Abstract
NOP2/Sun domain family member 2 (NSUN2) is upregulated in numerous types of tumors and may be implicated in multiple biological processes, including cell proliferation, migration and human tumorigenesis. However, little is known about how NSUN2 serves a role in these processes. In the present study, expression profiles of long noncoding RNAs (lncRNAs) and mRNAs were developed in NSUN2-deficient HepG2 cells by RNA-sequencing analysis. A total of 757 lncRNAs were differentially expressed, 392 of which were upregulated, and 365 were downregulated compared with wild-type HepG2 cells. Moreover, 212 lncRNAs were co-expressed with 368 target mRNAs. It was also observed that 253 pairs of lncRNAs and mRNAs exhibited negative correlations and that 290 pairs had positive correlations. Bioinformatics analysis indicated that these lncRNAs regulated by NSUN2 were associated with ‘signal transduction’, ‘extracellular exosome’ and ‘calcium ion binding’, and were enriched in ‘pathways in cancer’, ‘PI3K-Akt signaling pathway’ and ‘ECM-receptor interaction pathway’. These results illustrate the landscape and co-expression network of lncRNAs regulated by NSUN2 and provide invaluable information for studying the molecular function of NSUN2.
Keywords: NOP2/Sun domain family member 2, RNA methylation, long noncoding RNAs, RNA-sequencing, gene editing
Introduction
NOP2/Sun domain family member 2 (NSUN2), also known as Misu, is a type of RNA methyltransferase that is targeted by Myc and mediates Myc-induced cell proliferation and growth (1). Numerous studies have indicated that NSUN2 may be implicated in multiple biological processes, including cell proliferation, migration, testis differentiation, stem cell differentiation, and diseases such as intellectual disability and human cancer (1–4).
Previous studies have demonstrated that the NSUN2 gene is upregulated in several types of cancer, including esophageal, stomach, liver, pancreas, uterine cervix, prostate, kidney, bladder, thyroid, and breast cancers (5). High expression of NSUN2 leads to increased proliferation and metastasis of tumor cells (1,2), indicating that NSUN2 may be a valuable target for cancer therapy and a potential cancer diagnostic marker. Therefore, understanding NSUN2 function is extremely important in basic and clinical studies.
NSUN2 was recently reported to methylate various types of RNAs and modulate their functions. NSUN2 is able to stabilize p16 mRNA by methylating its 3′ untranslated region at A988 (6). Cyclin-dependent kinase 1 and p21 mRNAs are also methylated by NSUN2, which impacts the efficiency of their translation (7,8). NSUN2 may also methylate transfer RNAs (tRNAs) and microRNAs (miRNA) and influence their processing (9,10). However, it remains unknown whether lncRNAs are modulated by NSUN2.
lncRNAs, which are longer than 200 nucleotides (nt), have been identified as the most abundant non-protein-coding RNAs. Functional studies have revealed that lncRNAs are implicated in diverse biological processes and disease-associated pathways, including cell proliferation and differentiation, stem cell pluripotency, and tumorigenesis and metastasis (11,12). Multiple lines of evidence have suggested that the dysregulation of lncRNA is associated with cancer, indicating an important role of lncRNAs in tumorigenesis (12,13). Thus, it was speculated that NSUN2 may be implicated in tumorigenesis through its modulation of lncRNAs.
To investigate the effect of NSUN2 on lncRNA expression in cancer, a NSUN2-deficient HepG2 cell line was established with the clustered regularly interspaced short palindromic repeats/caspase 9 (CRISPR/Cas9) system, and expression profiles of lncRNAs were obtained by RNA-sequencing (RNA-seq) analysis.
Materials and methods
Cell line
The HepG2 cell line (a liver cancer cell line) was purchased from the American Type Culture Collection (cat. no. HB-8065; Manassas, VA, USA). Cells were maintained in Dulbecco's-modified Eagle's medium (DMEM, HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C with 5% CO2.
The NSUN2-deficient HepG2 cell line was established as previously described (14). In brief, two selection markers and a poly(A) signal were inserted into the first exon of the NSUN2 gene via CRISPR/Cas9-mediated homology-directed repair. Double marker selection was employed for screening the clonal cell lines for successful biallelic integration of the poly(A) signal. Finally, the efficiency of gene silencing was evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using primers specific for NSUN2 (Table I).
Table I.
Primers for reverse-transcription quantitative polymerase chain reaction.
| Gene symbol | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ACTB | AAGACCTGTACACCAACACAG | AGGGCAGTGATCTCCTTCT |
| NSUN2 | ATCTTGAGAAAATCGCCACAC | ATCATTCGCAATAACAAATCCCT |
| ENST00000623282 | TTTAACTGGACTCTTGGCACT | TGTTAAGCATACCCCTACCTG |
| NONHSAT194852.1 | GGAGTGTGTCAGTGTCCACC | GCACCAAATCTGCTTTCGCA |
| NONHSAT016752.2 | AGCACACAGGCATCTAGTGG | CAGGCTGCTCTTCCATTCCA |
| NONHSAT053044.2 | AACCTTAGGCAAGTTACGTT | GATAACTATGTGCTAGGCTCT |
| NONHSAT087855.2 | GGGACGGCTTCTCGGCAAT | TTCTGGGTGTATCCAGTTGTGC |
| ENST00000585065 | GAAGTTAGTCCCTGGGGTGTC | TGGGGCACAAATCCACATCT |
| NONHSAT180118.1 | CCAGTTCAGCCAGTACGTGT | CCTTTCCCTTTTAGATCCCTGC |
| ENST00000366365 | GTTGAGCCTGCCAAGTTGTG | ACGTAGGTCCTGTTTGCAGT |
| H19 | TTTGGTTACAGGACGTGGCA | CCTCGATCCCCTAAACCTCC |
| HIST1H4H | GAGGAGCTAAGCGTCATCGC | AGAAATTCGCTTGACACCGC |
| LOXL2 | CTCCACTGTACTGGCAACGA | GCGGTAGGTTGAGAGGATGG |
| PPARG | TCTCCGTAATGGAAGACCACT | AGGCTCCACTTTGATTGCACT |
| CGA | TTCGGATCCACAGTCAACCG | CACATCAGGAGCGGAATGGA |
| TGFA | GCGAGTGCCAGCAGAGAG | GCACGCAGCCAACACAATAC |
| FN1 | TTGCTCCTGCACATGCTTTG | TCGGGAATCTTCTCTGTCAGC |
| TNC | AAAGCGGGGAATGTTGGGAT | GCCTGTAAGCTTTTCCCAAGTG |
| TGFB1 | CGACTCGCCAGAGTGGTTAT | AGTGAACCCGTTGATGTCCA |
RNA library construction and sequencing
Total RNA of HepG2 cells and NSUN2-deficient HepG2 cells was extracted using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and quantified and qualified with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). For next generation sequencing, cDNA libraries were constructed using the NEBNext® Ultra™ RNA Library Prep kit for Illumina® (Illumina, Inc., San Diego, CA, USA), according to the manufacturer's protocol.
Then, the libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer's instructions (Illumina, Inc.). Sequencing was performed using a 2×150 bp paired-end configuration, and image analysis and base calling were conducted using HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina, Inc.) on the HiSeq instrument. The sequences were processed by Genewiz, Inc. (South Plainfield, NJ, USA).
Differential expression analysis
To remove technical sequences, including adapters, PCR primers, or fragments thereof, and quality of bases <20, pass filter data of fastq format were processed by Trimmomatic (v0.30) (15) to obtain high quality clean data.
Reference genome sequences (hg38) and gene model annotation files of humans were downloaded from ENSEMBL (http://asia.ensembl.org/info/data/ftp/index.html). Hisat2 (v2.0.1) was used to index the reference genome sequence (16). Finally, clean data were aligned to the reference genome using Hisat2 software (v2.0.1). lncRNAs were identified based on the NONCODE database (version: NONCODE 2016; http://www.bioinfo.org/NONCODE2016/).
To further analyze the expression levels of genes, Stringtie (version 1.3.0) (17) was used to count the number of fragments of each gene following Hisat2 comparison, which were normalized with the trimmed mean of M values method (18). The fragments per kilobase of exon model per million mapped reads (FPKM) value of each gene was calculated by the script.
Differential expression analysis was performed using the DESeq Bioconductor package (http://www-huber.embl.de/users/anders/DESeq), a model based on the negative binomial distribution. Fold-change was calculated based on the FPKM value. Following adjustment with the Benjamini and Hochberg method (19) for controlling the false discovery rate, genes with P<0.01 and log2 (fold-change) >2 were considered differentially expressed genes. For analysis of the differentially expressed lncRNAs, a more stringent standard (fold-change >4; P<0.01) was set to narrow the scope of candidate lncRNAs.
Prediction of target genes by lncRNAs
Target gene prediction was performed by evaluating trans- and cis-regulation. Trans-regulation predicts whether the database is the mRNA database of the species. First, Basic Local Alignment Search Tool (Magic-BLAST 1.3.0) was used to select complementary or similarity sequences (20). RNAplex (v0.2) was subsequently used to calculate the complementary energy between the two sequences, and sequences above the threshold were selected. RNAplex was designed to quickly identify possible hybridization sites for a query RNA in large RNA databases and is also useful for searching RNA-RNA interactions (21). Cis-regulation refers to an lncRNA acting on neighboring target genes. Genes with distances <10 kb upstream and downstream of lncRNAs were selected as targets for cis-regulation.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
GO::TermFinder comprises a set of object-oriented Perl modules for accessing GO information and evaluating and visualizing the collective annotation of a list of genes to GO terms (22). It was used to identify GO terms that annotate a list of enriched genes with P<0.05. KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs and chemical substances (http://en.wikipedia.org/wiki/KEGG) (23). KOBAS (v3.0) software (http://kobas.cbi.pku.edu.cn/index.php) was used to test the statistical enrichment of the differentially expressed genes in KEGG pathways. The background gene set comprised all genes expressed in HepG2 cells and NSUN2-deficient HepG2 cells, according to the FPKM value.
Construction of coexpression network
Genes enriched in KEGG pathways and their coexpressed lncRNAs were used to construct the coexpression network using Cytoscape software (v3.3.0) (24).
RT-qPCR analysis
Total RNA of HepG2 cells and NSUN2-deficient HepG2 cells was extracted using TRIzol® according to the manufacturer's instructions. Complementary DNAs were synthesized using the PrimeScript™ RT reagent kit plus gDNA Eraser (Takara, Kusatsu, Japan) and random primers. qPCR was performed using a CFX Connect™ real-time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the RNA level was quantified using a SYBR Green Master Mix (Vazyme, Piscataway, NJ, USA). The thermocycling conditions were: 95°C for 5 min, followed by 40 cycles of 94°C for 15 sec and 60°C for 30 sec. Comparative quantification of each RNA was normalized to the β-actin gene using the 2−∆∆Cq method (25). The specific primers used for amplification are presented in Table I.
Statistical analysis
All data were analyzed using SPSS software (version 20.0; IBM Corp., Armonk, NY, USA). Spearman's correlation test was used to estimate the coexpression relationship between lncRNAs and mRNAs (26,27). The RT-qPCR data are expressed as the mean ± standard error of the mean. Multigroup comparisons of the means were analyzed using one-way analysis of variance and post-hoc contrasts were performed using Tukey's test. P<0.05 was considered to indicate a statistically significant difference. Each group contained three samples, and all experiments were repeated at least three times.
Results
Differential expression of lncRNAs and mRNAs associated with NSUN2
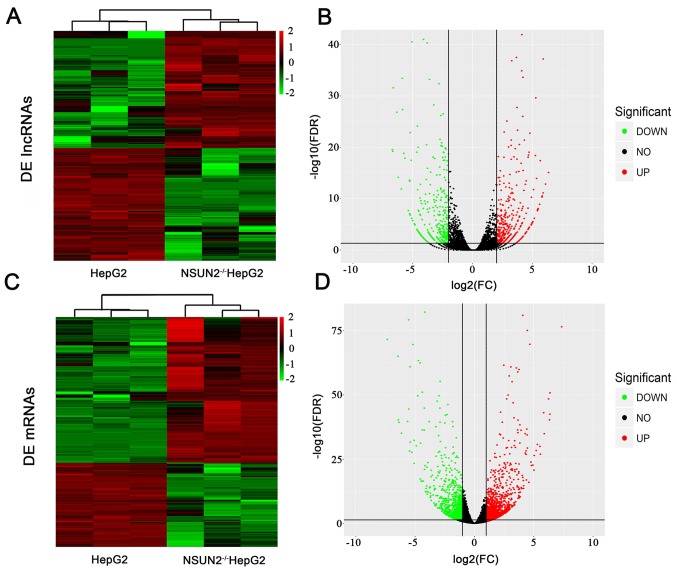
From the RNA-seq data, an average of 13.6 and 11.9 million clean reads were generated in NSUN2-deficient HepG2 cells and HepG2 cells. In NSUN2-deficient HepG2 cells and normal HepG2 cells, 94.8 and 95.5% of clean reads were uniquely mapped, respectively. A total of 757 lncRNAs were differentially expressed (fold-change >4; P<0.01) between NSUN2-deficient HepG2 cells and HepG2 cells, of which 392 lncRNAs were upregulated and 365 lncRNAs were downregulated (Fig. 1A and B; Table II).
Figure 1.
DE lncRNAs and mRNAs in NSUN2-deficient HepG2 cells. (A) Heatmap of the expression profiles of lncRNAs that exhibited significant expression changes. Red to green color indicates high to low expression levels, respectively. (B) Volcano plot of the P-values as a function of weighted fold-change for lncRNAs in NSUN2-deficient HepG2 cells and wild-type HepG2 cells. Dark dots represent lncRNAs not significantly differentially expressed (fold change <4; P>0.01) and red and green dots represent lncRNAs that are significantly differentially expressed (fold change ≥4; P<0.01). (C) Heatmap of the expression profiles of mRNAs that showed significant expression changes. Red to green color indicates high to low expression levels, respectively. (D) Volcano plot of the P-values as a function of weighted fold-change for lncRNAs in NSUN2-deficient HepG2 cells and wild-type HepG2 cells. Dark dots represent lncRNAs not significantly differentially expressed (fold change <4; P>0.01) (fold change <2; P>0.05) and red and green dots represent lncRNAs that are significantly differentially expressed (fold change ≥2; P<0.05). DE, differentially expressed; lncRNA, long noncoding RNA; NSUN2, NOP2/Sun domain family member 2; FDR, false discovery rate; FC, fold change.
Table II.
Top 10 downregulated and upregulated differentially expressed lncRNAs in NOP2/Sun domain family member 2-deficient HepG2 cells compared with normal controls.
| A, Downregulated | ||
|---|---|---|
| lncRNA | log2 fold change | P-value |
| NONHSAT061515.2 | −9.1672 | 8×10−210 |
| NONHSAT208991.1 | −7.89821 | 2.73×10−87 |
| NONHSAT053947.2 | −7.52574 | 1.94×10−49 |
| NONHSAT120697.2 | −7.35533 | 8.05×10−67 |
| NONHSAT175932.1 | −6.83085 | 7.51×10−46 |
| NONHSAT124573.2 | −6.70262 | 2.79×10−20 |
| NONHSAT186327.1 | −6.6746 | 6.06×10−20 |
| NONHSAT141130.2 | −6.62423 | 2.57×10−32 |
| ENST00000397381 | −6.48342 | 5.29×10−76 |
| NONHSAT053044.2 | −6.3331 | 1.46×10−27 |
| B, Upregulated | ||
| lncRNA | log2 fold change | P-value |
| NONHSAT179381.1 | 5.670156 | 3.91×10−18 |
| ENST00000443205 | 5.805754 | 4.68×10−11 |
| NONHSAT199040.1 | 5.831291 | 2.94×10−11 |
| ENST00000521369 | 5.905295 | 7.55×10−12 |
| NONHSAT202740.1 | 5.914346 | 6.53×10−38 |
| NONHSAT185459.1 | 6.106938 | 1.43×10−13 |
| NONHSAT060224.2 | 6.355958 | 8.03×10−16 |
| NONHSAT112221.2 | 6.63073 | 8.95×10−47 |
| NONHSAT103133.2 | 6.949018 | 1.39×10−53 |
| NONHSAT180405.1 | 7.830624 | 1.41×10−74 |
lncRNA, long noncoding RNA.
mRNA expression was also compared between NSUN2-deficient and wild-type cells. A total of 1,834 mRNAs were differentially expressed (fold-change >2; P<0.05) in NSUN2-deficient HepG2 cells compared with wild-type HepG2 cells (Fig. 1C and D; Table III). Of these genes, 1,163 mRNAs were upregulated and 671 mRNAs were downregulated in NSUN2-deficient HepG2 cells.
Table III.
Top 10 downregulated and upregulated differentially expressed mRNAs in NOP2/Sun domain family member 2-deficient HepG2 cells compared with normal controls.
| A, Downregulated | |||
|---|---|---|---|
| Gene | Gene symbol | log2 fold change | P-value |
| ENSG00000135373 | EHF | −7.352147492 | 2.47×10−72 |
| ENSG00000175874 | CREG2 | −7.111981449 | 5.31×10−88 |
| ENSG00000079308 | TNS1 | −6.554792085 | 4.72×10−153 |
| ENSG00000121797 | CCRL2 | −6.440975865 | 9.08×10−66 |
| ENSG00000161249 | DMKN | −6.403903015 | 4.81×10−41 |
| ENSG00000154997 | SEPT14 | −6.158418718 | 1.14×10−89 |
| ENSG00000127324 | TSPAN8 | −6.069707964 | 4.77×10−40 |
| ENSG00000178172 | SPINK6 | −5.636422774 | 8.06×10−36 |
| ENSG00000078098 | FAP | −5.556570307 | 6.05×10−189 |
| ENSG00000099998 | GGT5 | −5.544869611 | 2.19×10−40 |
| B, Upregulated | |||
| Gene | Gene symbol | log2 fold change | P-value |
| ENSG00000147246 | HTR2C | 5.306442373 | 1.95×10−31 |
| ENSG00000124253 | PCK1 | 5.524179098 | 1.08×10−30 |
| ENSG00000121904 | CSMD2 | 5.563322974 | 1.17×10−27 |
| ENSG00000164128 | NPY1R | 5.880433118 | 8.29×10−44 |
| ENSG00000105894 | PTN | 6.248807612 | 1.27×10−35 |
| ENSG00000163530 | DPPA2 | 6.323234879 | 3.53×10−49 |
| ENSG00000125740 | FOSB | 7.352207927 | 4.93×10−41 |
| ENSG00000170345 | FOS | 7.442578093 | 3.12×10−43 |
| ENSG00000006611 | USH1C | 7.895447434 | 1.58×10−51 |
| ENSG00000135346 | CGA | 10.37870244 | 2.89×10−77 |
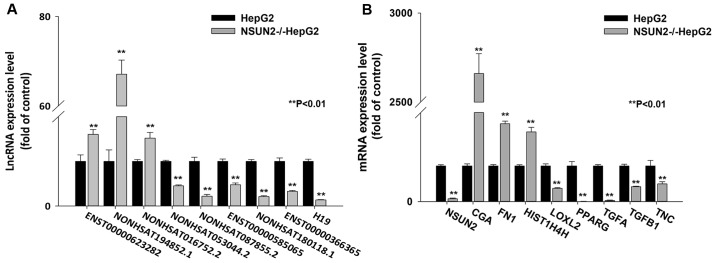
To further evaluate the reliability of the RNA-seq results, a total of 16 differentially expressed (DE) lncRNA and mRNA transcripts were randomly selected to validate the relative expression levels in the NSUN2-deficient HepG2 and wild-type HepG2 cells using RT-qPCR. The results demonstrated that all of the data for differential expression of lncRNAs and mRNAs were consistent with the RNA-seq results (Fig. 2A and B), indicating that the RNA-seq data were reliable.
Figure 2.
Verification of differentially expressed transcripts in NSUN2-deficient HepG2 cells. Relative expression levels of (A) lncRNAs and (B) mRNAs in NSUN2-deficient HepG2 cells using reverse transcription-quantitative polymerase chain reaction. The relative expression levels were normalized to β-actin gene expression. The results are expressed as the mean ± standard error of the mean. **P<0.01 vs. respective HepG2 group. NSUN2, NOP2/Sun domain family member 2; lncRNA, long noncoding RNA.
Correlation analysis of DE lncRNAs and target mRNAs
To better understand the functions of differentially expressed lncRNAs, target genes of 757 lncRNAs were predicted through cis and trans action. As a result, a total of 212 lncRNAs with 368 target mRNAs (rs>0.9; P<0.05) exhibited significant correlations (Table IV), among which the correlation of 253 mRNAs was negative and the correlation of 290 mRNAs was positive.
Table IV.
Top 20 significantly co-expressed lncRNAs and targeted mRNAs.
| lncRNA | log2 FC | Coef | Target gene | Gene name | log2 FC |
|---|---|---|---|---|---|
| ENST00000366365 | −2.23253 | −0.9868 | ENSG00000188732 | FAM221A | 2.2387726 |
| NONHSAT086754.2 | −2.73917 | −0.978 | ENSG00000152689 | RASGRP3 | 2.7018123 |
| NONHSAT017591.2 | 2.622214 | −0.978 | ENSG00000103381 | CPPED1 | −2.591183 |
| NONHSAT105304.2 | 3.80573 | −0.978 | ENSG00000125730 | C3 | −3.818577 |
| NONHSAT161933.1 | 3.905272 | −0.978 | ENSG00000134830 | C5AR2 | −3.92987 |
| NONHSAT031026.2 | 3.936562 | −0.978 | ENSG00000134830 | C5AR2 | −3.92987 |
| ENST00000564650 | 2.39005 | −0.9779 | ENSG00000188505 | NCCRP1 | −2.352221 |
| NONHSAT165291.1 | −2.28239 | −0.9765 | ENSG00000188732 | FAM221A | 2.2387726 |
| NONHSAT103857.2 | −2.07256 | −0.9765 | ENSG00000105650 | PDE4C | 2.1183298 |
| NONHSAT042028.2 | 3.698807 | −0.9747 | ENSG00000173698 | ADGRG2 | −3.652078 |
| NONHSAT000158.2 | −2.30185 | 0.978 | ENSG00000188112 | C6orf132 | −2.196761 |
| NONHSAT185304.1 | 2.199372 | 0.978 | ENSG00000008311 | AASS | 2.279951 |
| NONHSAT202599.1 | 2.73477 | 0.978 | ENSG00000121236 | TRIM6 | 2.8359967 |
| NONHSAT179528.1 | 2.426634 | 0.9824 | ENSG00000009950 | MLXIPL | 2.4755839 |
| NONHSAT018086.2 | 2.637192 | 0.9824 | ENSG00000139174 | PRICKLE1 | 2.6800691 |
| NONHSAT165633.1 | 2.144573 | 0.9845 | ENSG00000105650 | PDE4C | 2.1183298 |
| NONHSAT057058.2 | 2.729542 | 0.9845 | ENSG00000152689 | RASGRP3 | 2.7018123 |
| NONHSAT024073.2 | −3.80956 | 0.9868 | ENSG00000125730 | C3 | −3.818577 |
| NONHSAT058560.2 | 4.390725 | 0.9868 | ENSG00000241644 | INMT | 4.3854325 |
| NONHSAT159228.1 | 2.027489 | 0.9912 | ENSG00000066468 | FGFR2 | 2.0244554 |
lncRNA, long noncoding RNA.
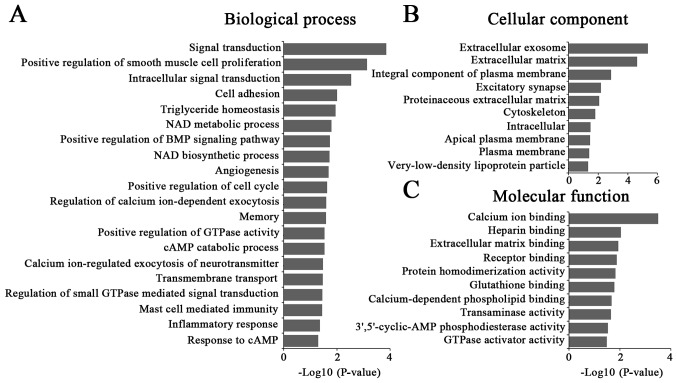
A total of 368 coexpressed mRNAs were selected for GO enrichment and KEGG pathway analysis. The results demonstrated that the most enriched GOs were involved in 42 biological processes, 18 cellular components and 23 molecular functions (Fig. 3). These dysregulated lncRNAs by NSUN2 were associated with ‘signal transduction’ (ontology: biological process), ‘extracellular exosome’ (ontology: cellular component) and ‘calcium ion binding’ (ontology: molecular function). KEGG pathway analysis demonstrated that the correlated mRNAs were primarily enriched in ‘pathways in cancer’ (hsa05200), ‘focal adhesion’ (hsa05032), the ‘PI3K-Akt signaling pathway’ (hsa04151) and ‘hematopoietic cell lineage’ (hsa04640) (Table V).
Figure 3.
Gene Ontology analysis of the significantly correlated mRNAs targeted by lncRNAs. (A) Biological processes, (B) molecular functions, and (C) cellular components enrichment analysis of the significantly correlated mRNAs targeted by lncRNAs. lncRNA, long noncoding RNA.
Table V.
Top 15 significantly enriched pathways for the significantly correlated mRNAs targeted by lncRNAs.
| Term | Count | P-value | Input |
|---|---|---|---|
| hsa05200: Pathways in cancer | 15 | 0.0009457 | FN1, WNT2B, LAMA4, GNB4, PGF, ARHGEF12, EGLN3, ARNT2, WNT7B, FGFR2, FLT3LG, PLCB1, ITGA2B, RASGRP3, JUN |
| hsa04921: Oxytocin signaling pathway | 8 | 0.0028716 | MYLK3, KCNJ12, PLCB1, CACNB2, CAMK4, CAMK1D, PPP1R12A, JUN |
| hsa04510: Focal adhesion | 9 | 0.0038045 | FN1, LAMA4, PGF, MYLK3, ITGA2B, THBS1, ARHGAP5, PPP1R12A, JUN |
| hsa04672: Intestinal immune network for IgA production | 4 | 0.0055313 | MAP3K14, TNFRSF13C, ICOSLG, CCL28 |
| hsa04810: Regulation of actin cytoskeleton | 9 | 0.005559 | FN1, ARHGEF4, ARHGEF12, LIMK1, MYLK3, ITGA2B, FGFR2, PIKFYVE, PPP1R12A |
| hsa04610: Complement and coagulation cascades | 5 | 0.0069775 | C1S, PROCR, C1R, THBD, C3 |
| hsa04512: ECM-receptor interaction | 5 | 0.0084585 | SV2B, FN1, LAMA4, THBS1, ITGA2B |
| hsa04640: Hematopoietic cell lineage | 5 | 0.0097028 | FLT3LG, CD37, IL6R, IL11, ITGA2B |
| hsa04611: Platelet activation | 6 | 0.011229 | MYLK3, ARHGEF12, ITGA2B, COL3A1, PLCB1, PPP1R12A |
| hsa05032: Morphine addiction | 5 | 0.0120466 | GNB4, GABRG3, PDE10A, PDE11A, PDE4C |
| hsa00561: Glycerolipid metabolism | 4 | 0.0123949 | DGKH, LPL, MGLL, AKR1B1 |
| hsa04151: PI3K-Akt signaling pathway | 11 | 0.0130241 | FN1, LAMA4, GNB4, PGF, IL6R, ITGA2B, THBS1, JAK3, FGFR2, IFNAR2, NR4A1. |
| hsa01100: Metabolic pathways | 28 | 0.0145589 | PTGES, KYNU, GGT5, QPRT, ETNPPL, GCNT3, SPTLC3, HKDC1, GALNT16, NMNAT1, UGDH, RDH10, AKR1B1, NDUFA9, AASS |
| hsa05146: Amoebiasis | 5 | 0.0171801 | FN1, SERPINB9, LAMA4, COL3A1, PLCB1 |
| hsa04530: Tight junction | 6 | 0.017745 | INADL, PARD6B, RAB3B, OCLN, MYH3, PRKCH |
Coexpression network of KEGG enriched mRNAs
The coexpression network comprised 128 nodes and 137 edges between 93 lncRNAs and 239 mRNAs. Among them, 70 pairs were positively correlated, and 324 pairs were negatively correlated (Fig. 4).
Figure 4.
Coexpression network of Kyoto Encyclopedia of Genes and Genomes enriched mRNAs and correlated lncRNAs. The coexpression network was established between the 93 significantly expressed lncRNAs and the 239 significantly differentially expressed mRNAs that had Spearman's correlation coefficients ≥0.90. The circles represent lncRNAs while the diamonds represent mRNAs. Red to green color indicates high to low expression levels, respectively. lncRNA, long noncoding RNA.
Discussion
NSUN2 is upregulated in numerous types of tumors and modulates the biological functions of multiple RNA species, including tRNA, mRNA, vault RNA, and miRNA (9,10,28,29). However, little is known about the association between NSUN2 and lncRNAs, and investigating this relationship may help further the understanding of the molecular role of NSUN2 in tumorigenesis. In the present study, the lncRNA expression profiles in NSUN2-deficient HepG2 cells and wild-type HepG2 cells were examined by genome-wide RNA-seq, and lncRNAs with statistically significant differential expression were identified. Integrated analyses of these lncRNAs, concentrating on target gene prediction, gene coexpression, GO and pathway analyses, were performed to elucidate their potential functions. As a result, a total of 757 lncRNAs were differentially expressed in response to NSUN2 knockout in HepG2 cells.
The biological role of lncRNAs is complicated due to their diverse and complex functions. Emerging evidence has indicated that lncRNAs may serve as signal, decoy, guide or scaffold molecules in the regulation of gene expression (30). Therefore, the function of lncRNAs may be assessed by analyzing the mRNAs that they regulate. The present study used cis and trans action to predict mRNAs targeted by DE lncRNAs. Furthermore, Spearman's correlation test was used to estimate the coexpression between lncRNAs and mRNAs. Ultimately, 212 DE lncRNAs were identified to be coexpressed with 368 target mRNAs, with 290 pairs positively correlated and 253 pairs negatively correlated. For example, the lncRNAs NONHSAT103857.2, NONHSAT001050.2, NONHSAT146065.2, ENSG00000130600 and NONHSAT 159228.1 were coexpressed with the Wnt7B, placental growth factor, epiregulin, insulin-like growth factor 2 and fibroblast growth factor receptor 2 (FGFR2) genes, which have been reported to be important regulators in tumors.
GO enrichment was employed to further examine the biological role of these DE lncRNAs. The coexpressed mRNAs were classified into hierarchical categories to determine gene regulatory networks based on the biological processes, cellular components and molecular functions. GO analysis revealed that these genes were significantly enriched in many cancer-associated biological processes, including cell proliferation, cell adhesion and angiogenesis. Furthermore, pathway analysis highlighted a number of pathways closely associated with carcinogenesis, including ‘pathways in cancer’, ‘focal adhesion’, ‘extracellular matrix (ECM)-receptor interaction’ and ‘PI3K-Akt signaling’.
Focal adhesions are large, dynamic protein complexes through which the cytoskeleton of a cell connects to the ECM. The focal adhesion and ECM-receptor pathways serve important roles in various biological processes such as cell proliferation, migration and angiogenesis, which are crucial for tumorigenesis (31–33).
The phosphatidylinositol-3-kinase-protein kinase B (PI3K-Akt) pathway is a signal transduction pathway that promotes survival and growth in response to extracellular signals. The PI3K-Akt pathway may be activated by a range of signals, including hormones, growth factors and components of the ECM, and further mediate various downstream responses, including survival, growth, cell proliferation and migration (34–37). The PI3K-Akt pathway also serves an important role in the regulation of angiogenesis (38,39), which is essential for embryonic development, tumor growth and metastasis. Recently, numerous studies have demonstrated dysfunction of the PI3K-Akt pathway in carcinogenesis. Various components of the PI3K-Akt pathway are frequently altered in different types of human cancer types, including breast, gastric, colon and hepatocellular carcinoma (HCC) (40–42). The PI3K pathway may transmit oncogenic signals to Akt, thus promoting tumorigenesis through a number of signaling pathways including the nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) pathway (43). Taken together, these results suggest that these dysregulated lncRNAs by NSUN2 deletion may be involved in tumor cell proliferation and angiogenesis by modulating focal adhesion, ECM-receptor interactions and PI3K-Akt signaling pathways.
A total of two well-studied lncRNAs, human urothelial carcinoma associated 1 (UCA1) and H19, were also demonstrated to be differentially expressed in the present study. Previous studies indicated that UCA1 and H19 may affect tumorigenesis by modulating the PI3K-Akt pathway. UCA1 is a known oncogene and is highly expressed in various types of cancer, including bladder cancer, gastric cancer, colorectal cancer and HCC (44–47). UCA1 is able to enhance the proliferation and metastasis of bladder cancer cells through the PI3K-Akt or Wnt signaling pathways (48). In HCC, UCA1 may serve an oncogenic role in promoting the proliferation and metastasis of HCC cells through the inhibition of miRNA-216b and activation of the FGFR1/extracellular signal-regulated kinases signaling pathway (47). Numerous studies have reported that NSUN2 is involved in regulating cell proliferation (1,7,49). In the present study, UCA1 was the most significantly downregulated lncRNA in NSUN2-deficient HepG2 cells, indicating that NSUN2 may affect cell proliferation by regulating the expression of UCA1.
As an important imprinting gene exclusively expressed from the maternal allele (50), H19 was also significantly downregulated in NSUN2-deficient HepG2 cells. The function of H19 is controversial. One study reported that H19 serves a role in the negative regulation of body weight and cell proliferation (51). Mutations in the H19 gene were also demonstrated to be associated with Beckwith-Wiedemann syndrome (52) and Wilms' tumorigenesis (53). However, increased H19 expression has been observed in various types of cancer, including colorectal cancer, esophageal cancer (54), lung cancer (55) and HCC (56). Downregulation of H19 lncRNA inhibits the migration and invasiveness of melanoma cells by inactivating the NF-κB and PI3K-Akt signaling pathways. On the other hand, overexpression of H19 lncRNA promotes invasion and autophagy via the PI3K/AKT/mammalian target of rapamycin pathways in trophoblast cells (57). In HCC, H19 overexpression might be a risk factor for tumor aggressiveness and poorer outcomes (56). Moreover, c-Myc, an oncogene that functions as a regulator of gene transcription, was also reported to directly induce H19 expression (55). A previous study demonstrated that NSUN2 was directly targeted by Myc and mediated Myc-induced cell proliferation and growth (1). The present study revealed that the H19 expression level was significantly decreased following NSUN2 deletion in HepG2 cells, indicating that NSUN2 may mediate the interaction between Myc and H19 lncRNA, further implicating NSUN2 in the regulation of tumor proliferation and metastasis.
It is noteworthy that the question of whether these lncRNAs are directly modulated by NSUN2 requires further experimental support. For example, an NSUN2-overexpression experiment may be performed to narrow the scope of candidate lncRNAs targeted by NSUN2. The application of technologies to investigate RNA-protein interaction, including RNA electrophoretic mobility shift assays and RNA binding protein immunoprecipitation assays, may help elucidate the molecular mechanisms behind NSUN2 modulation of these lncRNAs. Moreover, further research may address the functional significance of NSUN2 expression in other types of cancer cells in order to elucidate whether there is a common tumorigenic mechanism.
In conclusion, the present study revealed the expression patterns of lncRNAs in response to NSUN2 deletion in HepG2 cells. NSUN2 is an RNA m5C methyltransferase with important roles in tumor proliferation and metastasis, but its molecular function remains largely unknown. The results presented provide novel insight into the roles of lncRNAs associated with NSUN2 in carcinogenesis. Furthermore, a highly correlated expression pattern of lncRNAs induced by NSUN2 deficiency was observed, with coding genes that are involved in cancer development. These findings may be important for understanding the molecular function of NSUN2. Further studies are required to define the precise role of lncRNAs regulated by NSUN2 in tumorigenesis, in addition to the molecular mechanisms behind the regulation of lncRNAs and/or mRNAs by NSUN2. These will further our understanding of how the NSUN2 gene is involved in human diseases and cancer.
Acknowledgements
Not applicable.
Funding
This research was supported by the National Natural Science Foundation of China (grant nos. 81773013, 91540117), the National Key Research and Development Program in China (grant no. 2016YFC1303604), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (grant no. KYLX15_1376) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Animal Science and Veterinary Medicine).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZS performed the major experiments, and wrote and revised the manuscript. HC contributed to the experimental design, and revised the manuscript. SX, HX, YY and JO performed the experiments. XH, SC and ZY conducted the data analysis. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 2.Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang H, Dou Y, Liu W, Wang W, Du G, et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017;8:20751–20765. doi: 10.18632/oncotarget.10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M, et al. The mouse cytosine-5 RNA Methyltransferase NSun2 Is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol. 2013;33:1561. doi: 10.1128/MCB.01523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, Ogawa I, Takata T, Shimamoto F, Tatsuka M. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–671. doi: 10.1089/dna.2011.1446. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Liu Z, Yi J, Tang H, Xing J, Yu M, Tong T, Shang Y, Gorospe M, Wang W. The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3′-untranslated region of p16. Nat Commun. 2012;3:712. doi: 10.1038/ncomms1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing J, Yi J, Cai X, Tang H, Liu Z, Zhang X, Martindale JL, Yang X, Jiang B, Gorospe M, Wang W. NSun2 promotes cell growth via elevating cyclin-dependent kinase 1 translation. Mol Cell Biol. 2015;35:4043–4052. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, Wang W. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 Translation. J Cell Biochem. 2017;118:2587–2598. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, Han P, Luo Y, Zhang Z, Jiang B, et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34:3630–3641. doi: 10.1128/MCB.00243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Chang HY. Physiological roles of long noncoding RNAs: Insight from knockout mice. Trends Cell Biol. 2014;24:594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–9503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 13.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Han X, Yuan J, Geng T, Chen S, Hu X, Cui IH, Cui H. Biallelic insertion of a transcriptional terminator via the CRISPR/Cas9 system efficiently silences expression of protein-coding and non-coding RNA genes. J Biol Chem. 2017;292:5624–5633. doi: 10.1074/jbc.M116.769034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8:59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistical Soc. 1995;57:289–300. [Google Scholar]
- 20.Lobo I. Basic local alignment search tool (BLAST) J Mol Biol. 2012;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Tafer H, Hofacker IL. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 22.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO: TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:101–103, 119–128, 244–252.. [PubMed] [Google Scholar]
- 24.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Met Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Fu H, Cui J, Chen X. Differential lncRNA-mRNA co-expression network analysis revealing the potential regulatory roles of lncRNAs in myocardial infarction. Mol Med Rep. 2016;13:1195. doi: 10.3892/mmr.2015.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Yang H, Han L, Li F, Zhang T, Pang J, Feng X, Ren C, Mao S, Wang F. Long noncoding RNA expression profile changes associated with dietary energy in the sheep testis during sexual maturation. Sci Rep. 2017;7:5180. doi: 10.1038/s41598-017-05443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyenngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, Ewald AJ. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci USA. 2012;109:2595–2604. doi: 10.1073/pnas.1212834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong SJ, Wiberg M, Terenghi G, Kingham PJ. ECM molecules mediate both Schwann cell proliferation and activation to enhance neurite outgrowth. Tissue Eng. 2007;13:2863–2870. doi: 10.1089/ten.2007.0055. [DOI] [PubMed] [Google Scholar]
- 33.Cheresh DA, Stupack DG. Regulation of angiogenesis: Apoptotic cues from the ECM. Oncogene. 2008;27:6285–6298. doi: 10.1038/onc.2008.304. [DOI] [PubMed] [Google Scholar]
- 34.Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 36.Zheng H, Fu G, Dai T, Huang H. Migration of endothelial progenitor cells mediated by stromal cell-derived factor-1alpha/CXCR4 via PI3K/Akt/eNOS signal transduction pathway. J Cardiovasc Pharmacol. 2007;50:274. doi: 10.1097/FJC.0b013e318093ec8f. [DOI] [PubMed] [Google Scholar]
- 37.Brunet A, Datta SR, Greenberg ME. Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol. 2001;11:297–305. doi: 10.1016/S0959-4388(00)00211-7. [DOI] [PubMed] [Google Scholar]
- 38.Fang J, Ding M, Yang L, Liu L, Jiang B. PI3K/PTEN/AKT signalling regulates prostate tumor angiogenesis. Cell Signall. 2007;19:2487–2497. doi: 10.1016/j.cellsig.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karar J, Maity A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front Mol Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 41.Bellacosa A, De Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 42.Roy HK, Olusola BF, Clemens DL, Karolski WJ, Ratashak A, Lynch HT, Smyrk TC. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 43.Lee KB, Byun HJ, Park SH, Park CY, Lee SH, Rho SB. CYR61 controls p53 and NF-κB expression through PI3K/Akt/mTOR pathways in carboplatin-induced ovarian cancer cells. Cancer Lett. 2012;315:86–95. doi: 10.1016/j.canlet.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Wang T, Yuan J, Feng N, Li Y, Lin Z, Jiang Z, Gui Y. Hsa-miR-1 downregulates long non-coding RNA urothelial cancer associated 1 in bladder cancer. Tumour Biol. 2014;35:10075–10084. doi: 10.1007/s13277-014-2321-2. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, Zhou B, Chen JJ, Chen P. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Onco. 2015;17:640–646. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 46.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8. doi: 10.1016/j.gene.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. 2016;7:838–842. doi: 10.1002/wrna.1380. [DOI] [PubMed] [Google Scholar]
- 50.Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forné T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene networkcontrolling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 52.Debaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumour. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 54.Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, Ito K, Takagi H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480. [PubMed] [Google Scholar]
- 55.Barsytelovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 played different roles in overall and disease-free survival from hepatocellular carcinoma after curative hepatectomy: A preliminary analysis from gene expression omnibus. Dis Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J, Xia Y, Zhang H, Guo H, Feng K, Zhang C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed Pharmacother. 2018;101:691–697. doi: 10.1016/j.biopha.2018.02.134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.