Abstract
Objective:
This article describes a CDI outbreak in a long-term care (LTC) facility that used molecular typing techniques and whole-genome sequencing to identify widespread dissemination of the clonal strain in the environment which was successfully removed after terminal cleaning.
Setting:
This study was conducted in a long-term care facility in Texas.
Methods:
A recently hospitalized LTC patient was diagnosed with CDI followed shortly thereafter by 7 subsequent CDI cases. A stool specimen was obtained from each patient for culturing and typing. An environmental point-prevalence study of the facility was conducted before and after terminal cleaning of the facility to assess environmental contamination. Cultured isolates were typed using ribotyping, multilocus variant analysis, and whole-genome sequencing.
Results:
Stool samples were available for 5 of 8 patients; of these specimens, 4 grew toxigenic C. difficile ribotype 027. Of 50 environmental swab samples collected throughout the facility prior to the facility-wide terminal cleaning, 19 (38%) grew toxigenic C. difficile (most commonly ribotype 027, 79%). The terminal cleaning was effective at reducing C. difficile spores in the environment and at eradicating the ribotype 027 strain (P < .001). Using multilocus variance analysis and whole-genome sequencing, clinical and environmental strains were highly related and, in some cases, were identical.
Conclusion:
Using molecular typing techniques, we demonstrated reduced environmental contamination with toxigenic C. difficile and the eradication of a ribotype 027 clone. These techniques may help direct infection control efforts and decrease the burden of CDI in the healthcare system.
Clostridioides (formerly Clostridium) difficile, a gram-positive, spore-forming, anaerobic bacteria, is the most common cause of healthcare-associated infection in the United States.1 The spores of C. difficile are highly resistant to a wide range of disinfectants and can persist for long periods in many different environments. Previously, our group showed that C. difficile can be isolated from areas within and outside of the hospital regardless of proximity to a CDI patient.2 Thus, prevention and control of outbreaks or hyperendemic CDI likely requires an in-depth knowledge of the molecular epidemiology of the strains throughout the continuum of care. Thus, careful monitoring of patients outside of the acute-care hospital presents an important challenge for infection control.3,4 Outbreaks of CDI are recognized in hospitals and, to a lesser extent, in extended healthcare centers such as long-term care (LTC) facilities.5,6 The purposes of this study were (1) to describe a CDI outbreak in a LTC facility that used molecular typing techniques and whole-genome sequencing to identify the dissemination of the clonal strain in the environment and (2) to describe the effects of terminal cleaning.
Methods
Clinical setting and case definition
The outbreak occurred at a 146-bed LTC facility in Texas that provides nursing care and rehabilitation services to patients with chronic medical conditions. The facility had not previously had any outbreaks or recent cases of CDI.
During the outbreak investigation, a CDI case was defined as a resident with new onset of diarrhea as well as a positive diagnostic test for C. difficile toxins using a commercially available C. difficile toxin a/b EIA test.
Environmental decontamination and sampling
We hypothesized that environmental contamination outside the patient’s room contributed to the outbreak of cases throughout the facility. To test the hypothesis that C. difficile spores were likely disseminated throughout the facility, 50 surface samples of 930 cm2 (~1 square foot) were swabbed with presterilized cotton gauze moistened with sterile water. Samples were placed into a 50-mL presterilized tube and transported to the laboratory within 12 hours of collection. For each batch of 10 swabs, a negative control (ie, an unused swab) was included to ensure no cross contamination of swabs. To assess environmental contamination in patients’ rooms compared to communal areas in the facility, all patient rooms were swabbed (n = 30 swabs), with an additional 20 swabs taken from elsewhere in the facility. If the results of the initial environmental sampling confirmed the hypothesis of widespread spore dissemination in the environment, the entire facility was scheduled to undergo a terminal clean with a 10% bleach solution. The terminal clean was performed by trained facility staff with the guidance of the state health department. Environmental sampling of the facility was repeated 6 months later in the same patient rooms and communal areas to test for continued presence of C. difficile.
Microbiologic procedures
Stool and environmental samples were enriched in brain heart infusion (BHI) broth with 0.05% sodium taurocholate (Sigma Chemicals, St Louis, MO) and incubated anaerobically at 37°C for up to 3 days as previously described.2 From each sample, 10mL broth culture was centrifuged to concentrate the cells with the resulting pellet suspended in 100 μL of normal saline (0.85% NaCl), plated onto cycloserine cefoxitin fructose agar (CCFA, Anaerobic Systems, Morgan Hill, CA), and incubated anaerobically at 37°C for 40–48 hours (Forma Anaerobic System, Mode 1025/1029). Suspected colonies were tested using latex agglutination reagent (Oxoid, Hampshire, UK). Each batch of samples was processed with a positive and a negative control. The presence of toxin genes was assessed using multiplex polymerase chain reaction (PCR) to detect the presence of toxin A (tcdA), toxin B (tcdB), and tpi genes.7
Clostridioides difficile ribotyping
Fluorescent ribotyping was performed as previously described.8,9 Briefly, PCR ribotyping primers10 were synthesized with a fluorescent label (Life Technologies, Carlsbad, CA) and adjusted to 10 pmol/μL. A 25 μL PCR was performed using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) and the following conditions: 95°C (10 minutes); 35 cycles of 95°C (30 seconds), 55°C (30 seconds), and 72°C (1 minute 30 seconds); and a final extension of 72°C (10 minutes) (Eppendorf vapo-protect thermal cycler, Thermo Scientific, Waltham, MA). Amplicons were analyzed using an ABI3730xl DNA Analyzer and MapMaker 1000 ROX DNA sizing standard (BioVentures, Murfreesboro, TN). This technique does not distinguish between ribotypes 053 and 163, ribotypes 014 and 020, and ribotypes 078 and 126; therefore, these are reported as combined ribotypes (ie, 053-163, 014-020, and 078-126).
Clostridioides difficile multilocus variance analysis
The multilocus variance analysis (MLVA) typing of C. difficile isolates ribotype 027 was performed using a previously published fluorescent MLVA markers method11 described by Broukhanski et al.12,13 Briefly, amplicons are diluted in formamide, mixed with LIZ600 DNA ladder and run on an ABI 3130xl genetic analyzer (Applied Biosystems) for fragment analysis. Processing of capillary electrophoresis was done using BioNumerics version 7.6 software (Applied Maths, Austin, TX). Peak files from the genetic analyzer were normalized to the LIZ600 DNA ladder to determine amplicon size and to calculate the summed tandem-repeat difference (STRD). The copy number for each variable number of tandem repeats (VNTR) locus based on amplicon size was determined using previously published guidelines.12 Complexes containing >2 isolates whose MLVA genotypes generated an STRD <2 were defined as highly related isolates.4
Whole-genome sequencing
For whole-genome single-nucleotide polymorphism (SNP) analysis, cleaned sequence reads were mapped to the R20291 reference genome (GenBank accession no. FN545816) using the RedDog pipeline (https://github. com/katholt/RedDog) according to the developer’s previous guidelines. Briefly, Bowtie2 version 2.2.3 software14 was used for mapping, and SAMtools version 0.1.19 software15 was used for calling SNPs. Only high-quality SNPs were used for phylogenetic analyses. Phylogenetic trees were created in FigTree and edited using iTOL (https://itol.embl.de/). Transmission analyses were carried out as described previously.16 The ARG-ANNOT database was used to identify antimicrobial resistance genes.17
Results
Description of outbreak
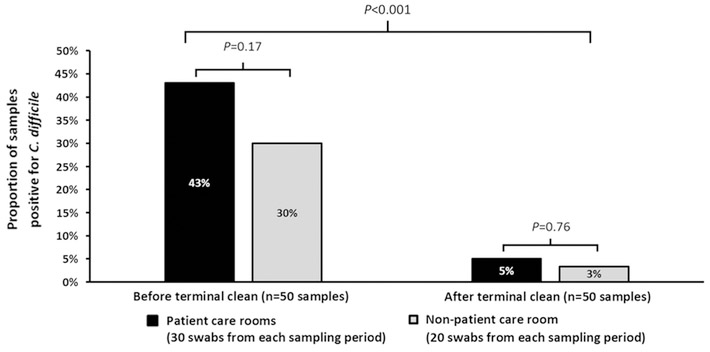
In September 2016, a 90-year-old male recently discharged from an acute-care facility was diagnosed with CDI after a new onset of diarrhea with abdominal pain. Despite terminal cleaning with a 10% bleach sporicidal solution in rooms with active cases, a total of 7 additional residents became symptomatic and tested positive for C. difficile toxins over the next 2 months. A description of the 8 patients who acquired CDI during the period at the facility is provided in Table 1. Stool specimens were available from 5 patients for C. difficile growth and typing. Of the 5 stool specimens, 4 grew toxigenic C. difficile: all 4 were ribotype 027. Also, 50 environmental swab samples were collected prior to the facility-wide terminal cleaning; 14 of these grew toxigenic C. difficile (28%). Of these 14 isolates, 11 (79%) were ribotype 027. Toxigenic C. difficile spores were disseminated throughout the facility; they were present in the patient care environment (43%) and in non-patient care areas (30%; P = 0.17). Environmental contamination was disseminated widely throughout the institution, including bed handrails, television remote control, doorway entrances, shower seat surface, wheelchair arms, toilet hand rails, bedside table, sink surfaces, physical therapy grip handrail, dining room table top, and communal shower chairs. These results prompted a facility-wide terminal cleaning, which was effective at decreasing contamination by C. difficile spores and eradicating the ribotype 027 strain from the environment (Fig. 1). After the terminal cleaning, the environmental swabbing of the facility was repeated. Of 50 environmental swab samples, 2 grew toxigenic C. difficile (1 swab each from a patient-care area and a non–patient care area). Neither of the 2 C. difficile isolates were ribotype 027. The MLST was not able to differentiate the isolates (all were ST-1); therefore, MLVA was conducted to provide more discrimination. The MLVA typing was successful in 11 environmental ribotype 027 isolates and 2 of the 4 ribotype 027 clinical isolates. Both clinical strains were highly related to each other, and 9 of the 11 environmental isolates were considered highly related to the clinical isolates (Table 2).
Table 1.
Description of Clostridioides difficile Infection (CDI) Cases
| Variable | No. |
|---|---|
| No. of patients | 8 |
| Males | 3 |
| Females | 5 |
| Age, y mean ± SD | 8 ±12 |
| Patients with CDI, no. (%) | 8 (100) |
| Treatment | |
| Vancomycin monotherapy | 1 |
| Vancomycin and metronidazole given sequentially | 3 |
| Vancomycin and metronidazole given together | 4 |
| Receiving a systemic non–C. difficile antibiotic at the time of CDI diagnosis | 3 |
| Expired within 60 d of onset, no. (%) | 2 (25) |
Note. SD, standard deviation.
Fig. 1.
Proportion of positive environmental samples before and after facility-wide terminal clean with 10% bleach solution. Plotted on the x-axis is the time point before and after terminal clean and the y-axis shows the percentage of samples that were positive for C. difficile. There was a significant difference in percentage of specimens positive for C. difficile between patient care rooms (black bars) and non-patient care areas at both time points. Terminal cleaning significantly reduced the C. difficile burden overall.
Table 2.
MLVA Typing of Ribotype 027 Clinical and Environmental Clostridioides difficile Isolates
| Source | Ribotype | A6Cd | B7Cd | G8Cd | C6Cd | E7Cd | STRD | StrainID |
|---|---|---|---|---|---|---|---|---|
| Clinical | F027 | 16 | 16 | 15 | 36 | 8 | Index | FCH 4 |
| Clinical | F027 | 17 | 16 | 15 | 0 | FCH 2 | ||
| Environ | F027 | 17 | 16 | 15 | 0 | LTC 10 | ||
| Environ | F027 | 17 | 16 | 15 | 0 | LTC 14 | ||
| Environ | F027 | 17 | 16 | 15 | 36 | 8 | 0 | LTC 15 |
| Environ | F027 | 16 | 16 | 15 | 0 | LTC 36 | ||
| Environ | F027 | 16 | 16 | 15 | 36 | 8 | 1 | LTC 39A |
| Environ | F027 | 17 | 16 | 15 | 1 | LTC 5A | ||
| Environ | F027 | 17 | 16 | 15 | 1 | LTC 7B | ||
| Environ | F027 | 36 | 8 | 0 | LTC 15B | |||
| Environ | F027 | 17 | 16 | 15 | 34 | 9 | 3 | LTC 19 |
| Environ | F027 | 16 | 16 | 15 | 36 | 8 | 4 | LTC 39 |
| Environ | F027 | 17 | 16 | 15 | 0 | LTC7B |
Note. Environ, environmental isolate.
To determine the extent to which the isolates were highly related, whole-genome sequencing was carried out. In total, 17 isolates were sequenced, including 3 clinical and 14 environmental isolates. Sequencing produced >10 million high-quality sequencing reads per isolate, which corresponded to >20 times the depth of coverage. Reads were mapped to the R20291 reference core genome, and SNPs were identified. All isolates were >99.999% genetically similar and only differed at 9 SNP sites, which included 4 intergenic mutations, 1 synonymous substitution, and 2 nonsynonymous substitutions. The 3 clinical strains varied at a total of 4 SNP sites; the clinical strain FCH-1 was different from another clinical strain FCH-2 by 2 SNPs and was different from clinical strain FCH-4 by 4 SNPs (Table 3). All of the strains were of the fluoroquinolone resistant 2 (FQR2) lineage.18 In addition to fluoroquinolone resistance, 2 strains (clinical strain FCH-4 and environmental strain LTC-7B) contained blaTEM-1D encoding a β-lactamase.
Table 3.
Whole-Genome Sequencing SNPs and Annotation
| Position | Gene | Product | Change | Ref | FCH-1 | LTC 15B |
LTC 15 |
LTC36 | LTC 36 |
LTC 39A |
FCH 2 |
LTC 10 |
LTC 15A |
LTC 44 |
LTC 5A |
LTC 7B |
FCH 4 |
LTC 14 |
LTC 14 |
LTC 19 |
LTC 39 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 120932 | CDR20291_0096 | DNA-directed RNA polymerase alpha chain | Synonymous | C | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 132939 | Intergenic | G | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | ||
| 132955 | Intergenic | C | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||
| 132958 | Intergenic | T | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | ||
| 132959 | Intergenic | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | ||
| 490091 | Intergenic | G | G | G | G | G | G | G | T | T | G | G | G | G | G | G | G | G | G | ||
| 490092 | Intergenic | T | T | T | T | T | T | T | C | C | T | T | T | T | T | T | T | T | T | ||
| 630219 | CDR20291_0515 | Hypothetical protein | Synonymous | G | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 685120 | Intergenic | A | A | A | A | A | A | A | G | G | G | G | G | G | A | A | A | A | A | ||
| 685442 | CDR20291_0565 | Two-component response regulator | Synonymous | C | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 737734 | CDR20291_0596 | Conserved hypothetical protein | Non-synonymous | G | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 904356 | CDR20291_0735 | Electron transfer flavoprotein beta-subunit | Non-synonymous | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| 1116866 | CDR20291_0901 | Putative membrane protein (pseudogene) | Non-synonymous | G | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 1138198 | CDR20291_0925 | Hypothetical protein | Non-synonymous | C | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 1411909 | Intergenic | G | G | G | G | G | G | G | G | G | G | G | G | G | T | G | G | G | G | ||
| 1547553 | CDR20291_1308 | Putative 5-nitroimidazole reductase | Non-synonymous | T | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 1592813 | Intergenic | A | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | ||
| 1662523 | CDR20291_1406 | Putative peptidyl-prolyl isomerase | Non-synonymous | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | T | C |
| 1713234 | CDR20291_1451 | Putative phage cell wall hydrolase | Non-synonymous | T | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 1865639 | CDR20291_1579 | TetR-family transcriptional regulator | Non-synonymous | T | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 1866352 | Intergenic | T | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||
| 1876233 | CDR20291_1593 | Putative arsenical pump membrane protein | Non-synonymous | A | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| 2061408 | CDR20291_1766 | transcription regulator (yobd protein) | Synonymous | C | C | C | C | C | C | A | C | C | C | C | C | C | C | C | C | C | C |
| 2076479 | Intergenic | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | ||
| 2160266 | CDR20291_1848 | Putative peptidase | Non-synonymous | T | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 2247604 | CDR20291_1924 | Putative uncharacterized protein | Non-synonymous | G | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 2297364 | CDR20291_1968 | Conserved hypothetical protein | Synonymous | A | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 2297662 | CDR20291_1968 | Conserved hypothetical protein | Non-synonymous | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| 2322226 | CDR20291_1987 | Putative FAD-binding subunit of xanthine dehydrogenase | Synonymous | G | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 2361948 | Intergenic | C | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||
| 2387569 | CDR20291_2036 | Putative membrane-associated protease | Non-synonymous | C | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 2576495 | Intergenic | C | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | ||
| 2582507 | Intergenic | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | ||
| 2976764 | CDR20291_2541 | UDP-N-acetylmuramoylalanine-D-glutamate ligase | Non-synonymous | G | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 3055365 | CDR20291_2602 | UDP-glucose 4-epimerase | Non-synonymous | T | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 3057112 | CDR20291_2605 | Hypothetical protein | Non-synonymous | T | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 3113416 | CDR20291_2644 | PTS system, phosphocarrier protein | Non-synonymous | G | G | G | G | G | G | G | G | G | G | G | G | G | A | G | G | G | G |
| 3164340 | CDR20291_2682 | Cell surface protein (S-layer precursor protein) | Non-synonymous | C | C | C | C | C | C | C | T | T | C | C | C | C | C | C | C | C | C |
| 3531583 | CDR20291_2969 | PTS system, IIabc component | Non-synonymous | C | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| 3715797 | Intergenic | T | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | ||
| 4104562 | CDR20291_3456 | Putative uncharacterized protein | Synonymous | G | G | G | G | G | G | G | G | G | G | G | G | G | G | A | A | G | G |
| 4112001 | CDR20291_3464 | Conjugative transposon protein | Non-synonymous | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | G |
| 4147900 | CDR20291_3497 | Hypothetical protein | Synonymous | A | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
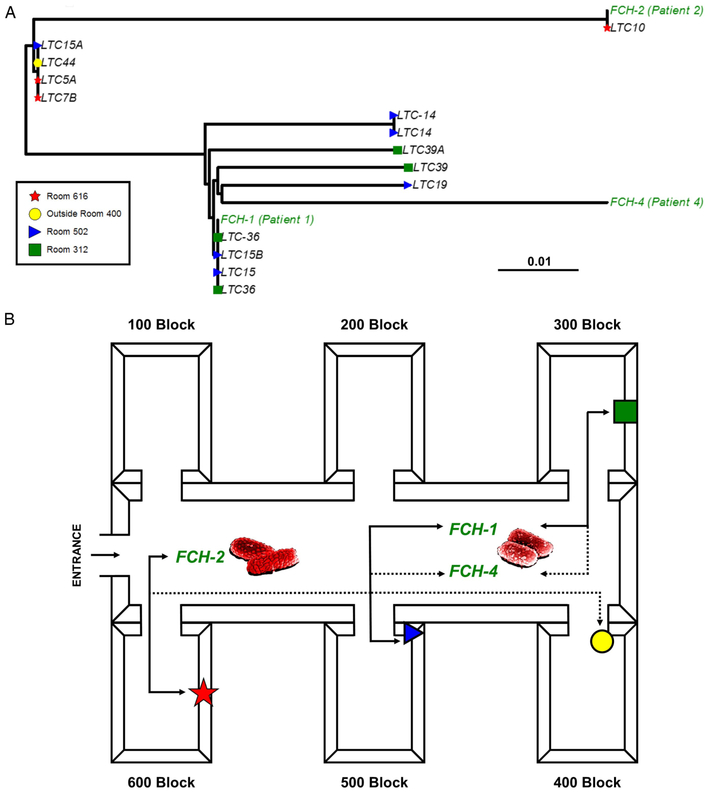
To demonstrate the overall genetic relatedness, a maximum likelihood phylogenetic tree was made from 44 SNP sites (Fig. 2A). As a proof of concept, 2 colonies each were isolated from 2 different isolates (LTC-36 and LTC-14), were sequenced in duplicate, and were identical. Certain environmental isolates were 100% genetically identical to the clinical isolates (Fig. 2A). Using epidemiological data with the sequencing results, transmission analyses were performed on all the isolates. Because chronology between clinical cases and environmental contamination was unclear, a conjectural transmission diagram was created from the isolation location and the SNP analysis (Fig. 2B). Because there was only a 2 SNP difference between FCH-1 and FCH-4, these strains were likely derived from a common source, either the 300 block or the 500 block of rooms. These data provide strong evidence that there was transmission between the patient and the LTC facility environment.
Fig. 2.
Maximum likelihood phylogeny and CDI transmission map. (A) The phylogenetic tree was made from 44 SNP sites and is midpoint rooted. Patient isolates are colored in green font and environmental samples in other colors based on location. The bar at the bottom of Fig. 2a (0.01) refers to degree of relatedness of isolates. (B) Using the epidemiological data with the phylogeny, a suspected transmission map was made of the long-term care facility. A solid line indicates an isolate shared a direct contact and was genetically identical to another isolate, whereas the dotted line indicates a suspected connection to another isolate. Red spore indicates a patient isolate. For example, clinical isolate FCH-2 was genetically identical to an environmental isolate from the 600 block of the LTC that was highly similar but not identical to an environmental isolate from the 400 block.
Discussion
Residents of LTC facilities have been recognized to be at risk for CDI due to a variety of factors including advanced age, administration of systemic antibiotics, and high rates of comorbid conditions.3 Hospitalized patients developing CDI are more likely to be discharged to LTC facilities than are other hospitalized patient populations.19 Two previous outbreaks of CDI due to ribotype 027 has been reported in residents of LTC facilities.20,21 However, whole-genome sequencing and subsequent molecular epidemiology of the outbreak were not conducted in these previous studies. In the current study, we used molecular typing techniques to identify a highly related, clonal outbreak of C. difficile ribotype 027. The clone was widely disseminated throughout the facility; it was present in patient rooms as well as other areas of the facility. The facility, guided by the results of an environmental contamination investigation, was able to eradicate the ribotype 027 from the environment and to reduce the contamination of C. difficile spores in the environment. The molecular typing was accomplished using high-throughput, cost-effective techniques to start (ribotyping) followed by more discriminatory typing techniques (MVLA and WGS) to confirm the relatedness of the strains. Using these tools, we were able to demonstrate clonal dissemination of ribotype 027 throughout the facility as well as successful eradication of the clone from the facility. Finally, using whole-genome sequencing, a potential route of transmission of isolates throughout the facility was generated and provided insight into the transmission of C. difficile between patients and the environment.
Although uncommon, use of environmental sampling to guide decontamination efforts has been reported.22 A tertiary-care referral center in England sampled 16 different sites within the hospital to guide terminal cleaning with either a sporicidal agent or aerial hydrogen peroxide. Although persistence in the environment was observed, a decrease in the number of positive C. difficile isolates was observed. A typing method was not used in this study. We have previously demonstrated that toxigenic C. difficile spores are present in the healthcare and non–healthcare environments, including shoe bottoms.2 It is possible that sites that underwent terminal cleaning became recontaminated from another source. This example demonstrates the value of typing in addition to culture to demonstrate removal of the original clone. For example, in our study, 2 positive C. difficile cultures were identified after terminal cleaning. Using ribotyping, we were able to demonstrate that these cultures were not from the original ribotype 027 clone present in the LTC facility.
This study has several limitations. Although we were able to demonstrate the effectiveness of our terminal clean to remove the ribotype 027 clone from the environment, we cannot infer the causality that this intervention interrupted the ongoing outbreak. However, because the isolates were identical between the environmental and clinical strains, it is possible. We were not able to perform MLVA analysis on 2 of the ribotype 027 clinical isolates, and we did not get deep enough sequence reads for a single ribotype 027 isolate for whole-genome sequencing analysis. Movement of patients throughout the facility made more detailed epidemiologic studies to correlate patient movement with spore dissemination difficult. This CDI outbreak was the first ever at this institution. Whether this guided decontamination technique would work at other healthcare settings with prior cases of CDI or higher rates of C. difficile contamination with more diverse ribotypes will require further study.
In conclusion, using a variety of molecular typing techniques, we were able to demonstrate reduced environmental contamination of toxigenic C. difficile spores and eradication of a ribotype 027 clone. Applying this technique to other healthcare centers may help to decrease the burden of CDI in the healthcare system.
Acknowledgments.
We would like to thank the University of Houston Seq-N-Edit core for help with sequencing. We would like to thank the University of Houston Center for Advanced Computing and Data Science, specifically Peggy Lindner, for assistance and support with the bioinformatics analyses.
Financial support. This work was supported by research grants from the Texas Department of State Health Services and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH NIAID grant no. 1UO1 AI-24290-01).
Footnotes
Conflicts of interest. All authors report no conflicts of interest.
References
- 1.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015;372:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam MJ, Walk ST, Endres BT, et al. Community environmental contamination of toxigenic Clostridium difficile. Open Forum Infect Dis 2017;4:ofx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra T, Goldstein EJ. Clostridium difficile infection in long-term care facilities: a call to action for antimicrobial stewardship. Clin Infect Dis 2015;60 Suppl 2:S72–S76. [DOI] [PubMed] [Google Scholar]
- 4.Curry SR, Muto CA, Schlackman JL, et al. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis 2013;57:1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziakas PD, Joyce N, Zacharioudakis IM, et al. Prevalence and impact of Clostridium difficile infection in elderly residents of long-term care facilities, 2011: a nationwide study. Medicine (Baltimore) 2016;95:e4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wust J, Sullivan NM, Hardegger U, Wilkins TD. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J Clin Microbiol 1982;16:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemee L, Dhalluin A, Testelin S, et al. Multiplex PCR targeting TPI (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 2004;42:5710–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinson JN, Broadaway S, Lohman E, et al. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. J Clin Microbiol 2015;53:1192–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam MJ, Anu A, Walk ST, Garey KW. Investigation of potentially pathogenic Clostridium difficile contamination in household environs. Anaerobe 2014;27:31–33. [DOI] [PubMed] [Google Scholar]
- 10.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 1999;175:261–266. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg RJ, Kuijper EJ, van Coppenraet LE, Claas EC. Rapid diagnosis of toxinogenic Clostridium difficile in faecal samples with internally controlled real-time PCR. Clin Microbiol Infect 2006; 12:184–186. [DOI] [PubMed] [Google Scholar]
- 12.Broukhanski G, Low DE, Pillai DR. Modified multiple-locus variable-number tandem-repeat analysis for rapid identification and typing of Clostridium difficile during institutional outbreaks. J Clin Microbiol 2011;49:1983–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broukhanski G, Simor A, Pillai DR. Defining criteria to interpret multilocus variable-number tandem repeat analysis to aid Clostridium difficile outbreak investigation. J Med Microbiol 2011;60:1095–1100. [DOI] [PubMed] [Google Scholar]
- 14.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar N, Miyajima F, He M, et al. Genome-based infection tracking reveals dynamics of Clostridium difficile transmission and disease recurrence. Clin Infect Dis 2016;62:746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta SK, Padmanabhan BR, Diene SM, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 2014;58:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 2013;45:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubberke ER, Butler AM, Reske KA, et al. Attributable outcomes of endemic Clostridium difficile-associated disease in nonsurgical patients. Emerg Infect Dis 2008;14:1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero DM, Nerandzic MM, Jury LA, Chang S, Jump RL, Donskey CJ. Clostridium difficile infection in a Department of Veterans Affairs long-term care facility. Infect Control Hosp Epidemiol 2011;32:513–515. [DOI] [PubMed] [Google Scholar]
- 21.Archbald-Pannone LR, Boone JH, Carman RJ, Lyerly DM, Guerrant RL. Clostridium difficile ribotype 027 is most prevalent among inpatients admitted from long-term care facilities. J Hosp Infect 2014;88:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yui S, Ali S, Muzslay M, Jeanes A, Wilson APR. Identification of Clostridium difficile reservoirs in the patient environment and efficacy of aerial hydrogen peroxide decontamination. Infect Control Hosp Epidemiol 2017;38:1487–1492. [DOI] [PubMed] [Google Scholar]