Abstract
Background
Traumatic brain injury (TBI) produces a series of pathological processes. Recent studies have indicated that autophagy pathway is persistently activated after TBI, which may lead to deterioration of nerve injury. Our preliminary work found miR-21-5p was upregulated in both in vivo and in vitro TBI models. MicroRNAs (miRNAs) could be loaded into exosomes to perform cell-to-cell interactions. This research aimed to evaluate the therapeutic effect of neuron-derived exosomes enriched with miR-21-5p on the TBI in vitro and to further explore the possible mechanisms.
Material/Methods
Brain extracts harvested from an rTBI mouse model were added to cultured HT-22 neurons to imitate the microenvironment of injured brain on in vitro cultured cells. Ultracentrifugation was performed to isolate exosomes. Transmission electron microscopy and Nano sight technology were used to examine exosomes. An in vitro model of TBI was established to study the effect of exosomal miR-21-5p on nerve injury and on neuronal autophagy regulation.
Results
The expression of miR-21-5p was increased in exosomes derived from HT-22 neurons after treatment with rTBI mouse brain extracts. Autophagy was activated in HT-22 neurons after scratch injury. Exosomal miR-21-5p produced a protective effect by suppressing autophagy in a TBI model in vitro. MiR-21-5p could directly target the Rab11a 3′UTR region to reduce its translation and further suppressed Rab11a-mediated neuronal autophagy.
Conclusions
The levels of miR-21-5p in neuronal exosomes increased from the acute to the chronic phase of TBI. Neuronal exosomes enriched with miR-21-5p can inhibit the activity of neuronal autophagy by targeting Rab11a, thus attenuating trauma-induced, autophagy-mediated nerve injury in vitro.
MeSH Keywords: Autophagy, Brain Injuries, Exosomes, MicroRNAs
Background
Traumatic brain injury (TBI) is one of the most common causes of mortality and morbidity worldwide, which makes it an important medical, public health, and societal problem [1,2]. More than 50 million people are newly diagnosed with TBI internationally each year and approximately half of the world’s population will have experience one or more TBIs during their lifetime [3–5].
TBI produces a complex series of pathological processes, which can be mainly divided into 2 stages. The primary insult is mainly caused by external impact, and leads to acute pathological changes, including brain contusions, intracerebral hemorrhage, and axonal shearing. The secondary pathological changes, including oxidative stress, Ca2+ overload, mitochondria injury, glutamate excitotoxicity, and neuroinflammation, leads to the further damage of nerve function [6,7]. In recent years, there have been more and more studies focus on the various molecular events after TBI. Autophagy is an evolutionarily conserved homeostatic process in which parts of the cytoplasm are fused within multi-membraned vesicles termed autophagosomes and then delivered to lysosomes for degradation [8,9]. Increased evidence suggests that autophagy plays an important role in the pathophysiologic process of both experimental and clinical TBI [10–13]. Recent studies indicated that autophagy pathway is persistently activated after TBI, which may lead to deterioration of nerve injury [14–17]. These findings suggest that the targeting trauma-induced neuronal autophagy might be a potential therapeutic method for the treatment of TBI.
MicroRNAs (miRNAs) are a class of single-stranded, non-coding endogenous RNA molecules that play important roles in the regulation of gene expression at post-transcription level [18]. Emerging evidences suggested dysregulation miRNAs were observed in TBI models [19–21]. Our preliminary work found that miR-21-5p was upregulated both in the brain cortex after TBI and cultured neurons after scratch injury [22–26]. MiR-21-5p has also been reported that it could regulate autophagy in the pathological processes of several diseases [27–30]. Bioinformatics analysis found Rab11a, which was involved in both early and late stages of autophagy, might be a target of miR-21-5p. These findings indicate that the expression of miR-21-5p may play vital roles in suppression autophagy.
Exosomes are small bioactive vesicles (40–100 nm) secreted by various cells, that can interact with other cells at the cell surface via a specific receptor or by mixing of their cargos with cellular contents after endocytosis. Proteins and nucleic acids can be loaded as cargos into exosomes, and can be transferred into target cells to perform cell-to-cell interactions and therefore affect many physiological and pathological processes [31,32]. Recent studies of exosomes have focused on their miRNA regulatory function in TBI [33]. In our previous study, we found for the first time that the miR-21-5p levels in the brain could increase after TBI, which could contribute to improving the neurological outcome of TBI [24–26]. But the specific mechanism of this process remains unclear. Based on these findings, we designed this research to evaluate the therapeutic effect of neuron-derived exosomal miR-21-5p on the TBI in vitro and to further explore the possible mechanisms of neuronal autophagy regulation induced by exosomal miR-21-5p.
Material and Methods
All experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA) and approved by the Tianjin Medical University Animal Care and Use Committee.
Controlled cortical impact-induced rTBI model
To clarify the role of neuronal exosomes on neurological outcome after TBI, we used an rTBI model, which has been shown to induce obvious neurological impairments [34,35]. Adult male C57BL/6 mice (age: 10–12 weeks, weight: 20–25 g) were purchased from the Chinese Academy of Military Science (Beijing, China). The mice were anesthetized with 4.6% isoflurane and positioned in a stereotaxic frame by using ear bars. After a midline scalp incision, a 3.0-mm craniotomy was performed centrally over the right parietal bone. The impounder tip of the injury device (eCCI, model 6.3; American Instruments, Richmond, VA, USA) was then extended to its full impact distance, positioned on the surface of the exposed dura mater, and reset to affect its surface.
The impact parameters were set at a velocity of 3.6 m/s and a deformation depth of 1.2 mm. Repetitive impact was performed for 4 times with 24-hour intervals [34]. Those mice with dural hernia were excluded from the group [22]. After each injury, the incision was stitched with interrupted 6-0 silk sutures and the mice were then placed in a well-heated cage at 37°C until they recovered consciousness. Mice from the control group experienced the same procedures except for the impact.
Preparation of brain extracts
To obtain the brain extracts after rTBI, the mice were euthanized by transcardiac perfusion with cold phosphate-buffered saline (PBS) at 3, 7, 14, or 21 days after the last brain injury (n=6 mice per group) [36]. The injured brains were dissected and isolated on ice. Brain tissue was homogenized by adding neurobasal medium containing 2% B27 and 1% glutamine (Thermo Fisher Scientific) at a concentration of 100 mg/mL. The homogenate was centrifuged at 12 000 g for 20 minutes at 4°C. The supernatant from brain tissue extracts was collected and stored at −80°C.
HT-22 cell line culture and treatment with rTBI brain extracts
HT-22 neurons were obtained from China Infrastructure of Cell Line Resources (Beijing, China). For the experiments, cells were cultured in DMEM/F12 culture medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific) in a 37°C incubator with 5% CO2. The purity of cultured cell was determined via immunofluorescence staining for microtubule-associated protein 2 (MAP-2).
HT-22 cells were then washed twice with PBS and cultured in neurobasal medium before treatment with the brain extracts. The brain extracts from rTBI or control group was added to the culture medium at a ratio of 1: 10 (extracts/culture medium). After 24-hour treatment, culture medium containing the brain extracts was removed, and the cells were washed with PBS twice to avoid any interference of FBS on the exosomes. HT-22 cells were cultured for another 48 hours in serum-free neurobasal medium before subsequent isolation of exosomes [36].
Exosome isolation, characterization, labelling and uptake
To isolate exosomes from the HT-22 cells, the cell culture supernatant was collected into 50 mL polypropylene tubes, and centrifuged at 300 g for 10 minutes to remove the free cells, 2000 g for 10 minutes to remove cell debris, 10 000g for 30 minutes to further remove the cell particles. Then it was filtered to remove dead cells and particles larger than 200 nm through a 0.22 mm filter (Millipore-Sigma). After that, an ultracentrifugation was performed at 100 000 g for 70 minutes to collect the exosomes. The supernatant was discarded, and the precipitates were stored for further experiments. All the above operations were completed at 4°C.
In order to identify the isolated exosomes, transmission electron microscopy (TEM, HT7700; Hitachi, Tokyo, Japan) was used measure the shape and size of particles in the precipitates. Exosomes from HT-22 cells were resuspended and the procedure of TEM observation was detailed previously [35]. Moreover, size distribution of HT-22 neuron derived exosomes was analyzed by using Nano sight technology (Particle Metrix, Meerbusch, Germany) according to the manufacturer’s protocols. The specific biomarkers for exosomes like CD9 and CD63, were detected by western blot analysis.
Exosomes were labelled with PKH67 dye (Sigma-Aldrich), according to the manufacturer’s instructions. In brief, exosome suspension in diluent C was mixed with 4 μL PKH67 and incubated for 10 minutes at room temperature. Then 20 mL chilled PBS was added to stop the labelling reaction. Labelled exosomes were ultra-centrifuged at 100 000 g for 70 minutes, washed with PBS, ultra-centrifuged again at 100 000 g and the precipitates were resuspended in PBS.
The uptake was performed by incubating HT-22 cell cultures with 100 μL of exosome solutions in a humid chamber for 12 hours (37°C, 5% CO2). A confocal microscope was performed to identify the uptake of exosomes.
Model of traumatic brain injury in vitro
Traumatic axonal injury is a consistent component of TBI, and it has been recognized as a major pathology of TBI [37]. To study the impact of traumatic injury on neurons in vitro, a scratch injury model was performed as we reported [25]. By using a 10 mL pipette tip, cultured HT22 neurons were scratched across the cell surface vertically and horizontally with a 4 mm space between each line, and floated cells were washed away by culture medium. This scratch injury was used in TBI model for neurons in vitro, since scratch injury activated the primary damage of neurons and then affect the entire neurons.
Target prediction and luciferase reporter assay
For the dysregulated miR-21-5p, target predication was performed with Target Scan (http://www.targetscan.org/). To identify whether miR-21-5p directly targeted Rab11a mRNA in HT22 neurons, luciferase reporter assay was performed [38]. Luciferase reporter constructs were made by ligating 3′UTR fragments of Rab11a containing the predicted binding sites (TargetScan, http://www.targetscan.org/cgi-bin/targetscan/mmu_71/view_gene.cgi?rs=ENSMUST00000169058.2&taxid=10090&members=&showcnc=0&shownc=0&showncf1=&showncf2=&subset=1#miR-21-5p) into the luciferase reporter vector pGL3. Briefly, the fragment was amplified by polymerase chain reaction (PCR) from mouse genomic DNA. The pGL3-Rab11a-3′UTR construct was then inserted into the pGL3 control vector containing SV40 promoter (Promega, Madison, WI, USA), using XbaI enzyme. Additionally, the mutant type (Mut) of luciferase reporter was generated by deleting the original binding site for miR-21-5p.
For the luciferase assay, HT22 neurons were cultured in 96-well plates. The wild type (WT) or mutant (Mut) Rab11a-3′UTR was co-transfected with 200 pmol miR-21-5p mimic or scrambled oligonucleotides (Gene Pharma) in neurons with Lipofectamine-3000. After incubated for 48 hours, the cells were harvested and the luciferase activity was detected with a dual-luciferase reporter system (Promega, Madison, WI, USA) according to the manufacturer’s protocols.
Transfection of the HT-22 neurons
To study the role of miR-21-5p, miR-21-5p mimic and negative control (NC) mimic (Gene Pharma, Shanghai, China) were transfected into HT-22 neurons as previously described (Table 1) [25,39]. Briefly, the miR-21-5p mimic or NC mimic was diluted to an accurate concentration of 20 mM. After that, 5 mL miR-21-5p mimic or NC mimic were mixed with 5 mL Lipofectamine-3000 (Thermo Fisher Scientific) in 500 mL serum-free DMEM/F12 and incubated for 20 minutes at room temperature. This transfection solution was washed twice with PBS and then added to the culture plates. After 48 hours of transfection, the neurons were prepared for the further experiment.
Table 1.
Sequences of miR-21-5p mimic and Rab11a siRNA.
| Sense (5′-3′) | |
|---|---|
| miR-21-5p mimic | UAGCUUAUCAGCUGAUGUUGA |
| Mimic negative control | UUCUCCGAACGUGUCACGUTT |
| Rab11a siRNA | AATGTCAGACAGACGCGAAAA |
| Control siRNA | UUCUCCGAACGUGUCACGUTT |
To further clarify the effect of miR-21-5p on autophagy through targeting Rab11a, a recombinant vector that overexpresses Rab11a was used (GenePharma, Shanghai, China), and for specific gene knockdown on Rab11a mRNA, the vectors of siRNA targeting Rab11a (si-Rab11a) or scramble siRNA Control (si-Control) was used (GenePharma, Shanghai, China) (Table 1). They were transfected into HT22 neurons with Lipofectamine-3000 as described above.
Immunofluorescence staining
To identify the cultured cells and measure the autophagy levels, immunofluorescence staining was performed. The cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature. Then cells were treated with 3% bovine serum albumin (BSA) for 30 minutes at 37°C to block nonspecific staining. They were incubated overnight at 4°C with primary antibodies: MAP-2 at 1: 200 (Abcam, Cambridge, United Kingdom), LC3 at 1: 200 (Cell Signaling Technology, Danvers, MA, USA). The next day, they were rinsed with PBS, and then incubated for 1 hour at room temperature with secondary antibodies. The nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, Abcam). Then the cells were evaluated under an OLYMPUS microscope (cellSens system) and an OLYMPUS confocal microscope (FV 1200).
Real time PCR
Total RNA was extracted from the cultured parental and treated HT-22 neurons, neuronal exosomes with TRIzol reagent (Thermo Fisher Scientific). The RNA concentration and quality were evaluated by Nanodrop Spectrophotometer (ND-2000; Thermo Fisher Scientific). cDNA generation and quantitative real-time (RT)-PCR were performed with the Hairpin-it miR-21-5p/mRNA RT-PCR Quantitation kit (Gene Pharma) with corresponding primers (Table 2) according to the manufacturer’s protocols. U6 served as the internal control for miR-21-5p, and GAPDH was used as the internal control for Rab11a. The Ct was acquired using the CFX Connect RT-PCR system (Bio-Rad, Hercules, CA, USA). The data were analyzed with the 2−ΔΔCt formula.
Table 2.
List of the primer sequence for quantitative RT-PCR.
| Gene | Primer sequence, 5′–3′ | |
|---|---|---|
| Forward | Reverse | |
| miR-21-5p | ACGTTGTGTAGCTTATCAGACTG | AATGGTTGTTCTCCACACTCTC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Rab11a | CCTGGTCCCACAGATACCAC | CTCAGACCTGGGAAATGGAC |
| GAPDH | GCCAAGGCTGTGGGCAAGGT | TCTCCAGGCGGCACGCAGA |
Western blot analysis
The Western blot analysis was performed at 24 hours after scratch injury and other treatments. An 8% SDS acrylamide gel was used for detecting Rab11a (1: 1000; Cell Signaling Technology). SDS-polyacrylamide gel (10%) was used for detecting Beclin-1 at 1: 1000 and P62 at 1: 1000; both from Cell Signaling Technology, SDS-polyacrylamide gels (12%) were used for detecting CD9 at 1: 2000, CD63 at 1: 1000, both from Abcam; and cleaved caspase-3 at 1: 1000, Bcl-2 at 1: 1000, LC3B at 1: 1000, and Rab11a at 1: 1000, all from Cell Signaling Technology. β-Actin at 1: 1000 from Cell Signaling Technology was used as the internal control. For densitometry, the ChemiDoc XRS+ Imaging System (Bio-Rad) was used. Measurement of mean pixel density of each band was performed with Quantity One software (Bio-Rad). The information of the antibodies mentioned above are detailed in Table 3.
Table 3.
List of the antibodies.
| Antibody | Manufacturer | Catalogue | Application | Dilution |
|---|---|---|---|---|
| LC3B | CST | 3868 | IF | 1: 200 |
| MAP-2 | Abcam | Ab5392 | IF | 1: 200 |
| CD9 | Abcam | Ab92726 | WB (25 kDa) | 1: 2000 |
| CD63 | Abcam | Ab193349 | WB (26 kDa) | 1: 1000 |
| LC3B | CST | 3868 | WB (14, 16 kDa) | 1: 1000 |
| P62 | CST | 23214 | WB (62 kDa) | 1: 1000 |
| Beclin-1 | CST | 4122 | WB (60 kDa) | 1: 1000 |
| Cleaved Caspase3 | CST | 9662 | WB (19kDa) | 1: 1000 |
| Bcl-2 | CST | 3498 | WB (26 kDa) | 1: 1000 |
| β-actin | CST | 3700 | WB (45 kDa) | 1: 1000 |
| Rab11a | CST | 2413 | WB (25 kDa) | 1: 1000 |
IF – immunofluorescence staining; WB – Western blotting.
Statistical analysis
All data represent the results of at least 3 independent experiments and are expressed as means ± standard deviation (SD). Statistical comparisons were performed using one-way ANOVA followed by least-significant difference post hoc analyses or Student’s t-tests. P values less than 0.05 were considered significant.
Result
The expression of miR-21-5p were increased in HT-22 neurons and their exosomes after treatment with rTBI mouse brain extracts
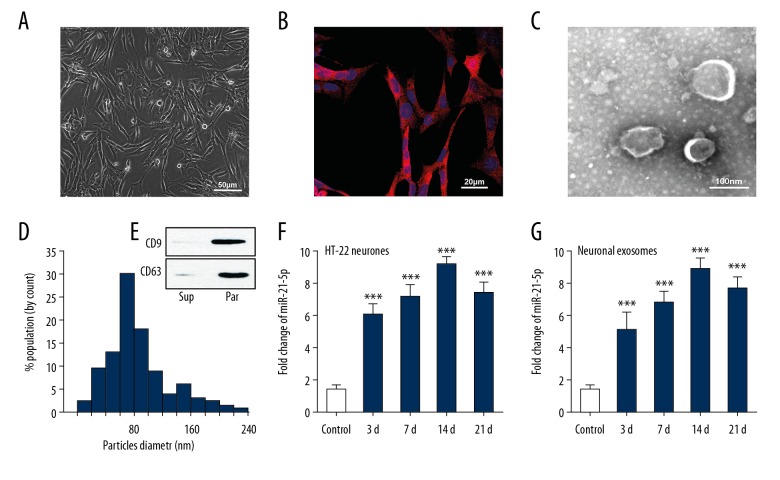
In order to detect the expression of miR-21-5p in neuronal exosomes after TBI, we first cultured HT-22 cells – a common mouse hippocampal neuron cell line. Pure HT-22 neurons were characterized by immunofluorescence staining for MAP-2 (Figure 1A, 1B), following which an rTBI mouse model was prepared. Cultured HT-22 cells were treated with extracts of the injured brain tissue at 3, 7, 14, and 21 days after the last brain injury. In order to identify the exosomes produced by HT-22 neurons, we isolated the neuron-generated particles from supernatants of the culture medium. TEM was used to observe the isolated exosomes, which were found double membrane particles ranging from 40–100 nm in size (Figure 1C). By using Nano Sight technology, the particle diameter was found to be heterogeneous, and that the peak diameter was 80.0±0.7 nm (Figure 1D). Moreover, the expression of CD9 and CD63 were abundant in the particles (Figure 1E). These results indicated that these particles were mainly composed of exosomes. Total RNA was then extracted from HT-22 cells and their exosomes, and the expression of miR-21-5p was detected by RT-PCR. Results showed that the expression of miR-21-5p were increased in both HT-22 neurons and neuronal exosomes from all rTBI groups (Figure 1F, 1G).
Figure 1.
Identification of exosomes and expression of miR-21-5p in HT-22 neurons and their exosomes after treatment with rTBI mouse brain extracts. (A) Cultured HT-22 neurons under the transmission light microscope. (B) Immunofluorescence staining for MAP-2 to identify neurons. (C) TEM image of exosomes isolated from the HT-22 neuron culture medium. The exosomes presented a round shape and double-layer membrane, with a size range of 40-100 nm. (D) The size distribution of the neuron-derived particles determined by Nano sight technology. The peak diameter of the particles was 80.8 ± 1.9 nm. (E) Immunoblot analysis showed that expressions of CD9 and CD63 were higher in the neuron-derived particles (par) than in the supernatant (sup), suggesting that these particles were mainly composed of exosomes. (F, G) Real-time PCR results reconfirmed that expression of miR-21-5p were upregulated in both HT-22 neurons and neuronal exosomes from all rTBI groups (n=6/group). *** P<0.001 versus control group. rTBI – repetitive traumatic brain injury; MAP-2 – microtubule-associated protein 2; TEM – transmission electron microscopy
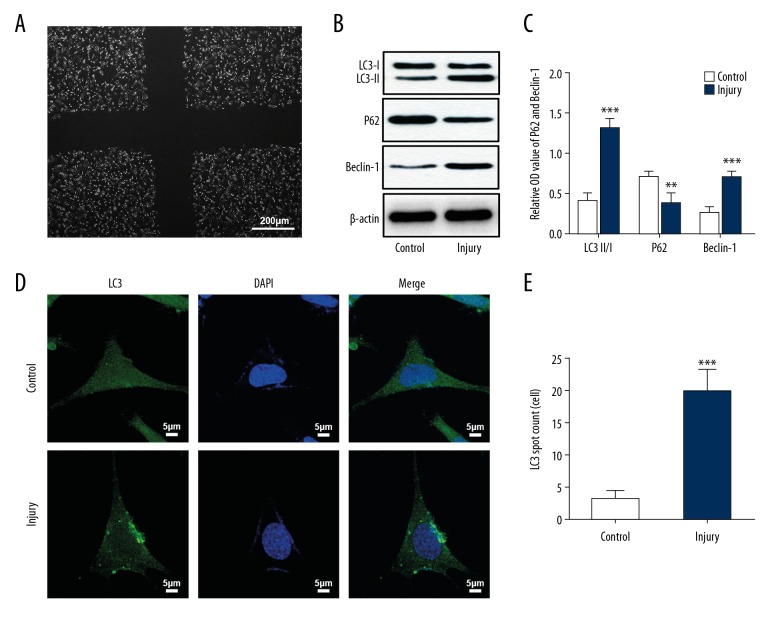
Neuronal autophagy was upregulated in the TBI models in vitro
To clarify the role of autophagy in TBI pathology, scratch-injury was used in cultured HT22 neurons., following which they served as in vitro models of TBI (Figure 2A). Expressions of LC3, P62, and Beclin-1 were detected by using western blot analyses in HT-22 neurons after injury. Results showed that, compared with the sham group, there was an increase in the expressions of LC3 and Beclin-1 and a decrease in the expression of P62 in the injured neurons (Figure 2B, 2C). Immunofluorescence staining also revealed that LC3 was highly expressed in the injured group (Figure 2D, 2E). This indicated that scratch injury can upregulate neuronal autophagy. Regulation of the activated neuronal autophagy might be a way to protect neurons from trauma-induced injury.
Figure 2.
(A–E) Neuronal autophagy was upregulated in the TBI models in vitro. (A) Cultured HT22 neurons with a scratch injury under the transmission light microscope. (B, C) Immunoblot analysis showed that LC3 II/I and Beclin-1 expressions were upregulated in cultured HT22 neurons after scratch injury, while P62 expression was downregulated after scratch injury. (D) Immunofluorescence staining showed upregulation of LC3 puncta in HT22 neurons after scratch injury. ** P<0.01, *** P<0.001 versus control group. TBI – traumatic brain injury.
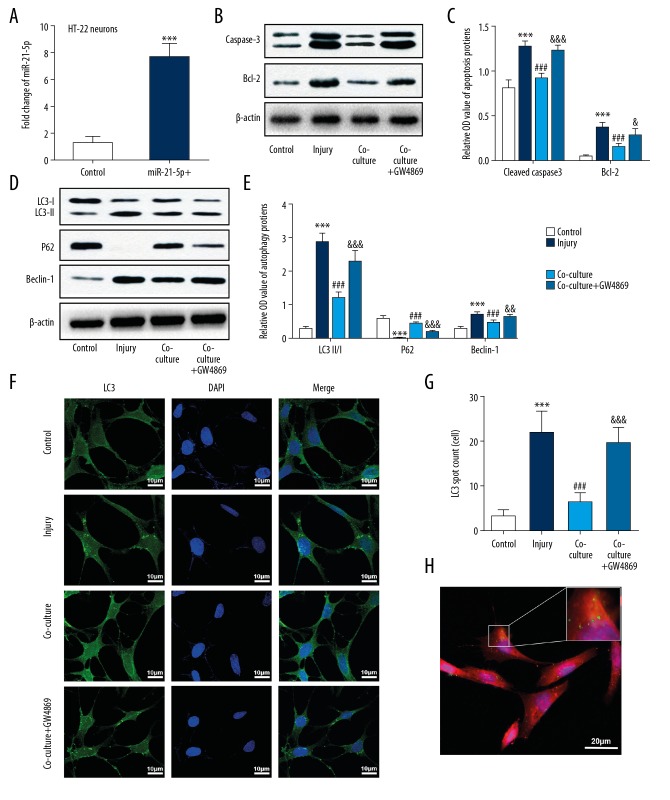
Exosomes from miR-21-5p-overexpressing HT-22 neurons inhibited scratch injury-induced neuronal autophagy and exerted a protective effect in vitro
To determine the effects of miR-21-5p-overexpressing HT-22 neurons and their exosomes on nerve injury and neuronal autophagy after TBI in vitro, neuron apoptosis and autophagy were evaluated. We first transfected miR-21-5p mimics into the cultured HT-22 neurons. Transfection efficiency of miR-21-5p was identified by RT-PCR, miR-21-5p levels were significantly upregulated following transfection (Figure 3A). The injured neurons were then co-cultured with miR-21-5p-overexpressing neurons, or with miR-21-5p-overexpressing neurons conditioned with GW4869, which inhibits the generation of exosomes. We measured levels of LC3, Beclin-1, P62, cleaved caspase-3, and Bcl-2 via western blot analysis, and further examined LC3 levels via immunofluorescence staining. Result suggested that expressions of apoptosis mediators (cleaved caspase-3 and Bcl-2) were increased in the injury group, and were significantly attenuated in the co-cultured group (Figure 3B, 3C). Moreover, increases in autophagy-related proteins (LC3, Beclin-1) and decrease in P62 expression in the injury group were remarkably attenuated by co-culture treatment (Figure 3D–3G). Similarly, results of immunofluorescence staining also suggested that LC3 was more highly expressed in the injury group, and was decreased by co-culture treatment. These results suggested that co-culture of injured neurons with miR-21-5p-overexpressing neurons might inhibit trauma-induced autophagy and exert protective effects to the injured neurons. However, compared with the co-culture group, changes in protein levels of LC3, Beclin-1, P62, cleaved caspase-3, and Bcl-2 were alleviated following GW4869 treatment, indicating that an exosome inhibitor may reverse the inhibition of autophagy and the protective effects induced by co-culture with miR-21-5p-overexpressing neurons (Figure 3B–3G).
Figure 3.
Exosomes from miR-21-5p-overexpressing HT-22 neurons inhibited scratch injury-induced neuronal autophagy and exerted a protective effect in vitro. (A) Real-time PCR analyses showed that miR-21-5p levels were significantly upregulated in the cultured HT-22 neurons after transfection with an miR-21-5p mimic. (B, C) Immunoblot analysis showed that expressions of apoptosis-related proteins (cleaved caspase 3, Bcl-2) were increased in the scratch-injured neurons, and were reduced after co-cultured with miR-21-5p-overexpressing neurons. This inhibitory effect was alleviated by GW4869 treatment. (D, E) Immunoblot analysis of autophagy-related proteins (LC3, P62, Beclin-1) revealed that autophagy was inducted in the cultured neurons after scratch injury and was inhibited after co-cultured with miR-21-5p-overexpressing neurons. This inhibitory effect was alleviated by GW4869 treatment. (F, G) Immunofluorescence staining showed upregulation of LC3 puncta in HT22 neurons after scratch injury, which decreased after treatment with co-cultured with miR-21-5p-overexpressing neurons. Similarly, the inhibitory effect was alleviated by GW4869 treatment. (H) Immunofluorescence staining revealed that the labeled exosomes were taken into neurons. *** P<0.001 versus control group; ### P<0.001 versus injury group; & P<0.05, && P<0.01,&&&P < 0.001 versus co-culture group.
In order to confirm that exosomes can be transferred among the neurons, isolated exosomes were labeled with PKH67 and incubated with HT-22 neurons. Immunofluorescence staining experiments showed that the labeled exosomes were transferred into the HT-22 neurons (Figure 3H). All these results indicate that a co-culture with miR-21-5p-overexpressing neurons produces a protective effect by suppressing neuronal autophagy in scratch-injured neurons. These effects were probably achieved via neuronal exosomes.
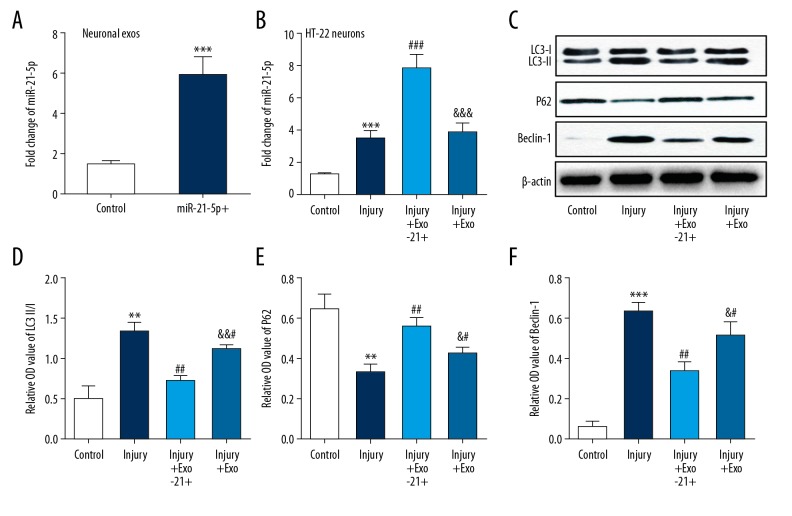
Exosomal miR-21-5p inhibited the neuronal autophagy by increasing miR-21-5p expression in the injured neurons
RT-PCR analyses showed that the expression of miR-21-5p was increased in exosomes harvested from miR-21-5p mimics transfected HT-22 neurons (Figure 4A). To clarify the effect of exosomal miR-21-5p on scratch injury-induced neuronal autophagy, the injured neurons were treated with normal exosomes or miR-21-5p-overexpressing exosomes. Results found that scratch injury increased miR-21-5p expression in neurons, and that such expression was significantly upregulated following treatment with miR-21-5p-overexpressing exosomes. There was no significant change in miR-21-5p expression in injured neurons following treatment with normal exosomes (Figure 4B). Then the neuronal autophagy-related proteins including LC3, Beclin-1, and P62 were measured by western bolt analysis. Results demonstrated that treatment with neuronal exosomes decreased the level of LC3 and Beclin-1 while increasing the level of P62, especially in miR-21-5p-overexpressing exosomes treated group (Figure 4C–4F). These data suggested that suppression effect of autophagy in injured neurons was mainly caused by exosomal miR-21-5p.
Figure 4.
Exosomal miR-21-5p inhibited the neuronal autophagy by increasing miR-21-5p expression in the injured neurons. (A) Real-time PCR showed that expression of miR-21-5p was increased in exosomes harvested from miR-21-5p mimics transfected HT-22 neurons. (B) Real-time PCR showed that miR-21-5p expression was increased in HT22 neurons after scratch injury, and further increasing significantly after treatment with miR-21-5p-overexpressing exosomes. (C–F) Immunoblot analysis of autophagy-related proteins (LC3, P62, Beclin-1) showed that autophagy was induced in the cultured HT-22 neurons after scratch injury. Activation of autophagy was inhibited by neuronal exosomes, especially miR-21-5p-overexpressing exosomes. ** P<0.01, *** P<0.001 versus control group; ## P<0.01, ### P<0.001 versus injury group; & P<0.05, && P<0.01 versus injury+ Exo-21+ group.
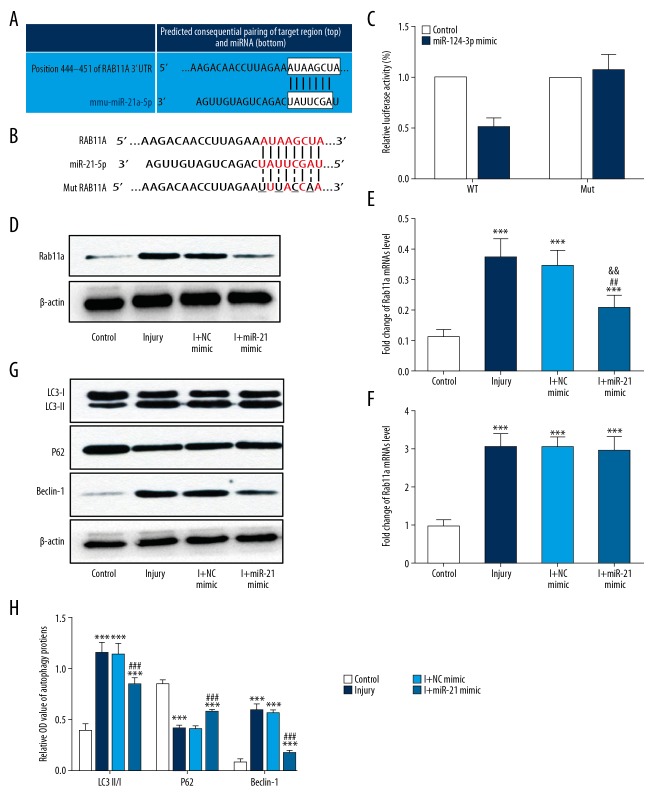
MiR-21-5p targeted Rab11a via repressing translation in HT-22 neurons
To evaluate the mechanism underlying the suppression of scratch injury-induced neuronal autophagy by miR-21-5p, potential target molecule was predicted by using bioinformatics analysis (http://www.targetscan.org/). Results showed that Rab11a, a protein associated with the process of autophagy, may be a target of miR-21-5p (Figure 5A). Then a dual-luciferase reporter assay was performed to identify whether miR-21-5p could directly target the 3′UTR of Rab11a mRNA. The fragment of the 3′UTR of Rab11a mRNA containing the putative miR-21-5p binding site or its mutant 3′UTR was cloned into luciferase reporter vector pGL3 to construct pGL3-Rab11a-3′UTR WT or pGL3-Rab11a-3′UTR Mut, and then co-transfected with miR-21-5p mimics or scrambled oligonucleotides into HT-22 neurons (Figure 5B). Results suggested that relative luciferase activity was remarkably downregulated in the neurons co-transfected with pGL3-Rab11a-3′UTR WT with miR-21-5p, while no difference was found in the neurons co-transfected with pGL3-Rab11a-3′UTR Mut with miR-21-5p, compared with the relative luciferase activity observed in the control group (Figure 5C). All of the data revealed that miR-21-5p can directly target the 3′UTR of Rab11a.
Figure 5.
miR-21-5p targeted Rab11a via repressing translation in HT-22 neurons. (A) The potential binding sites for miR-21-5p in the Rab11a 3′UTR, which were predicted by using bioinformatics analysis (http://www.targetscan.org/). (B) The WT (Rab11a WT 3′UTR) and mutant type (Rab11a mut 3′UTR) luciferase reporter constructs exhibited intact and mutated seed sequences (underlined), respectively, in the miR-21-5p binding site. (C) The relative luciferase activity of the WT and mut reporter constructs, which were co-transfected with either the miR-21–5p mimic or scrambled oligonucleotides. Data indicated that miR-21-5p inhibited the luciferase activity of the WT, but not the mut type. (D, E) Immunoblot analysis showed that Rab11a expression was upregulated in the cultured HT22 neurons after scratch injury. Overexpression of miR-21-5p suppressed the protein expression of Rab11a, and no apparent difference was observed in I+ NC mimic group, relative to the levels observed in injury group. (F) No obvious differences in Rab11a mRNA expression were observed among the injury group and transfected groups. (G, H) Immunoblot analysis of autophagy-related proteins (LC3, P62, Beclin-1) showed that neuronal autophagy was inhibited in the I+miR-21-5p mimic group, and no apparent difference was observed in the I+ NC mimic group, relative to the levels observed in injury group. *** P<0.001 versus control group; ## P<0.01, ### P<0.001 versus injury group. WT – wild-type; Mut – mutant-type; NC – negative control.
To further identify the effect of miR-21-5p on Rab11a expression, we respectively transfected miR-21-5p mimic and negative control mimic into HT22 neurons. Overexpression of miR-21-5p remarkably suppress the protein expression of Rab11a, and no apparent difference was observed in the negative control mimic group, relative to the levels observed in injury group (Figure 5D, 5E). Moreover, there were no obvious changes in levels of Rab11a mRNA expression among the injury group and transfected groups (Figure 5F). The result indicated that miR-21-5p suppressed the translation of Rab11a, rather than its transcription. In addition, we also examined the expression of autophagy-related proteins to evaluate the direct effect of miR-21-5p on autophagy in injured neurons. As expected, our results indicated that autophagy was inhibited in injured neurons transfected with the miR-21-5p mimic, except the negative control mimic group (Figure 5G, 5H). These results demonstrated that miR-21-5p had induced autophagy inhibition in the injured neurons, possibly by targeting Rab11a.
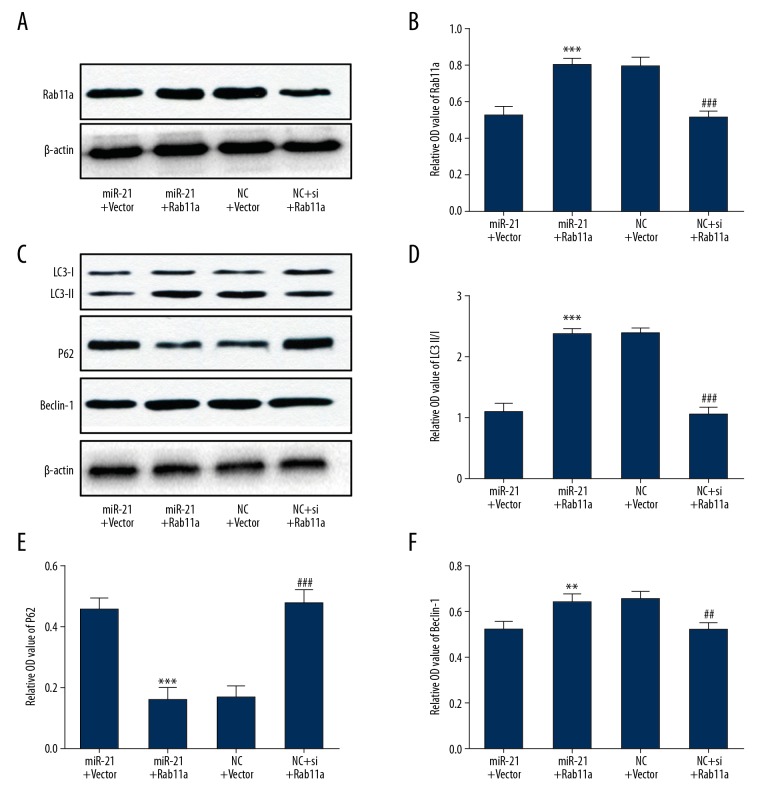
Overexpression of miR-21-5p mainly suppressed Rab11a-mediated neuronal autophagy
To further clarify the contribution of Rab11a to the suppression of neuronal autophagy, a functional rescue experiment was carried out by overexpressing Rab11a in injured HT-22 neurons. MiR-21-5p mimic and Rab11a overexpression plasmid were co-transfected into the neurons prior to scratch-injury. Moreover, we also transfected si-Rab11a and si-negative control into the HT-22 neurons. Western blot analyses showed that overexpression of miR-21-5p suppressed Rab11a protein expression, and co-transfection of Rab11a overexpression plasmid reversed this inhibitory effect (Figure 6A, 6B). In addition, normal neuronal expression of Rab11a was silenced by si-Rab11a (Figure 6A, 6B). More importantly, our results demonstrated that the inhibitory effects of miR-21-5p overexpression on autophagic activation were remarkably blocked by Rab11a overexpression, and that silencing of Rab11a via siRNA mimicked the effects of miR-21-5p in scratch injured neurons (Figure 6C–6F). These data indicated that Rab11a, as the target of miR-21-5p, plays a pivotal role in the process of scratch injury-induced neuronal autophagy.
Figure 6.
Overexpression of miR-21-5p mainly suppressed Rab11a-mediated neuronal autophagy. (A, B) Immunoblot analysis showed that overexpression of Rab11a reversed the miR-21-5p-induced inhibition of Rab11a, and that silencing of Rab11a by siRNA suppressed Rab11a expression in the scratch-injured neurons. (C–F) Overexpression of Rab11a blocked miR-21-5p-induced inhibition of neuronal autophagy, and silencing of Rab11a by siRNA mimicked the effects of miR-21-5p in the scratch-injured neurons. ** P<0.01, *** P<0.001 versus miR-21-5p+vector group; ## P<0.01, ### P<0.001 versus NC+vector group. NC – negative control.
Discussion
In this study, we confirmed that miR-21-5p levels in neuronal exosomes increase from the acute to chronic phase of TBI at first. Then we investigated whether neuronal exosomes enriched with miR-21-5p have a protective effect on a TBI model in vitro and we also explored the possible mechanism. The results showed as follows: i) neuronal autophagy was activated in the HT-22 neurons after scratch injury; ii) exosomes from miR-21-5p-overexpressing HT-22 neurons significantly alleviated nerve injury by suppressing autophagy in TBI model in vitro; iii) miR-21-5p loaded as a cargo in the exosomes plays the key role in the inhibition of neuronal autophagy; iv) miR-21-5p could specifically binds with the 3′UTR of Rab11a mRNA to modulate its translation; v) the functional rescue experiment showed the overexpression of Rab11a abrogated the inhibition effect of miR-21-5p on neuronal autophagy post-TBI, suggesting that upregulated miR-21-5p mainly suppressed Rab11a-mediated neuronal autophagy. Taken together, our results indicated that neuron derived exosomal miR-21-5p may represent a therapeutic target for intervention in TBI-induced neuronal autophagy.
Exosomes are extracellular micro-vesicles that play important roles in intercellular communication by transporting functional cargos such as RNA, lipids, and proteins from one cell to another [40]. In the central nervous system (CNS), exosomes have been proved to be released from different types of cells, such as neurons, astrocytes, microglia, and oligodendrocytes [41–45]. Accumulating evidence demonstrates that exosomes in the central nervous system exert a number of roles, such as intercellular communication, maintenance of myelination, synaptic plasticity, trophic support of neurons and so on, suggesting they take part in pathologic changes in many CNS disorders [32,42,44,46,47]. Neuron derived exosome is a type of the exosomes derived from CNS, it has been reported to have a utility as diagnostic biomarkers in Alzheimer disease (AD), frontotemporal dementia, and dementia with lewy bodies [48–51]. Recent studies suggest that neuron derived exosomes may play a vital role in the clearance of Aβ. Neuronal exosomes can receive APP from early endosomes after cleavage into Aβ peptides, which may then be secreted from the cells in exosomes [52,53]. Other studies report that neuronal exosomes may transmit Aβ and tau to neighboring cells and other brain regions, causing the propagation of these toxic cargos [48,54,55]. These findings indicate neuronal exosome as a promising therapeutic target of Alzheimer disease. Although neuronal exosomes have been studied in chronic neurologic diseases, especially in neurodegenerative diseases, little attention has been paid to their functions in acute neurologic diseases, such as TBI.
Therefore, we focused on studying the impact of miRNAs loaded in the neuron derived exosomes on injured neurons in the present study. Using the rTBI mouse model, we harvested the brain extracts, which were added to the cultured HT-22 neurons, to imitate the microenvironment of injured brain on in vitro cultured cells. Thus, the neuronal exosomes under TBI condition could be separately collected from brain-derived exosomes for further investigation of their roles in TBI. We confirmed that miR-21-5p levels are increased in exosomes released from HT-22 neurons exposed to TBI brain extracts at 3, 7, 14, and 21 days after the last brain injury. MiR-21-5p has been proved to act as a protective role in several diseases of the CNS [27,30,56]. In our previous work, we identified that the level of miR-21-5p in brain was increased after TBI [26]. Our previous studies also suggested that miR-21-5p could contribute to improving the nerve injury and neurologic outcome of TBI both in vivo and in vitro [24,25]. Moreover, we found miR-21-5p may alleviate blood-brain barrier damage in brain microvascular endothelial cells [22]. In this present study, we confirmed that miR-21-5p exert an anti-apoptosis effect on scratch injured neurons via their transfer by neuronal exosomes. Thus, these results suggested that the increased miR-21-5p in neuronal exosomes confer a neuroprotective effect in TBI models in vitro.
Autophagy is a self-catabolic process for the degradation of superfluous or aberrant cytoplasmic components, representing an essential cell survival mechanism. Autophagy has been confirmed to be critical for various biological activities. Recent studies have suggested that neuronal autophagy is significantly activated in response to trauma and participate in the pathophysiological processes of TBI [14,16,17,57]. However, the role of neuronal autophagy in the pathological process of TBI remains uncertain. Previous studies suggested that trauma induced neuronal autophagy as a protective process at early stage after TBI, autophagic activation may eliminate aberrant cellular components and protect neurons against from trauma-induced injury [14,58]. More recent reports have suggested that dysregulated autophagy is associated with neuronal death, inhibition of excessive autophagy can attenuate brain injury and improve neurological outcomes after TBI [12,33]. A series of microRNAs have been proved involving in the different autophagy cascades by acting on different targets [59–61]. Emerging studies demonstrated the possible relationship between miR-21 and autophagy in different diseases. It was reported that miR-21 may promote the proliferation, migration and invasion of NSCLC A549 cells by regulating autophagy activity of NSCLC A549 cells [62]. Another study indicated miR-21 promoted ECM degradation through inhibiting autophagy in human degenerated NP cells [63]. Furthermore, treatment with antimiR-21 could promote the autophagy activation and increase chemosensitivity of leukemia cells [64]. In the present study, we first identified that miR-21-5p-overexpressing HT-22 neurons could suppress scratch injury-induced neuronal autophagy. Then we proved that neuron derived exosomal miR-21-5p play the key role in the inhibition of neuronal autophagy, thus exert neuroprotective effect in scratch injured neurons. These findings indicate that increased miR-21-5p expression in neuron derived exosomes may inhibit neuronal autophagy and influence the pathological progression of TBI.
Furthermore, by using bioinformatics analysis, Rab11a was found to be a potential target gene of miR-21-5p. Rab GTPases, including more than 60 members in the human genome, serve as multifaceted organizers in almost all membrane and vesicle trafficking processes [65]. Rab11 was a member of Rab GTPases family. Recent studies found that Rab11 was involved in both early and late stages of autophagy and play an important role in the maturation process of autophagosomes [66–68]. It was reported that Rab11, involving in the interaction between multivesicular bodies and autophagic pathway, was required for autophagosome in K652 cells [69]. Several studies revealed that recycling endosomes (RE) might be a membrane source for autophagosome formation and Rab11 was thought to take part in vesicle trafficking from REs to the sites of autophagosome formation [70,71]. Another study suggested that Rab11 may promote the fusion of endosomes and autophagosomes by removing Hook from mature late endosomes [72]. Rab11a, a member of Rab11 subfamily, has been shown to be critical to the process of autophagy. One recent report indicated that Rab11a-positive membrane was a primary direct platform for canonical autophagosome formation, loss of Rab11a impaired the recruitment and assembly of the autophagic machinery [73]. In the present study, we subsequently confirmed that miR-21-5p binds directly to the 3′UTR region of Rab11a mRNA via a luciferase reporter assay. In addition, overexpression of Rab11a abrogated the neuroprotective effects of miR-21-5p on neuronal injury post-TBI, further supporting the notion that miR-21-5p protects against TBI-induced neuronal injury by suppressing Rab11a-mediated neuronal autophagy. Altogether, these findings suggest that increased levels of miR-21-5p in neuronal exosomes can be transferred to the injured neurons to regulate excessive neuronal autophagy by suppressing Rab11a, thereby conferring protective effects against neuronal injury post-TBI in vitro.
Conclusions
In summary, our results demonstrate that miR-21-5p levels in neuronal exosomes increase from the acute to chronic phase of TBI. Furthermore, our findings indicate that neuronal exosomes enriched with miR-21-5p can inhibit the activity of Rab11a-mediated neuronal autophagy signaling by targeting Rab11a, thus attenuating trauma-induced, autophagy-mediated nerve injury in vitro. Our study also suggests that treatment with neuronal exosomes enriched with miR-21-5p represents a promising therapeutic strategy for the treatment of nerve injury after TBI. Neuronal exosomes manipulated with miRNA may thus represent a novel therapy for TBI and other neurologic diseases.
Footnotes
Source of support: This research was supported by grants from the National Natural Science Foundation of China (grant no. 81772060, 81471252), Tianjin Science Foundation (grant no. 16JCYBJC27200, 16JCQNJC11000, 16JCYBJC26900, 18ZXDBSY00090), Tianjin Medical University General Hospital Youth Cultivation Foundation (grant no. ZYYFY2016001)
References
- 1.Johnson WD, Griswold DP. Traumatic brain injury: A global challenge. Lancet Neurol. 2017;16:949–50. doi: 10.1016/S1474-4422(17)30362-9. [DOI] [PubMed] [Google Scholar]
- 2.Majdan M, Plancikova D, Brazinova A, et al. Epidemiology of traumatic brain injuries in Europe: A cross-sectional analysis. Lancet Public Health. 2016;1:e76–83. doi: 10.1016/S2468-2667(16)30017-2. [DOI] [PubMed] [Google Scholar]
- 3.Gao GY, Jiang JY. Chinese Head Trauma Data Bank: Effect of gender on the outcome of patients with severe traumatic brain injury. J Neurotrauma. :2012. doi: 10.1089/neu.2011.2134. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Jiang JY Chinese Head Trauma Study Collaboration. Head trauma in China. Injury. 2013;44:1453–57. doi: 10.1016/j.injury.2012.08.045. [DOI] [PubMed] [Google Scholar]
- 5.Maas AIR, Menon DK, Adelson PD, et al. Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 2017;16:987–1048. doi: 10.1016/S1474-4422(17)30371-X. [DOI] [PubMed] [Google Scholar]
- 6.Levin H, Smith D. Traumatic brain injury: Networks and neuropathology. Lancet Neurol. 2013;12:15–16. doi: 10.1016/S1474-4422(12)70300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlosberg D, Benifla M, Kaufer D, Friedman A. Blood-brain barrier breakdown as a therapeutic target in traumatic brain injury. Nat Rev Neurol. 2010;6:393–403. doi: 10.1038/nrneurol.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky DJ. Cell biology: Regulated self-cannibalism. Nature. 2004;431:31–32. doi: 10.1038/431031a. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Kroemer G. Snapshot: Macroautophagy. Cell. 2008;132:162e1–e3. doi: 10.1016/j.cell.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y, Zhang MY, Wang T, et al. IL-33/ST2L signaling provides neuroprotection through inhibiting autophagy, endoplasmic reticulum stress, and apoptosis in a mouse model of traumatic brain injury. Front Cell Neurosci. 2018;12:95. doi: 10.3389/fncel.2018.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin CJ, Chen TH, Yang LY, Shih CM. Resveratrol protects astrocytes against traumatic brain injury through inhibiting apoptotic and autophagic cell death. Cell Death Dis. 2014;5:e1147. doi: 10.1038/cddis.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar C, Zhao Z, Aungst S, et al. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy. 2014;10:2208–22. doi: 10.4161/15548627.2014.981787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Wang H. Autophagy in traumatic brain injury: A new target for therapeutic intervention. Front Mol Neurosci. 2018;11:190. doi: 10.3389/fnmol.2018.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CL, Chen S, Dietrich D, Hu BR. Changes in autophagy after traumatic brain injury. J Cereb Blood Flow Metab. 2008;28:674–83. doi: 10.1038/sj.jcbfm.9600587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Liu A, Zhang J, et al. miR-23b improves cognitive impairments in traumatic brain injury by targeting ATG12-mediated neuronal autophagy. Behav Brain Res. 2018;340:126–36. doi: 10.1016/j.bbr.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Zhao M, Wang Y, et al. Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochem Biophys Res Commun. 2017;482:1141–47. doi: 10.1016/j.bbrc.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Au AK, Aneja RK, Bayir H, et al. Autophagy biomarkers Beclin 1 and p62 are increased in cerebrospinal fluid after traumatic brain injury. Neurocrit Care. 2017;26:348–55. doi: 10.1007/s12028-016-0351-x. [DOI] [PubMed] [Google Scholar]
- 18.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–9. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Sun T, Liu Z, et al. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS One. 2014;9:e103948. doi: 10.1371/journal.pone.0103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez B, Peplow PV. MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen Res. 2017;12:1749–61. doi: 10.4103/1673-5374.219025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meissner L, Gallozzi M, Balbi M, et al. Temporal profile of microRNA expression in contused cortex after traumatic brain injury in mice. J Neurotrauma. 2016;33:713–20. doi: 10.1089/neu.2015.4077. [DOI] [PubMed] [Google Scholar]
- 22.Ge X, Han Z, Chen F, et al. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150–57. doi: 10.1016/j.brainres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Ge X, Huang S, Gao H, et al. MiR-21-5p alleviates leakage of injured brain microvascular endothelial barrier in vitro through suppressing inflammation and apoptosis. Brain Res. 2016;1650:31–40. doi: 10.1016/j.brainres.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Ge XT, Lei P, Wang HC, et al. MiR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:6718. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Z, Chen F, Ge X, et al. MiR-21 alleviated apoptosis of cortical neurons through promoting PTEN-Akt signaling pathway in vitro after experimental traumatic brain injury. Brain Res. 2014;1582:12–20. doi: 10.1016/j.brainres.2014.07.045. [DOI] [PubMed] [Google Scholar]
- 26.Lei P, Li Y, Chen X, et al. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 27.Ji W, Jiao J, Cheng C, Shao J. MicroRNA-21 in the pathogenesis of traumatic brain injury. Neurochem Res. 2018;43:1863–68. doi: 10.1007/s11064-018-2602-z. [DOI] [PubMed] [Google Scholar]
- 28.Su C, Yang X, Lou J. Geniposide reduces alpha-synuclein by blocking microRNA-21/lysosome-associated membrane protein 2A interaction in Parkinson disease models. Brain Res. 2016;1644:98–106. doi: 10.1016/j.brainres.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Xie W, Yang SY, Zhang Q, et al. Knockdown of microRNA-21 promotes neurological recovery after acute spinal cord injury. Neurochem Res. 2018;43:1641–49. doi: 10.1007/s11064-018-2580-1. [DOI] [PubMed] [Google Scholar]
- 30.Yao X, Wang Y, Zhang D. MicroRNA-21 confers neuroprotection against cerebral ischemia-reperfusion injury and alleviates blood-brain barrier disruption in rats via the MAPK signaling pathway. J Mol Neurosci. 2018;65:43–53. doi: 10.1007/s12031-018-1067-5. [DOI] [PubMed] [Google Scholar]
- 31.Shi M, Sheng L, Stewart T, et al. New windows into the brain: Central nervous system-derived extracellular vesicles in blood. Prog Neurobiol. 2019;175:96–106. doi: 10.1016/j.pneurobio.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson AG, Gray E, Heman-Ackah SM, et al. Extracellular vesicles in neurodegenerative disease – pathogenesis to biomarkers. Nat Rev Neurol. 2016;12:346–57. doi: 10.1038/nrneurol.2016.68. [DOI] [PubMed] [Google Scholar]
- 33.Jiang M, Wang H, Jin M, et al. Exosomes from MiR-30d-5p-ADSCs reverse acute ischemic stroke-induced, autophagy-mediated brain injury by promoting M2 microglial/macrophage polarization. Cell Physiol Biochem. 2018;47:864–78. doi: 10.1159/000490078. [DOI] [PubMed] [Google Scholar]
- 34.Gao H, Han Z, Bai R, et al. The accumulation of brain injury leads to severe neuropathological and neurobehavioral changes after repetitive mild traumatic brain injury. Brain Res. 2017;1657:1–8. doi: 10.1016/j.brainres.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 35.Huang S, Ge X, Yu J, et al. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018;32:512–28. doi: 10.1096/fj.201700673R. [DOI] [PubMed] [Google Scholar]
- 36.Xin H, Li Y, Buller B, et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–64. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greer JE, Hanell A, McGinn MJ, Povlishock JT. Mild traumatic brain injury in the mouse induces axotomy primarily within the axon initial segment. Acta Neuropathol. 2013;126:59–74. doi: 10.1007/s00401-013-1119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 39.Han Z, Ge X, Tan J, et al. Establishment of lipofection protocol for efficient miR-21 transfection into cortical neurons in vitro. DNA Cell Biol. 2015;34:703–9. doi: 10.1089/dna.2015.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osier N, Motamedi V, Edwards K, et al. Exosomes in acquired neurological disorders: New insights into pathophysiology and treatment. Mol Neurobiol. 2018;55:9280–93. doi: 10.1007/s12035-018-1054-4. [DOI] [PubMed] [Google Scholar]
- 41.Bianco F, Perrotta C, Novellino L, et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009;28:1043–54. doi: 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budnik V, Ruiz-Canada C, Wendler F. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci. 2016;17:160–72. doi: 10.1038/nrn.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faure J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642–48. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Kramer-Albers EM, Bretz N, Tenzer S, et al. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics Clin Appl. 2007;1:1446–61. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- 45.Potolicchio I, Carven GJ, Xu X, et al. Proteomic analysis of microglia-derived exosomes: Metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol. 2005;175:2237–43. doi: 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- 46.Coleman BM, Hill AF. Extracellular vesicles – their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol. 2015;40:89–96. doi: 10.1016/j.semcdb.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Rajendran L, Bali J, Barr MM, et al. Emerging roles of extracellular vesicles in the nervous system. J Neurosci. 2014;34:15482–89. doi: 10.1523/JNEUROSCI.3258-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiandaca MS, Kapogiannis D, Mapstone M, et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015;11:600–7e1. doi: 10.1016/j.jalz.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gamez-Valero A, Beyer K, Borras FE. Extracellular vesicles, new actors in the search for biomarkers of dementias. Neurobiol Aging. 2019;74:15–20. doi: 10.1016/j.neurobiolaging.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Goetzl EJ, Kapogiannis D, Schwartz JB, et al. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016;30:4141–48. doi: 10.1096/fj.201600816R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ngolab J, Trinh I, Rockenstein E, et al. Brain-derived exosomes from dementia with Lewy bodies propagate alpha-synuclein pathology. Acta Neuropathol Commun. 2017;5:46. doi: 10.1186/s40478-017-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Yuan M, Liu N, Wang X, et al. The mechanism of exosomes function in neurological diseases: A progressive review. Curr Pharm Des. 2018;24:2855–61. doi: 10.2174/1381612824666180903113136. [DOI] [PubMed] [Google Scholar]
- 53.Yuyama K, Sun H, Usuki S, et al. A potential function for neuronal exosomes: Sequestering intracerebral amyloid-beta peptide. FEBS Lett. 2015;589:84–88. doi: 10.1016/j.febslet.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 54.Jaunmuktane Z, Mead S, Ellis M, et al. Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature. 2015;525:247–50. doi: 10.1038/nature15369. [DOI] [PubMed] [Google Scholar]
- 55.Kapogiannis D, Boxer A, Schwartz JB, et al. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer’s disease. FASEB J. 2015;29:589–96. doi: 10.1096/fj.14-262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Wang Y, Lv Q, et al. MicroRNA-21 overexpression promotes the neuroprotective efficacy of mesenchymal stem cells for treatment of intracerebral hemorrhage. Front Neurol. 2018;9:931. doi: 10.3389/fneur.2018.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun L, Gao J, Zhao M, et al. A novel cognitive impairment mechanism that astrocytic p-connexin 43 promotes neuronic autophagy via activation of P2X7R and down-regulation of GLT-1 expression in the hippocampus following traumatic brain injury in rats. Behav Brain Res. 2015;291:315–24. doi: 10.1016/j.bbr.2015.05.049. [DOI] [PubMed] [Google Scholar]
- 58.Ban BK, Jun MH, Ryu HH, et al. Autophagy negatively regulates early axon growth in cortical neurons. Mol Cell Biol. 2013;33:3907–19. doi: 10.1128/MCB.00627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korkmaz G, le Sage C, Tekirdag KA, et al. MiR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–76. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 60.Menghini R, Casagrande V, Marino A, et al. MiR-216a: A link between endothelial dysfunction and autophagy. Cell Death Dis. 2014;5:e1029. doi: 10.1038/cddis.2013.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu H, Wang F, Hu S, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179–86. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Li S, Zeng X, Ma R, Wang L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018;16:2038–45. doi: 10.3892/etm.2018.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang WJ, Yang W, Ouyang ZH, et al. MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. Biomed Pharmacother. 2018;99:725–34. doi: 10.1016/j.biopha.2018.01.154. [DOI] [PubMed] [Google Scholar]
- 64.Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, et al. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets. 2013;14:1135–43. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 65.Rivero-Rios P, Gomez-Suaga P, Fernandez B, et al. Alterations in late endocytic trafficking related to the pathobiology of LRRK2-linked Parkinson’s disease. Biochem Soc Trans. 2015;43:390–95. doi: 10.1042/BST20140301. [DOI] [PubMed] [Google Scholar]
- 66.Lamb CA, Longatti A, Tooze SA. Rabs and GAPs in starvation-induced autophagy. Small GTPases. 2016;7:265–69. doi: 10.1080/21541248.2016.1220779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Longatti A, Lamb CA, Razi M, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–75. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards P, Didszun C, Campesan S, et al. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington’s disease. Cell Death Differ. 2011;18:191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–50. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 70.Knaevelsrud H, Soreng K, Raiborg C, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–49. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puri C, Renna M, Bento CF, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–99. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szatmari Z, Kis V, Lippai M, et al. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell. 2014;25:522–31. doi: 10.1091/mbc.E13-10-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puri C, Vicinanza M, Ashkenazi A, et al. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev Cell. 2018;45:114–31e8. doi: 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]