Abstract
CYP2C9 and CYP2C19 are highly polymorphic pharmacogenes; however, clinically actionable genetic variability in drug metabolism due to these genes has been limited to a few common alleles. The identification and functional characterization of less-common open reading frame sequence variation might help to individualize therapy with drugs that are substrates for the enzymes encoded by these genes. The present study identified seven uncharacterized variants each in CYP2C9 and CYP2C19 using next-generation sequence data for 1013 subjects, and functionally characterized the encoded proteins. Constructs were created and transiently expressed in COS-1 cells for the assay of protein concentration and enzyme activities using fluorometric substrates and liquid chromatography– tandem mass spectrometry with tolbutamide (CYP2C9) and (S)-mephenytoin (CYP2C19) as prototypic substrates. The results were compared with the SIFT, Polyphen, and Provean functional prediction software programs. Cytochrome P450 oxidoreductase (CPR) activities were also determined. Positive correlations were observed between protein content and fluorometric enzyme activity for variants of CYP2C9 (P < 0.05) and CYP2C19 (P < 0.0005). However, CYP2C9 709G>C and CYP2C19 65A>G activities were much lower than predicted based on protein content. Substrate intrinsic clearance values for CYP2C9 218C>T, 343A>C, and CYP2C19 337G>A, 518C>T, 556C>T, and 557G>A were less than 25% of wild-type allozymes. CPR activity levels were similar for all variants. In summary, sequencing of CYP2C9 and CYP2C19 in 1013 subjects identified low-frequency variants that had not previously been functionally characterized. In silico predictions were not always consistent with functional assay results. These observations emphasize the need for high-throughput methods for pharmacogene variant mutagenesis and functional characterization.
Introduction
Pharmacogenomics (PGx) is the study of the role of genetic variation in variability in drug response phenotypes (Weinshilboum, 2003). Individual response to drug therapy varies widely, with genetic factors possibly accounting for 20%–30% of that variation (Ingelman-Sundberg and Rodriguez-Antona, 2005). While genetic variants contribute to variability in function for genes encoding drug-metabolizing enzymes, drug transporters, drug receptors, and signaling molecules, genes encoding drug-metabolizing enzymes have received the most attention for clinical implementation (Ingelman-Sundberg and Rodriguez-Antona, 2005; Weinshilboum and Wang, 2017). Significant associations of nonsynonymous single-nucleotide polymorphisms (SNPs) in these genes with drug treatment outcomes are reported regularly (Sim et al., 2013). Genetic heterogeneity in genes encoding drug-metabolizing enzymes contributes to population heterogeneity in drug response by influencing both pharmacokinetics and pharmacodynamics (Dresser et al., 2000; Kim et al., 2008).
Cytochrome P450 (P450) enzymes in families 1–3 metabolize 70%–80% of all clinically used drugs that undergo phase I metabolism (Ingelman-Sundberg et al., 2007; Sim et al., 2013). Nearly 40%–45% of those drugs are cleared by oxidative metabolism catalyzed by CYP2C9, CYP2C19, and CYP2D6 (Kirchheiner et al., 2004). The genes encoding these P450s have numerous polymorphisms, and those polymorphisms may have significant impact on the clinical effects of drugs with narrow therapeutic indices. Therefore, the US Food and Drug Administration has adopted black box warnings for several narrow therapeutic index drugs, recommending PGx testing before prescribing these drugs. Similarly, many drugs with potentially promising efficacy never reach the market because of adverse drug reactions or large variation in efficacy. It is possible that PGx variants may explain the large variation in response to drug treatment and predict adverse drug reactions (Friedman et al., 1999; Murphy, 2000).
Advanced sequencing technologies continue to identify variants in the drug-metabolizing enzyme genes that encode allozymes with poorly characterized or unknown clinical significance. Next-generation sequencing (NGS) studies of large populations such as the 100,000 Genomes Projects (Mark et al., 2017) have generated DNA sequence data for PGx genes. Several years ago, the Mayo Clinic instituted the “Right Drug, Right Dose, Right Time—Using Genomic Data to Individualize Treatment Protocol” (i.e., the RIGHT protocol) in collaboration with the National Human Genome Research Institute, the Electronic Medical Records and Genomics Network, and the Pharmacogenomics Research Network (Bielinski et al., 2014). That project was designed to investigate the clinical implementation of preemptive PGx testing and provide clinicians with point-of-care PGx-guided prescribing information. Specifically, 1013 subjects gave permission for the sequencing of their DNA for 84 pharmacogenes among which CYP2C19, CYP2C9, VKORC1, CYP2D6, and SLCO1B1 are incorporated into the electronic health record (Ji et al., 2016).
The RIGHT study identified a series of rare variants in the open reading frames (ORFs) of CYP2C9 and CYP2C19. Those variants either lacked functional characterization or had not been previously reported. The standard strategy to identify the functional significance of variants begins with the preparation of recombinant allozymes using site-directed mutagenesis followed by expression in human cell lines, measurement of protein concentrations, and determination of enzyme activity using prototypic substrates (Weinshilboum et al., 1999; Wang et al., 2003, 2005; DeLozier et al., 2005; Dai et al., 2014a,b, 2015; Niinuma et al., 2014; Hu et al., 2015). Computational methods have also been developed in an attempt to predict the effect of genetic variation in encoded amino acid sequence on protein stability and enzyme activity, but their validation and accuracy remains unclear (Flanagan et al., 2010).
The present study was designed to determine the functional effect of rare variants found in both CYP2C9 and CYP2C19 in the RIGHT sequence data. We used a standard approach to prepare recombinant variant allozymes and studied the effect of those variants on protein level and enzyme activity using both fluorescent probe substrates and prototypic substrates. The functional consequences of many of the variants could not be accurately predicted by current algorithms. Study of these variants showed functional effects that might indicate clinical utility with regard to variation in drug response phenotypes. If these results can be generalized, they indicate that, ultimately, DNA sequencing will be preferable to genotyping for the clinical implementation of pharmacogenomic variants, and they also strongly support the need for high-throughput functional assays and accurate predictive algorithms to make it possible to achieve the optimal reduction in adverse drug reactions and the optimal increase in drug efficacy that pharmacogenomics promises.
Materials and Methods
Study Subjects.
The RIGHT Study enrolled 1013 participants and sequenced 84 pharmacogenes for each subject. This study was conducted according to the Declaration of Helsinki and was reviewed and approved by the Mayo Clinic Institutional Review Board. The subjects were 86% non-Hispanic Caucasians, and 53% were women with 96.7% Caucasian, 1% Asian, 0.6% African American, 0.1% American Indian/Alaskan Native, 0.5% other race, and 1.1% unknown or chose not to disclose their ethnicity (Bielinski et al., 2014; Ji et al., 2016).
CYP2C9 and CYP2C19 Gene Sequencing.
PGx gene capture and NGS of DNA were conducted in the Personalized Genomics Laboratory and Clinical Genome Sequencing Laboratory, Mayo Clinic, Rochester, MN. The Pharmacogenomics Research Network sequencing (PGRN-Seq, version 1.0) capture reagent panel was used to sequence 84 pharmacogenes and covered 968 kb that included approximately 2 kb upstream of and downstream from the coding regions of these genes (Ji et al., 2016). The KAPA HTP Library Preparation Kit (Kapa Biosystems, Inc., Wilmington, MA) and Bioo Scientific NEXTflex barcode adapters (Bioo Scientific Corporation, Austin, TX) were used for library preparation and precapture pooling. Samples were sequenced using the Illumina HiSeq2500 Sequencing System in the rapid run mode by using the TruSeq Rapid SBS Kit (Illumina, San Diego, CA) with the 200-cycle and 2 × 101 pair end reads capability (Ji et al., 2016). FASTQC (Babraham Institute, Cambridge, UK) was used to assess raw read quality. Files were aligned to the hg19 reference genome using NovoAlign (VN:V2.07.13; Novocraft Technologies, Selangor, Malaysia). Single-nucleotide variants were identified by CLC’s Neighborhood Quality calling method.
Specific regions around known CYP2C9 and CYP2C19 alleles were examined, and Sanger sequencing was used to confirm observed variants. Each variant was confirmed 10 times. The NGS workbench, which is an internally developed Mayo Clinic program that facilitates results interpretation, was used to review quality metrics and manually annotate novel or ambiguous sequence alterations (Bielinski et al., 2014).
CYP2C9 and CYP2C19 cDNA Expression Constructs.
Human CYP2C9 cDNA and human CYP2C19 cDNA clones in the eukaryotic expression vector pCMV6-XL5 were obtained from OriGene Technologies, Inc. (Rockville, MD). Site-directed mutagenesis was then performed using the QuikChange Lightning Kit (Agilent Technologies, Santa Clara, CA) to create expression constructs for each of the variants to be studied. Sequences of the primers used to perform site-directed mutagenesis are listed in the Supplemental Material (see Supplemental Tables 1 and 2). The sequences of the human CYP2C9 cDNA and CYP2C19 cDNA clones and all variant constructs were also confirmed by Sanger sequencing.
Expression of CYP2C9 and CYP2C19 Variant Proteins in COS-1 Cells.
COS-1 African green monkey kidney cells do not express CYP2C9 or CYP2C19, and as a result were selected for use in our expression studies. Specifically, the cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. At 24 hours before transfection, the cells were plated at a density of 1.25 × 106 per T75 flask. Subsequently, the cells were transfected with plasmids carrying CYP2C9 or CYP2C19 cDNA using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After 6 hours of incubation at 37°C, the culture medium was replaced with Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, and the cells were incubated for an additional 66 hours. Each expression clone was homozygous for the nucleotide change. The cells were then washed with PBS, followed by trypsinization and pelleting in PBS (2000 rpm centrifugation for 10 minutes at 4°C) for S9 fractionation.
Preparation of S9 Fractions of Cell Pellets.
Cells were resuspended in 300 µl of 0.25 M sucrose. After sonication for 20 seconds using a probe sonicator and centrifugation at 2500 rpm for 5 minutes, the supernatant was transferred to a new tube and centrifuged at 9000 rpm for 10 minutes. The resulting supernatant was transferred to a microcentrifuge tube. The DC Protein Assay Kit (Bio Rad, Hercules, CA) was used to measure the protein content of the S9 fractions.
Western Blot Analysis.
Quantitative Western blot analyses were performed using CYP2C9 or CYP2C19 S9 fractions. Proteins were separated by SDS-PAGE prior to transfer to polyvinylidene fluoride membranes. The membranes were incubated with rabbit polyclonal CYP2C9 antibody (Abcam) or CYP2C19 antibody (Sigma, St. Louis, MO) at 1:1000 or 1:250 dilution, respectively. ACTB protein was measured using mouse monoclonal ACTB antibody (Sigma), and its expression was used as a loading control. The expression of each variant was normalized with regard to that of the wild type (WT). Proteins were detected using the SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, Rockford, IL), and images were captured on X-ray films. Quantification of protein density on X-ray films was performed with the National Institutes of Health ImageJ software program (https://imagej.nih.gov/ij/download.html).
Protein Degradation Experiments.
In some experiments, cells were treated with 10 μM 3-methyladenine [(3MA); Selleckchem, Houston, TX] for 48 hours or 10 μM carbobenzoxy-L-leucyl-L-leucyl-L-leucinal (MG132), a proteasome inhibitor (Selleckchem), for 8 hours to determine whether CYP2C9 and CYP2C19 variant allozymes might be degraded by either autophagy (3MA) or by a proteasome-mediated process (MG132) in COS-1 cells (Wang et al., 2004, 2005; Ji et al., 2007; Pereira et al., 2010; Liu et al., 2017). The cells were then collected for S9 fractionation.
Enzyme Assays.
CYP2C9 and CYP2C19 enzyme activities were measured using a modification of the Vivid assay (Thermo Fisher Scientific, Carlsbad, CA). In this assay, blocked dye substrates, benzyloxy-methyl-fluorescein and 7-(ethoxymethoxy)-3-cyanocouamrin for the CYP2C9 and CYP2C19 enzymes, respectively, were metabolized into fluorescent products in an aqueous solution. In our experiments, we incubated CYP2C9 and CYP2C19 S9 fractions at protein concentrations of 4.8 and 3.9 mg/100 μl, respectively, with an NADPH regeneration system consisting of glucose 6-phosphate and glucose-6-phosphate dehydrogenase and reaction buffer, which were included in the kit, at room temperature in triplicate for 20 minutes in a 96-well plate. The enzyme reaction was initiated by the addition of a mix of NADP+ and appropriate Vivid substrate concentrations [2 μM of Vivid benzyloxy-methyl-fluorescein substrate for CYP2C9 and 10 μM of Vivid 7-(ethoxymethoxy)-3-cyanocouamrin substrate for CYP2C19, which were provided in the kit]. Immediately (less than 2 minutes) after initiation of the enzyme reaction, fluorescent products were measured at intervals of 15 minutes spectrophotometrically at excitation/emission wavelengths of 490/520 nm for CYP2C9 and 415/460 nm for CYP2C19 substrates, against Vivid fluorescent standard concentrations of 500, 250, 125, 62.5, 15.625, 7.8125, and 0 nM in duplicate wells. Relative enzyme activities were measured compared with WT activity as 100%. Enzyme activity of all variant allozymes of CYP2C9 and CYP2C19 were conducted on the same day at the same time alongside their respective WT enzymes.
CYP2C9 and CYP2C19 enzyme kinetic parameters were characterized with tolbutamide and (S)-mephenytoin, respectively, as substrates. Specifically, S9 fractions of CYP2C9 and CYP2C19 enzyme suspensions were incubated in a 2 ml microcentrifuge tube maintained at 37°C in a shaker bath. Each incubation mixture (100 µl) contained S9 fraction (0.4 mg/ml protein final concentration), NADPH (1 mM), magnesium chloride (5 mM), and potassium phosphate buffer (50 mM pH 7.5, CYP2C9) or Tris buffer (100 mM pH 7.4, CYP2C19). Identical S9 incubations in which NADPH was omitted served as controls. After preincubation of the S9 fractions with the reaction buffer (NADPH+/−) for 2 minutes, the metabolic process was initiated by adding tolbutamide (CYP2C9) or (S)-mephenytoin (CYP2C19) for final concentrations of 1000, 500, 100, 50, and 10 µM. Reactions were terminated after 30-minute incubations by mixing with ice-cold methanol (2:1, v/v) containing 282 nM (hydroxy tolbutamide-d9) or 354 nM [(+/−)-4-hydroxy mephenytoin-d3] internal standards, and were then vortexed for 1 minute before centrifugation at 14,000g for 15 minutes. The resultant supernatant was collected for liquid chromatography– tandem mass spectrometry (LC-MS/MS) analysis.
Cytochrome P450 oxidoreductase (CPR) activity was measured in the S9 fraction of cells in which CYP2C9 and CYP2C19 variant allozymes had been expressed using a colorimetric Cytochrome P450 Reductase Activity Assay Kit (Abcam). This assay uses the oxidation of NADPH catalyzed by cytochrome P450 reductase to cause the conversion of a nearly colorless probe substrate into a brightly colored product with an absorbance at a 460-nm optical density wavelength. A glucose 6-phosphate standard curve preparation of 0, 2, 4, 6, 8, and 10 nmol/well in a 96-well plate was used. To subtract any extraneous reductase activity in the samples, all variant samples were assayed in parallel with the presence and absence of 0.1 mM diphenyleneiodonium chloride, an inhibitor of NADPH-dependent flavoprotein, subtracting any residual activity detected with the inhibitor present. The optical density was measured using a spectrophotometer in kinetic mode for 30 minutes. The CPR activity was calculated between two time points in the linear range. The rate of color formation is directly proportional to CPR activity. Activity was expressed as milliunits per milligram of total protein. One unit of CPR activity is equal to the amount of cytochrome P450 reductase that will generate 1.0 µmol of reduced substrate per minute by oxidizing 1.0 µmol NADPH to β-NADP+ at pH 7.7 at 25°C.
Drug Analysis Using LC-MS/MS Assay.
Tolbutamide, hydroxy tolbutamide, hydroxy tolbutamide-d9, (S)-mephenytoin, (S)-4-hydroxy mephenytoin, and (+/−)-4-hydroxy mephenytoin-d3 (used for internal standard) were obtained from Toronto Research Chemicals (Toronto, ON, Canada). High-performance liquid chromatography–grade methanol and water were purchased from EM Science (Gibbstown, NJ). Formic acid (minimum 95%), DMSO, β-nicotinamide adenine dinucleotide phosphate reduced form (NADPH), potassium phosphate dibasic, potassium phosphate monobasic, and magnesium chloride were purchased from Sigma. PBS was purchased from Invitrogen. Tris HCL buffer 10X solution was purchased from Sigma. Sodium hydroxide solution was purchased from Sigma Aldrich. Deionized and distilled water was used to prepare buffer solution.
Stock solutions of 1 mg/ml of tolbutamide, hydroxy tolbutamide, hydroxyl tolbutamide-d9, (S)-mephenytoin, (S)-4-hydroxy mephenytoin, and (+/−)-4-hydroxy mephenytoin-d3 were prepared in methanol and stored at −20°C. The concentrations of 20X stock solutions were 2.9, 14.6, 29.1, 58.2, 146, 291, 1455, and 2910 nM for hydroxy tolbutamide and 3.6, 17.8, 35.6, 71.1, 178, 356, 1789, and 3558 nM for (S)-4-hydroxy mephenytoin, which were prepared by diluting the 1 mg/ml stock solutions with 1:1 methanol:water and were stored at −20°C. Standard samples and Quality Control’s [hydroxy tolbutamide (8.7, 218, and 2182 nM) and (S)-4-hydroxy mephenytoin (10.7, 267, and 2668 nM) concentrations] were prepared in assay buffer containing 0.4 mg of protein (bovine serum albumin). Incubations of triplicate standard curves, Quality Control’s, and sample mixtures were performed at 37°C in a total volume of 100 μl. Interday and Intraday variability for CYP2C9 and CYP2C19 standard curves was <25% for their respective lowest standard. The concentration ranges of samples for CYP2C9 and CYP2C19 were 0–178 and 0–694 nM, respectively.
The separation of tolbutamide, hydroxy tolbutamide, hydroxy tolbutamide-d9, (S)-mephenytoin, (S)-4-hydroxy mephenytoin, and (+/−)-4-hydroxy mephenytoin-d3 was achieved with a precolumn filter (Column Saver, MAC-MOD Analytical, Inc., Chadds Ford, PA) and a Waters Select HSS T3 column (2.1 × 100 mm, XP, 2.5 μm) (Waters Corporation, Milford, MA) by gradient elution utilizing the following profile: 2–6 minutes for 75% A and 25% B, 6–8 minutes for 5% A and 95% B, and 8–10 minutes for 75% A and 25% B, where solvent A was water containing 0.1% formic acid and solvent B was methanol containing 0.1% formic acid. The flow rate was 0.2 ml/min. After sample injection (20 μl), the column effluent was diverted to waste for 3 minutes, after which the flow was switched to the mass spectrometer.
Metabolites of tolbutamide and (S)-mephenytoin were monitored using a modification of a previously published LC-MS/MS assay (Peng et al., 2015). The LC-MS/MS system used to perform the assays consisted of a Shimadzu liquid chromatograph (Wood Dale, IL) with two LC-10ADvp pumps (flow rate 0.200 ml/min), and an SIL-10ADvp autoinjector (injection volume 20 μl) coupled to a Quattro Micro mass spectrometer fitted with an electrospray ionization (ESI) probe (Waters Corporation). Hydroxy tolbutamide detection was accomplished using multiple reactions monitoring in positive ESI mode with a parent ion of 287.1 mass-to-charge ratio (m/z) and daughter ion of 74.1 m/z (dwell = 0.1 second, cone = 28 V, and collision energy = 13 eV). Internal standard (hydroxy tolbutamide-d9) detection was performed using multiple reactions monitoring in positive ESI mode with a parent ion of 296.1 m/z and daughter ion of 83.2 m/z (dwell = 0.1 second, cone = 30 V, and collision energy = 14 eV). (S)-4-hydroxy mephenytoin detection was accomplished with a parent ion of 235.1 m/z and daughter ion of 150.2 m/z (dwell = 0.1 seconds and cone = 19 eV). Internal standard [(+/−)-4-hydroxy mephenytoin-d3] detection was performed using multiple reactions monitoring in positive ESI mode with a parent ion of 238.2 m/z and daughter ion of 150.2 m/z (dwell = 0.1 second and cone = 19 eV). The source temperature, desolvation temperature, and cone and desolvation gas flows were 120°C, 350°C, and 650 and 25 l/h, respectively. Mass spectrometry data were collected for 10 minutes after injection. Spectra and chromatograms were processed using the MassLynx version 3.5 software (Waters Corporation). Metabolism data were acquired using a full scan function (mass spectrometry scan) over the range of potential metabolites (50–450 m/z). Once the metabolite masses were determined, a daughter ion scan was performed to determine and confirm the structure of the metabolite.
In Silico Variant Sequence Prediction Analysis.
The Polyphen version 2 (Adzhubei et al., 2010), PROVEAN (Choi and Chan, 2015), and SIFT software programs (Sim et al., 2012)—three commonly used, publically available variant effect prediction tools—were used to predict the functional impact of the novel variants identified across CYP2C9 and CYP2C19. The web server versions of these tools were used under their default settings.
Statistics.
Protein expression data were analyzed using GraphPad Prism7 software (GraphPad Software, La Jolla, CA). Data are displayed as mean ± S.E.M. Protein expression was analyzed using ANOVA followed by appropriate post hoc tests for multiple comparisons. A value of P < 0.05 was considered statistically significant. Enzyme activity was analyzed using one-way ANOVA, with mean comparison for each variant genotype against the wild-type control performed using Dunnet’s test. Corresponding P values were multiplicity adjusted and reported as such. Associations between protein content and enzyme activity were evaluated using Pearson correlations, with two-sided t test P values reported. Enzyme kinetics parameters were modeled using nonlinear regression based on the Michaelis-Menten equation. All P values less than 0.05 were considered statistically significant. Analyses were performed using GraphPad Prism version 7.
Results
Variants in CYP2C9 and CYP2C19.
Sequencing of the DNA from the 1013 subjects enrolled in the RIGHT Study identified seven heterozygous variants in the CYP2C9 gene and seven variants in the CYP2C19 gene (Table 1) that are not included in current clinical algorithms or guidelines for the metabolic phenotypes of these enzymes. The Genome Aggregation Database [(gnomAD); https://gnomad.broadinstitute.org/] and the SNP database [(dbSNP); https://www.ncbi.nlm.nih.gov/projects/SNP/], were searched for previous reports of these variants and the results from the gnomAD and dbSNP are listed in Table 2. For CYP2C9, variant 218C>T was found in our cohort with a heterozygous CYP2C9*2 allele. CYP2C9 variants 229C>A and 709G>C are variants that had not been reported in the gnomAD or dbSNP. For CYP2C19, variant 65A>G was found together with heterozygous CYP2C19*17, variant 337G>A was found with heterozygous CYP2C19*2 and CYP2C19*17, variants 518C>T and 578A>G were found with heterozygous CYP2C19*2, and variants 556C>T and 557G>A were found with homozygous CYP2C19*2. Variant 815A>G is a variant that had not been reported in the gnomAD or dbSNP data banks.
TABLE 1.
CYP2C9 and CYP2C19 variants cDNA and encoded amino acid changes
| Gene | cDNA Change | Amino Acid Change (Protein) | Comment |
|---|---|---|---|
| CYP2C9 | 218C>T | Pro73Leu | Found with heterozygous CYP2C9 *2 |
| CYP2C9 | 229C>A | Leu77Met | |
| CYP2C9 | 343A>C | Ser115Arg | |
| CYP2C9 | 707delA | Asn236Thrfs*5 | |
| CYP2C9 | 709G>C | Val237Leu | |
| CYP2C9 | 791T>C | Ile264Thr | |
| CYP2C9 | 801C>T | Phe267Phe | |
| CYP2C19 | 65A>G | Gln22Arg | Found with heterozygous CYP2C19 *17 |
| CYP2C19 | 337G>A | Val113Ile | Found with heterozygous CYP2C19 *2 and *17 |
| CYP2C19 | 518C>T | Ala173Val | Found with heterozygous CYP2C19 *2 |
| CYP2C19 | 556C>T | Arg186Cys | Found with homozygous CYP2C19 *2 |
| CYP2C19 | 557G>A | Arg186His | Found with homozygous CYP2C19 *2 |
| CYP2C19 | 578A>G | Gln193Arg | Found with heterozygous CYP2C19 *2 |
| CYP2C19 | 815A>G | Glu272Gly |
TABLE 2.
Data search results for CYP2C9 and CYP2C19 variants in the gnomAD data bank
The database search results were obtained from gnomAD (https://gnomad.broadinstitute.org/) and dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/) in August 2018.
| Gene | cDNA Change | dbSNP Data Bank |
gnomAD Data Bank (Minor Allele Frequency) | |
|---|---|---|---|---|
| rsID | Validation | |||
| CYP2C9 | 218C>T | rs762081829 | Not done | 5.69 × 10−05 |
| CYP2C9 | 229C>A | Not present | Not recorded | |
| CYP2C9 | 343A>C | rs771237265 | 1 | 1.01 × 10−04 |
| CYP2C9 | 707delA | rs549167718 | 2 | 4.099 × 10−06a |
| CYP2C9 | 709G>C | Not present | Not recordedb | |
| CYP2C9 | 791T>C | rs761895497 | 1 | 1.09 × 10−05 |
| CYP2C9 | 801C>T | rs149158426 | 1, 2, 3 | 8.55 × 10−04 |
| CYP2C19 | 65A>G | rs144928727 | 3 | 1.22 × 10−05 |
| CYP2C19 | 337G>A | rs145119820 | 1, 2, 3 | 2.24 × 10−04 |
| CYP2C19 | 518C>T | rs61311738 | 1, 2, 3 | 4.66 × 10−03 |
| CYP2C19 | 556C>T | rs183701923 | 1, 2, 3 | 9.74 × 10−05 |
| CYP2C19 | 557G>A | rs140278421 | 1, 2, 3 | 1.08 × 10−04 |
| CYP2C19 | 578A>G | Not present | 4.06 × 10−06 | |
| CYP2C19 | 815A>G | Not present | Not recorded | |
rsID, reference SNP identification; 1, validated by frequency or genotype data (minor alleles were observed in at least two chromosomes); 2, SNP was sequenced in the 1000 genome project; 3, validated by multiple, independent submissions to the reference SNP cluster.
Without frameshift.
Other mutations to Ile, Phe, Val, and Ala recorded at this position.
Changes in Gene Expression by Novel CYP2C9 and CYP2C19 Variants.
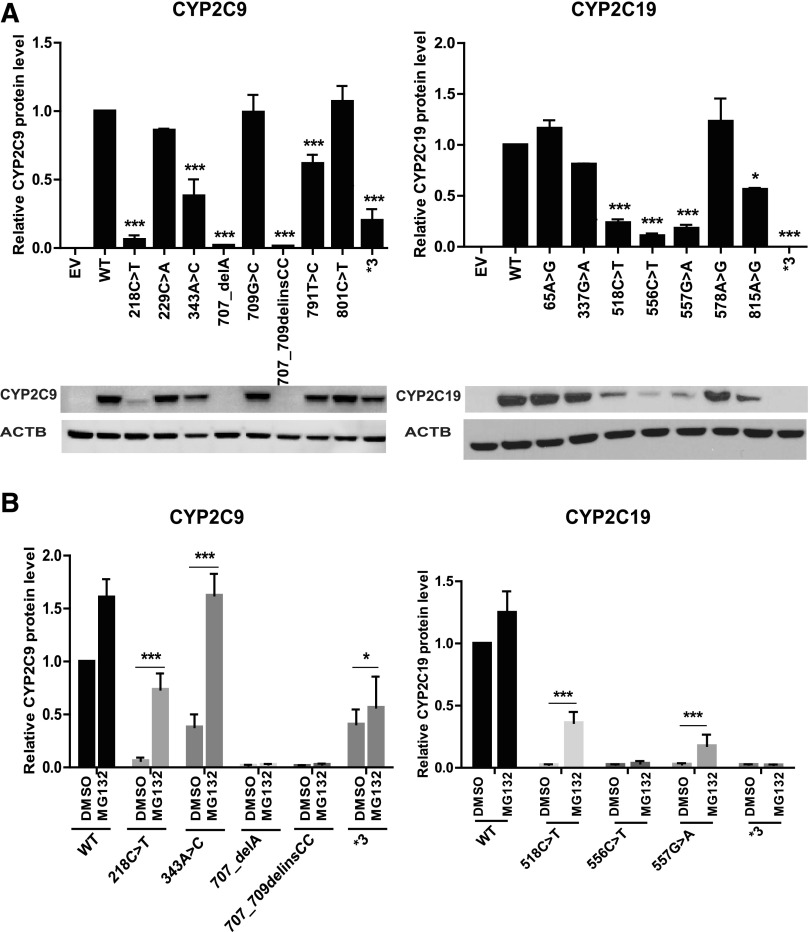
As a first step in our functional analysis, we sought to determine if the variants found in both CYP2C9 and CYP2C19 would influence protein expression and the enzyme activity of allozymes encoded by the variant sequences. Each expression clone was homozygous for the nucleotide change. Quantitative western blot analysis of protein expression showed reduced expression of CYP2C9 protein for three of the identified CYP2C9 variants (229C>A, 709G>C, and 801C>T) (Fig. 1A). In a similar fashion, quantitative western blot analysis of protein expression showed reduced expression of CYP2C19 protein for four of the identified CYP2C19 variants (65A>G, 337 G>A, 578A>G, and815A>G), as well as in loss of function variants CYP2C9*3 (Andersson et al., 2012; Prieto-Pérez et al., 2013; Lee et al., 2014) and CYP2C19*3 (Scott et al., 2011, 2013) (Fig. 1A).
Fig. 1.
(A) Quantitative western blot analysis of newly identified variants in CYP2C9 and CYP2C19 (*P ≤ 0.05; ***P < 0.001 vs. WT). (B) Effect of MG132 treatment on the protein levels of CYP2C9 and CYP2C19. All values are mean ± S.E.M. for three separate independent assays. ANOVA was performed to compare gene expression, followed by multiple comparisons tests for individual comparisons when significant effects were detected. *P ≤ 0.05; ***P ≤ 0.001.
We next set out to determine the mechanism(s) responsible for the decreased protein levels of CYP2C9 and CYP2C19 variant allozymes. Accelerated protein degradation is a common mechanism that can contribute to decreased levels of protein, either as a result of ubiquitin-proteasome or autophagy-mediated degradation (Li et al., 2008; Deng et al., 2016). Therefore, we tested the effect of an autophagy inhibitor (3MA) and a proteasome inhibitor (MG132) on protein expression levels of selected variant allozymes with levels that were decreased to at least 50% or less compared with the WT allozymes (Fig. 1B). Strikingly, three variants for CYP2C9 and two variants for CYP2C19 displayed significantly increased protein expression compared with baseline after treatment with MG132. Although it was not statistically significant, we also noticed that both CYP2C9 and CYP2C19 WT protein levels increased in response to exposure to MG132—suggesting that the proteasome may play a role in basal levels of protein expression for these enzymes. These observations suggest that several of the variants for both enzymes undergo accelerated proteasome-mediated degradation. Finally, protein levels for none of the variant allozymes were significantly affected by treatment with 3MA (data not shown).
Changes in Enzyme Activity in Novel CYP2C9 and CYP2C19 Variants.
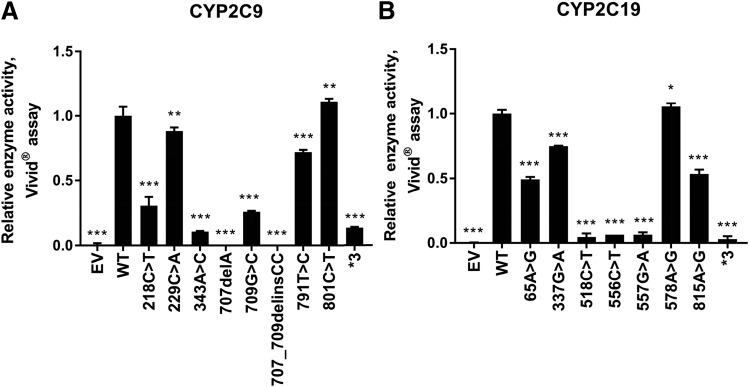
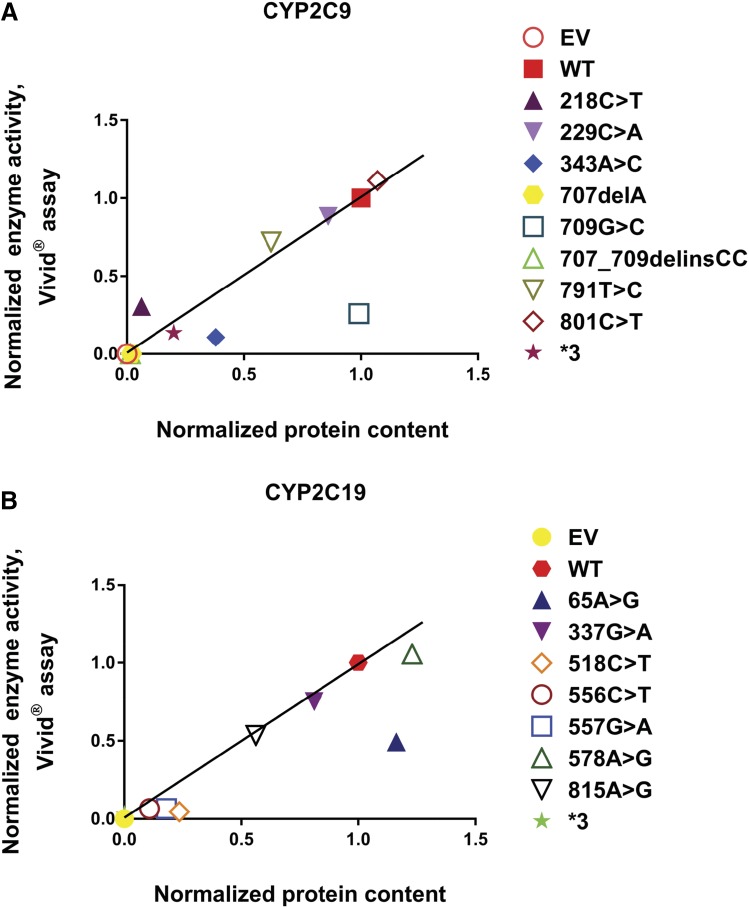
Enzyme activity levels for these same preparations were determined using the fluorogenic Vivid substrates, and those results are shown in Fig. 2. These results were generally comparable with the results of the protein expression studies (Fig. 1A) with some notable exceptions. For example, CYP2C9 709G>C and CYP2C19 65A>G displayed significantly reduced enzyme activity, but their protein level was similar to that of the WT allozyme. The reduced activity could be due to changes in the nucleotide sequence near the active site affecting the enzyme activity more than the protein level. Overall, there was a significant positive correlation between levels of enzyme activity and protein content for both CYP2C9 (P = 0.0015; r2 = 0.693) and CYP2C19 (P = 0.0003; r2 = 0.825), respectively, as shown graphically in Fig. 3.
Fig. 2.
(A) Relative CYP2C9 Vivid enzyme activity. Bar graphs show normalized Vivid enzymatic activity of the CYP2C9 WT and variant proteins (nanomolar fluorescent product formed per minute per milligram of total protein); error bars depict S.D. of the mean in three independent activity assays. (B) Relative CYP2C19 Vivid enzyme activity. Bar graphs show normalized Vivid enzymatic activity of the CYP2C19 WT and variant proteins (nanomolar fluorescent product formed per minute per milligram of total protein); error bars depict S.D. of the mean in three experimental samples. Enzyme activity was analyzed using one-way ANOVA, with mean comparison for each variant genotype against the WT control performed using Dunnet’s test. Corresponding P values were multiplicity adjusted and reported as such. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Fig. 3.
(A) CYP2C9 Vivid enzyme activity vs. protein content: correlation of recombinant protein quantity vs. Vivid enzyme activity for CYP2C9 variant allozymes. The normalized protein quantities are plotted on the horizontal axis and the normalized Vivid enzyme activities are plotted on the vertical axis. R2 = 0.693 and P = 0.0015. (B) CYP2C19 Vivid enzyme activity vs. protein content: correlation of recombinant protein quantity vs. Vivid enzyme activity for CYP2C19 variant allozymes. The normalized protein quantities are plotted on the horizontal axis and the normalized Vivid enzyme activities are plotted on the vertical axis. R2 = 0.825 and P = 0.0003. Associations between protein content and enzyme activity were evaluated using Pearson correlations, with two-sided t test P values reported. The solid line illustrates the line of identity for the association between normalized activity and protein content.
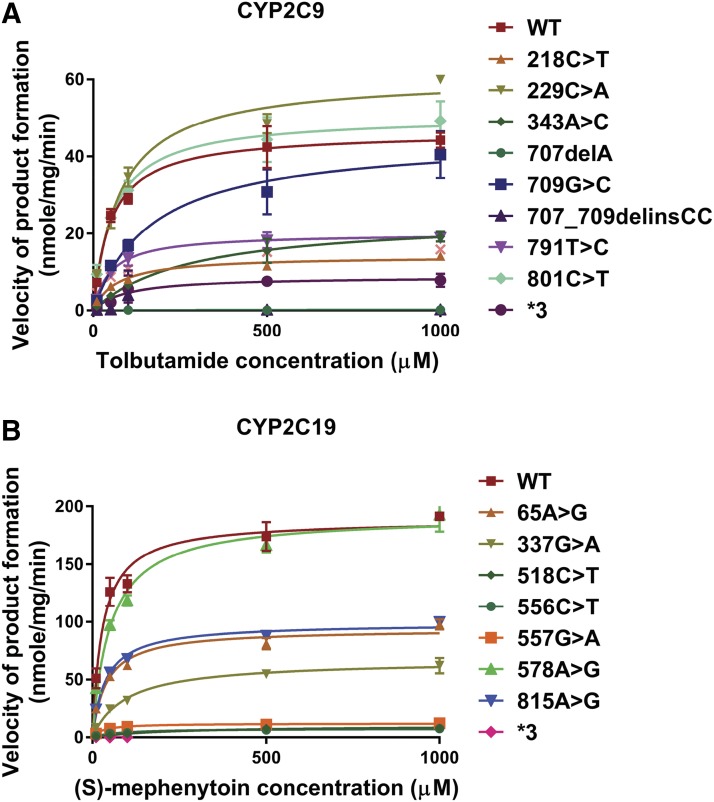
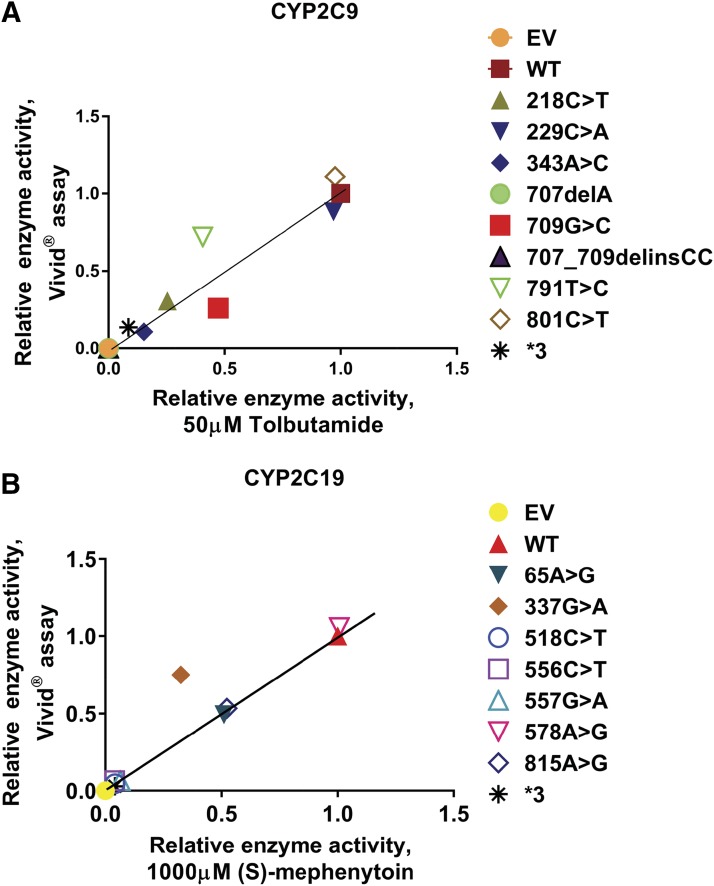
The enzyme activities of CYP2C9 and CYP2C19 variant allozymes expressed in COS-1 cells were also determined with the prototypic substrates tolbutamide and (S)-mephenytoin, respectively. The concentration ranges of product formation in samples for CYP2C9 and CYP2C19 were 0–178 and 0–694 nM, respectively. Michaelis-Menten plots for tolbutamide (CYP2C9) and (S)-mephenytoin (CYP2C19) are shown in Fig. 4. Kinetic parameters for recombinant CYP2C9 variant allozymes as well as empty vector, WT, and CYP2C9*3 as negative controls, are listed in Table 3. Similarly, kinetic parameters for recombinant CYP2C19 variants together with empty vector, WT, and CYP2C9*3 as negative control are given in Table 4. For CYP2C9, we found that the catalytic activities of variants were less than 25% for 218C>T, 343A>C, 707delA, and 707_709delinsCC; 25%–50% (intermediate activity) for variants 709G>C and 791T>C; and 50%–100% (equal or similar to WT) for 229C>A and 801C>T when compared with that of the WT allozyme. Similarly, the catalytic activities of CYP2C19 variants were less than 25% for 337G>A, 518C>T, 556C>T, and 557G>A; 25%–50% (intermediate activity) for variants 65A>G and 815A>G; and 50%–100% (equal or similar) for 578A>G when compared with that of the WT allozyme. There was a significant positive correlation between enzyme activities determined using the Vivid reagent and the velocity of product formation at 50 μM drug concentration for CYP2C9 (P = <0.0001; r2 = 0.907) and 1000 μM drug concentration for CYP2C19 (P = <0.0001; r2 = 0.898), respectively, as shown in Fig. 5.
Fig. 4.
(A) Michaelis-Menten curves of the enzymatic activities of the recombinant WT and 9 variant CYP2C9 allozymes (including CYP2C9*3) toward tolbutamide (each point represents the mean ± S.D. of three separate experiments). (B) Michaelis-Menten curves of the enzymatic activities of the recombinant WT and eight variant CYP2C19 proteins (including CYP2C19*3) toward (S)-mephenytoin (each point represents the mean ± S.D. of three separate experiments). Enzyme kinetics was modeled using nonlinear regression based on the Michaelis-Menten equation.
TABLE 3.
Enzyme kinetic properties of recombinant WT and mutant CYP2C9 proteins for tolbutamide hydroxylation and in silico functional prediction of CYP2C9 variants
These data represent the mean ± S.D. of three independently performed catalytic assays. Concordance is defined as agreement of the kinetic assay result with at least two of three in silico software results.
| cDNA |
Vmax ± S.D. × 10−3 |
Km ± S.D. |
CLINT × 10−4 [Relative CLINT (%WT)] |
Polyphen |
SIFT |
PROVEAN |
Enzyme Activity in Relation to WT |
Concordance
(Yes/No) |
|
|---|---|---|---|---|---|---|---|---|---|
| Vivid* |
MS* |
||||||||
| nmol/mg per minute | µM | µl/mgprotein per minute | % | % | |||||
| WT | 13.5 ± 0.64 | 51 ± 10.51 | 2.71 ± 0.41 | ||||||
| 218C>T | 4.11 ± 0.22 | 67.8 ± 16.1 | 0.62 ± 0.11 (23)** | Probably damaging | Damaging | Deleterious | 31 | 23 | Yes |
| 229C>A | 17.7 ± 0.61 | 76.2 ± 7.49 | 2.33 ± 0.16 (86) | Probably damaging | Damaging | Neutral | 88 | 86 | No |
| 343A>C | 7.12 ± 0.79 | 283 ± 49.2 | 0.26 ± 0.04 (9.6)** | Probably damaging | Damaging | Deleterious | 11 | 10 | Yes |
| 707_delAa | 0.42 ± 0.04 | 141 ± 183 | Possibly damaging | N/A | N/A | 0 | 0 | N/A | |
| 709G>C | 13.9 ± 3.36 | 219 ± 127 | 0.73 ± 0.24 (29)** | Benign | Tolerated | Neutral | 26 | 27 | No |
| 707_709delinsCCa | 0.06 | N/A | N/A | N/A | |||||
| 791T>C | 5.83 ± 0.41 | 50.1 ± 5.62 | 1.17 ± 0.09 (43.6)* | Possibly damaging | Tolerated | Deleterious | 72 | 43 | Yes |
| 801C>T | 14.7 ± 0.46 | 56.5 ± 10.6 | 2.66 ± 0.42 (98.1) | Tolerated | Neutral | 111 | 98 | Yes | |
| *3 | 2.80 ± 0.81 | 149 ± 157 | 0.32 ± 0.24 (10.3)** | 1 | 10 | ||||
CLINT, intrinsic clearance; Km, Michaelis constant; MS, mass spectrometry; N/A, not applicable.
The kinetic parameters for tolbutamide hydroxylation of CYP2C9 variants 707_delA and 707_709delinsCC could not be determined because the amount of produced metabolite was at or below the detection limit at the lower substrate concentrations.
P < 0.05; **P < 0.01 compared with CYP2C9 WT.
TABLE 4.
Enzyme kinetic properties of recombinant wild-type and mutant CYP2C19 proteins for (S)-mephenytoin hydroxylation and in silico functional prediction of CYP2C19 variants
The kinetic parameters for (S)-mephenytoin hydroxylation of variants 518C>CT, 556C>CT, and *3 could not be determined because the amount of produced metabolite was at or below the detection limit at the lower substrate concentrations. These data represent the mean ± S.D. of three independently performed catalytic assays. Concordance is defined as agreement of the kinetic assay result with at least two of three in silico software results.
| cDNA |
Vmax ± S.D. × 10−3 |
Km ± S.D. |
CLINT × 10−4 (%WT) |
Polyphen |
SIFT |
PROVEAN |
Enzyme Activity in Relation to WT |
Concordance Yes/No | |
|---|---|---|---|---|---|---|---|---|---|
| Vivid* |
MS* |
||||||||
| nmol/mg per minute | µM | µl/mg protein per minute | % | % | |||||
| WT | 67.2 ± 2.51 | 30.1 ± 7.79 | 23.4 ± 6.27 | ||||||
| 65A>G | 33.3 ± 0.74 | 40.3 ± 0.86 | 8.25 ± 0.36 (36.2) | Benign | Tolerated | Deleterious | 49 | 36 | No |
| 337G>A | 23.6 ± 2.75 | 96.6 ± 23.1 | 2.49 ± 0.28 (10.7)* | Benign | Tolerated | Neutral | 75 | 11 | No |
| 518C>T | 3.65 ± 0.25 | 205 ± 11.8 | 0.18 ± 0.02 (0.7)* | Probably Damaging | Tolerated | Deleterious | 5 | 1 | Yes |
| 556C>T | 3.03 ± 0.36 | 148 ± 69.5 | 0.23 ± 0.09 (1.6)* | Probably Damaging | Damaging | Deleterious | 7 | 2 | Yes |
| 557G>A | 4.33 ± 0.34 | 30.5 ± 7.12 | 1.45 ± 0.20 (6.5)* | Probably Damaging | Tolerated | Deleterious | 7 | 7 | Yes |
| 578A>G | 68.5 ± 4.69 | 51.5 ± 7.69 | 13.4 ± 1.06 (57.9) | Benign | Tolerated | Neutral | 106 | 58 | Yes |
| 815A>G | 35.2 ± 0.11 | 38.4 ± 2.47 | 9.19 ± 0.54 (39.7) | Benign | Damaging | Deleterious | 53 | 40 | Yes |
| *3 | 0.02 ± 0.01 | 4.82 ± 0.00 | 0.05 ± 0.02 (0.2)* | 3 | N/A | ||||
CLINT, intrinsic clearance; Km, Michaelis constant; MS, mass spectrometry; N/A, not applicable.
P < 0.05 compared with CYP2C19 WT.
Fig. 5.
(A) Correlation analysis between enzyme activities determined by high-throughput assay vs. velocity of product formation determined by mass spectrometric assay for CYP2C9 variant allozymes. Relative velocity values of hydroxy tolbutamide formation (nanomoles of product formed per milliliter per minute) at 50 μM tolbutamide concentration are plotted on the horizontal axis and relative enzyme activity values determined by Vivid assay (nanomolars of fluorescent product formed per minute per milligram of total protein) are plotted on the vertical axis. R2 = 0.907 and P = < 0.0001. (B) Correlation analysis between enzyme activities determined by high-throughput assay vs. velocity of product formation determined by mass spectrometric assay for CYP2C19 variants. Relative velocity values of (S)-4-hydroxy mephenytoin formation (nanomoles of product formed per milliliter per minute) at 1000 μM (S)-mephenytoin concentration are plotted on the horizontal axis and relative Vivid enzyme activity values determined by Vivid assay (nanomolars of fluorescent product formed per minute per milligram of total protein) are plotted on the vertical axis. R2 = 0.898 and P ≤ 0.0001. Associations between protein content and enzyme activity were evaluated using Pearson correlations, with two-sided t test P values reported. The solid line illustrates the line of identity for the association between enzyme activities determined by high-throughput assay and intrinsic clearance determined by mass spectrometric assay.
The CPR activities in the majority of the cells transfected with CYP2C9 variant allozymes were similar to those for the CYP2C9 WT allozymes, as expected. In a similar fashion, cells transfected with CYP2C19 variant allozymes showed CPR activities similar to those observed in cells transfected with the CYP2C19 WT allozymes, as shown graphically in the Supplemental Material (see Supplemental Fig. 1, A and B, respectively).
In Silico Variant Sequence Prediction Analysis.
The impact of CYP2C9 and CYP2C19 variant sequence on enzyme activity was predicted in silico using Polyphen, SIFT, and Provean (Adzhubei et al., 2010; Sim et al., 2012; Choi and Chan, 2015) (Tables 3 and 4, respectively). The three programs predicted no effects (benign/tolerated/neutral) for 1/7 CYP2C9 and 2/7 CYP2C19 variants and loss of function (probably damaging, damaging, and deleterious) for 2/7 CYP2C9 and 1/7 CYP2C19 variants. Concordance, defined as agreement of the kinetic assay result with at least two of three in silico software results, was observed for 9 of 14 variants. Predictions differed among the programs for CYP2C9 variants 229C>A and 791T>C and CYP2C19 variants 65A>G, 518C>T, 557G>A, and 815A>G. Furthermore, there were differences between programs and functional assay results, for example, CYP2C9 variant 709G>C, which had low catalytic activity (<50% of WT), was predicted by Polyphen, SIFT, and Provean (Adzhubei et al., 2010; Sim et al., 2012; Choi and Chan, 2015) to be benign, tolerated, and neutral, respectively. Similarly, CYP2C19 variant 65A>G, which had low catalytic activity (<50% of WT), was predicted by Polyphen, SIFT, and Provean (Adzhubei et al., 2010; Sim et al., 2012; Choi and Chan, 2015) to be benign, tolerated, and deleterious, respectively.
Discussion
PGx will be the first aspect of clinical genomics to achieve broad clinical implementation, eventually touching virtually all patients (Weinshilboum and Wang, 2017). In the Mayo Clinic RIGHT1K study, NGS was performed for 1013 participants to identify sequence variation in 84 pharmacogenes (Bielinski et al., 2014). We identified six nonsynonymous ORF variants in the CYP2C9 gene and seven in the CYP2C19 gene. Expression of these variants after transfection into COS-1 cells most often yielded differing protein levels for variant allozymes compared with the WT sequence. Using fluorometric probe and prototypical substrates to assess the impact of these genetic polymorphisms on biologic function, we observed substantial variability in enzyme activity among the variants that generally correlated with protein expression levels. We also observed a significant correlation between protein expression levels and Vivid enzyme activities for CYP2C9 and CYP2C19. These results suggest decreased protein level may be the major factor responsible for the decreased enzyme activity that we observed. This conclusion is consistent with protein degradation as a result of misfolding due to alteration in the amino acid sequence that leads to decreased protein levels for the variant enzyme—as has been observed in the past for variant allozymes as a result of nonsynonymous SNPs (Wang et al., 2003, 2005; Li et al., 2008).
We also studied the impact of these polymorphisms on enzyme kinetics using the clinically relevant prototypic drug substrates tolbutamide and (S)-mephenytoin for CYP2C9 and CYP2C19, respectively. Three of seven CYP2C9 variant allozymes had low catalytic activity (<25%), two of seven had intermediate activity (25%–50%), and two of seven had similar activity (50%–100%) to WT. Similarly, four of seven CYP2C19 variant allozymes had low catalytic activity (<25%), two of seven had intermediate activity (25%–50%), and one of seven had similar activity (50%–100%) to WT. In addition, we found a significant correlation between our Vivid fluorometric high-throughput assay and enzyme kinetics of the prototypical substrates.
The COS-1 cell expression system used in this study to test recombinant enzymes does not constitutively express P450 enzymes, but does sufficiently express CPR and cytochrome b5 enzymes to support P450 activities (Gonzalez and Korzekwa, 1995). Oxidation and reduction reactions supported by P450s require interactions between the P450 with CPR flavoprotein and NADPH. Changes in the level of CPR could potentially affect drug disposition (Backes and Kelley, 2003). Because cytochrome b5 is not necessary in systems that coexpress CPR and CYP2C9 or CYP2C19 (Yamazaki et al., 2002), and the COS-1 expression system yields small amounts of protein, it was not measured in this study. Since CPR is important and differences in expression may alter CYP2C9 and CYP2C19 activity, we measured CPR enzyme activity in all of the CYP2C9 and CYP2C19 variant samples. The observation of similar CPR activity among the recombinant samples suggests that variation in CPR levels is likely a minor factor in affecting P450 enzyme activity. While we cannot rule out variation in the nature of the interaction between the P450s that we studied and CPR due to sequence variation in the P450s, our results strongly suggest that protein degradation is a major factor for this variation in intrinsic clearance.
In silico predictions have been widely applied to genetic variation in structure and function of proteins. When we compared our enzyme kinetic results with in silico predictions, we found several differences between our functional results and the in silico predictions. For example, the CYP2C9 variant 709G>C was predicted to be benign, whereas functional studies with a prototypic substrate showed only 27% of the WT enzyme activity. Polyphen 2, SIFT, and Provean are only three of a large number of programs designed to predict the effects of nonsynonymous ORF SNPs, but our results—and those of others (Flanagan et al., 2010; Min et al., 2016)—support the importance of functional studies as the gold standard for the functional assessment of variants that alter encoded amino acid sequence.
Gotoh (1992) proposed six putative substrate recognition sites (SRSs) in mammalian P450s, including SRS-1 (B-C loop), SRS-2 (F-helix), SRS-3 (G-helix), SRS-4 (I-helix), SRS-5 (β3 area), and SRS-6 (C-terminal β-strand region 4 β5) (Gotoh, 1992). These SRSs constitute about 76–79 amino acids and afford the structural and functional basis for drug and enzyme alignment (Gotoh, 1992). Furthermore, nonsynonymous mutations appeared to be more frequent within SRSs. Polymorphisms within SRS regions could cause protein structure conformational changes (Wester et al., 2004) and disrupt hydrophobic interactions with drug substrates (Straub et al., 1994). Earlier studies indicate that even single mutations at key residues result in significant geometrical alterations to substrate binding regions (Negishi et al., 1996). Thus, variant nucleotides may cause conformational changes in the enzyme, alter substrate binding affinity at the active site, and cause differences in enzyme activity. In our experiments, CYP2C9 variant 218C>T occurs in SRS region 1, variant 343A>C occurs in SRS 3 region, and variants 707delA, 709G>C, and 707_709delinsCC occur in SRS 2 and SRS 3. Each of these alterations in amino acid sequence results in a significant reduction (<25%) in enzyme activity when compared with the WT sequence. Similarly, CYP2C19 variant 337G>A occurs in SRS 1 and showed only ∼10% of the CYP2C19 WT activity for (S)-mephenytoin hydroxylation, but ∼80% activity in our fluorometric assay, which might be explained by the substrate specificity of these enzymes. Obviously, the role of genetic variants in altering the physiochemical and structural properties of substrate binding sites and substrate affinity in different P450-substrate interactions requires further study. However, in vivo studies in animals and humans are difficult, since many of these variants are rare and testing them in vivo would be expensive and time consuming. Therefore, the incorporation of the results of functional studies, such as those described here, into pharmacogenomic variant analysis for clinical purposes is essential to predict patient phenotypes and to allow for the use of clinical decision support tools that are currently being built into electronic health records.
The standard method to determine the effect of genetic variation on enzyme function involves cDNA expression in COS/yeast/insect/human cells, followed by characterization of enzyme kinetics. This approach has both strengths and limitations. The strengths include the fact that enzyme kinetic studies using prototypic substrates are clinically relevant and the results are closer to biologic enzyme activity than many other characterization studies. The limitations of this approach include the fact that these studies are laborious and time consuming, making their use in high-throughput assays difficult. Given the overwhelming number of new variants being identified, the time-consuming nature of standard methods of functional characterization, and the limited accuracy of computational in silico predictions, other approaches such as CRISPR Cas9 techniques (Guo et al., 2018), which are used to create genetic variants and recombinants, as well as techniques such as massively parallel single-nucleotide mutagenesis (Haller et al., 2016) are being tested for functional genomic applications. Similarly, high-throughput screening methods should be developed to make it possible to rapidly screen variants for their functional effects (Stresser et al., 2000; Kariv et al., 2001; Trubetskoy et al., 2005; Rainville et al., 2008; Cheng et al., 2009; Alden et al., 2010).
In summary, we have identified 13 nonsynonomous ORF variants and one ORF synonymous variant in two important, clinically actionable pharmacogenes, CYP2C9 and CYP2C19. Functional studies of these variants showed impaired metabolism by the encoded allozymes that may be due, at least in part, to effects on protein stability, resulting in decreased metabolism of the drug substrate. These results serve to emphasize the need for high-throughput functional genomic methods to address the tidal wave of novel variants discovered as pharmacogenomics moves from genotyping common variants that have been previously identified and functionally tested to preemptive sequencing. The move to preemptive NGS in ever larger population cohorts will identify ever larger numbers of variants with functional effects. For example, the Mayo Clinic has expanded the initial RIGHT protocol to an additional 10,085 patients, while variants identified in the 100,000 Genomes Project and those already present in gnomAD represent only two of the larger population cohorts that have become available. The application of standard approaches to study the functional effects of pharmacogene variants discovered during these studies will not be practically possible. Therefore, novel robust high-throughput methods are needed to identify nucleotide alterations that affect enzyme function, prepare recombinant enzymes that incorporate those alterations, and rapidly characterize the functional effects on enzyme activity resulting from those alterations. If the present results can be generalized, they suggest that, ultimately, DNA sequencing will be preferable to genotyping for the clinical implementation of pharmacogenomic variants, and they also support the need for high-throughput functional assays and highly accurate predictive algorithms if we are to achieve the optimal decrease in adverse drug reactions and the optimal increase in drug efficacy that PGx promises.
Abbreviations
- CPR
cytochrome P450 oxidoreductase
- dbSNP
single-nucleotide polymorphism database
- ESI
electrospray ionization
- gnomAD
Genome Aggregation Database
- LC-MS/MS
liquid chromatography– tandem mass spectrometry
- 3MA
3-methyladenine
- MG123
carbobenzoxy-L-leucyl-L-leucyl-L-leucinal
- m/z
mass-to-charge ratio
- NGS
next-generation sequencing
- ORF
open reading frame
- P450
cytochrome P450
- PGx
pharmacogenomics
- RIGHT, Right Drug, Right Dose, Right Time
Using Genomic Data to Individualize Treatment
- SNP
single-nucleotide polymorphism
- SRS
substrate Recognition Sites
- WT
wild type
Authorship Contributions
Participated in research design: Reid, Wang, Weinshilboum, Devarajan, Moon.
Conducted experiments: Devarajan, Moon.
Performed data analysis: Devarajan, Moon.
Wrote or contributed to the writing of the manuscript: Devarajan, Moon, Ho, Larson, Black, Bielinski, Neavin, Moyer, Scherer, Wang, Weinshilboum, Reid.
Footnotes
This work was supported in part by the Mayo Cancer Center [Support Grant CA 15083, (grant #T32 GM072474 to D.R.N.)]; the National Institutes of Health [Grants U19 GM61388, R01 GM28157, R01 GM125633, and U01 HG06379]; the Pharmacogenomics Program of the Mayo Clinic Center for Individualized Medicine; the Mayo Clinic Robert D. and Patricia E. Kern Center for the Science of Healthcare Delivery; and the Mayo Clinic Center for Individualized Medicine.
J.L.B. has licensed intellectual property to the companies AssureX Health and OneOme. In addition, he has stock ownership in OneOme. R.M.W. and L.W. are cofounders and stockholders in OneOme.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alden PG, Plumb RS, Jones MD, Rainville PD, Shave D. (2010) A rapid ultra-performance liquid chromatography/tandem mass spectrometric methodology for the in vitro analysis of pooled and cocktail cytochrome P450 assays. Rapid Commun Mass Spectrom 24:147–154. [DOI] [PubMed] [Google Scholar]
- Andersson ML, Eliasson E, Lindh JD. (2012) A clinically significant interaction between warfarin and simvastatin is unique to carriers of the CYP2C9*3 allele. Pharmacogenomics 13:757–762. [DOI] [PubMed] [Google Scholar]
- Backes WL, Kelley RW. (2003) Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther 98:221–233. [DOI] [PubMed] [Google Scholar]
- Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, Ryu E, Targonski PV, Van Norstrand MD, Hathcock MA, et al. (2014) Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time—using genomic data to individualize treatment protocol. Mayo Clin Proc 89:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Sohl CD, Guengerich FP. (2009) High-throughput fluorescence assay of cytochrome P450 3A4. Nat Protoc 4:1258–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP. (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield M, Davies J, Dennys M, Elbahy L, Fowler T, Hill S, Hubbard T, Jostins L, Maltby N, Mahon-Pearson J, McVean G, Nevin-Ridley K, Parker M, Parry V, Rendon A, Riley L, Turnbull C, Woods K. (2017) The 100,000 Genomes Project Protocol, figshare.
- Dai DP, Wang SH, Geng PW, Hu GX, Cai JP. (2014a) In vitro assessment of 36 CYP2C9 allelic isoforms found in the Chinese population on the metabolism of glimepiride. Basic Clin Pharmacol Toxicol 114:305–310. [DOI] [PubMed] [Google Scholar]
- Dai DP, Wang SH, Li CB, Geng PW, Cai J, Wang H, Hu GX, Cai JP. (2015) Identification and functional assessment of a new CYP2C9 allelic variant CYP2C9*59. Drug Metab Dispos 43:1246–1249. [DOI] [PubMed] [Google Scholar]
- Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, Yang LP, Qian JC, Wang ZS, Zhu GH, et al. (2014b) CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J 14:85–92. [DOI] [PubMed] [Google Scholar]
- DeLozier TC, Lee SC, Coulter SJ, Goh BC, Goldstein JA. (2005) Functional characterization of novel allelic variants of CYP2C9 recently discovered in Southeast Asians. J Pharmacol Exp Ther 315:1085–1090. [DOI] [PubMed] [Google Scholar]
- Deng M, Yang X, Qin B, Liu T, Zhang H, Guo W, Lee SB, Kim JJ, Yuan J, Pei H, et al. (2016) Deubiquitination and activation of AMPK by USP10. Mol Cell 61:614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser GK, Spence JD, Bailey DG. (2000) Pharmacokinetic-pharmacodynamic consequences and clinical relevance of cytochrome P450 3A4 inhibition. Clin Pharmacokinet 38:41–57. [DOI] [PubMed] [Google Scholar]
- Flanagan SE, Patch AM, Ellard S. (2010) Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers 14:533–537. [DOI] [PubMed] [Google Scholar]
- Friedman MA, Woodcock J, Lumpkin MM, Shuren JE, Hass AE, Thompson LJ. (1999) The safety of newly approved medicines: do recent market removals mean there is a problem? JAMA 281:1728–1734. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Korzekwa KR. (1995) Cytochromes P450 expression systems. Annu Rev Pharmacol Toxicol 35:369–390. [DOI] [PubMed] [Google Scholar]
- Gotoh O. (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90. [PubMed] [Google Scholar]
- Guo X, Chavez A, Tung A, Chan Y, Kaas C, Yin Y, Cecchi R, Garnier SL, Kelsic ED, Schubert M, et al. (2018) High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR-Cas9 in yeast. Nat Biotechnol 36:540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller G, Alvarado D, McCall K, Mitra RD, Dobbs MB, Gurnett CA. (2016) Massively parallel single-nucleotide mutagenesis using reversibly terminated inosine. Nat Methods 13:923–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GX, Pan PP, Wang ZS, Yang LP, Dai DP, Wang SH, Zhu GH, Qiu XJ, Xu T, Luo J, et al. (2015) In vitro and in vivo characterization of 13 CYP2C9 allelic variants found in Chinese Han population. Drug Metab Dispos 43:561–569. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Rodriguez-Antona C. (2005) Pharmacogenetics of drug-metabolizing enzymes: implications for a safer and more effective drug therapy. Philos Trans R Soc Lond B Biol Sci 360:1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526. [DOI] [PubMed] [Google Scholar]
- Ji Y, Moon I, Zlatkovic J, Salavaggione OE, Thomae BA, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. (2007) Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J Pharmacol Exp Ther 322:529–540. [DOI] [PubMed] [Google Scholar]
- Ji Y, Skierka JM, Blommel JH, Moore BE, VanCuyk DL, Bruflat JK, Peterson LM, Veldhuizen TL, Fadra N, Peterson SE, et al. (2016) Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next-generation DNA sequencing and a customized CYP2D6 genotyping cascade. J Mol Diagn 18:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariv I, Fereshteh MP, Oldenburg KR. (2001) Development of a miniaturized 384-well high throughput screen for the detection of substrates of cytochrome P450 2D6 and 3A4 metabolism. J Biomol Screen 6:91–99. [DOI] [PubMed] [Google Scholar]
- Kim KA, Park PW, Hong SJ, Park JY. (2008) The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther 84:236–242. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, Brockmöller J. (2004) Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 9:442–473. [DOI] [PubMed] [Google Scholar]
- Lee HI, Bae JW, Choi CI, Lee YJ, Byeon JY, Jang CG, Lee SY. (2014) Strongly increased exposure of meloxicam in CYP2C9*3/*3 individuals. Pharmacogenet Genomics 24:113–117. [DOI] [PubMed] [Google Scholar]
- Li F, Wang L, Burgess RJ, Weinshilboum RM. (2008) Thiopurine S-methyltransferase pharmacogenetics: autophagy as a mechanism for variant allozyme degradation. Pharmacogenet Genomics 18:1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ho MF, Schaid DJ, Scherer SE, Kalari K, Liu M, Biernacka J, Yee V, Evans J, Carlson E, et al. (2017) Breast cancer chemoprevention pharmacogenomics: deep sequencing and functional genomics of the ZNF423 and CTSO genes. NPJ Breast Cancer 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Nie M, Zhang A, Wen J, Noel SD, Lee V, Carroll RS, Kaiser UB. (2016) Computational analysis of missense variants of G protein-coupled receptors involved in the neuroendocrine regulation of reproduction. Neuroendocrinology 103:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. (2000) Current pharmacogenomic approaches to clinical drug development. Pharmacogenomics 1:115–123. [DOI] [PubMed] [Google Scholar]
- Negishi M, Iwasaki M, Juvonen RO, Sueyoshi T, Darden TA, Pedersen LG. (1996) Structural flexibility and functional versatility of cytochrome P450 and rapid evolution. Mutat Res 350:43–50. [DOI] [PubMed] [Google Scholar]
- Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, Hiratsuka M. (2014) Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J 14:107–114. [DOI] [PubMed] [Google Scholar]
- Peng Y, Wu H, Zhang X, Zhang F, Qi H, Zhong Y, Wang Y, Sang H, Wang G, Sun J. (2015) A comprehensive assay for nine major cytochrome P450 enzymes activities with 16 probe reactions on human liver microsomes by a single LC/MS/MS run to support reliable in vitro inhibitory drug-drug interaction evaluation. Xenobiotica 45:961–977. [DOI] [PubMed] [Google Scholar]
- Pereira NL, Aksoy P, Moon I, Peng Y, Redfield MM, Burnett JC, Jr, Wieben ED, Yee VC, Weinshilboum RM. (2010) Natriuretic peptide pharmacogenetics: membrane metallo-endopeptidase (MME): common gene sequence variation, functional characterization and degradation. J Mol Cell Cardiol 49:864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Pérez R, Ochoa D, Cabaleiro T, Román M, Sánchez-Rojas SD, Talegón M, Abad-Santos F. (2013) Evaluation of the relationship between polymorphisms in CYP2C8 and CYP2C9 and the pharmacokinetics of celecoxib. J Clin Pharmacol 53:1261–1267. [DOI] [PubMed] [Google Scholar]
- Rainville PD, Wheaton JP, Alden PG, Plumb RS. (2008) Sub one minute inhibition assays for the major cytochrome P450 enzymes utilizing ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 22:1345–1350. [DOI] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, Roden DM, Klein TE, Shuldiner AR, Clinical Pharmacogenetics Implementation Consortium (2011) Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther 90:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR, Clinical Pharmacogenetics Implementation Consortium (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40:W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SC, Kacevska M, Ingelman-Sundberg M. (2013) Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J 13:1–11. [DOI] [PubMed] [Google Scholar]
- Straub P, Lloyd M, Johnson EF, Kemper B. (1994) Differential effects of mutations in substrate recognition site 1 of cytochrome P450 2C2 on lauric acid and progesterone hydroxylation. Biochemistry 33:8029–8034. [DOI] [PubMed] [Google Scholar]
- Stresser DM, Blanchard AP, Turner SD, Erve JCL, Dandeneau AA, Miller VP, Crespi CL. (2000) Substrate-dependent modulation of CYP3A4 catalytic activity: analysis of 27 test compounds with four fluorometric substrates. Drug Metab Dispos 28:1440–1448. [PubMed] [Google Scholar]
- Trubetskoy OV, Gibson JR, Marks BD. (2005) Highly miniaturized formats for in vitro drug metabolism assays using Vivid® fluorescent substrates and recombinant human cytochrome P450 enzymes. J Biomol Screen 10:56–66. [DOI] [PubMed] [Google Scholar]
- Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. (2005) Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc Natl Acad Sci USA 102:9394–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sullivan W, Toft D, Weinshilboum R. (2003) Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics 13:555–564. [DOI] [PubMed] [Google Scholar]
- Wang L, Yee VC, Weinshilboum RM. (2004) Aggresome formation and pharmacogenetics: sulfotransferase 1A3 as a model system. Biochem Biophys Res Commun 325:426–433. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R. (2003) Inheritance and drug response. N Engl J Med 348:529–537. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Szumlanski CL. (1999) Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol 39:19–52. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Wang L. (2017) Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc 92:1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-Å resolution. J Biol Chem 279:35630–35637. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, et al. (2002) Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr Purif 24:329–337. [DOI] [PubMed] [Google Scholar]