Abstract
Background
Hypercoagulation and neutrophilia are described in several cancers, however, whether they are involved in lung carcinogenesis is currently unknown. Emodin is the main bioactive component from Rheum palmatum and has many medicinal values, such as anti-inflammation and anticancer. This study is to investigate the contributions of neutrophils to the effects of emodin on hypercoagulation and carcinogenesis.
Methods
The effects of emodin on neutrophil phenotypes were assessed by cell proliferation, morphological changes, phagocytosis and autophagy in vitro. The anti-coagulation and cancer-preventing actions of emodin were evaluated in the urethane-induced lung carcinogenic model. The expressions of Cit-H3 and PAD4 in lung sections were assessed by immunohistochemistry, CD66b+ neutrophils were distinguished by immunofluorescence, and cytokines and ROS were examined with ELISA. The neutrophils-regulating and hypercoagulation-improving efficacies of emodin were confirmed in a Lewis lung cancer allograft model. The related targets and pathways of emodin were predicted by network pharmacology.
Results
In vitro, emodin at the dose of 20 µM had no effect on cell viability in HL-60N1 but increased ROS and decreased autophagy and thus induced apoptosis in HL-60N2 with the morphological changes. In the urethane-induced lung carcinogenic model, before lung carcinogenesis, urethane induced obvious hypercoagulation which was positively correlated with lung N2 neutrophils. There were the aggravated hypercoagulation and lung N2 neutrophils after lung carcinoma lesions. Emodin treatment resulted in the ameliorated hypercoagulation and lung carcinogenesis accompanied by the decreased N2 neutrophils (CD66b+) in the alveolar cavity. ELISA showed that there were more IFN-γ, IL-12 and ROS and less IL-6, TNF-α and TGF-β1 in the alveolar cavity in the emodin group than those in the control group. Immunohistochemical analysis showed that emodin treatment decreased Cit-H3 and PAD4 in lung sections. In the Lewis lung cancer allograft model, emodin inhibits tumor growth accompanied by the attenuated coagulation and intratumor N2 neutrophils. Network pharmacology indicated the multi-target roles of emodin in N2 neutrophil activation.
Conclusions
This study suggests a novel function of emodin, whereby it selectively suppresses N2 neutrophils to prevent hypercoagulation and lung carcinogenesis.
Keywords: Emodin, Neutrophil phenotypes, NETs, Hypercoagulation, Carcinogenesis
Background
Due to being diagnosed at advanced stages, lung cancer is the leading cause of cancer-related death [1]. In spite of revised and aggressive approaches to therapy, the 5-year survival rate for advanced stage lung cancer has not improved significantly [2]. As is the case for other types of cancer, the normal lung cells spend several years transforming into early precursor lesions and finally into lung cancer, which provides us with several opportunities for preventing or reversing the progression of precancerous lesions [3]. Undoubtedly, the application of chemopreventive agents may decrease the cancer incidence, as well as treatment cost, and contribute to the improvement of patient survival and quality of life [4]. The herb-derived compounds display an unparalleled potential for the proposed development of preventive pharmaceutics [5]. Emodin is a natural anthraquinone present in various Chinese medicinal herbs and is known as the main bioactive component in Rheum palmatum which has been used for over 2000 years in China [6]. Pharmacological research indicated that emodin has a variety of functions, including anti-tumor and anti-inflammation [7, 8]. Previous studies have shown that emodin possesses anti-proliferative function in various cancer cells in vitro and in vivo [9, 10]. A recent study revealed that emodin could also exert tumor preventive efficacy in DMBA-induced oral carcinogenesis [11]. Furthermore, the injury preventive action of emodin was confirmed in several conditions, such as pancreatitis [12], ulcerative colitis [13], intestinal mucosal injury [14] and lung injury [15]. These findings strongly suggest that emodin may protect organ injury against tissue carcinogenesis, however, the different conditions and the involved mechanism need to be further investigated.
Recently, neutrophils, which are the most important effectors of innate immunity, have been shown to have a crucial role in tumor initiation and progression by releasing cytokines, chemokines, ECM remodeling enzymes, and so on [16]. Neutrophils accumulate in many kinds of human and murine tumors and participate in nearly all steps of tumor progression [17, 18]. In clinical practice, the neutrophils denote heterogeneous populations in response to inflammatory stimuli, Fridlender et al. proposed the paradigm of anti-tumoral “N1 neutrophils” versus pro-tumoral “N2 neutrophils” based on neutrophil diversity and plasticity [19]. Most recently, it has been known that neutrophil extracellular traps (NETs) are an important effector mechanism by which neutrophils elaborate response to numerous stimuli [20]. Cancer patients are commonly affected by thrombotic events induced by tumor-related platelet activation [21]. Several studies have suggested an important role of NETs in cancer progression and tumor-associated thrombosis [22–24]. These studies have indicated the complex relationship among neutrophils, hypercoagulation and carcinogenesis. It was reported that spontaneous intestinal tumorigenesis correlated with the accumulation of low density neutrophils with a pro-tumorigenic N2 phenotype, and that hypercoagulation and neutrophilia are frequent incidents in cancer patients [25]. It was also reported that emodin promoted polymorphonuclear neutrophil apoptosis in pancreatitis-associated acute lung injury [26] and ameliorated diesel exhaust particles-induced impairment of vascular and cardiac homeostasis [27]. However, to date, whether hypercoagulation involved in lung carcinogenesis and whether the cancer preventive efficacy of emodin is associated with its antithrombosis are currently unknown. In this study, we explored the relationship between hypercoagulation and carcinogenesis and investigated the contributions of neutrophils to the effects of emodin on hypercoagulation and carcinogenesis.
Materials and methods
Materials
Emodin (purity > 98%) was purchased from Xi-an Helin Biological Engineering Co. (Xi-an, Shanxi, China). Urethane, phorbol 12-myristate 13-acetate (PMA), 4′,6-diamidino-2-phenylindole (DAPI) and 2′,7′-dichlorodihydrofluorescein (DCFH-DA) diacetate were purchased from Sigma Chemical Co (St.Louis. MO. USA). Ly6G microbeads, CD66b microbeads and Ly6G monoclonal antibody (Ly6G mAb) were from Miltenyi. Antibodies including anti-CD66b, anti-histone Cit-H3, anti-PAD4, FITC-conjugated anti-mouse CD66b were obtained from BD Pharmingen. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG polyclonal antibody, peroxidase substrate DAB (3,3′-diaminobenzidine) were obtained from Nichirei Bioscience (Tokyo, Japan). Annexin V-FITC Apoptosis kit and mouse quantitative ELISA kits (IFN-γ, ROS, IL-12, TNF-α, IL-6 and TGF-β1) were obtained from R&D Systems. Four coagulation kits were obtained from Nanjing jiancheng bioengineering institute. Standard rodent chow was purchased from Henan Provincial Medical Laboratory Animal Center (Zhengzhou, China), License No. SCXK (YU) 2015-0005, Certificate No. 41000100002406.
Cell culture and assays
The human acute promyelocytic leukemia (HL-60) cells and the Lewis lung carcinoma (LLC) cells from ATCC were purchased from the Chinese Academy of Sciences and were grown in RPMI1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) in a humidified atmosphere containing 5% CO2 and 95% air at 37 °C. To obtain N1-like neutrophils (HL-60N1), HL-60 cells were seeded at 2 × 105 cells/mL in medium supplemented with 10 μM all-trans retinoic acid (ATRA) for 7 days in a flask [28], changing the medium after 3 days. To obtain N2-like neutrophils (HL-60N2), HL-60N1 cells were stimulated with 100 ng/mL of TGF-β1 for 3 h prior to further assays [19]. HL-60N1 (CD66b−) and HL-60N2 (CD66b+) cells were magnetically separated by anti-CD66b microbeads (Miltenyi).
HL-60N1 or HL-60N2 cells were seeded in 6-well plate and incubated for 1 h with the indicated concentrations of emodin and then challenged with 50 nM PMA for 3 h. Cell proliferation was examined by MTT according to our previous method [29]. Cells were analyzed by Laser holographic cell imaging and analysis system (HoloMonitor M4, Phiab, Sweden). The phagocytic function of neutrophils was detected by neutral red uptake [30] and E. coli phagocytosis [31]. Cell autophagic proteins were tested by western blot. Cell apoptosis was examined by the binding of ANXV-FITC to phosphatidylserine using an automated cell counter and analysis system (Nexcelom Cellometer X2, Nexcelom, USA). ROS was detected by DCFH-DA (2′,7′-dichlorodihydrofluorescein diacetate) using a fluorescence photometer. NET formation was observed by DAPI staining [32].
Western blot
The intracellular protein was extracted from HL-60N1 (CD66b−) and HL-60N2 (CD66b+) cells in cell lysis buffer. Equal amounts of protein were separated via 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, electroblotted onto nitrocellulose membranes, and probed with antibodies against P62, LC3-B, and β-actin. Antibody binding was detected via enhanced chemiluminescence according to the manufacturer’s instructions (Pierce, Rockford, IL). Band density was quantified using ImageJ software (NIH, Bethesda, MD, USA) and normalized to the corresponding control group.
Animals
Ten-week-old female ICR mice were obtained from Henan Provincial Medical Laboratory Animal Center. All mice were housed in individually ventilated cages (lights on 7:00 AM to 7:00 PM). Animals were fed standard rodent chow and water. All animal procedures were approved by the Animal Experimentation Ethics Committee of Henan University (permission number HUSAM 2016-288), and all procedures were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals and the Regulation of Animal Protection Committee to minimize suffering and injury.
Urethane-induced lung carcinogenesis model
Urethane (600 mg/kg body weight), alone or in combination with Ly6G mAb (500 µg/mouse), was injected intraperitoneally (i.p.) into ICR mice (thirty mice per group) once a week for 4, 8 or 10 weeks, according to our previous protocol [33]. Following the first urethane injection, mice received emodin (10 mg/kg/day) via intragastric administration once a day for 4, 8 or 17 weeks. At 5, 9 and 18 weeks after the first urethane injection, the blood from the retro-orbital sinuses was collected in anticoagulant tubes for platelet counts by a blood cell analyzer, and platelet-rich and platelet-free plasma were extracted for measurement of coagulation parameters (PT, APTT, PAgT and FIB) using a coagulation analyzer. The mice were sacrificed under anesthesia with pentobarbital sodium (45 mg/kg), the alveolar fluid was collected by inserting a cannula into the trachea with three sequential injections of 1 mL PBS, supernatant was used for cytokine assay (IFN-γ, IL-12, ROS, TNF-α, IL-6 and TGF-β1) after centrifugation, and the centrifuged cells were resuspended in 0.9% sterile saline for total cell counts and added into a magnetic cell sorting column for neutrophile collection based on anti-LY6G-coated beads (providing > 90% purity, as assessed by Wright-Giemsa stain) Neutrophil immunophenotypes were analyzed by FITC-conjugated anti-mouse CD66b staining using an automated cell counter and analysis system (Nexcelom Cellometer X2, Nexcelom, USA).
To examine the bleeding time (BT), the mice were anesthetized using intraperitoneal injection of pentobarbital sodium (45 mg/kg). A 5-mm tail tip was cut off and the bleeding time was recorded when bleeding had stopped for more than 30 s.
To observe NETs, the neutrophils were incubated in RPMI1640 medium in 96-well plates for 30 min, then stimulated with 100 nM PMA for 4 h, fixed in 4% paraformaldehyde, stained by DAPI and analyzed under a wide-field fluorescent microscope (Olympus BX60) with MetaMorph software.
A part of each lung was preserved in 10% buffered formalin and routinely embedded in paraffin. For lung NETs, lung sections were stained by immunohistochemistry (anti-mouse-Cit-H3, anti-mouse-PAD4) according to our previous method [34]. The total immunohistochemical score was calculated by the intensity score and proportion score by excluding the primary antibody and IgG matched serum, respectively as positive and negative controls.
For H&E staining, each lung section was systematically scanned under a microscope, and five successive fields were graded using a lesion score (LS) based on the area of involved lesions: grade 0, no lesions; grade 1, lesions occupy less than 10% of the scanned field; grade 2, lesions occupy 10–30% of the scanned field; grade 3, lesions occupy 30–50% of the scanned field; grade 4, lesions occupy more than 50% of the scanned field.
IFN-γ, IL-12, ROS, TNF-α, IL-6 and TGF-β1 were determined by ELISA kits, according to the manufacturer’s protocols. The results were calculated from linear curves obtained using the Quantikine kit standards.
Tumor allograft model
LLC cells were used for tumor allograft experiments. 200 µL saline containing 1 × 106 cells was injected subcutaneously into the lateral axilla of mice to establish tumor allografts. One day after tumor inoculation, in vitro HL-60-drived HL-60N1 or HL-60N2 cells (2 × 106 cells in 200 µL saline), were injected intravenously into mice once a week for 3 weeks; simultaneously, mice received emodin (10 mg/kg once a day via intragastric administration) or DNase I (200 U/mouse once a week via intraperitoneal administration) for 3 weeks (ten mice per group). Tumor size was monitored twice a week with calipers and calculated as the length × width2/2. On the 22nd day after tumor inoculation, the mice were euthanized; the blood for blood cell counts measurement of coagulation parameters (PT, APTT, PAgt and FIB). The tumors were removed and weighed. Tumor neutrophils were magnetically separated by anti-LY6G-coated beads, and their expressions of MPO and CD66 were examined by western blot. CD66+ cells were also counted by an automated cell counter and analysis system.
The regulatory mechanism of emodin on neutrophils
The gene expression profiles GSE43254 was obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43254), up- and down-regulated genes related to tumor-associated neutrophils (N2) were identified using GEO2R, and the human structures of these differential proteins were collected from the protein data bank (PDB) for docking. The chemical structure of emodin was obtained from PubChem and the docking exercise was conducted using the online software systemsDock (http://systemsdock.unit.oist.jp) with the auto-movement of nonspecified protein structures. Docking scores over 6 were regarded as the potential targets for emodin. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed for the potential targets using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov) and the online software Omicshare (http://www.omicshare.com). The protein–protein interaction (PPI) among these potential targets was constructed using the STRING database (https://string-db.org/cgi/input.pl) and the hub genes were identified using Cytoscape.
Statistical analyses
The data was statistically analyzed using GraphPad Prism, Version 5.0 (San Diego, CA, USA) and presented as the mean ± SD. The differences between the two groups were evaluated using a t-test. A P value of less than 0.05 was considered statistically significant.
Results
Emodin selectively regulates neutrophils in vitro
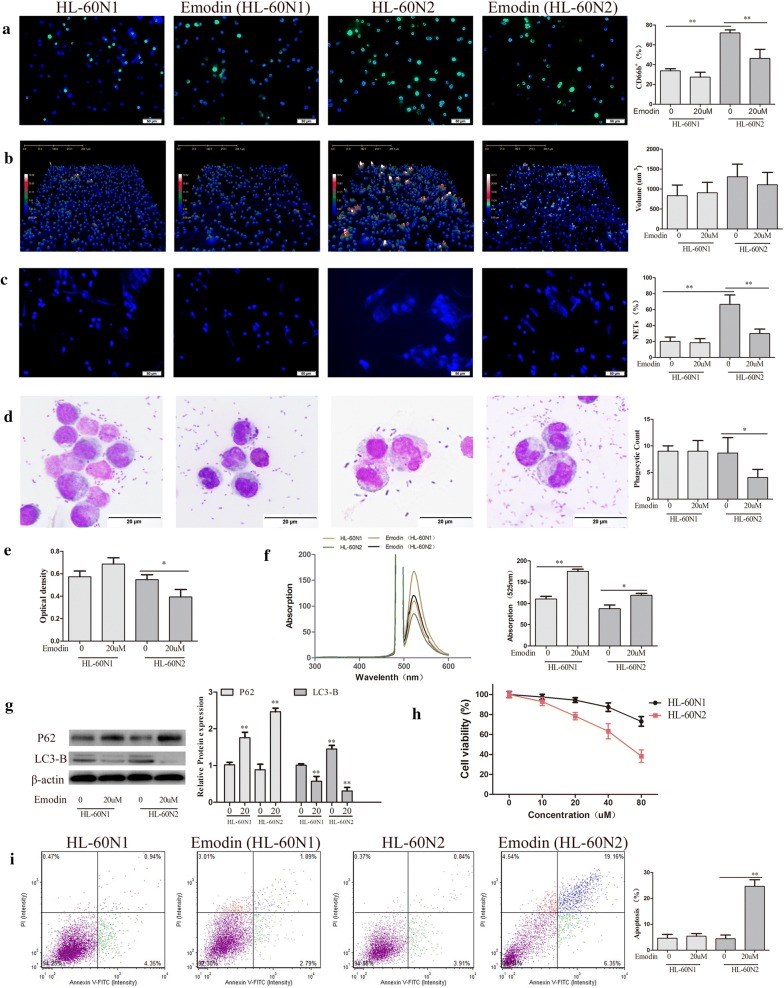
HL-60 cells have high plasticity and are widely used in the study of phenotype and function of neutrophils [35]. To explore the effect of emodin on neutrophils, we induced the differentiation of HL-60 cells into N1-like neutrophils (HL-60N1) and N2-like neutrophils (HL-60N2). Compared to HL-60N1, HL-60N2 cells had high levels of surface CD66b expression (Fig. 1a) with the morphological changes (Fig. 1b) and were easy to form NETs (Fig. 1c). There was no difference in E.coli phagocytosis (Fig. 1d) and neutral red uptake (Fig. 1e) between HL-60N1 and HL-60N2, however, HL-60N2 had a reduction in ROS (Fig. 1f) and an increase in autophagy (Fig. 1g). When administered at the dose (20 μM) that has no effect on cell viability in HL-60N1 (Fig. 1h), emodin increased ROS and decreased autophagy (Fig. 1f, g) and thus induced apoptosis (Fig. 1i) in HL-60N2 with the morphological changes (Fig. 1b). In addition, emodin treatment also decreased NETs in HL-60N2 cells (Fig. 1c). The experiments were repeated 3 times and the similar results were obtained.
Fig. 1.
Emodin selectively regulates N2 neutrophils in vitro. a CD66b expression in HL-60-differentiated N1 and N2 neutrophils analyzed by FITC-conjugated anti-mouse CD66b staining (n = 5, ×40). b Morphological changes N1 and N2 neutrophils analyzed by Laser holographic cell imaging and analysis system (n = 5, ×40). c NET formation analyzed by DAPI staining (n = 5, ×40). d Neutrophil phagocytosis detected by the Giemsa staining (n = 5, ×100). e Neutral red uptake examined with Microplate reader (n = 5). f ROS detected by DCFH-DA (n = 5). g Cell autophagy-associated proteins examined by Western blot (n = 3). h Cell viability examined by MTT (n = 5). i Cell apoptosis detected by Annexin V-FITC apoptosis kit (n = 5). The data present mean ± SD, the experiments were repeated 3 times, and statistical significance was determined by a t-test. *P < 0.05, **P < 0.01
Urethane-induced lung carcinogenesis was positively correlated with the neutrophil-associated coagulation
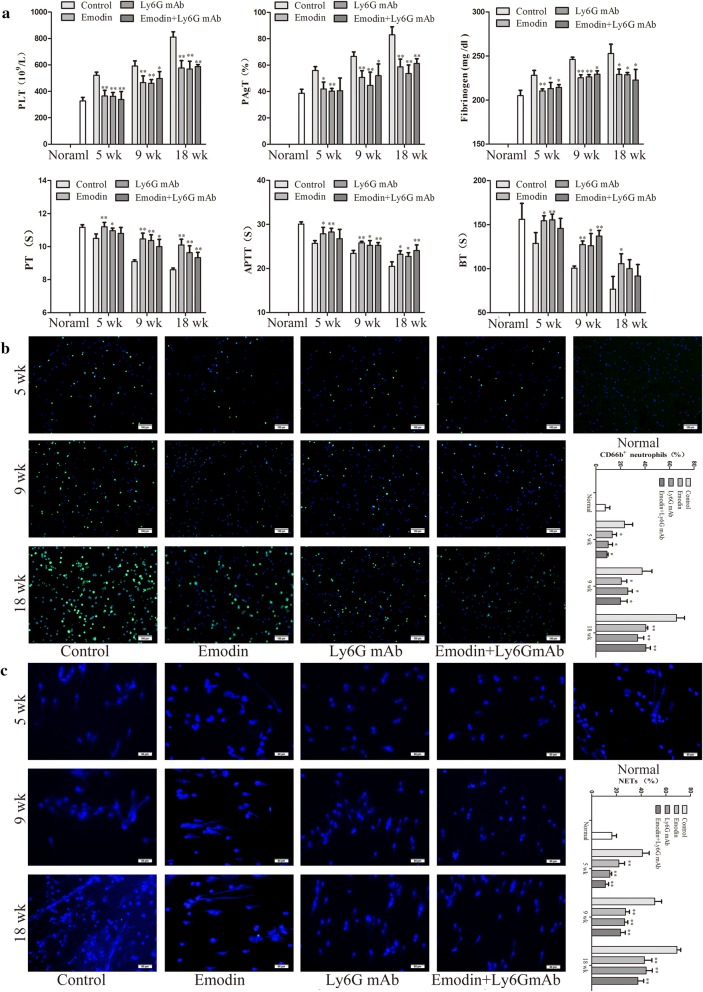
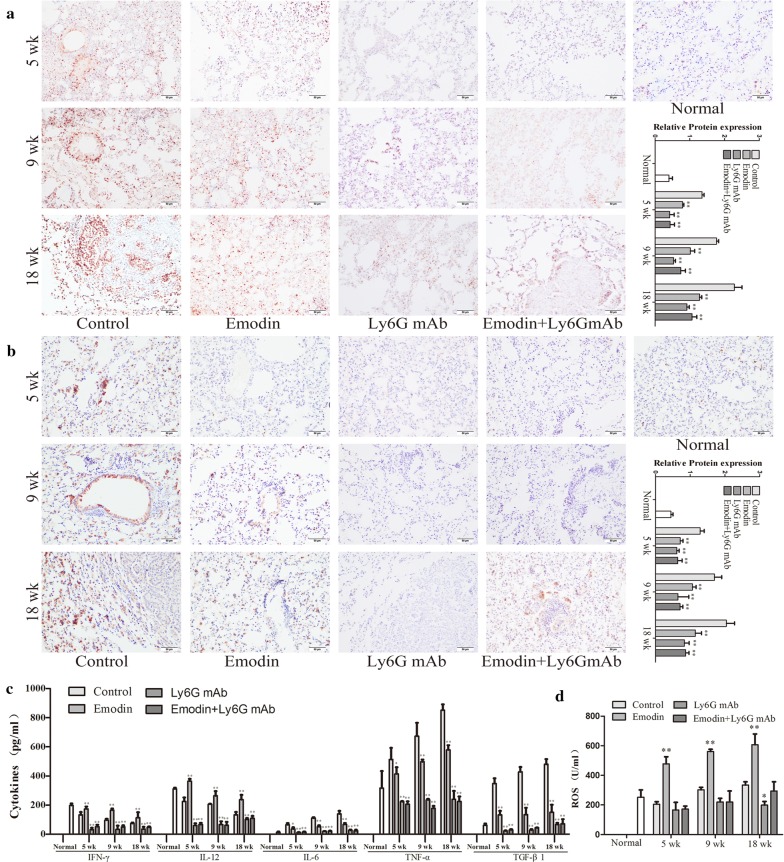
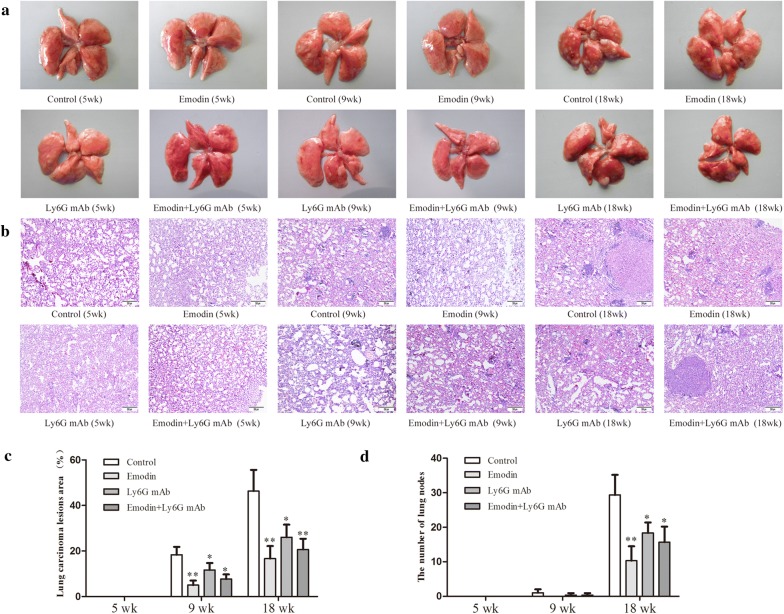
Urethane-induced mouse lung cancer model is used for studying basic lung tumor biology and finding new tumor intervention strategies. In this study, urethane at the dose of 600 mg/kg body weight was injected intraperitoneally (i.p.) once a week. At 5 weeks, prior to lung carcinogenesis, the four urethane injections led to obvious hypercoagulation in control group, presenting as an increase in platelet counts, platelet aggregation, fibrinogen (FIB) production, and a reduction in prothrombin time (PT), activated partial thromboplastin time (APTT) and bleeding time (BT) (Fig. 2a) which were positively correlated with N2 neutrophils (CD66b+ cells) and NETs (Fig. 2b, c). Immunohistochemistry confirmed NET formation in lung tissues, as indicated by staining for histone Cit-H3 (Fig. 3a) and peptidyl-arginine deiminase 4 (PAD4) (Fig. 3b). At 9 weeks, the eight urethane injections resulted in visible lung carcinoma lesions (Fig. 4b) under a microscope accompanied by the aggravated hypercoagulation, N2 neutrophils and NETs (Fig. 2), but no lung cancer nodes were visible to the naked eye (Fig. 4a). At 18 weeks, ten urethane injections led to the development of lung cancer nodes visible to the naked eye (Fig. 4a) accompanied by further aggravated hypercoagulation, N2 neutrophils and NETs (Fig. 2). The number of lung cancer nodes was 28.2 ± 5.2 (Fig. 4d), regardless of the heterogeneity of tumor histology in the experimental group. ELISA showed that the levels of IFN-γ, IL-12 and ROS decreased and the levels of TNF-α, IL-6 and TGF-β1 increased in alveolar cavities in control mice compared to normal mice (Fig. 3c, d). As expected, neutrophil depletion using the Ly6G mAb following urethane injection significantly inhibited lung carcinoma lesions and carcinogenesis accompanied by the decreased hypercoagulation, N2 neutrophils and NETs (Figs. 2, 3, 4). The experiments were repeated 3 times and the similar results were obtained.
Fig. 2.
Emodin suppresses N2 neutrophiles to attenuate hypercoagulation in a time-dependent manner during urethane-induced lung carcinogenesis. a Hypercoagulation indicated by platelet counts, platelet aggregation, fibrinogen (FIB), prothrombin time (PT), activated partial thromboplastin time (APTT) and bleeding time (BT) (n = 6). b CD66b expression in alveolar neutrophils (n = 5, ×20). c NET formation in alveolar neutrophils analyzed by DAPI staining (n = 5, ×40). The data present Mean ± SD, the experiments were repeated 3 times, and statistical significance was determined by a t-test. *P < 0.05, **P < 0.01 vs control
Fig. 3.
Emodin attenuates NETs and changes cytokines and ROS during urethane-induced lung carcinogenesis. a NETs in lung sections indicated by Cit-H3 examined by immunohistochemistry (n = 5, 40 ×). b NETs in lung sections indicated by PAD4 examined by immunohistochemistry (n = 5, 40 ×). c Cytokine levels in alveolar cavity examined by ELISA (n = 6). d ROS in alveolar cavity examined by ELISA (n = 6). The data present Mean ± SD, the experiments were repeated 3 times, and statistical significance was determined by a t-test. *P < 0.05, **P < 0.01 vs control
Fig. 4.
Emodin prevents lung carcinoma lesions and carcinogenesis during urethane-induced lung carcinogenesis. a The whole lung in naked eye (n = 10). b HE staining of lung tissues (n = 10, ×10). c The area of involved lesions in HE staining (n = 10). d The number of nodes in lung surface (n = 10). The data present mean ± SD, the experiments were repeated 3 times, and statistical significance was determined by a t-test. *P < 0.05, **P < 0.01 vs control
Emodin regulates neutrophils to prevent hypercoagulation and urethane-induced lung carcinogenesis
To prove the preventive effects of emodin on hypercoagulation and lung carcinogenesis, the mice received emodin after the first urethane injection. At five weeks, compared to the control group, emodin treatment resulted in a reduction in coagulation accompanied by the decreased N2 neutrophils and NETs (Fig. 2a–c). The NET preventive efficacy of emodin was further verified in immunohistochemical staining of histone Cit-H3 and peptidyl-arginine deiminase 4 (PAD4) (Fig. 3a, b). At 9 and 18 weeks, respectively, there were slighter carcinoma lesions and less lung cancer nodes in emodin group than those in control group (Fig. 4), which were in line with the decreased hypercoagulation, N2 neutrophils and NETs (Fig. 2) and the improved cytokine levels indicated by the increased IFN-γ, IL-12 and ROS and the decreased TNF-α, IL-6 and TGF-β1 (Fig. 3c, d). Neutrophil depletion using the Ly6G mAb significantly attenuated the preventive effects of emodin on hypercoagulation and lung carcinogenesis (Figs. 2, 3, 4). The experiments were repeated 3 times and the similar results were obtained.
Emodin exhibits neutrophil-regulating capacity in a Lewis lung cancer allograft model
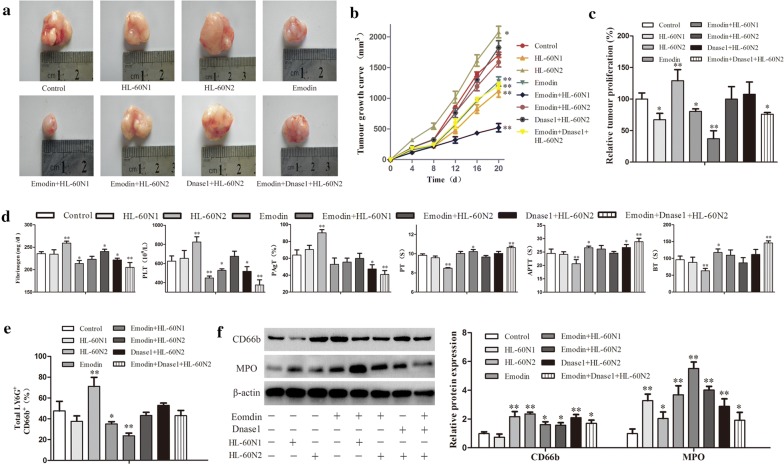
To further prove the relativity of emodin-ameliorated hypercoagulation and lung carcinogenesis to N2 neutrophils, we established an allograft model of Lewis lung cancer and used emodin alone or in combination with HL-60N1 or HL-60N2 cells to treat the model mice. The transfer of HL-60N2 cells promoted tumor growth, whereas the transfer of HL-60N1 cells or the degradation of NETs with DNAse I prevented tumor growth (Fig. 5a–c). Emodin alone suppressed tumor growth by 20%, but synergistically prevented tumor growth in combination with HL-60N1 cells or DNAse I and attenuated the tumor-promoting efficacy of HL-60N2 cells (Fig. 5a–c), which was consistent with hypercoagulation that was promoted by HL-60N2 cells and attenuated by emodin and DNAse I (Fig. 5d). CD66b+ cell count (Fig. 5e) and western blot (Fig. 5f) further confirmed the N2 neutrophil-dependent effect of emodin on hypercoagulation and lung carcinogenesis. The experiments were repeated 3 times and the similar results were obtained.
Fig. 5.
Emodin exhibits neutrophil-regulating capacity in a Lewis lung cancer allograft model. a Tumor appearance. b The curve of tumor growth (n = 10). c The relative tumor proliferation (n = 10). d Hypercoagulation indicated by platelet counts, platelet aggregation, fibrinogen (FIB), prothrombin time (PT), activated partial thromboplastin time (APTT) and bleeding time(BT)(n = 6). e CD66b+ cells in intratumor neutrophils analyzed using fluorescent cell count (n = 5). f The protein expression in intratumor neutrophils examined by western blot (n = 3). The data present Mean ± SD, the experiments were repeated 3 times, and statistical significance was determined by a t-test. *P < 0.05, **P < 0.01 vs control
Emodin regulates neutrophils by targeting multi-protein network
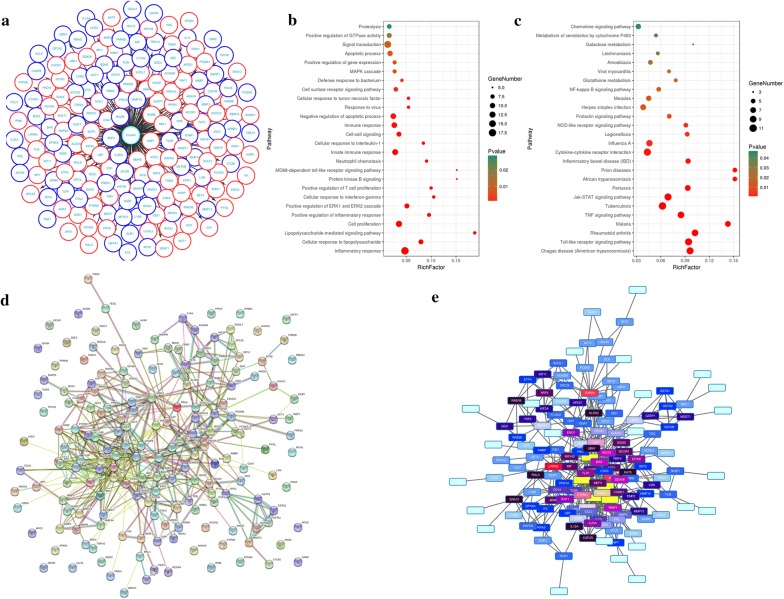
We queried 853 up-regulated genes (LogFC ≥ 1.5, P < 0.05) and 1023 down-regulated genes (LogFC ≤ − 1.5, P < 0.05) related to N2 neutrophils and obtained 237 targeting information (Fig. 6a). 177 of the potential targets with docking score > 6.0 (pKd/pKi) were selected for GO and KEGG analyses (Table 1). The hypergeometric distribution count > 2 and P < 0.05 were set as threshold criteria to identify the functional gene ontology and pathway. GO enrichment analysis indicated that the potential targets of emodin were primarily associated with the “inflammatory response”, “signal transduction”, “cell proliferation”, “innate immune response” and “negative regulation of apoptotic process” terms (Fig. 6b). KEGG enrichment analysis revealed that the potential targets of emodin were significantly enriched in the “Toll-like receptor signaling pathway”, “Jak-STAT signaling pathway” and “Cytokine-cytokine receptor interaction” terms (Fig. 6c). The PPI network (Fig. 6d) identified 4 key genes (IL-10, TLR-4, START3 and CCL2), which were hub genes for emodin (Fig. 6e).
Fig. 6.
Emodin regulates N2 neutrophils by targeting multi-protein network. a The potential targets of emodin with docking score > 6.0. b GO enrichment analysis performed by DAVID and visualized by Omicshare. c KEGG enrichment analysis performed by DAVID and visualized by Omicshare. d The PPI network constructed by the STRING database. e The hub genes identified by Cytoscape
Table 1.
177 of the potential targets with docking score > 6.0
| Protein name | PDB ID | Docking scores (pKd/pKi) | Protein name | PDB ID | Docking scores (pKd/pKi) | Protein name | PDB ID | Docking scores (pKd/pKi) |
|---|---|---|---|---|---|---|---|---|
| Up-regulated genes | ||||||||
| CCL3 | 3FPU | 7.775 | IL23A | 3D85 | 6.835 | MMP3 | 1HY7 | 6.668 |
| IL10 | 2ILK | 7.354 | NT5C | 2I7D | 6.826 | ISG15 | 1Z2M | 6.664 |
| BDH2 | 2AG5 | 7.183 | CCL8 | 1ESR | 6.824 | TPRKB | 3ENP | 6.659 |
| GGCT | 3CRY | 7.093 | EFNA5 | 1SHX | 6.824 | MMP12 | 1Y93 | 6.512 |
| GSTM2 | 2C4J | 7.062 | PRPS1 | 2H06 | 6.824 | PAOX | 1W6G | 6.509 |
| DCXR | 3D3W | 6.981 | MTSS1 | 2D1L | 6.822 | ACY1 | 1Q7L | 6.431 |
| CBR2 | 1CYD | 6.941 | GAS6 | 1H30 | 6.81 | JAK3 | 3LXL | 6.417 |
| ICAM1 | 1IAM | 6.941 | CCL2 | 1DOK | 6.8 | JMJD6 | 3K2O | 6.414 |
| AMBP | 1BIK | 6.929 | PRDX1 | 3HY2 | 6.799 | LY86 | 3M7O | 6.389 |
| SDC4 | 1OBY | 6.929 | DDIT4 | 3LQ9 | 6.79 | TLR1 | 2Z7X | 6.375 |
| C1QB | 2WNV | 6.922 | LMNA | 1IFR | 6.775 | CUTA | 1KR4 | 6.354 |
| CD69 | 3HUP | 6.92 | RRAS | 2FN4 | 6.775 | PIM1 | 1XWS | 6.342 |
| TRPV2 | 2ETB | 6.915 | BST2 | 3NWH | 6.761 | EVL | 1QC6 | 6.34 |
| NPC2 | 1NEP | 6.907 | CD14 | 1WWL | 6.751 | TLR6 | 3A79 | 6.31 |
| UCK2 | 1UJ2 | 6.903 | DAPP1 | 1FAO | 6.747 | GATM | 1JDW | 6.302 |
| GSTA2 | 1ML6 | 6.9 | IL12A | 1F45 | 6.746 | PCSK9 | 2QTW | 6.219 |
| LAG3 | 2FO1 | 6.89 | SOCS3 | 2HMH | 6.742 | APOM | 2WEW | 6.213 |
| BAX | 2G5B | 6.888 | TREX1 | 2IOC | 6.731 | GLB1 | 3G46 | 6.207 |
| GSTA1 | 1K3Y | 6.881 | ARF2 | 1J2 J | 6.73 | ISG20 | 1WLJ | 6.203 |
| TLR4 | 2Z62 | 6.871 | MEFV | 2WL1 | 6.709 | RGS10 | 2IHB | 6.192 |
| NR1D1 | 1A6Y | 6.87 | THYN1 | 3EOP | 6.685 | VDR | 1IE9 | 6.159 |
| CBR3 | 2HRB | 6.869 | LTC4S | 2UUI | 6.681 | CCND1 | 2W96 | 6.144 |
| APBB3 | 2DYQ | 6.864 | CASP8 | 1QTN | 6.678 | MMP14 | 3C7X | 6.135 |
| LXN | 2BO9 | 6.855 | MMP13 | 830C | 6.677 | TNNI2 | 1A2X | 6.118 |
| PRL | 3D48 | 6.855 | IL2RG | 2B5I | 6.676 | |||
| GRHPR | 2GCG | 6.841 | MIF | 3DJH | 6.673 | |||
| Down-regulated genes | ||||||||
| STAT3 | 1BG1 | 7.945 | RNF4 | 2XEU | 6.841 | KIF11 | 1X88 | 6.645 |
| NCOR1 | 3KMZ | 7.085 | AGFG1 | 2OLM | 6.837 | DCK | 1P5Z | 6.641 |
| NCOA4 | 1T5Z | 7.084 | FUS | 1SVF | 6.837 | AHSP | 1Z8U | 6.629 |
| ACADM | 1EGD | 7.073 | ANXA3 | 1AXN | 6.836 | NR1H2 | 1PQ9 | 6.624 |
| KAT5 | 2OU2 | 7.065 | RBM12 | 2EK1 | 6.834 | FCHO2 | 2V0O | 6.593 |
| SDF2 | 3MAL | 7.065 | IPMK | 2IEW | 6.832 | NQO2 | 2QWX | 6.592 |
| TDG | 1WYW | 7.021 | SCP2 | 2SAS | 6.824 | RGS3 | 3FBK | 6.584 |
| MYLK | 2O5G | 7.005 | OGT | 1MGT | 6.82 | AQP1 | 1J4N | 6.582 |
| TOP2A | 1ZXM | 7.002 | HVCN1 | 3A2A | 6.816 | MMP8 | 1I76 | 6.484 |
| ITK | 3MIY | 6.98 | PDE6D | 1KSH | 6.81 | MKL1 | 2V52 | 6.459 |
| UBA1 | 3CMM | 6.971 | LRRK2 | 2ZEJ | 6.809 | PTPRE | 2JJD | 6.438 |
| DDX58 | 3LRR | 6.966 | CDK9 | 3MI9 | 6.808 | GET4 | 2WPV | 6.414 |
| NCF1 | 1KQ6 | 6.944 | GGA1 | 1J2J | 6.8 | XDH | 3NRZ | 6.407 |
| PYGL | 3DDS | 6.936 | IL6RA | 1N26 | 6.797 | TESC | 1NJK | 6.398 |
| RAP2A | 1KAO | 6.935 | MGST1 | 2H8A | 6.786 | GNA12 | 1ZCA | 6.384 |
| GRK6 | 2ACX | 6.924 | PPOX | 2IVD | 6.784 | YKT6 | 3KYQ | 6.377 |
| CD47 | 2JJS | 6.923 | SRSF1 | 3BEG | 6.773 | ARAF | 8ABP | 6.346 |
| KIF2A | 2GRY | 6.922 | SNX17 | 3LUI | 6.772 | SGOL1 | 3FGA | 6.336 |
| RAB28 | 2HXS | 6.919 | CSF3R | 1CD9 | 6.759 | PPM1A | 1A6Q | 6.331 |
| IRF3 | 3A77 | 6.916 | MAFG | 3A5T | 6.756 | HIP1R | 1R0D | 6.319 |
| SUMO1 | 2UYZ | 6.912 | RALA | 1U8Z | 6.756 | LBP | 2HPS | 6.317 |
| GP1BA | 1P9A | 6.903 | NCK1 | 2CI9 | 6.742 | RXRB | 1H9U | 6.283 |
| PDIA3 | 2H8L | 6.896 | PADI4 | 2DEW | 6.738 | GDI1 | 2BCG | 6.27 |
| VPS54 | 3N1E | 6.887 | STIP1 | 1ELW | 6.738 | PTPRC | 1YGR | 6.268 |
| ATG3 | 2DYT | 6.883 | EXOC2 | 1UAD | 6.719 | ENO1 | 2AL1 | 6.225 |
| NPY | 1R9N | 6.881 | TIMP2 | 2E2D | 6.719 | WNK1 | 3FPQ | 6.169 |
| SPC25 | 2FTX | 6.878 | EP300 | 3BIY | 6.707 | CAT | 1Q23 | 6.105 |
| RAF1 | 1C1Y | 6.87 | PRG2 | 1H8U | 6.701 | PGAM1 | 1YFK | 6.07 |
| AP2S1 | 2VGL | 6.859 | TGM3 | 1VJJ | 6.701 | DDX47 | 3BER | 6.062 |
| FIS1 | 2PQR | 6.855 | ETFA | 1O97 | 6.694 | PSIP1 | 2B4J | 6.019 |
| GPX1 | 2V1M | 6.854 | DOK1 | 2V76 | 6.679 | ANXA1 | 1HM6 | 6.014 |
| DAPK2 | 2A2A | 6.853 | UCHL5 | 3A7S | 6.665 | GSTP1 | 2A2R | 6.007 |
| RHOU | 2Q3H | 6.853 | ALDH2 | 1O04 | 6.655 | RAB1B | 3NKV | 6.007 |
Discussion
Neutrophils are categorized into two subsets, high-density neutrophils (HDNs) and low-density neutrophils (LDNs), characterized by distinct morphology, phenotype and function [36]. HDNs are a homogenous population of mature neutrophils and can acquire a cytotoxic phenotype to exert anti-tumorigenic (N1) function in response to a variety of stimuli [37]. In contrast, LDNs have a larger volume than HDNs, and comprise a heterogeneous population of immature and mature neutrophils and can display pro-inflammatory and immunosuppressive properties to exert pro-tumorigenic (N2) function [38]. One way to classify HDNs and LDNs is by their sedimentation properties in density gradients [39]. In fact, both of HDNs and LDNs contain diverse cell populations with different functions. In this study, we use N2 cell surface marker-CD66b and NET formation to classify HDNs and LDNs based on the literature [25, 40]. We found that urethane induced obvious hypercoagulation which was positively correlated with lung N2 neutrophils and NETs before lung carcinogenesis, and that there were the aggravated hypercoagulation and lung N2 neutrophils as well as NETs after lung carcinoma lesions, whereas neutrophil depletion resulted in the decreased hypercoagulation and lung carcinogenesis, indicating a loop of N2 activation-NETs-hypercoagulation-carcinogenesis during urethane induces lung cancer. We suppose that neutrophil phenotypes are dynamic and can be modulated by small molecular compounds, our results showed that emodin treatment significantly attenuated lung N2 neutrophils and NETs inconsistent with the decreased hypercoagulation and carcinogenesis, suggesting a relativity of emodin-ameliorated hypercoagulation and lung carcinogenesis to N2 neutrophils.
Cancer patients often exhibit hypercoagulation, which is one of the most common causes of cancer-related death [41–43]. A recent study suggested that neutrophils were essential for the initiation and propagation of cancer-associated thrombosis by releasing NETs, indicating a novel link between neutrophils and carcinogenesis, as well as cancer-associated hypercoagulation [44, 45]. Neutrophils are the first immune cell type to be recruited to the inflammation site and exert the first line defense against a variety of pathogens [18]. Abundant evidence suggest that N2 neutrophils are an immunosuppressive subset of neutrophils that acts as a key regulator at the various stages of tumor development [39]. Considering the anti-tumorigenic function of neutrophils (N1), we suppose that regulating of N2 to N1, rather than complete depletion of neutrophils, is a better approach for tumor prevention. In this study, we found that emodin at the optimal dose selectively suppressed N2 neutrophils in vitro. The relativity of emodin-ameliorated hypercoagulation and lung carcinogenesis to N2 neutrophils was also further proved in Lewis lung cancer allograft model, in which emodin did not attenuate significantly tumor growth but prevented synergistically hypercoagulation and tumor development in combination with HL-60N1 cells or DNase I, which tested our hypothesis and also suggested the dual roles of hypercoagulation as an initiator and a promoter in carcinogenesis. Although emodin has been extensively investigated for cancer prevention and treatment, there is insufficient information related to the mechanism by which emodin exerts its anticancer properties in different cancers [46]. It has been proved that emodin can modulate mitochondrial apoptosis pathway, mitotic catastrophe and P2Y receptors in different cancers [47]. Xing et al. revealed that there were significant changes in many endogenous metabolites after emodin exposure concerning oxidative stress and disturbances in amino acid and energy metabolism [48]. In this study, we try hard to explain the mechanism by which emodin regulates N2 neutrophils to attenuate hypercoagulation and lung carcinogenesis. Network pharmacology indicated that the potential targets of emodin regulating N2 neutrophils were primarily associated with the “inflammatory response”, “signal transduction”, “cell proliferation”, “innate immune response” and “negative regulation of apoptotic process, indicating a function of emodin targeting multi-protein network, such as “Toll-like receptor signaling pathway”, “Jak-STAT signaling pathway” and “Cytokine-cytokine receptor interaction”, whereby it regulates neutrophil phenotypes to maintain system homeostasis for cancer prevention. Further mechanism studies are necessary to verify its medicinal applications.
Cancer chemoprevention is an important prophylactic strategy for reduction of cancer-related death [49]. Despite substantial progress and successful precedent, such as the use of HPV vaccine for prevention of cervical cancer, cancer chemoprevention has faced many challenges and needs to be used more broadly [50, 51]. One misunderstanding is that the molecular targets of chemopreventive agents must coincide with the targets of anti-cancer therapies [52]. It is very important for medical professionals to fully appreciate the benefits of cancer chemoprevention and the need for the development of precise strategies, as well as for understanding the mechanistic aspects of these strategies, in order to prevent progression of precancer lesions into advanced stage cancer [53]. Therefore, the focus of this study was not on tumor killing efficacy of emodin but on effect of emodin on neutrophil phenotypes. Here we propose a specific mechanism by which emodin can delay or reverse the procession of early precancer lesions. Although lacking a precise explanation of mechanism by which emodin regulates N2 neutrophils, our research provides a causative role for coagulation in lung carcinogenesis. With cancer being a global issue and understanding the need for activation of patient’s innate antineoplastic agents, emodin may provide an efficient opportunity as a chemoprevention agent targeting multi-protein network.
Conclusions
In summary, N2 neutrophils play an important role in hypercoagulation and cancer progression, our findings suggest several potential clinical implications. First, hypercoagulation is the result and cause of carcinogenesis. Second, emodin can selectively suppress N2 to maintain N1 neutrophils for hypercoagulation and cancer prevention. Third, the combination of emodin and N1 neutrophils synergistically prevent hypercoagulation and carcinogenesis and can be a novel strategy for preventing cancer and cancer-associated thrombosis.
Authors’ contributions
GD conceived and designed the experiments. GD, ZL and YL wrote the paper and plotted the results. GD, ZL, HL and QT analyzed the data. GD, HL and QT contributed reagents, materials and analysis tools. All authors took part in performing experiments. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by Animal Experimentation Ethics Committee of Henan University (permission number HUSAM 2016-288), and all procedures were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals and the Regulation of Animal Protection Committee.
Funding
This study was supported by Natural Science Foundation of Henan Province of China (No. 182300410310) and National Natural Science Foundation of China (No. 81472745).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- NET
neutrophil extracellular traps
- PMA
phorbol 12-myristate 13-acetate
- DAPI
4′,6-diamidino-2-phenylindole
- DCFH-DA
2′,7′-dichlorodihydrofluorescein
- PLT
platelet
- PAgT
platelet aggregation test
- FIB
fibrinogen
- PT
prothrombin time
- APTT
activated partial thromboplastin time
- BT
bleeding time
- HDN
high-density neutrophil
- LDN
low-density neutrophil
Contributor Information
Zibo Li, Email: 963235757@qq.com.
Yukun Lin, Email: 15736880141@163.com.
Shuhui Zhang, Email: 15537873819@163.com.
Lin Zhou, Email: 13137591738@163.com.
Guixi Yan, Email: 17637616880@163.com.
Yuehua Wang, Email: 13137596876@163.com.
Mengdi Zhang, Email: zhangmengdi166565@163.com.
Mengqi Wang, Email: 13783602198@163.com.
Haihong Lin, Phone: 86-0371-23880680, Email: linhaihong369@126.com.
Qiaozhen Tong, Email: qztong88@126.com.
Yongjian Duan, Email: dyj9062@163.com.
Gangjun Du, Phone: 86-0371-23880680, Email: 10200029@vip.henu.edu.cn.
References
- 1.Melloni G, Muriana P, Bandiera A. Prognostic role of liver X receptor-alpha in resected stage II and III non-small-cell lung cancer. Clin Respir J. 2018;12(1):241–246. doi: 10.1111/crj.12522. [DOI] [PubMed] [Google Scholar]
- 2.Hirashima T, Suzuki H, Okamoto N. Important factors for achieving survival of five years or more in non-small cell lung cancer patients with distant metastasis. Oncol Lett. 2014;8(1):327–334. doi: 10.3892/ol.2014.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki H, Suzuki A, Tatematsu T. Prognosis of recurrent non-small cell lung cancer following complete resection. Oncol Lett. 2014;7(4):1300–1304. doi: 10.3892/ol.2014.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano D, Lazzeroni M, Bonanni B. Cancer chemoprevention: Much has been done, but there is still much to do. State of the art and possible new approaches. Mol Oncol. 2015;9(5):1008–1017. doi: 10.1016/j.molonc.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ancuceanu RV, Istudor V. Pharmacologically active natural compounds for lung cancer. Altern Med Rev. 2004;9(4):402–419. [PubMed] [Google Scholar]
- 6.Dong X, Fu J, Yin X. Emodin: a review of its pharmacology, toxicity and pharmacokinetics. Phytother Res. 2016;30(8):1207–1218. doi: 10.1002/ptr.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Zhang J, Zhang H. Anti-inflammatory effect of emodin on lipopolysaccharide induced keratitis in Wistar rats. Int J Clin Exp Med. 2015;8(8):12382–12389. [PMC free article] [PubMed] [Google Scholar]
- 8.Chun W, Jun Y, Bei L. Anti-tumor activity of emodin against human chronic myelocytic leukemia K562 cell lines in vitro and in vivo. Eur J Pharmacol. 2010;627(1–3):33–41. doi: 10.1016/j.ejphar.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Xu Y, Zhao Z. Emodin suppresses proliferation, migration and invasion in ovarian cancer cells by down regulating ILK in vitro and in vivo. OncoTargets Ther. 2017;10:3579–3589. doi: 10.2147/OTT.S138217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sun Y, Wang X, Zhou Q. Inhibitory effect of emodin on migration, invasion and metastasis of human breast cancer MDA-MB-231 cells in vitro and in vivo. Oncol Rep. 2014;33(1):338–346. doi: 10.3892/or.2014.3585. [DOI] [PubMed] [Google Scholar]
- 11.Manimaran A, Manoharan S. Tumor Preventive Efficacy of Emodin in 7,12-Dimethylbenz Anthracene-Induced Oral Carcinogenesis: a Histopathological and Biochemical Approach. Pathol Oncol Res. 2017;24(1):19–29. doi: 10.1007/s12253-017-0205-7. [DOI] [PubMed] [Google Scholar]
- 12.Cai Baochang. Emodin attenuates calcium overload and endoplasmic reticulum stress in AR42 J rat pancreatic acinar cells. Mol Med Rep. 2013;9(1):267–272. doi: 10.3892/mmr.2013.1773. [DOI] [PubMed] [Google Scholar]
- 13.Luo S, Deng X, Liu Q. Emodin ameliorates ulcerative colitis by the flagellin-TLR5 dependent pathway in mice. Int Immunopharmacol. 2018;59:269–275. doi: 10.1016/j.intimp.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Ning JW, Zhang Y, Yu MS. Emodin alleviates intestinal mucosal injury in rats with severe acute pancreatitis via the caspase-1 inhibition. Hepatobiliary Pancreat Dis Int. 2017;16(4):431–436. doi: 10.1016/S1499-3872(17)60041-9. [DOI] [PubMed] [Google Scholar]
- 15.Xiao M, Zhu T, Zhang W. Emodin Ameliorates LPS-Induced Acute Lung Injury, Involving the Inactivation of NF-κB in Mice. Int J Mol Sci. 2014;15(11):19355–19368. doi: 10.3390/ijms151119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell DR, Huttenlocher A. Neutrophils in the tumor microenvironment. Trends Immunol. 2016;37(1):41–52. doi: 10.1016/j.it.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Jia Y, Wang N, et al. The clinical significance of tumor-infiltrating neutrophils and neutrophil-to-CD8 + lymphocyte ratio in patients with resectable esophageal squamous cell carcinoma. J Transl Med. 2014;12(1):7. doi: 10.1186/1479-5876-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navegantes KC, Gomes RS, Czaikoski PG, et al. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J Transl Med. 2017;15(1):36. doi: 10.1186/s12967-017-1141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridlender ZG, Sun J, Kim S. Polarization of tumor-associated neutrophil (TAN) phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell. 2009;16(3):183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs TA, Abed U, Goosmann C. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cedervall J, Hamidi A, Olsson AK. Platelets, NETs and cancer. Thromb Res. 2018;164(Suppl 1):S148. doi: 10.1016/j.thromres.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 22.Demers M, Wagner D. Neutrophil extracellular traps: a new link to cancer-associated thrombosis and potential implications for tumor progression. Oncoimmunology. 2013;2(2):e22946. doi: 10.4161/onci.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsson AK, Cedervall J. NETosis in Cancer-platelet-neutrophil crosstalk promotes tumor-associated pathology. Front Immunol. 2016;7:1243–1252. doi: 10.3389/fimmu.2016.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cools-Lartigue J, Spicer J, Najmeh S. Neutrophil extracellular traps in cancer progression. Cell Mol Life Sci. 2014;71(21):4179–4194. doi: 10.1007/s00018-014-1683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silvia G, Andrea C, Elena Z. Coagulation induced by C3aR-dependent NETosis drives protumorigenic neutrophils during small intestinal tumorigenesis. Nat Commun. 2016;7(7):11037. doi: 10.1038/ncomms11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui H, Li S, Xu C. Emodin alleviates severe acute pancreatitis-associated acute lung injury by decreasing pre-B-cell colony-enhancing factor expression and promoting polymorphonuclear neutrophil apoptosis. Mol Med Rep. 2017;16(4):5121–5128. doi: 10.3892/mmr.2017.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemmar A, Al Dhaheri R, Alamiri J. Diesel Exhaust particles induce impairment of vascular and cardiac homeostasis in mice: ameliorative effect of emodin. Cell Physiol Biochem. 2015;36(4):1517–1526. doi: 10.1159/000430315. [DOI] [PubMed] [Google Scholar]
- 28.Almzaiel AJ, Billington R, Smerdon G. Effects of hyperbaric oxygen treatment on antimicrobial function and apoptosis of differentiated HL-60 (neutrophil-like) cells. Life Sci. 2013;93(2–3):125–131. doi: 10.1016/j.lfs.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Guo Z, Ma X. The M2 macrophages induce autophagic vascular disorder and promote mouse sensitivity to urethane-related lung carcinogenesis. Dev Comp Immunol. 2016;59:89–98. doi: 10.1016/j.dci.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Ates G, Vanhaecke T, Rogiers V, et al. Assaying cellular viability using the neutral red uptake assay. Methods Mol Biol. 2017;1601:19. doi: 10.1007/978-1-4939-6960-9_2. [DOI] [PubMed] [Google Scholar]
- 31.Qiu W, Liu S, Chen J. The primary culture of carp (Cyprinus carpio) macrophages and the verification of its phagocytosis activity. Vitro Cell Dev Biol Anim. 2016;52(1):1–10. doi: 10.1007/s11626-015-9942-7. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama T, Saitoh H. Tunicamycin-induced neutrophil extracellular trap (NET)-like structures in cultured human myeloid cell lines. Cell Biol Int. 2015;39(3):355–359. doi: 10.1002/cbin.10396. [DOI] [PubMed] [Google Scholar]
- 33.Ma X, Deng J, Cao N. Lasting glycolytic stress governs susceptibility to urethane-induced lung carcinogenesis in vivo and in vitro. Toxicol Lett. 2016;240(1):130–139. doi: 10.1016/j.toxlet.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Meng M, Geng S, Du Z. Berberine and cinnamaldehyde together prevent lung carcinogenesis. Oncotarget. 2017;8(44):76385–76397. doi: 10.18632/oncotarget.20059. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Hauert AB, Martinelli S, Marone C. Differentiated HL-60 cells are a valid model system for the analysis of human neutrophil migration and chemotaxis. Int J Biochem Cell Biol. 2002;34(7):838–854. doi: 10.1016/s1357-2725(02)00010-9. [DOI] [PubMed] [Google Scholar]
- 36.Eruslanov EB. Phenotype and function of tumor-associated neutrophils and their subsets in early-stage human lung cancer. Cancer Immunol Immunother. 2017;66(8):997–1006. doi: 10.1007/s00262-017-1976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagiv JY, Michaeli J, Assi S. Phenotypic diversity and plasticity in circulating neutrophil sub populations in cancer. Cell Rep. 2015;10(4):562–573. doi: 10.1016/j.celrep.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Hu Y, Gu F. Phenotypic and clinical characterization of low density neutrophils in patients with advanced lung adenocarcinoma. Oncotarget. 2017;8(53):90969–90978. doi: 10.18632/oncotarget.18771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishalian I, Granot Z, Fridlender ZG. The diversity of circulating neutrophils in cancer. Immunobiology. 2016;222(1):82–88. doi: 10.1016/j.imbio.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Kanamaru R, Ohzawa H, Miyato H. Low density neutrophils (LDN) in postoperative abdominal cavity assist the peritoneal recurrence through the production of neutrophil extracellular traps (NETs) Sci Rep. 2018;8(1):632. doi: 10.1038/s41598-017-19091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi JL, Chung JW, Ahn MJ. Hypercoagulability and mortality of patients with stroke and active cancer: the OASIS-CANCER study. J Stroke. 2017;19(1):77–87. doi: 10.5853/jos.2016.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu C, Chen R, Chen W. Thrombocytosis is a significant indictor of hypercoagulability, prognosis and recurrence in gastric cancer. Exp Therap Med. 2014;8(1):125. doi: 10.3892/etm.2014.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guglietta S, Rescigno M. Hypercoagulation and complement: connected players in tumor development and metastases. Semin Immunol. 2016;28(6):578–586. doi: 10.1016/j.smim.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Cedervall J, Zhang Y, Huang H. Neutrophil extracellular traps accumulate in peripheral blood vessels and compromise organ function in tumor-bearing animals. Can Res. 2015;75(13):2653–2662. doi: 10.1158/0008-5472.CAN-14-3299. [DOI] [PubMed] [Google Scholar]
- 45.Demers M, Krause DS, Schatzberg D. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc Natl Acad. Sci USA. 2012;109(32):13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrimali D, Shanmugam MK, Kumar AP. Targeted abrogation of diverse signal transduction cascades by emodin for the treatment of inflammatory disorders and cancer. Cancer Lett. 2013;341(2):139–149. doi: 10.1016/j.canlet.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Li L, Guan R. Emodin inhibits ATP-induced proliferation and migration by suppressing p2y receptors in human lung adenocarcinoma cells. Cell Physiol Biochem. 2017;44:1337–1351. doi: 10.1159/000485495. [DOI] [PubMed] [Google Scholar]
- 48.Xing YX, Li MH, Tao L. Anti-cancer effects of emodin on HepG2 cells as revealed by 1H-NMR based metabolic profiling. J Proteome Res. 2018;17(5):1943–1952. doi: 10.1021/acs.jproteome.8b00029. [DOI] [PubMed] [Google Scholar]
- 49.See HT, Cheung YB, Yong F, et al. Acceptance of prophylactic surgery and chemoprevention of cancer in Singapore. Ann Acad Med. 2005;34(3):238–242. [PubMed] [Google Scholar]
- 50.Tsai Horng-Jyh. Clinical cancer chemoprevention: from the hepatitis B virus (HBV) vaccine to the human papillomavirus (HPV) vaccine. Taiwanese J Obstet Gynecol. 2015;54(2):112–115. doi: 10.1016/j.tjog.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Hutubessy R, Levin A, Wang S. A case study using the United Republic of Tanzania: costing nationwide HPV vaccine delivery using the WHO Cervical Cancer Prevention and Control Costing Tool. Bmc Medicine. 2012;10(1):136. doi: 10.1186/1741-7015-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walczak K, Marciniak S, Grażyna R. Cancer chemoprevention-selected molecular mechanisms. Postepy Higieny I Medycyny Doswiadczalnej. 2017;71(1):149. doi: 10.5604/01.3001.0010.3799. [DOI] [PubMed] [Google Scholar]
- 53.Steele VE, Kelloff GJ. Development of cancer chemopreventive drugs based on mechanistic approaches. Mutat Res Fundam Mole Mech Mutagenesis. 2005;591(1–2):16–23. doi: 10.1016/j.mrfmmm.2005.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.