Abstract
The human single‐stranded DNA binding Protein 2 (SSBP2) is a tumor suppressor implicated in multiple cancer forms. The SSBP2 and related SSBP3/SSBP4 proteins are predicted to be intrinsically disordered excepted for their highly conserved N‐terminal LUFS (LUG/LUH, Flo8, and SSBP/SSDP) domain. LUFS domains are found in a number of proteins including some transcriptional co‐repressors. Although LUFS domains contain an N‐terminal Lis homology (LisH) motif that typically forms a stable dimer, no 3D structure of any LUFS domain is available. Here, we report a crystal structure of the LUFS domain of human SSBP2 at 1.52 Å resolution. We show that the SSBP2 LUFS domain forms a homo‐tetramer and reveal how an alpha‐helix C‐terminal to the LisH motif mediates SSBP2 tetramerization (dimerization of dimers). Conservation of the tetramerization interface among LUFS domains suggests that other LUFS domains may also form tetramers in similar manners.
Keywords: single‐stranded DNA binding Protein 2; SSBP proteins; tumor suppressor; LUFS (LUG/LUH, Flo8 and SSBP/SSDP) domain; Lis homology (LisH) motif; tetramerization; X‐ray crystallography
Short abstract
PDB Code(s): 6IWV
Introduction
The human single‐stranded DNA binding Protein 2 (SSBP2) gene was isolated as a candidate myeloid leukemia suppressor from a critical region of loss in chromosome 5q13‐14.1 The single‐stranded DNA binding proteins (SSBP or SSDP), were first identified based on induced differentiation of avian chondrocytes in culture, which was shown to selectively bind pyrimidine‐rich elements within the α2(I) collagen gene promotor.2 SSBP2 is a tumor suppressor implicated in acute myelogenous leukemia,1, 3, 4 acute preB lymphoblastic leukemia,5 prostate cancer,6 suggesting that disruption of the SSBP2‐regulated pathways may be an infrequent but critical step in malignant transformation of multiple tissues. One of the key biochemical functions of SSBP2 is to interact with and stabilize the transcriptional adaptor protein LIM domain‐binding protein 1 (LDB1),7, 8 which regulates transcriptional activities in multiple systems, including red blood cell development and Wnt signaling.9, 10
The highly conserved N terminus of SSBP2 contains a LUFS (LUG/LUH, Flo8, single‐strand DNA‐binding protein) domain.11, 12 Database search with SSBP2 N‐terminal 100 amino acids revealed 35% and 30% sequence identity to the Arabidopsis LUG and the yeast Flo8, suggesting that the LUFS domains are evolutionarily ancient and may have a conserved function.7, 11, 12 The N terminal part of LUFS domain (Residues 18–50 for SSBP2) harbors a Lis homology (LisH) motif, which can be found in over 100 eukaryotic proteins.13 LisH motifs are believed to be involved in microtubule dynamics and organization, cell migration, and chromosome segregation; several of them are associated with genetic diseases.14, 15, 16, 17 LisH motifs typically form dimers (e.g., that of Lis1, which results in lissencephaly [smooth brain])13; that of FOP, a centrosomal protein),18 but some tetramerize (e.g., that of TBL1, a subunit of a HDAC recruiting co‐repressor).19 The sequence character of LUFS is that it contains P‐X‐GFL‐XX‐WW‐X‐VFWD sequence C‐terminal to a LisH motif [Fig. 1(A)].7 Fiedler et al. also found that Drosophila SSDP(1–92) forms a stable tetramer in solution by using the size‐exclusion chromatography‐coupled to multi‐angle light scattering (SEC‐MALS) method.10
Figure 1.

Biochemical characterization of N‐terminal LUFS domain of SSBP2. (A) Evolutionary conservation of the LUFS domain among SSBP2/SSBP3/SSBP4, LEUNG/LUH, Flo8, MSS11, and SOMA proteins.20, 21, 22, 23 The double slash in SOMA sequence represents a 32‐residue hydrophilic loop. Hs, Homo sapiens; At, Arabidopsis thaliana; Ca, Candida albicans; Nf, Neosartorya fumigate. (B) Limited proteolysis analysis of SSBP2 fragments. One subtilisin‐resistant fragment can be obtained through limited proteolysis of SSBP2(1–94) (left) or SSBP2(10–94) (right), which is about 7 kDa, red box marked. The marked ratios are the molar ratios of subtilisin over SSBP2(1–94) or SSBP2(10–94) used in limited proteolysis. The SDS‐PAGE gel was stained with Coomassie brilliant blue. (C) SEC‐MALS analysis of SSBP2(1–94), SSBP2(10–77) and the F68D/L71A/Y72A triple mutant of SSBP2(1–94) (SSBP2(1‐94)Mut for short). Molecular weights measured by SEC‐MALS are shown. (D) Alignment of SEC chromatograms of SSBP2(1–94) (in red), SSBP2(10–77) (in green), and SSBP2(1–94)Mut (in blue). Calculated molecular weights for SSBP2(1–94) tetramer, SSBP2(10–77) tetramer, and SSBP2(1–94) dimer are labeled.
Here, we report the crystal structure of the N‐terminal LUFS domain of human SSBP2 at 1.52 Å resolution. Our crystallographic and SEC studies suggest that the SSBP2 LUFS domain is sufficient for forming tetramers, using a tetramerization scheme distinct from these of other LisH motif contained proteins.
Results and Discussion
Biochemical characterization of the highly conserved N‐terminal domain of SSBP2
SSBP proteins (SSBP2, SSBP3, and SSBP4) contain a highly conserved N‐terminal domain (Residues 1–94 for human SSBP2) (Supporting Information Fig. S1). We overexpressed and purified various human SSBP2 fragments including SSBP2(1–94) and SSBP2(10–94). Since these SSBP2 fragments did not yield any useful crystal in our crystallization trials, we performed limited proteolysis analysis of SSBP2(1–94) and SSBP2(10–94) fragments. We found that both SSBP2(1–94) and SSBP2(10–94) are sensitive to subtilisin treatment [Fig. 1(B)], suggesting a protease‐sensitive C‐terminal tail in both SSBP2 fragments that is likely structurally flexible. Analysis of intrinsically disordered regions using the GeneSilico MetaDisorder server24 (http://iimcb.genesilico.pl/metadisorder/) suggested that human SSBP2 is intrinsically disordered except for the LUFS domain (Residues 10–77) (Supporting Information Fig. S2). Indeed, purified SSBP2(10–77) is well‐behaved, resistant to protease treatment and can be crystallized.
Purified SSBP2 proteins were checked for their oligomerization states by size‐exclusion chromatography‐coupled multi‐angle light scattering (SEC‐MALS). Our SEC and SEC‐MALS data are in agreement with a previous report that the Drosophila SSDP(1–92) protein fragment forms a homo‐tetramer.10 Importantly, our SEC‐MALS data demonstrate that the SSBP2 LUFS domain (SSBP2 Residues 10–77) is sufficient for tetramerization [Fig. 1(C,D)].
Crystal structure of the LUFS domain of human SSBP2
We determined crystal structure of the LUFS domain of human SSBP2 (Residues 10–77) using the single‐wavelength anomalous dispersion (SAD) method and refined the structure at 1.52 Å resolution (Fig. 2 and Table 1). There are two SSBP2 molecules in each asymmetric unit which form a homodimer [Fig. 2(A)], and two dimers related by a crystallographic two‐fold axis form a tetramer via a conserved hydrophobic interface [Fig. 2(B)]. Each SSBP2(10–77) subunit consists of three helices: α1 (Residues 16–32), α2 (Residues 36–45), and α3 (Residues 60–73). The Helices α1 and α2 correspond to the LisH motif and mediate homodimerization by forming a four‐helix bundle in each crystallographic asymmetric unit (Supporting Information Fig. S3), whereas four α3 helices from respective SSBP2 molecules are responsible for forming a homo‐tetramer [Fig. 2(B)].
Figure 2.
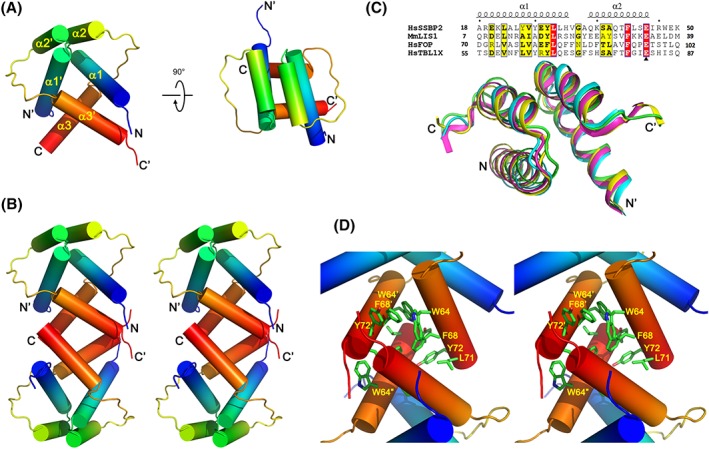
Crystal structure of the LUFS domain of human SSBP2. (A) Cartoon representation of SSBP2(10–77) dimer observed in each asymmetric unit, in two orthogonal orientations. Each SSBP2(10–77) subunit is colored in gradient, with blue to red representing the peptide chain from N‐ to C‐terminus. (B) Cartoon representation the overall structure of SSBP2(10–77) tetramer in stereo. The two dimers forming each tetramer are related by a “horizontal” two‐fold axis in this orientation. (C) Sequence alignment and structural superposition of different LisH motifs: these of SSBP2 (in green), LIS1 (in magenta, PDB code:1UUJ), FOP (in cyan, PDB code: 2D68) and TBL1X (in yellow, PDB code:2XTC). (D) Key residues involved in the tetramerization of the SSBP2 LUFS domain, shown in stereo.
Table 1.
Data Collection and Refinement Statistics
| Data collection | SSBP2(10–77) native | SSBP2(10–77) Au derivative |
|---|---|---|
| Space group | P6322 | P6322 |
| Cell dimensions | ||
| a, b, c (Å) | 116.91, 116.91, 42.35 | 117.26, 117.26, 42.71 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Wavelength (Å) | 1.00 | 1.02 |
| Resolution (Å) | 20–1.52 (1.55–1.52) | 50–1.85 (1.88–1.85) |
| R merge (%) | 5.9 (155.5) | 6.9 (154.4) |
| I/σI | 94.73 (1.25) | 68.46 (2.17) |
| CC1/2 | 1.003 (0.626) | 0.996 (0.741) |
| Completeness (%) | 99.8 (99.9) | 100 (100) |
| Redundancy | 22.8 (10.4) | 21.8 (16.1) |
| Refinement | ||
| Resolution (Å) | 20–1.52 | |
| No. reflections | 25,214 | |
| R work/R free (%) | 14.57/18.06 | |
| No. atoms | 1204 | |
| No. of water | 129 | |
| Average B factors (Å2) | ||
| Protein | 38.49 | |
| H2O | 50.91 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | |
| Bond angles (°) | 1.247 | |
| Ramachandran plot (%) | ||
| Most favorable | 98.29 | |
| allowed | 1.71 | |
| Outliers | 0 |
The LisH motif comprising two antiparallel SSBP2 helices α1 and α2 is shared in the related proteins including Lis1, FOP, and TBL1, and shows similar 3D structures [Fig. 2(C)]. The homodimer interface is mainly maintained by hydrophobic interactions formed by Leu22, Val26, Tyr29, Leu30, Ala35, and Phe42, which are conserved among LisH motif proteins [Supporting Information Fig. S3(A)]. In addition to hydrophobic contacts, the dimer is stabilized by hydrogen bonds formed between Glu45, one of the most conserved residues of the LisH motif, and Gln36, Lys37 and Ser38 of the other chain of the dimer [Supporting Information Fig. S3(A)]. In addition to the LisH motif, the helix α3 also forms part of the dimer interface by interacting with the helix α1 of the other SSBP2 subunit in the asymmetric unit. In this α1/α3 dimer interface, a hydrophobic interface adjoin the LisH four‐helix bundle is formed by Ala18 and Leu22 in helix α1, as well as Phe60, Leu61, Trp65, and Trp69 from helix α3 [Supporting Information Fig. S3(B)]. The α1/α3 dimer interface is also stabilized by two pairs of hydrogen bonds formed between α1/α3 sidechains, these of Arg19/His62 and Gln17/Trp69, and another hydrogen bond formed between the mainchain carbonyl of Ala18 and the sidechain of Trp65.
The SSBP2 LUFS domain tetramerization interface formed by the α3 helix
The tetramerization interface of the SSBP2 LUFS domain between two dimers related by a crystallographic two‐fold axis is formed predominantly by hydrophobic residues in the C‐terminal helix α3, including Phe60, Leu61, Trp64, Trp65, Val67, Phe68, Leu71, and Tyr72. Among these hydrophobic residues, Phe60, Leu61, Trp65, and Val67 seem to be involved in both dimerization and tetramerization, whereas the highly conserved Trp64 and three residues in the C‐terminal half of Helix α3 (Phe68, Leu71, and Tyr72) may only contribute to SSBP2 tetramerization [Fig. 2(D)]. To validate biochemical relevance of the tetramerization interface, we generated SSBP2(1–94) triple mutant (F68D/L71A/Y72A). Our SEC and SEC‐MALS analysis demonstrates that while SSBP2(1–94) and SSBP2(10–77) form tetramers in solution, purified SSBP2(1–94; F68D/L71A/Y72A) mutant protein forms a dimer instead of tetramer, in strong support of our structural observation [Fig. 1(C,D)].
A search for similarly folded proteins using the Dali server25 (http://ekhidna2.biocenter.helsinki.fi/dali/) results in TBL1, Cstf‐50, Topless/TPR, and Lis1. Among these proteins, Lis1 and Cstf‐50 form dimers but not tetramers.13, 26 Interestingly, the LisH motif–containing protein TBL1 also forms a tetramer, using a tetramerization interface distinct from that of SSBP2 – through the base of its LisH four‐helix bundle (Supporting Information Fig. S4).19 Key residues on the TBL1 tetramerization interface are not conserved in SSBP2, and no similar crystal packing exist in our SSBP2 LUFS crystal lattice. In the meantime, Topless/TPR forms a tetramer via the CTLH‐CRA domain C‐terminal to its LisH motif (Supporting Information Fig. S4).27 Therefore, the SSBP2 tetramerization interface is distinct from these known LisH motif–containing protein tetramerization interfaces. Nonetheless, it is important to note that residues on our SSBP2 tetramerization interface (hydrophobic residues in Helix α3) are highly conserved among LUFS domains [Figs. 1(A) and 2(D)]. This strongly suggests that the homo‐tetramerization structural features observed in our SSBP2 LUFS crystal structure are likely conserved for other LUFS domains.
Materials and Methods
Cloning, expression, and purification
The SSBP2 gene was amplified from plasmid #28090 (Addgene). SSBP2 was cloned into a pET28a vector (Novagen) and expressed in Escherichia coli BL21 (DE3) to produce an N‐terminal hexa‐histidine tagged protein with a Tobacco etch virus (TEV) cleavage site for removal of the His‐tag. Cells were grown in LB medium with 30 μg/mL kanamycin at 37°C until OD600 = 0.6, when protein expression was induced with 0.2 mM isopropyl beta‐d‐1‐thiogalactopyranoside (IPTG) for 16–18 h at 18°C. Cells pellets were resuspended in lysis buffer (50 mM Tris pH 8, 400 mM NaCl, 20 mM imidazole pH 8, 2 mM DTT) and sonicated. The cell lysate were cleared by centrifugation (26,000g for 1 h) and the supernatant was loaded on Ni‐NTA affinity column (GE Healthcare), and washed with lysis buffer. The protein samples were subsequently eluted with lysis buffer containing 300 mM imidazole pH 8. After cleavage of the His‐tag at 4°C overnight, the proteins were loaded onto a HiTrap Q column (GE Healthcare) equilibrated with buffer A (20 mM Tris pH 8, 20 mM NaCl, 2 mM DTT) and the protein was eluted with a 0–100% gradient of 20 mM Tris pH 8, 1 M NaCl in 20 column volumes. Then the proteins were further purified on a HiLoad Superdex 75 10/300 gel filtration column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris pH 8, 100 mM NaCl, 2 mM DTT. The final protein sample was concentrated to 15 mg/mL, flash‐frozen in liquid nitrogen, and stored at −80°C until use.
Limited proteolysis analysis
SSBP2(1–94) and SSBP2(10–94) were digested with subtilisin at different molar ratios of subtilisin:SSBP2 (1:1000, 1:500, 1:200, and 1:100). The incubation buffer is 20 mM Tris pH 8.0, 100 mM NaCl. Samples were incubated at room temperature for 1 h. After that, protein loading buffer was added and boiled at 100°C for 5 min for sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE).
Crystallization and data collection
SSBP2(10–77) crystals were grown at room temperature by hanging drop vapor diffusion by mixing 1 μL of the protein solution and 1ul of solution containing 0.1 M Bis–Tris pH 6.3, 1.9 M ammonium sulfate. The crystal has a Matthews coefficient (V M) of 2.64 and a solvent content of 53.51%. Crystals were transferred stepwise into 0.1 M Bis–Tris pH 6.3, 2 M sodium malonate and allowed to stabilize for at least 24 h before frozen with liquid nitrogen. Various heavy‐atom chemicals were added to SSBP2 crystals soaking drops to search for the isomorphous derivatives. A gold derivative was successfully obtained when crystals were soaked in 10 mM Potassium dicyanoaurate (I) (KAu[CN]2) (Hampton Research) overnight at room temperature.
Data sets of native SSBP2 crystals and its gold derivative were collected from the Beamline 821 at ALS (Advanced Light Source). These data sets were integrated and scaled using HKL3000.28 A 1.52 Å data set of native SSBP2 crystals and a 1.85 Å data set of gold derivative were used in the structure determination.
Structure determination and refinement
The SSBP2 structure was determined by the single‐wavelength anomalous dispersion (SAD) method,with a KAu(CN)2 derivative, using Autosol in PHENIX.29 And autobuilding was done with AutoBuild in PHENIX29 and REFMAC.30 Model building was performed with the graphics software COOT31 and CCP4.30 All structural pictures were generated using PyMOL.32 Crystallographic statistics are shown in Table 1.
Accession numbers
The structure factors and co‐ordinates have been deposited in the Protein Data Bank under the accession code 6IWV.
Supporting information
Figure S1. Sequence alignment of full length SSBP proteins from different species. Apparently, the N‐terminal domains are the most conserved region of SSBP2/3/4 proteins. Abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Pt, Pan troglodytes; Bt, Bos taurus; Oc, Oryctolagus cuniculus; Ml, Myotis lucifugus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Dm, Drosophila melanogaster.
Figure S2. The protein disorder tendency prediction of the full length human SSBP2, using the GeneSilico MetaDisorder server (http://iimcb.genesilico.pl/metadisorder/). The X‐axis represents residues 1–361 of the full‐length human SSBP2 protein, whereas the Y‐axis is the disorder tendency score for each residue in the context of the SSBP2 sequence. The regions with the disorder tendency higher than 0.5 have a high tendency to be structurally disordered. The results by two different prediction methods, MetaDisorder and MetaDisorder MD2, are shown in cyan and orange, respectively.
Figure S3. Details of the SSBP2 LisH‐mediated dimerization interface. (A) The four‐helix bundle interface formed among LisH motif helices α1, α2, α1’, and α2’. (B) The interface formed among helicesα1, α3, α1’, and α3’.
Figure S4. Comparison of the SSBP2 (A), TBL1X [(B); PDB code: 2XTC], and TPR [(C); PDB code:5NQS] tetramerization schemes. The tetramer interfaces are highlighted by dashed gray boxes. Key residues involved in the tetramerization, shown in stick.
Acknowledgments
We appreciate assistance in data collection from staff of beamline 8.2.1 and 8.2.2, ALS. We also thank Xiaoxia Yu for help on the SEC‐MALS experiment (Institute of Biophysics, CAS). This work was supported by the National Laboratory of Biomacromolecules, and National Natural Science Foundation of China Grant nos. 31570794 and 31629002, the Chinese Academy of Sciences Pilot Strategic Science and Technology Projects B grant XDB08010303, to X.Y. and W.X., respectively.
Contributor Information
Xiao‐Xue Yan, Email: snow@ibp.ac.cn.
Wenqing Xu, Email: wxu@uw.edu.
REFERENCES
- 1. Castro P, Liang H, Liang JC, Nagarajan L (2002) A novel, evolutionarily conserved gene family with putative sequence‐specific single‐stranded DNA‐binding activity. Genomics 80:78–85. [DOI] [PubMed] [Google Scholar]
- 2. Bayarsaihan D, Soto RJ, Lukens LN (1998) Cloning and characterization of a novel sequence‐specific single‐stranded‐DNA‐binding protein. Biochem J 331:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang H, Samanta S, Nagarajan L (2005) SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene 24:2625–2634. [DOI] [PubMed] [Google Scholar]
- 4. Wang Y, Klumpp S, Amin HM, Liang H, Li J, Estrov Z, Zweidler‐McKay P, Brandt SJ, Agulnick A, Nagarajan L (2010) SSBP2 is an in vivo tumor suppressor and regulator of LDB1 stability. Oncogene 29:3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, Morton CC (2008) Novel SSBP2‐JAK2 fusion gene resulting from a t(5;9)(q14.1;p24.1) in pre‐B acute lymphocytic leukemia. Genes Chromosomes Cancer 47:884–889. [DOI] [PubMed] [Google Scholar]
- 6. Liu JW, Nagpal JK, Sun W, Lee J, Kim MS, Ostrow KL, Zhou S, Jeronimo C, Henrique R, Van Criekinge W, Moon CS, Califano JA, Trink B, Sidransky D (2008) ssDNA‐binding protein 2 is frequently hypermethylated and suppresses cell growth in human prostate cancer. Clin Cancer Res 14:3754–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Meyel DJ, Thomas JB, Agulnick AD (2003) Ssdp proteins bind to LIM‐interacting co‐factors and regulate the activity of LIM‐homeodomain protein complexes in vivo. Development 130:1915–1925. [DOI] [PubMed] [Google Scholar]
- 8. Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ (2007) Single‐stranded DNA‐binding proteins regulate the abundance of LIM domain and LIM domain‐binding proteins. Genes Dev 21:942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Love PE, Warzecha C, Li L (2014) Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet 30:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fiedler M, Graeb M, Mieszczanek J, Rutherford TJ, Johnson CM, Bienz M (2015) An ancient Pygo‐dependent Wnt enhanceosome integrated by Chip/LDB‐SSDP. Elife 4:e09073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conner J, Liu Z (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci U S A 97:12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci U S A 101:11494–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim MH, Cooper DR, Oleksy A, Devedjiev Y, Derewenda U, Reiner O, Otlewski J, Derewenda ZS (2004) The structure of the N‐terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 12:987–998. [DOI] [PubMed] [Google Scholar]
- 14. Umeda M, Nishitani H, Nishimoto T (2003) A novel nuclear protein, Twa1, and Muskelin comprise a complex with RanBPM. Gene 303:47–54. [DOI] [PubMed] [Google Scholar]
- 15. Pancoast M, Dobyns W, Golden JA (2005) Interneuron deficits in patients with the Miller–Dieker syndrome. Acta Neuropathol 109:400–404. [DOI] [PubMed] [Google Scholar]
- 16. Popovici C, Zhang B, Grégoire MJ, Jonveaux P, Lafage‐Pochitaloff M, Birnbaum D, Pébusque MJ (1999) The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood 93:1381–1389. [PubMed] [Google Scholar]
- 17. Emes RD, Ponting CP (2001) A new sequence motif linking lissencephaly, Treacher Collins and oral–facial–digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet 10:2813–2820. [DOI] [PubMed] [Google Scholar]
- 18. Mikolajka A, Yan X, Popowicz GM, Smialowski P, Nigg EA, Holak TA (2006) Structure of the N‐terminal domain of the FOP (FGFR1OP) protein and implications for its dimerization and centrosomal localization. J Mol Biol 359:863–875. [DOI] [PubMed] [Google Scholar]
- 19. Oberoi J, Fairall L, Watson PJ, Yang JC, Czimmerer Z, Kampmann T, Goult BT, Greenwood JA, Gooch JT, Kallenberger BC, Nagy L, Neuhaus D, Schwabe JW (2011) Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol 18:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, Chen J, Liu H (2006) The Flo8 transcription factor is essential for Hyphal development and virulence in Candida albicans . Mol Biol Cell 17:295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Su C, Li Y, Lu Y, Chen J (2009) Mss11, a transcriptional activator, is required for hyphal development in Candida albicans . Eukaryot Cell 8:1780–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin CJ, Sasse C, Gerke J, Valerius O, Irmer H, Frauendorf H, Heinekamp T, Straßburger M, Tran VT, Herzog B, Braus‐Stromeyer SA, Braus GH (2015) Transcription factor SomA is required for adhesion, development and virulence of the human pathogen Aspergillus fumigatus . PLoS Pathog 11:e1005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozlowski LP, Bujnicki JM (2012) MetaDisorder: a meta‐server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm L, Laakso LM (2016) Dali server update. Nucleic Acids Res 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreno‐Morcillo M, Minvielle‐Sébastia L, Mackereth C, Fribourg S (2011) Hexameric architecture of CstF supported by CstF‐50 homodimerization domain structure. RNA 17:412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin‐Arevalillo R, Nanao MH, Larrieu A, Vinos‐Poyo T, Mast D, Galvan‐Ampudia C, Brunoud G, Vernoux T, Dumas R, Parcy F (2017) Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression. Proc Natl Acad Sci U S A 114:8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. [DOI] [PubMed] [Google Scholar]
- 29. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. [DOI] [PubMed] [Google Scholar]
- 31. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA: DeLano Scientific, 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of full length SSBP proteins from different species. Apparently, the N‐terminal domains are the most conserved region of SSBP2/3/4 proteins. Abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Pt, Pan troglodytes; Bt, Bos taurus; Oc, Oryctolagus cuniculus; Ml, Myotis lucifugus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Dm, Drosophila melanogaster.
Figure S2. The protein disorder tendency prediction of the full length human SSBP2, using the GeneSilico MetaDisorder server (http://iimcb.genesilico.pl/metadisorder/). The X‐axis represents residues 1–361 of the full‐length human SSBP2 protein, whereas the Y‐axis is the disorder tendency score for each residue in the context of the SSBP2 sequence. The regions with the disorder tendency higher than 0.5 have a high tendency to be structurally disordered. The results by two different prediction methods, MetaDisorder and MetaDisorder MD2, are shown in cyan and orange, respectively.
Figure S3. Details of the SSBP2 LisH‐mediated dimerization interface. (A) The four‐helix bundle interface formed among LisH motif helices α1, α2, α1’, and α2’. (B) The interface formed among helicesα1, α3, α1’, and α3’.
Figure S4. Comparison of the SSBP2 (A), TBL1X [(B); PDB code: 2XTC], and TPR [(C); PDB code:5NQS] tetramerization schemes. The tetramer interfaces are highlighted by dashed gray boxes. Key residues involved in the tetramerization, shown in stick.


