine phosphate, CB1954)114. Bacterial heat inactivation abolished these differential drug modification responses, suggesting that enzymatic activities, and not structural components, are implicated in this phenomenon. These findings suggest that bacteria often directly metabolize chemotherapeutic drugs and change their efficacy (Fig. 3).
Cancer management through bacteriotherapy
While much recent research has focused on how bacteria can contribute to disease in the gut, especially cancer, these microorganisms could also potentially be enlisted for cancer prevention, detection or drug delivery. For example, probiotics or microorganisms associated with health benefits have long been used to attempt to resolve gut dysbiosis with ‘good’ bacteria, although often with limited clinical evidence of their effectiveness64. Lactic acid bacteria, with demonstrated beneficial effects as probiotics115,116, are beginning to be explored in the context of CRC117,118. As mentioned previously, abundances of these microorganisms are often reduced in the biota of CRC patients. Capsule delivery of probiotics (B. longum, Lactobacillus acidophilus and Enterococcus faecalis) in CRC patients produced increased microbial diversity and lower abundance of Fusobacterium with increased representation of Enterococcus on the intestinal mucosal tissue of patients compared to placebo-treated patients119. Studies in colitis and enteric infectious diseases have yielded limited evidence suggesting that such multi-species probiotics have greater efficacy relative to that of single-strain probiotics120,121. However, the vast majority of studies involving probiotics as cancer therapies have focused on single-species probiotics117,122–124 and a rigorous comparison of the efficacy of single- versus multispecies probiotics is needed. Such comparisons would allow for the evaluation of tradeoffs between multiple species with multiple cancer targets and potential intra-probiotic antagonism between species. Although probiotics are mainly viewed as preventive agents, some Lactobacillus spp. are used to mitigate the GI toxicity associated with anticancer drugs109. The proposed mechanisms of action are wide-ranging from providing antioxidants to improving the function of the immune response64. However, even when they alter microbial community composition, probiotics do not always offer protection against cancer, and in some models many even enhance tumourigenesis125. This again emphasizes the ultimate importance of understanding each individual’s unique microenvironments when designing interventions.
Microbial replacement therapy or faecal microbial transplantation (FMT) consists of repopulating the lower GI tract of patients with GI pathologies using stools from healthy subjects. This intervention has emerged as a ‘natural’ and promising therapy for patients with relapsing C. difficile infection, irritable bowel syndrome and IBD126. In the case of recurrent C. difficile infection, FMT has shown a 90% clinical success rate127. Following FMT, donor and recipient bacteria strains coexist for more than 90 days, although optimal ecosystem transfer and duration may be dependent on donor-recipient compatibility128. With the growing popularity of FMT, access to donor stools are made easier by nonprofit companies such as OpenBiome, which provides stools from healthy subjects packaged into pills for researchers implicated in clinical trials, as the oral mode of delivery is easier than faecal infusion using colonoscopy, endoscopy or enema. However, as opposed to C. difficile infection, where the disease-causing agent has been identified and is trackable, IBD and CRC are complex diseases resulting from polymicrobial interactions between microorganisms, the host and the environment that evolve over a long period of time. In addition, the presence of a microbial biofilm in these patients may limit FMT efficacy77,78 as these communities are typically resilient to intervention. Indeed, studies on FMT as a remission-inducing approach for IBD has resulted in conflicting reports on its efficacy and safety, and further investigation is needed129,130. The effectiveness of FMT in treating CRC is likewise not yet clear. While the idea of utilizing FMT as a preventive measure65–67, following surgery for remission maintenance or during treatment is intriguing, there is little data to support such incorporation of FMT for cancer management and more studies are urgently needed to evaluate the value of ecosystem transfer in cancer.
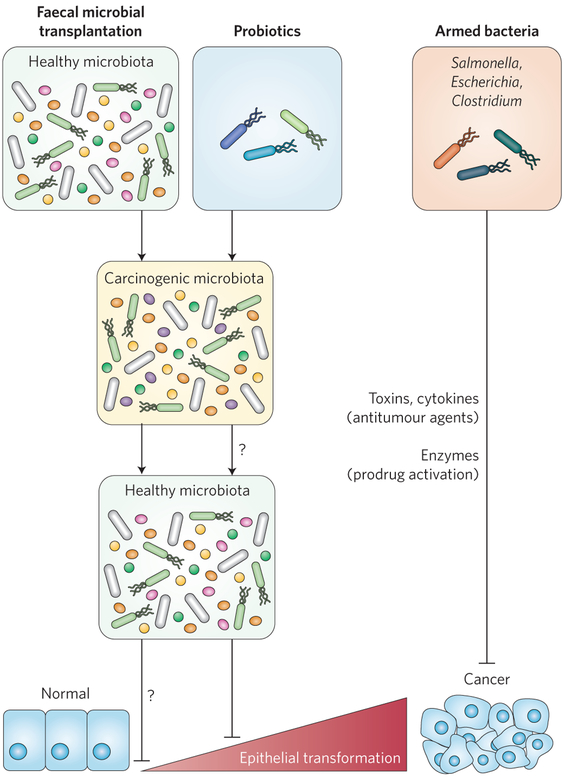
Bacteria could also be directly enlisted in the fight against cancer, either by acting as detecting agents or in the shuttle delivery of specific therapeutics. The fact that bacteria such as Salmonella, Escherichia, Clostridium and Listeria can migrate and penetrate tumour tissues makes them potentially valuable tools for cancer management. The ability of bacteria to detect tumours is probably due to their response to microorganism-specific signals that come from the tumour microenvironment131. Administration of attenuated Salmonella typhimurium VNP20009 showed antitumour efficacy in preclinical models132. In addition, using a ‘tumour-on-a-chip’ system, Panteli et al. showed that attenuated Salmonella engineered to express and release the fluorescent protein ZsGreen was able to detect microscopic solid tumours133. Moreover, E. coli engineered to sense a glucose gradient were able to penetrate deep into microfluidic tumours134, suggesting that these engineered bacteria could be ‘armed’ to deliver therapeutics deep into neoplastic tissues. The armed bacteria could deliver a battery of compounds, ranging from toxins and host signalling molecules (such as TRAIL (TNF-related apoptosis-inducing ligand), Fas, cytokines) to enzymes that selectively activate prodrugs in the tumour tissues, thereby avoiding systemic toxicity135. However, although the phase I trial demonstrated a safety profile for S. typhimurium VNP20009 in patients with metastatic melanoma, no tumour regression was observed in these patients, possibly due to limited accumulation of the bacterium in the tumours136. Nevertheless, similar to oncolytic viruses137, exploiting the unique properties of bacteria may yet prove to be a powerful tool in combatting carcinogenesis (Fig. 4).
Figure 4 ∣. Cancer management through the use of bacteriotherapy.
Potential strategies to enlist bacteria to prevent or treat carcinogenesis include FMT, probiotics and armed bacteria. In FMT, the carcinogenic microbiota from patients is ‘replaced’ by a new, healthy microbiota to eliminate carcinogenic activities. The introductions of probiotics may result in a ‘rebalanced’ microbiota with less potential to cause cancer. Bacteria could also be engineered to deliver specific cargo, such as cell death signalling molecules, toxins or enzymes, to selectively activate antitumour prodrugs in the tumour tissues. Whether probiotic intake results in a healthy microbiota or whether healthy microbiota prevents development of carcinogenesis (question marks) is unclear.
Conclusions
The exciting conceptual models reviewed suggest that the traditional screening and treatment of cancer could be greatly enhanced by monitoring microbial composition and function and manipulating the microbiome. For example, future diagnostics could screen for microbial genes as biomarkers of increased cancer risk. For cancer prevention, the manipulation of the microbiome through microbial replacement, probiotics and/or diet could promote a microbiome that minimized inflammation and carcinogenic activities, thereby reducing cancer risk. The remarkable advances in our understanding of how bacteria interact with host immune response suggest that treatment could be personalized through monitoring the microbiome to determine how individual patients are likely to metabolize individual drugs or ‘pair well’ with given anticancer drugs to enhance efficacy while reducing toxicity. This holistic concept is supported by the recent demonstration of microbiome–cytokine interaction in humans138.
Although there exists a great deal of optimism in the potential for mining the microbiome for cancer management drugging this ecosystem for therapeutic purposes will likely be very challenging. There are probably more genes within the human microbiome than human genes, and while it is enticing to think about new targets and biology within this microbiome, the vast increase in complexity when considering both human and microbial targets makes this a daunting mission. In addition, the literature reports time and time again that the taxonomic composition of the gut microbiome of an individual is highly distinct and, at least in some studies, stable in the face of differences in diet139. For example, a recent study swapped the diet of a rural cohort in Africa and African Americans and found minor changes in microbial community composition, but large changes in mucosal markers of cancer risk, some of which are probably microbial in origin140. This intriguing study suggests that how microorganisms contribute to cancer risk may be determined more by the environments an individual creates for their microbial communities, rather than simple individual differences in community composition. Consistent with this idea, a recent study comparing vegans to non-vegans found robust differences in the metabolome but very few differences in microbial community composition141.
If what we feed our microorganisms and the resulting metabolites they produce is of greater importance to cancer risk than community composition, then studies that examine microbial differences between cancer and healthy subjects using metatranscriptomics or metabolic profiles may generate a deeper understanding of bacterial roles in tumourigenesis than 16S rRNA or metagenomics studies. Moreover, if the environment of the microbial community, rather than just community composition, needs to be changed to impact risk, this may limit the potential of interventions that rely on probiotics to engineer a healthier gut. It may be that features of the microenvironment beyond just the bacteria themselves are transferred in a faecal transfer procedure, and that removing bacteria from their environmental context also strips them of their beneficial behaviours. In addition, interactions between viruses and bacteria are important components of host homeostasis142,143 and efforts in engineering defined bacterial cocktails to promote health may prove to be of limited efficacy. These important questions of what the requirements of engineering a protective gut microflora are will unquestionably receive much attention over the coming years.
In summary, attempts to utilize the microbiome to improve cancer detection, progression and therapy have tremendous potential, but substantial efforts in basic and clinical research that will allow us to better understand the complexity of host–environment interactions will probably be needed before the translational potential of the microbiome is fully realized.
Acknowledgements
C. Jobin acknowledges support from NIH (RO1DK073338, RO1 AT08623 and R21 CA195226) and the University of Florida Department of Medicine Gatorade Fund.
Footnotes
Competing interests
The authors declare no competing financial interest.
References
- 1.Garrett WS Cancer and the microbiota. Science 348, 80–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwabe RF & Jobin C The microbiome and cancer. Nat. Rev. Cancer 13, 800–812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh J-K & Weiderpass E Infection and cancer: global distribution and burden of diseases. Ann. Glob. Health 80, 384–392 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Biedermann L et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS ONE 8, e59260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan XC et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevers D et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mottawea W et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 7, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymond F et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 10, 707–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Keefe SJ Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 13, 691–706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieleman LA et al. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect. Immun. 68, 5107–5113 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuss SK et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334, 249–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B et al. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346, 861–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones MK et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346, 755–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan XC & Huttenhower C Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology 146, 1437–1448 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Grice EA & Segre JA The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet 13, 151–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson GP, Lee SM & Mazmanian SK Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 14, 20–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sender R, Fuchs S & Milo R Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Fujimura KE & Lynch SV Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 17, 592–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scher JU & Abramson SB The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol 7, 569–578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang WH & Hazen SL The contributory role of gut microbiota in cardiovascular disease. J. Clin. Invest 124, 4204–4211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilg H & Kaser A Gut microbiome, obesity and metabolic dysfunction. J. Clin. Invest 121, 2126–2132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wlodarska M, Kostic AD & Xavier RJ An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 17, 577–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF & Pimentel-Nunes P Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev. Esp. Enferm. Dig 107, 659–671 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Vipperla K & O’Keefe SJ The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr. Clin. Pract 27, 624–635 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Parsonnet J et al. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl J. Med 325, 1127–1131 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Wroblewski LE, Peek RM & Wilson KT Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin. Microbiol. Rev 23, 713–739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsugawa H et al. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe 12, 764–777 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Hatakeyama M Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15, 306–316 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Koeppel M, Garcia-Alcalde F, Glowinski F, Schlaermann P & Meyer TF Helicobacter pylori infection causes characteristic DNA damage patterns in human cells. Cell Rep. 11, 1703–1713 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Liu X et al. A systematic review on the association between the Helicobacter pylori vacA i genotype and gastric disease. FEBS Open Bio. 6, 409–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palframan SL, Kwok T & Gabriel K Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front. Cell. Infect. Microbiol 2, 92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jo HJ et al. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 21, 364–374 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Helicobacter and Cancer Collaborative Group. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut 49, 347–353 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamangar F et al. Opposing risks of gastric cardia and noncardia gastric adenocarcinomas associated with Helicobacter pylori seropositivity. J. Natl. Cancer Inst 98, 1445–1452 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Hansen S, Melby KK, Aase S, Jellum E & Vollset SE Helicobacter pylori infection and risk of cardia cancer and non-cardia gastric cancer. A nested case-control study. Scand. J. Gastroenterol 34, 353–360 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Ye W et al. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J. Natl. Cancer Inst 96, 388–396 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Islami F & Kamangar F Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev. Res 1, 329–338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie F-J et al. Helicobacter pylori infection and esophageal cancer risk: an updated meta-analysis. World J. Gastroenterol 19, 6098–6107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wistuba II & Gazdar AF Gallbladder cancer: lessons from a rare tumour. Nat. Rev. Cancer 4, 695–706 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Scanu T et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17, 763–774 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Aran D et al. Widespread parainflammation in human cancer. Genome Biol. 17, 145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum S Why cancer and inflammation? Yale J. Biol. Med 79, 123–130 (2006). [PMC free article] [PubMed] [Google Scholar]
- 43.Coussens LM & Werb Z Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colotta F, Allavena P, Sica A, Garlanda C & Mantovani A Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Itzkowitz SH & Yio X Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am. J. Physiol. Gastrointest. Liver Physiol 287, G7–G17 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Honda K & Littman DR The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi J & Chang EB The gut microbiota and inflammatory bowel diseases. Transl. Res 179, 38–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen-Vercoe E & Jobin C Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol. Lett. 162, 54–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilg H & Moschen AR Food, immunity, and the microbiome. Gastroenterology 148, 1107–119 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Tan J et al. The role of short-chain fatty acids in health and disease. Adv. Immunol 121, 91–119 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Rivera-Chávez F et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh V et al. Microbiota-dependent hepatic lipogenesis mediated by stearoyl CoA desaturase 1 (SCD1) promotes metabolic syndrome in TLR5-deficient mice. Cell Metab. 22, 983–996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belcheva A et al. Gut microbial metabolism drives transformation of MSH2- deficient colon epithelial cells. Cell 158, 288–299 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Winter SE, Lopez CA & Bäumler AJ The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14, 319–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louis P, Hold GL & Flint HJ The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol 12, 661–672 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Wynendaele E et al. Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 64, 40–48 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Pevsner-Fischer M et al. Role of the microbiome in non-gastrointestinal cancers. World J. Clin. Oncol 7, 200–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marchesi JR et al. Towards the human colorectal cancer microbiome. PLoS ONE 6, e20447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flemer B et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut http://doi.org/bx3c (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakatsu G et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 6, 8727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Q et al. Gut microbiome development along the colorectal adenoma- carcinoma sequence. Nat. Commun 6, 6528 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Gao Z, Guo B, Gao R, Zhu Q & Qin H Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol 6, 20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu Y et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 6, 26337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu Y, Michelle Luo T, Jobin C & Young HA Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 309, 119–127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zackular JP, Rogers MAM, Ruffin MT & Schloss PD The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res 7, 1112–1121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narayanan V, Peppelenbosch MP & Konstantinov SR Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev. Res 7, 1108–1111 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Zeller G et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol 10, 766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasolli E, Truong DT, Malik F, Waldron L & Segata N Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput. Biol 12, e1004977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhernakova A et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodrich JK et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arthur JC et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun 5, 4724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moschen AR et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 19, 455–469 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Zackular JP et al. The gut microbiome modulates colon tumorigenesis. mBio 4, e00692–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagao-Kitamoto H et al. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol. Gastroenterol. Hepatol 2, 468–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baxter NT, Zackular JP, Chen GY & Schloss PD Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2, 20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhan Y et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 73, 7199–7210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dejea CM et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA 111, 18321–18326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swidsinski A, Weber J, Loening-Baucke V, Hale LP & Lochs H Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin.Microbiol 43, 3380–3389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Macfarlane S & Dillon JF Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol 102, 1187–1196 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Babbar N & Gerner EW Targeting polyamines and inflammation for cancer prevention. Recent Results Cancer Res. 188, 49–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson CH et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21, 891–897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomkovich S et al. Human colorectal cancer-associated biofilms promote tumorigenesis in susceptible mice. Gastroenterology 150, S77 (2016). [Google Scholar]
- 83.Hay M, Thomas DW, Craighead JL, Economides C & Rosenthal J Clinical development success rates for investigational drugs. Nat. Biotechnol 32, 40–51 (2014). [DOI] [PubMed] [Google Scholar]
- 84.ElRakaiby M et al. Pharmacomicrobiomics: the impact of human microbiome variations on systems pharmacology and personalized therapeutics. OMICS 18, 402–414(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pardoll DM The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller JFAP & Sadelain M The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer Cell 27, 439–449 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Khan H, Gucalp R & Shapira I Evolving concepts: immunity in oncology from targets to treatments. J. Oncol 2015, 847383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nicodemus CF Antibody-based immunotherapy of solid cancers: progress and possibilities. Immunotherapy 7, 923–939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gelao L, Criscitiello C, Esposito A, Goldhirsch A & Curigliano G Immune checkpoint blockade in cancer treatment: a double-edged sword cross-targeting the host as an “innocent bystander”. Toxins 6, 914–933 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abdel-Wahab N, Shah M & Suarez-Almazor ME Adverse events associated with immune checkpoint blockade in patients with cancer: a systematic review of case reports. PLoS ONE 11, e0160221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vétizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dubin K et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun 7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mazmanian SK, Liu CH, Tzianabos AO & Kasper DL An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Chu H et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sivan A et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zitvogel L, Ayyoub M, Routy B & Kroemer G Microbiome and anticancer immunosurveillance. Cell 165, 276–287 (2016). [DOI] [PubMed] [Google Scholar]
- 97.McCarthy EF The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J 26, 154–158 (2006). [PMC free article] [PubMed] [Google Scholar]
- 98.Shirota H & Klinman DM Recent progress concerning CpG DNA and its use as a vaccine adjuvant. Expert Rev. Vaccines 13, 299–312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iida N et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muggia FM, Dimery I & Arbuck SG Camptothecin and its analogs An overview of their potential in cancer therapeutics. Ann. N. Y. Acad. Sci 803, 213–223 (1996). [DOI] [PubMed] [Google Scholar]
- 101.Nagar S & Blanchard RL Pharmacogenetics of uridine diphosphoglucuronosyltransferase (UGT) 1A family members and its role in patient response to irinotecan. Drug Metab. Rev 38, 393–409 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Wallace BD et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roberts AB, Wallace BD, Venkatesh MK, Mani S & Redinbo MR Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 84, 208–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wallace BD et al. Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol 22, 1238–1249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paci A et al. Review of therapeutic drug monitoring of anticancer drugs part 1--cytotoxics. Eur. J. Cancer 50, 2010–2019 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Frank M et al. TLR signaling modulates side effects of anticancer therapy in the small intestine. J. Immunol 194, 1983–1995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Touchefeu Y et al. Systematic review: the role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis - current evidence and potential clinical applications. Aliment. Pharmacol. Ther 40, 409–421 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Ferreira MR, Muls A, Dearnaley DP & Andreyev HJ Microbiota and radiation-induced bowel toxicity: lessons from inflammatory bowel disease for the radiation oncologist. Lancet Oncol. 15, 139–147 (2014). [DOI] [PubMed] [Google Scholar]
- 109.Ciorba MA, Hallemeier CL, Stenson WF & Parikh PJ Probiotics to prevent gastrointestinal toxicity from cancer therapy: an interpretive review and call to action. Curr. Opin. Support Palliat. Care 9, 157–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emadi A, Jones RJ & Brodsky RA Cyclophosphamide and cancer: golden anniversary. Nat. Rev. Clin. Oncol 6, 638–647 (2009). [DOI] [PubMed] [Google Scholar]
- 111.Viaud S et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chaney SG, Campbell SL, Bassett E & Wu Y Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol. Hematol 53, 3–11 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Nicholson JK et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Lehouritis P et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep 5, 14554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Masood MI, Qadir MI, Shirazi JH & Khan IU Beneficial effects of lactic acid bacteria on human beings. Crit. Rev. Microbiol 37, 91–98 (2011). [DOI] [PubMed] [Google Scholar]
- 116.Rafter JJ The role of lactic acid bacteria in colon cancer prevention. Scand. J. Gastroenterol 30, 497–502 (1995). [DOI] [PubMed] [Google Scholar]
- 117.Yu A-Q & Li L The potential role of probiotics in cancer prevention and treatment. Nutr. Cancer 68, 535–544 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Zhong L, Zhang X & Covasa M Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J. Gastroenterol 20, 7878–7886 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gao Z et al. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol. Med. Rep 12, 6119–6127 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Chapman CMC, Gibson GR & Rowland I In vitro evaluation of single- and multi-strain probiotics: inter-species inhibition between probiotic strains, and inhibition of pathogens. Anaerobe 18, 405–413 (2012). [DOI] [PubMed] [Google Scholar]
- 121.Chapman CMC, Gibson GR & Rowland I Health benefits of probiotics: are mixtures more effective than single strains? Eur. J. Nutr 50, 1–17 (2011). [DOI] [PubMed] [Google Scholar]
- 122.So SS, Wan ML & El-Nezami H Probiotics-mediated suppression of cancer. Curr. Opin. Oncol 29, 62–72 (2017). [DOI] [PubMed] [Google Scholar]
- 123.Khan AA, Khurshid M, Khan S & Alshamsan A Gut microbiota and probiotics: current status and their role in cancer therapeutics. Drug Dev. Res 74, 365–375 (2013). [Google Scholar]
- 124.dos Reis SA et al. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr. Res 37, 1–19 (2017). [DOI] [PubMed] [Google Scholar]
- 125.Arthur JC et al. VSL#3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci. Rep 3, 2868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smits LP, Bouter KE, de Vos WM, Borody TJ & Nieuwdorp M Therapeutic potential of fecal microbiota transplantation. Gastroenterology 145, 946–953 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Kassam Z, Lee CH, Yuan Y & Hunt RH Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol 108, 500–508 (2013). [DOI] [PubMed] [Google Scholar]
- 128.Li SS et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352, 586–589 (2016). [DOI] [PubMed] [Google Scholar]
- 129.Grinspan AM & Kelly CR Fecal microbiota transplantation for ulcerative colitis: not just yet. Gastroenterology 149, 15–18 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Kelly CR et al. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 149, 223–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Forbes NS Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Luo X et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol. Res. 12, 501–508 (2001). [DOI] [PubMed] [Google Scholar]
- 133.Panteli JT, Forkus BA, Van Dessel N & Forbes NS Genetically modified bacteria as a tool to detect microscopic solid tumor masses with triggered release of a recombinant biomarker. Integr. Biol. (Camb) 7, 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Panteli JT & Forbes NS Engineered bacteria detect spatial profiles in glucose concentration within solid tumor cell masses. Biotechnol. Bioeng 113, 2474–2484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Van Dessel N, Swofford CA & Forbes NS Potent and tumor specific: arming bacteria with therapeutic proteins. Ther. Deliv 6, 385–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Toso JF et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol 20, 142–152 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zwiebel JA Cancer gene and oncolytic virus therapy. Semin. Oncol 28, 336–343 (2001). [DOI] [PubMed] [Google Scholar]
- 138.Schirmer M et al. Linking the human gut microbiome to inflammatory cytokine production capacity. 167, 1–21 (2016). [DOI] [PubMed] [Google Scholar]
- 139.Winglee K & Fodor AA Intrinsic association between diet and the gut microbiome: current evidence. Nutr. Diet. Suppl 7, 69–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O’Keefe SJD et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun 6, 6342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu GD et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65, 63–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang J-Y et al. Enteric viruses ameliorate gut inflammation via Toll-like receptor 3 and Toll-like receptor 7-mediated interferon-β production. Immunity 44, 889–900 (2016). [DOI] [PubMed] [Google Scholar]
- 143.Manrique P et al. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113, 10400–10405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vogtmann E & Goedert JJ Epidemiologic studies of the human microbiome and cancer. Br. J. Cancer 114, 237–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Meurman JH Oral microbiota and cancer. J. Oral Microbiol 2, 5195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang L & Ganly I The oral microbiome and oral cancer. Clin. Lab. Med 34, 711–719 (2014). [DOI] [PubMed] [Google Scholar]
- 147.Guerrero-Preston R et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget 7, 51320–51334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pushalkar S et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 12, 144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schmidt BL et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE 9, e98741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Flynn KJ, Baxter NT & Schloss PD Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere 1, e00102–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michaud DS & Izard J Microbiota, oral microbiome, and pancreatic cancer. Cancer J 20, 203–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Farrell JJ et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ahn J, Chen CY & Hayes RB Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 23, 399–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hosgood HD et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen 55, 643–651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gui Q-F, Lu H-F, Zhang C-X, Xu Z-R & Yang Y-H Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet. Mol. Res 14, 5642–5651 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Yu G et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 17, 163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yan X et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 5, 3111–3122 (2015). [PMC free article] [PubMed] [Google Scholar]
- 158.Hieken TJ et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci. Rep 6, 30751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Urbaniak C et al. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82, 5039–5048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xuan C et al. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 9, e83744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lakritz JR et al. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget 6, 9387–9396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Roderburg C & Luedde T The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes 5, 441–445 (2014). [DOI] [PubMed] [Google Scholar]
- 163.Huang Y et al. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J. Clin. Pathol 57, 1273–1277 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rocha M et al. Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut 54, 396–401 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dapito DH et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer cell 21, 504–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang H-L et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J. Hepatol 57, 803–812 (2012). [DOI] [PubMed] [Google Scholar]