Abstract
Hereditary spherocytosis (HS) is a common heterogeneous type of inherited hemolytic anemia characterized by jaundice and splenomegaly. Diagnosis of HS in neonates is considered unreliable, and is generally based on positive family history, spherocytes in peripheral smears, increased osmotic fragility, and jaundice. In the present study, routine laboratory tests, next-generation sequencing, and Sanger sequencing were applied to diagnose a neonatal patient with Coombs-negative hemolytic jaundice. The neonate had no family history of HS; however, spherocytes were observed in peripheral smears, and the patient exhibited Coombs-negative and severe hemolytic jaundice, normal mean corpuscular hemoglobin concentration (MCHC) and mean corpuscular volume (MCV), normal glucose-6-phosphate dehydrogenase activity, negative thalassemia genetic mutation screening results, and negative autoimmune antibody tests. Novel compound heterozygous mutations in the spectrin-α, erythrocytic 1 (SPTA1) gene (c.3897-1G>C and c.5029G>A) were identified. The SPTA1 c.3897-1G>C mutation in intron 27-1, which disrupted the consensus splice site, was inherited from his asymptomatic mother, and the SPTA1 c.5029G>A (p.Gly1677Arg) mutation in trans with the SPTA1 c.3897-1G>C mutation was inherited from his asymptomatic father. Sanger sequencing of mRNA reverse transcribed into cDNA identified a deletion of the first 10 nucleotides of exon 28, confirming the splicing mutation. In conclusion, the present study reports a rare case of autosomal-recessive HS with a severe clinical phenotype, but normal MCHC and MCV.
Keywords: hereditary spherocytosis, anemia, jaundice, autosomal recessive, α-spectrin
Introduction
Hereditary spherocytosis (HS) is a group of heterogeneously inherited hemolytic anemic disorders, characterized by the presence of spherical-shaped spherocytes on peripheral blood film, jaundice and splenomegaly (1). HS is the most frequent cause of inherited chronic hemolysis in North America and northern Europe, with prevalence rates of 1/5,000 and 1/2,000, respectively. HS is usually associated with quantitative or qualitative abnormalities in red blood cell membrane proteins, including ankyrin, band 3, β-spectrin, spectrin-α, erythrocytic 1 and protein 4.2, encoded by ANK1, SLC4A1, SPTB, SPTA1 and EPB42, respectively (2). Splenectomy is widely considered as the standard surgical treatment in moderate and severe forms of HS (3). Newborn infants who have HS may develop anemia, extreme hyperbilirubinemia, bilirubin-induced neurologic dysfunction and kernicterus (4,5). Exact diagnosis of HS is considered unreliable around birth. However, if the early diagnosis of HS is recognized, appropriate treatment provided at the beginning of congenital hemolysis may prevent adverse outcomes in neonates with HS, caused by either de novo or compound heterozygous mutations, particularly for those with no family history of HS (6,7).
Deficiencies in ankyrin, band 3, spectrins or protein 4.2 account for 50–60, 15–20, 20 and <5% of patients with HS, respectively (8). Ethnic differences may exist: For instance in Japan, ankyrin defects account for only 5–10% of patients with HS, whereas protein 4.2 defects account for 45–50% (9). The major structural component of the skeletal network is the spectrin tetramer, which is formed by α-spectrin and β-spectrin (10). Spectrins are the central components of a complex spectrin-actin scaffold at the inner surface of the erythrocyte membrane. The C-terminal of α-spectrin and the N-terminal of β-spectrin associate to form heterodimers (αβ), and two heterodimers associate at the N-terminal of α-spectrin and the C-terminal of β-spectrin to form functional tetramers. The tetramers constitute the filaments of this network, and attach to the cellular membrane via interactions with various transmembrane proteins (11). Deficiency of α-spectrin is only clinically apparent in the homozygous or compound heterozygous state, and these patients have severe clinical manifestations. SPTA1 mutations cause HS in association with recessive forms of inheritance, and the presence of two null SPTA1 alleles is speculated to be lethal (12,13).
In the present study, a neonate with severe Coombs-negative hemolytic jaundice but no family history of HS was observed. The neonate was further clinically diagnosed with HS based on the following features: Spherocytes on the blood film, increased osmotic fragility, normal G6PD activity, normal hemoglobin in electrophoresis, negative thalassemia genetic mutation screening results and negative autoimmune antibody tests. Normal mean corpuscular hemoglobin concentration (MCHC) and mean corpuscular volume (MCV) were observed. The molecular basis of HS was elucidated via next-generation sequencing (NGS) of relevant genes. Novel compound heterozygous mutations in the SPTA1 gene (c.3897-1G>C and c.5029G>A), which were inherited from his asymptomatic mother and father, respectively, were identified to be responsible for his clinical phenotype.
Materials and methods
Subjects
A one-month old neonate with suspected HS and his asymptomatic parents (age, 30 years) were asked to participate in the study in February 2017. Written informed consent to use their blood samples and consent for publication were obtained from the parents, and this study was formally approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). All procedures were performed in accordance with the approved guidelines.
The available clinical characteristics of the neonate, along with hematological data, osmotic fragility, glucose-6-phosphate dehydrogenase (G6PD) activity, thalassemia genetic mutation screening and autoimmune antibody tests are summarized in Table I.
Table I.
Blood test results.
| Hematological parameters | Result | Reference |
|---|---|---|
| RBC | 3.77×1012/lb | 4.3–5.8×1012/l |
| MCV | 99.7 fl | 82–100 fl |
| MCH | 33.4 pg | 27–34 pg |
| MCHC | 33.5 g/dl | 31.6–35.4 g/dl |
| TBIL | 114.5 µmol/la | 3.4–20.5 µmol/l |
| IBIL | 104.7 µmol/la | ≤13.3 µmol/l |
| BRD | 9.8 µmol/la | 0–6.8 µmol/l |
| G6PD | 4,415 U/l | >1,100 U/l |
Elevated
Decreased. RBC, red blood cell; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; TBIL, total bilirubin; IBIL, indirect bilirubin; BRD, bilirubin direct; G6PD, glucose-6-phosphate dehydrogenase.
Ampliseq NGS panel design
A custom NGS panel was designed on the Ion AmpliSeq Designer website (http://www.ampliseq.com; Ion AmpliSeq Designer; Thermo Fisher Scientific, Inc., Waltham, MA, USA) to sequence all exons and adjacent introns of the ANK1, SLC4A1, SPTB, SPTA1 and EPB42 genes, with coverage of 96.71, 96.53, 99.54, 100 and 100%, respectively. The uncovered region was amplified and sequenced by Sanger sequencing.
NGS
Ion Torrent adapter-ligated DNA libraries were built according to the Ampliseq™ Library Preparation kit 2.0 96Lv (Thermo Fisher Scientific, Inc.) using 10 ng of input DNA from peripheral blood mononuclear cells (PBMCs), quantitated by Qubit 2.0 (Invitrogen; Thermo Fisher Scientific, Inc.) in two primer-pools with 118 and 115 amplicons, respectively. The polymerase chain reaction (PCR) products of the two pools were mixed together, with samples barcoded using Ion Xpress Barcodes 1–16 (Thermo Fisher Scientific, Inc.), followed by purification using AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA) and quantification by Ion Library TaqMan™ Quantitation kit (Thermo Fisher Scientific, Inc.). Prepared libraries were pooled in equal amount and diluted to 100 pM. Subsequently, 2 µl of pooled library were enriched using a Ion One-Touch Two (OT2) system with the Ion Personal Genome Machine™ (PGM) Hi-Q™ OT2 kit (Thermo Fisher Scientific, Inc.), followed by enrichment of Ion Sphere Particles on the Ion OneTouch Enrichment System apparatus (Thermo Fisher Scientific, Inc.). Enriched products were sequenced using the Ion Torrent PGM Hi-Q™ Sequencing kit using the Ion 316™ chip v2 (Thermo Fisher Scientific, Inc.).
Bioinformatics analysis
Raw data, following removal of adapter sequences, were aligned to the hg19 human reference genome. Coverage analysis and variant identification were performed on the Ion Torrent Server software version 4.4.2 (Thermo Fisher Scientific, Inc.), with default parameters. Variant annotation was performed with the Ion Reporter 5.0 software (Thermo Fisher Scientific, Inc.), according to the nomenclature recommended by the Human Genome Variation Society (http://www.hgvs.org), to classify variants as single nucleotide variants, multiple nucleotide variants or indels. All identified variants and regions with <20X coverage depth were visually verified with the Integrative Genomics Viewer v2.3.8 (Broad Institute, Cambridge, MA, USA). The clinical significance of annotated variants was assessed according to The 1000 Genomes Project (1000G; http://www.internationalgenome.org/), the Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php), the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/), ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/variation) and InterVar (http://wintervar.wglab.org/) databases (14). Functional prediction of missense mutations was performed using multiple software packages, including Sorting Intolerant From Tolerant (SIFT; http://sift.jcvi.org/www/SIFT_BLink_submit.html), MutationTaster (http://www.mutationtaster.org/) and Polymorphism Phenotyping (PolyPhen)-2 (http://genetics.bwh.harvard.edu/pph2/) (15–17).
Sanger sequencing
Sanger sequencing was performed as previously described (18). Briefly, genomic DNA was extracted from PBMCs with the QIAamp DNA blood mini kit (Qiagen GmbH, Hilden, Germany). Coding exons and splice junctions of relevant genes were amplified for identified mutations, failed amplicons or uncovered regions. RNA was extracted from PBMCs with TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA at 42°C for 1 h for sequencing. Sanger sequencing was performed bi-directionally on an ABI 3500 Dx Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.). NM_003126.2 was used as the reference transcript of the SPTA1 gene. The following primers were used in the study: SPTA1 c.3897-1G>C: Forward, 5′-CAAGGGATTGTTATCATTAGGTG-3′ and reverse, 5′-AAGGGGGAAGAAATCAGTGA-3′; SPTA1 c.5029G>A: Forward, 5′-CTACCTCTATCGCTCCCACAT-3′ and reverse, 5′-AACTGCTCTTCACTTCCTCCA-3′; RT PCR primers: Forward, 5′-GTTCAGGCTCTTCAGCGACG-3′ and reverse, 5′-CAAATCGTCCCGTTTCTTCAT-3′. PCR was performed using I-5™ 2X High-Fidelity Master Mix (MCLAB, South San Francisco, CA, USA) in a 50 µl volume including 19 µl water, 25 µl master mix, 2 µl forward primer, 2 µl reverse primer and 2 µl DNA or cDNA. The PCR was performed as following: Initial denaturation at 98°C for 2 min, followed by 35 cycles of 98°C for 10 sec, 62°C for 30 sec, 72°C for 3 sec, and a final extension at 72°C for 1 min.
Protein structure analysis
Protein structure analysis of the SPTA1 c.5029G>A (p.Gly1677Arg) mutation was performed using the HOPE software (version 1.1.1; http://www.cmbi.ru.nl/hope/) (19). The sequence relative to SPTA1 was downloaded from UniProtKB (https://www.uniprot.org/uniprot/P02549).
Results
Clinical features of the neonate with HS
This male neonate was delivered vaginally at 35 weeks of gestation (G2P2G35+4) following a normal pregnancy. No family members had a history of anemia or jaundice. The neonate weighed 5,100 g, and his Apgar scores were 9 and 10 (1 and 10 min, respectively), with no dysmorphic features. At 30 h, jaundice was observed, with a total serum bilirubin of 304.4 µmol/l (transcutaneous jaundice) and hemoglobin (Hb) of 118 g/l; phototherapy was initiated. On day 8, the neonate was transferred to the affiliated hospital of the present study due to jaundice and neonatal anemia, with an Hb of 85 g/l. The blood type of the parents was A Rh(+). The clinical characteristics of the neonate on day 8 are presented in Table I. Spherocytes were observed in peripheral smears, Coombs test was negative, G6PD activity was normal, no abnormal Hb was observed in electrophoresis, α- and β-thalassemia genetic mutation screen results were negative and autoimmune antibody tests were negative. Osmotic fragility was also increased. Discrepancies occurred in the patient between alterations in MCHC, MCV and clinical phenotype. The neonate had normal MCHC, MCV, and MCHC/MCV ratio, but received various blood transfusions in the three subsequent months due to severe anemia. The 31-year old father and 32-year old mother were asymptomatic.
NGS output and coverage
The sequencing of coding exons and adjacent introns of five HS-associated genes on the PGM achieved an average output of 425,997 mapped reads (98.9% on target). In summary, 100% of targeted amplicons were covered at least once, 99.14% amplicons were covered at least 20 times, and 97.42% amplicons were covered at least 100 times. The mean uniformity of base coverage was 95.09% in this panel. The average read depth was 1,453 fold.
Mutation detection and sanger sequencing validation
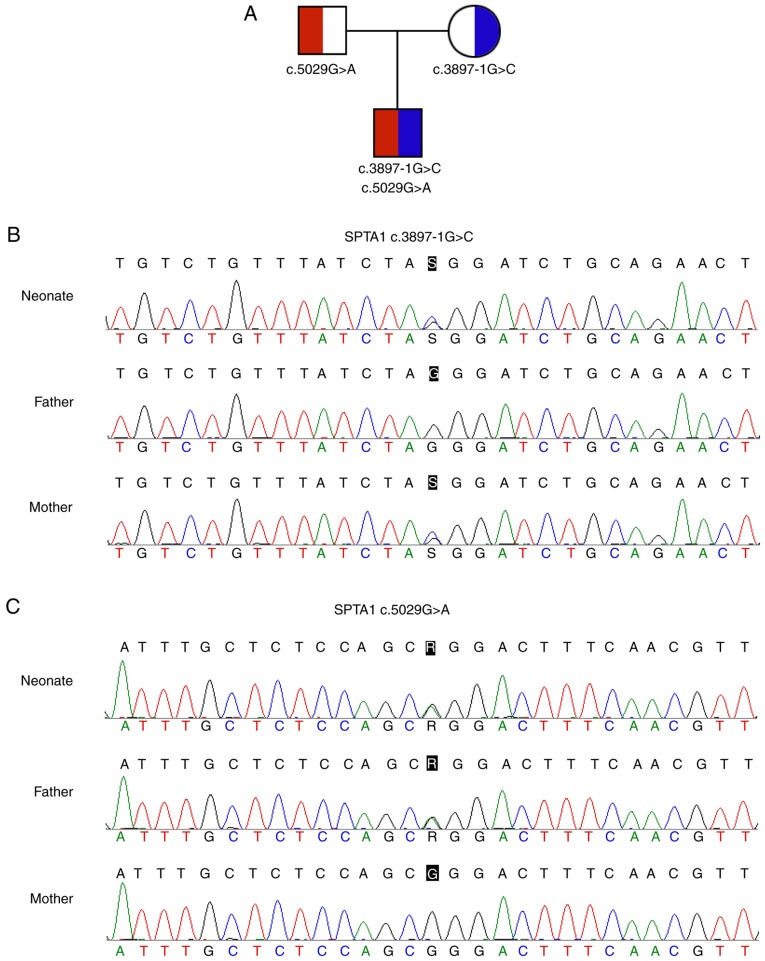
A total of 51 variants in the neonate were found and processed to discover pathogenic mutations. Following annotation and filtration, novel compound heterozygous mutations in the SPTA1 gene (c.3897-1G>C and c.5029G>A) were identified. The SPTA1 c.3897-1G>C (chr1:158615385, hg19) in intron 27-1, which may disrupt the consensus splice site, was inherited from his asymptomatic mother. The SPTA1 c.5029G>A (p.Gly1677Arg, chr1:158607983, hg19) in trans with the SPTA1 c.3897-1G>C mutation was inherited from his asymptomatic father (Fig. 1).
Figure 1.
Compound heterozygous mutations in the SPTA1 gene. (A) Family tree of a Chinese family with a neonate with hereditary spherocytosis. Square and circle denote male and female, respectively. (B) SPTA1 c.3897-1G>C maternally transmitted mutation. (C) SPTA1 c.5029G>A paternally transmitted mutation. SPTA1, spectrin-α, erythrocytic 1.
Functional prediction of the mutation
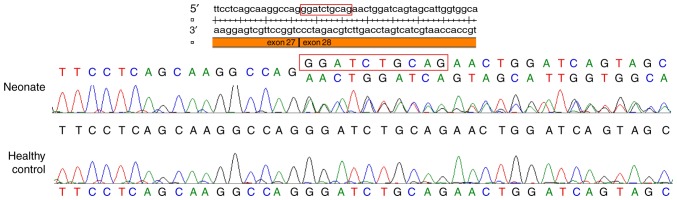
Sanger sequencing of mRNA reverse transcribed into cDNA identified a deletion of the first 10 nucleotides of exon 28, and confirmed that the SPTA1 c.3897-1G>C in intron 27-1 disrupted the consensus splice site (Fig. 2). This mutation was not found in the 1000G, EVS, ExAC or HGMD databases.
Figure 2.
Sanger sequencing of mRNA reverse transcribed into cDNA from peripheral blood mononuclear cells. A deletion of the first 10 nucleotides of exon 28 was observed in the neonate.
The SPTA1 c.5029G>A (p.Gly1677Arg) mutation was not found in the 1000G, EVS or HGMD databases, whereas four carriers (one in a South Asian and three in Non-Finnish Europeans) were found in the ExAC database, with an allele frequency of 3.328×10−05, indicating a very low frequency in the population. This variant was predicted to be ‘probably damaging’ in PolyPhen, ‘deleterious’ in SIFT, ‘disease causing’ in MutationTaster, and ‘likely pathogenic’ in InterVar, with evidence of PM2 [absent from controls in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium (20–22)], PM3 (detected in trans with a pathogenic variant), PP2 (missense variant in a gene that has a low rate of benign missense variation and in which missense variants are a common mechanism of disease) and PP3 (multiple lines of computational evidence support a deleterious effect on the gene or gene product) (23). The SPTA1 c.5029G>A (p.Gly1677Arg) mutation may function in trans with SPTA1 c.3897-1G>C in intron 27-1, causing HS in the neonate.
Protein structure analysis of the SPTA1 c.5029G>A mutation
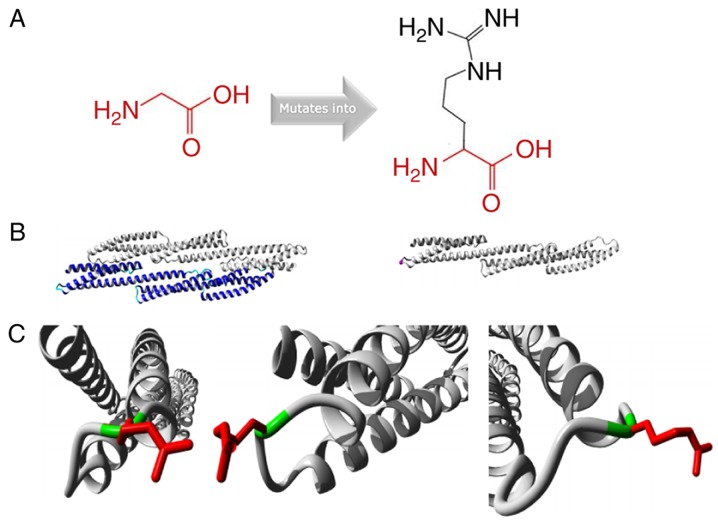
Protein structure analysis of the SPTA1 c.5029G>A (p.Gly1677Arg) mutation was performed using the HOPE approach (http://www.cmbi.ru.nl/hope/) (19). The mutant residue was larger compared with the wild-type residue. The wild-type residue charge was neutral, and the mutant residue charge was positive. The wild-type residue was more hydrophobic compared with the mutant residue (Fig. 3A). The mutation was located within a stretch of residues that was repeated in the protein; this repeat was termed Spectrin 16. The wild-type residue was a glycine, which is the most flexible of all residues (19). This flexibility may be necessary for the function of the protein, and mutation of this glycine may abolish this function (Fig. 3B). The mutated residue was located in a domain that was important for binding of other molecules, and was in contact with residues in a domain that was also important for binding. The mutation may disturb the interaction between these two domains and negatively affect the function of the protein. Moreover, the torsion angles for this residue were unusual. Only glycine was flexible enough to generate these torsion angles, and mutation into another residue may force the local backbone into an incorrect conformation and disrupt the local structure (Fig. 3C).
Figure 3.
Protein structure analysis of SPTA1 c.5029G>A (p.Gly1677Arg). (A) Schematic structures of the original (left) and the mutant (right) amino acid. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black. (B) Left is an overview of the protein in ribbon-presentation. The protein is colored by element: α-helix, blue; β-strand, red; turn, green; 3/10 helix, yellow; and random coil, cyan. Other molecules in the complex are colored grey when present. Right is an overview of the mutant protein in ribbon-presentation. The protein is colored grey and the side chain of the mutated residue is colored magenta (pink spheres). (C) Close-up of the mutation. The protein is colored grey, the side chains of the wild-type and the mutant residue are presented and colored green and red, respectively. The structure is presented in three different angles.
Discussion
HS is a heterogeneous disorder, and the majority of cases are inherited in an autosomal-dominant fashion with an unambiguous family history. The phenotype of HS in neonates ranges from asymptomatic to hydrops fetalis (6,13). HS is the third most common underlying hemolytic anemia, following G6PD and ABO hemolytic disease, among neonates listed in the USA Kernicterus Registry (6). Moreover, HS is the leading cause of Coombs-negative hemolytic anemia, requiring blood transfusion in the first months of life (6). Neonates with HS may present with extreme hyperbilirubinemia, thus a timely diagnosis may prevent adverse outcomes in neonates with HS (24).
For neonates with Coombs-negative hemolytic jaundice and a negative family history, the presence of spherocytes on blood film, an increased MCHC and a decreased MCV suggest a diagnosis of HS (25). Christensen et al (26), and Christensen and Sheffield (27), speculated that an MCHC of 36.0 g/dl or more could alert caregivers to the possibility of HS. However, the neonate described in the present study exhibited normal MCHC, MCV and MCHC/MCV ratio. Hemolytic jaundice and anemia were the principal signs of HS, and blood transfusions were performed almost monthly to maintain the Hb concentration in the blood. Neonates with normal MCHC and MCV have also been reported previously. For example, Yaish et al (25) described a neonate with α-spectrin deficient HS confirmed by SDS-PAGE, and the neonate had Coombs-negative spherocytic hemolytic jaundice in addition to normal MCHC and MCV. These results suggested that for patients with normal MCHC, MCV or MCHC/MCV ratio, additional tests may be required. NGS is widely used in the diagnosis of hereditary hemolytic anemia, including HS (28–31). Moreover, the mutation spectrum and genetic characteristics in the Chinese population have been identified by exome sequencing (31). Additionally, the eosin-5′-maleimide binding test, performed using flow cytometry, has been also recommend for HS diagnosis (32).
The SPTA1 gene is located at 1q22-23, and α-spectrin is an integral part of the erythrocyte cytoskeleton, which is linked to the surface of the plasma membrane through ankyrin and band 3. Homozygous or compound heterozygous mutations in the SPTA1 gene may be involved in the pathogenesis of recessive spectrin-deficient HS, and heterozygous individuals may still produce sufficient α-spectrin to balance β-spectrin and maintain the erythrocyte cytoskeleton (33,34). Missense and splicing mutations account for the majority of cases.
In this case, novel compound heterozygous mutations in the SPTA1 gene (c.3897-1G>C and c.5029G>A) were identified in the neonate. The SPTA1 c.3897-1G>C mutation was not found in the 1000G, EVS, ExAC or HGMD databases. Sanger sequencing of the mRNA reverse transcribed into cDNA identified a deletion of the first 10 nucleotides of exon 28, and the ninth and tenth nucleotides of exon 28 were AG. This result indicated that the SPTA1 c.3897-1G>C mutation in the consensus splice site led to a new AG splice acceptor site at the ninth and tenth nucleotides of exon 28, resulting in a deletion of the first 10 nucleotides of exon 28.
The SPTA1 c.5029G>A mutation resulted in a glycine-to-arginine substitution at amino acid position 1,677 of α-spectrin. Various types of software predicted that this missense mutation would have deleterious effects. Moreover, protein structure analysis implied that this mutation may negatively affect the function of α-spectrin as follows. First, the wild-type residue charge was neutral, whereas the mutant residue was positive. Additionally, the wild-type residue was more hydrophobic compared with the mutant residue, and the mutated residue was located in a domain important for the binding of other molecules and in contact with residues in a domain important for binding. The mutation may disrupt the interaction between these two domains. Only glycine is flexible enough to generate these torsion angles, and mutation into arginine may force the local backbone into an incorrect conformation and disrupt the local structure. Thus, SPTA1 c.5029G>A (p.Gly1677Arg) in trans with the SPTA1 c.3897-1G>C mutation may have caused HS in this neonate.
In summary, novel compound heterozygous mutations in the SPTA1 gene (c.3897-1G>C and c.5029G>A) may be responsible for the clinical manifestations of the described neonatal patient with HS. In the present study. NGS provided a rapid and powerful approach for the genetic diagnosis of HS.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81500925).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
AL, YL and QH designed the study. XW and YL performed the experiments and drafted the manuscript. AL and QH cared for the patient. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent to use blood samples from the subjects was obtained from the parents. The present study was formally approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). All procedures were performed in accordance with the approved guidelines.
Patient consent for publication
Written consent for publication from all subjects was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Da Costa L, Galimand J, Fenneteau O, Mohandas N. Hereditary spherocytosis, elliptocytosis, and other red cell membrane disorders. Blood Rev. 2013;27:167–178. doi: 10.1016/j.blre.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Shen N, Huang M, Lu Y, Hu Q. Novel hereditary spherocytosis-associated splice site mutation in the ANK1 gene caused by parental gonosomal mosaicism. Haematologica. 2018;103:e219–e222. doi: 10.3324/haematol.2017.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iolascon A, Andolfo I, Barcellini W, Corcione F, Garçon L, De Franceschi L, Pignata C, Graziadei G, Pospisilova D, Rees DC, et al. Recommendations regarding splenectomy in hereditary hemolytic anemias. Haematologica. 2017;102:1304–1313. doi: 10.3324/haematol.2016.161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson L, Bhutani VK, Karp K, Sivieri EM, Shapiro SM. Clinical report from the pilot USA kernicterus registry (1992 to 2004) J Perinatol. 2009;29(Suppl 1):S25–S45. doi: 10.1038/jp.2008.211. [DOI] [PubMed] [Google Scholar]
- 5.Gallagher PG. Disorders of red cell volume regulation. Curr Opin Hematol. 2013;20:201–207. doi: 10.1097/MOH.0b013e32835f6870. [DOI] [PubMed] [Google Scholar]
- 6.Christensen RD, Yaish HM, Gallagher PG. A pediatrician's practical guide to diagnosing and treating hereditary spherocytosis in neonates. Pediatrics. 2015;135:1107–1114. doi: 10.1542/peds.2014-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andres O, Eber S, Speer CP. Early postnatal diagnosis of hereditary spherocytosis by combining light microscopy, acidified glycerol lysis test and eosin-5′-maleimide binding assay. Ann Hematol. 2015;94:1959–1964. doi: 10.1007/s00277-015-2491-z. [DOI] [PubMed] [Google Scholar]
- 8.An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367–375. doi: 10.1111/j.1365-2141.2008.07091.x. [DOI] [PubMed] [Google Scholar]
- 9.Yawata Y, Kanzaki A, Yawata A, Doerfler W, Ozcan R, Eber SW. Characteristic features of the genotype and phenotype of hereditary spherocytosis in the Japanese population. Int J Hematol. 2000;71:118–135. [PubMed] [Google Scholar]
- 10.Speicher DW, DeSilva TM, Speicher KD, Ursitti JA, Hembach P, Weglarz L. Location of the human red cell spectrin tetramer binding site and detection of a related ‘closed’ hairpin loop dimer using proteolytic footprinting. J Biol Chem. 1993;268:4227–4235. [PubMed] [Google Scholar]
- 11.Lam VQ, Antoniou C, Rolius R, Fung LW. Association studies of erythroid alpha-spectrin at the tetramerization site. Br J Haematol. 2009;147:392–395. doi: 10.1111/j.1365-2141.2009.07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narla J, Mohandas N. Red cell membrane disorders. Int J Lab Hematol. 2017;39(Suppl 1):S47–S52. doi: 10.1111/ijlh.12657. [DOI] [PubMed] [Google Scholar]
- 13.Perrotta S, Gallagher PG, Mohandas N. Hereditary spherocytosis. Lancet. 2008;372:1411–1426. doi: 10.1016/S0140-6736(08)61588-3. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter. 2013;7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Zhu Y, Shen N, Peng J, Wang C, Liu H, Lu Y. Mutation analysis of a Chinese family with oculocutaneous albinism. Oncotarget. 2016;7:84981–84988. doi: 10.18632/oncotarget.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auer PL, Reiner AP, Wang G, Kang HM, Abecasis GR, Altshuler D, Bamshad MJ, Nickerson DA, Tracy RP, Rich SS, et al. Guidelines for large-scale sequence-based complex trait association studies: Lessons learned from the NHLBI exome sequencing project. Am J Hum Genet. 2016;99:791–801. doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng-Bradley X, Flicek P. Applications of the 1000 genomes project resources. Brief Funct Genomics. 2017;16:163–170. doi: 10.1093/bfgp/elw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen RD, Nussenzveig RH, Yaish HM, Henry E, Eggert LD, Agarwal AM. Causes of hemolysis in neonates with extreme hyperbilirubinemia. J Perinatol. 2014;34:616–619. doi: 10.1038/jp.2014.68. [DOI] [PubMed] [Google Scholar]
- 25.Yaish HM, Christensen RD, Agarwal A. A neonate with Coombs-negative hemolytic jaundice with spherocytes but normal erythrocyte indices: A rare case of autosomal-recessive hereditary spherocytosis due to alpha-spectrin deficiency. J Perinatol. 2013;33:404–406. doi: 10.1038/jp.2012.67. [DOI] [PubMed] [Google Scholar]
- 26.Christensen RD, Henry E. Hereditary spherocytosis in neonates with hyperbilirubinemia. Pediatrics. 2010;125:120–125. doi: 10.1542/peds.2009-0864. [DOI] [PubMed] [Google Scholar]
- 27.Sheffield MJ, Christensen RD. Evaluating neonatal hyperbilirubinemia in late preterm Hispanic twins led to the diagnosis of hereditary spherocytosis in them, and in their sibling and in their mother. J Perinatol. 2011;31:625–627. doi: 10.1038/jp.2010.212. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal AM, Nussenzveig RH, Reading NS, Patel JL, Sangle N, Salama ME, Prchal JT, Perkins SL, Yaish HM, Christensen RD. Clinical utility of next-generation sequencing in the diagnosis of hereditary haemolytic anaemias. Br J Haematol. 2016;174:806–814. doi: 10.1111/bjh.14131. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Yi B, Mu K, Shen N, Zhu Y, Hu Q, Lu Y. Identification of a novel de novo ANK1 R1426* nonsense mutation in a Chinese family with hereditary spherocytosis by NGS. Oncotarget. 2017;8:96791–96797. doi: 10.18632/oncotarget.18243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Jia S, Dewan RK, Liao N. Novel mutations in patients with hereditary red blood cell membrane disorders using next-generation sequencing. Gene. 2017;627:556–562. doi: 10.1016/j.gene.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Wang R, Yang S, Xu M, Huang J, Liu H, Gu W, Zhang X. Exome sequencing confirms molecular diagnoses in 38 Chinese families with hereditary spherocytosis. Sci China Life Sci. 2018;61:947–953. doi: 10.1007/s11427-017-9232-6. [DOI] [PubMed] [Google Scholar]
- 32.Farias MG. Advances in laboratory diagnosis of hereditary spherocytosis. Clin Chem Lab Med. 2017;55:944–948. doi: 10.1515/cclm-2016-0738. [DOI] [PubMed] [Google Scholar]
- 33.Wichterle H, Hanspal M, Palek J, Jarolim P. Combination of two mutant alpha spectrin alleles underlies a severe spherocytic hemolytic anemia. J Clin Invest. 1996;98:2300–2307. doi: 10.1172/JCI119041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nussenzveig RH, Christensen RD, Prchal JT, Yaish HM, Agarwal AM. Novel α-spectrin mutation in trans with alpha-spectrin causing severe neonatal jaundice from hereditary spherocytosis. Neonatology. 2014;106:355–357. doi: 10.1159/000365586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.