Abstract
Background:
Salmonella and Shigella spp. are 2 of the most frequent and deadly enteric bacterial pathogens recorded worldwide. In developing countries Salmonella infections are responsible for many deaths annually and these mortality rates are prone to increase due to the emergence of resistance to antibiotics. In this overall scenario new alternative therapeutic approaches are needed.
Methods:
For the first time, we investigated the activity of 3 commercial bacteriophage cocktails (INTESTI, Septaphage, PYO) against a collection of contemporary Salmonella spp. (n = 30) and Shigella spp. (n = 20) strains isolated in Switzerland. Phage susceptibility was determined by implementing the spot test.
Results:
The overall susceptibility of Salmonella spp. to INTESTI and Septaphage was 87% and 77%, respectively. With regard to Shigella spp., the overall susceptibility to INTESTI and Septaphage was 95% and 55%, respectively. PYO was observed to be active against only 10% of Salmonella spp. but against 95% of Shigella spp.
Conclusions:
Our results seem promising, especially for the INTESTI biopreparation against Salmonella enterica infections. Nevertheless, such speculation should be supported by further in vivo studies to confirm efficacy and safety of the cocktails. We also emphasize the importance of large in vitro screening analyses aimed to assess the activity of such biopreparations against contemporary multidrug-resistant strains that are emerging worldwide.
Keywords: commercial, bacteriophages, Salmonella, Shigella, cocktails
INTRODUCTION
Salmonella and Shigella spp. are the most frequently found and deadly enteric bacterial pathogens. For instance, each year 500,000 cases of diarrheal shigellosis and about 1.2 million cases of nontyphoidal salmonellosis with 380 deaths are recorded in the United States [1–4]. Moreover, in developing countries Salmonella infections are responsible for 1 million deaths annually and these mortality rates are likely to increase due to the emergence of resistance to commonly implemented antibiotics [5, 6]. In this overall scenario, new alternative and cost-effective therapeutic approaches are needed.
Bacteriophages are highly species-specific self-propagating viruses that can infect and lyse bacteria. Their employment is part of the standard medical practice in countries of the former Soviet Union, whereas in Western nations the use of phage therapy is unfamiliar, and this has led to a lack of studies analyzing efficacy and possible alternatives to antibiotics [7, 8].
Numerous in vitro and in vivo reports exploring both lytic activity and clinical effectiveness to control Salmonella infections are available. However, such analyses have exclusively used monophages and focused on reducing contamination of food stuffs or intestinal colonization in food animals [9–13]. With regard to Shigella, Mai et al tested a phage cocktail (ShigActiveTM) in a mice model obtaining encouraging results [14].
To our knowledge, data regarding the in vitro activity of bacteriophage cocktails against large collections of Salmonella and Shigella spp. strains are still lacking. In this study, for the first time, we explored the in vitro activity of 3 commercially available bacteriophage cocktails currently implemented in the country of Georgia to treat human intestinal infections.
METHODS
The following cocktails of sterile-filtrate phage lysates of different bacterial species were tested: PYO Bacteriophage, INTESTI Bacteriophage (Eliava Biopreparations, Tbilisi, Georgia; concentration of 105-6 Plaque Forming Units, PFU/mL), and Septaphage (Biochimpharm, Tbilisi, Georgia; 105 PFU/mL). PYO targets Escherichia coli, Proteus spp., Pseudomonas aeruginosa, Staphylococcus spp., and Streptococcus spp., whereas INTESTI and Septaphage target over 12 gastrointestinal pathogens, such as Shigella, Salmonella, Proteus, Staphylococcus, Pseudomonas spp. and different serovars of enteropathogenic E. coli. PYO is used to treat purulent skin and surgical, oral, enteral, and gynecological infections, whereas INTESTI and Septaphage are implemented for intestinal infections [15]. Notably, INTESTI is the only molecularly well-characterized phage cocktail [16].
The collection of strains tested during the present study included contemporary Salmonella (n = 30) and Shigella spp. (n = 20) isolated from human infections which occurred in Switzerland. Species identification (ID) was routinely obtained using the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; Bruker). The ID confirmation and further typing were performed at the National Reference Laboratory for Enteropathogenic Bacteria and Listeria (Institute for Food Safety and Hygiene, Zurich, Switzerland). The antibiotic susceptibility profiles were obtained by disc-diffusion tests [17]. Most Salmonella spp. strains were pan-susceptible to tested antibiotics (ampicillin, ceftriaxone, cotrimoxazole, chloramphenicol, nalidixic acid, and ciprofloxacin), whereas only ceftriaxone was always active in vitro against isolates of Shigella spp. (Supplementary Table 1).
Phage susceptibility was determined with the spot test with double agar overlay method [18]. Briefly, 100 μl of a 0.5 McFarland bacterial suspension was mixed in a brain heart infusion (BHI) agarose matrix (0.6%), which was then distributed to solidify on a standard BHI agar plate. Then, 10 μl of each phage-suspension was spotted on the plate and incubated overnight. The day after, lysis zones were quantified [18]. Specifically, strains showing confluent lysis (complete clearing: ++++), semi-confluent lysis (clearing throughout, but with faint hazy background: +++), opaque lysis (turbidity throughout the cleared zone: ++), and taches vierges (individual clear or opaque plaques: +) were defined as susceptible to the phage compounds tested. Strains showing no activity (no clearing: R) were defined as resistant. For all strains (n = 50) susceptibility tests were performed in duplicate and on distinct days.
RESULTS AND DISCUSSION
As shown in Table 1, the overall susceptibility of Salmonella spp. to INTESTI and Septaphage was 86.7% (of which 23/30 were +++ or ++++) and 76.7% (none of which were +++ or ++++), respectively (examples in Supplementary Figure 1). With regard to Shigella spp., the overall susceptibility to INTESTI and Septaphage was 95% (of which 9/20 were +++ or ++++) and 55% (of which 3/20 were +++ or ++++), respectively. This data is promising, but we should note that the spot test can lead to an overestimation of the susceptibility as a consequence of the lysis-from-without phenomenon [19].
Table 1.
Summary of the susceptibility of the Salmonella and Shigella spp. strains to the 3 commercial bacteriophage cocktails
| Phage Cocktails | Strain groups | Results of the spot test (%) a |
||||
|---|---|---|---|---|---|---|
| R | + | ++ | +++ | ++++ | ||
| PYO Bacteriophage (Eliava) | Overall strains (n = 50) | 56.0 | 4.0 | 24.0 | 12.0 | 4.0 |
| Salmonella spp. (n = 30) | 90.0 | 3.3 | 0.0 | 3.3 | 3.3 | |
| Shigella spp. (n = 20) | 5.0 | 5.0 | 55.0 | 30.0 | 5.0 | |
| INTESTI Bacteriophage (Eliava) | Overall strains (n = 50) | 10.0 | 6.0 | 20.0 | 36.0 | 28.0 |
| Salmonella spp. (n = 30) | 13.3 | 3.3 | 6.7 | 33.3 | 43.3 | |
| Shigella spp. (n = 20) | 5.0 | 10.0 | 40.0 | 40.0 | 5.0 | |
| Septaphage (Biochimpharm) | Overall strains (n = 50) | 32.0 | 42.0 | 20.0 | 0.0 | 6.0 |
| Salmonella spp. (n = 30) | 23.3 | 53.3 | 23.3 | 0.0 | 0.0 | |
| Shigella spp. (n = 20) | 45.0 | 25.0 | 15.0 | 0.0 | 15.0 | |
a Strains were defined as susceptible to the bacteriophages when confluent lysis (ie, complete clearing: ++++), semi-confluent lysis (ie, clearing throughout but with faint hazy background: +++), opaque lysis (ie, turbidity throughout the cleared zone: ++), taches vierges (ie, a few individual plaques: +) were recorded. Strains showing no activity (ie, no clearing “R”) were defined as resistant.
We did not expect any activity for PYO against our strains because, according to the manufacturer, this preparation should not contain lytic phages against Salmonella spp. and Shigella spp. However, we were surprised to note that this cocktail was active against 10% (of which 2/30 were +++ or ++++) of Salmonella spp. and, more importantly, against 95% (of which 7/20 were +++ or ++++) of Shigella spp. This could be explained by the presence of bacteriophages unable to selectively differentiate Salmonella and Shigella spp. from E. coli (all 3 being phylogenetically closely related bacterial species, especially the latter 2 [20]) that might share several common phage targets [21]. Moreover, taking into account the lysis-from-without phenomenon where a high multiplicity of infection can lead to bacterial death without infection, we are aware that by exclusively using the spot test, our susceptibility results might be slightly overestimated [19].
In conclusion, we showed the distinct spectrum and lytic activity of commercial bacteriophage cocktails targeting Salmonella and Shigella species. In particular, Septaphage proved to be active, though overall weakly, against 68% of the tested strains, whereas INTESTI exhibited a strong response against 90% of our isolates. Therefore, our results seem promising, especially for the latter biopreparation against Salmonella enterica infections. Nevertheless, such speculation should be supported by further animal studies together with human clinical trials in order to confirm efficacy and safety of cocktails. We also emphasize the importance of large in vitro screening analyses aimed to assess the activity of such biopreparations against contemporary multidrug-resistant strains emerging worldwide [2, 22, 23]. The sum of these steps, if successful, could lead to the maturation—also in Western countries—of an alternative approach for the treatment of bacillary dysenteries and salmonellosis.
ACKNOWLEDGMENTS
We are grateful to Dr. Cédric Hirzel for obtaining the commercial phage cocktails during his trip in Georgia. This work was supported by Swiss National Science Foundation grant No. 153377 to AE. Odette J. Bernasconi is a PhD student (2015-2018) supported by the Hans Sigrist Foundation (Bern, Switzerland).
Supplementary Table 1.
Characteristics of the 30 Salmonella and 20 Shigella spp. strains and susceptibility to 3 commercial bacteriophage cocktails
| No. | ID strain | Species | Source | Detection Month/Year | Susceptibility according to CLSI |
Bacteriophage Susceptibility a |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CRO | SXT | CHL | NAL | CIP | INTESTI | Septaphage | PYO | |||||
| 1 | 6301.21 | S. enteritidis | Stool | 08/16 | S | S | S | S | S | S | ++++ | + | R |
| 2 | 6301.22 | S. enteritidis | Stool | 08/16 | S | S | S | S | S | S | R | + | R |
| 3 | 6301.23 | S. enterica subsp. enterica 4,12:i | Stool | 08/16 | S | S | S | S | S | S | R | + | R |
| 4 | 6212.52 | S. enteritidis | Stool | 08/16 | S | S | S | S | S | S | R | + | R |
| 5 | 6212.46 | S. enterica subsp. enterica 4,12:i | Stool | 08/16 | R | S | S | S | S | S | R | + | R |
| 6 | 6212.47 | S. enteritidis | Stool | 08/16 | S | S | S | S | S | S | +++ | + | R |
| 7 | 6211.59 | S. enterica subsp. enterica 6,7:y:- | Stool | 08/16 | S | S | S | S | S | S | ++++ | ++ | +++ |
| 8 | 6211.25 | S. enteritidis | Stool | 08/16 | S | S | S | S | S | S | +++ | + | R |
| 9 | 5804.66 | S. paratyphi A | Blood culture | 04/15 | S | S | S | S | R | I | +++ | + | R |
| 10 | 6102.20 | S. typhimurium | Urine | 01/16 | S | S | S | S | S | S | +++ | R | R |
| 11 | 6103.32 | S. typhimurium | Stool | 02/16 | S | S | S | S | S | S | ++++ | + | R |
| 12 | 6107.71 | S. typhimurium | Stool | 03/16 | S | S | S | S | S | S | ++++ | ++ | R |
| 13 | 6007.27 | S. panama | Stool | 11/15 | S | S | S | S | S | S | ++++ | + | R |
| 14 | 5804.47 | S. paratyphi B | Stool | 04/15 | S | S | S | S | S | S | +++ | + | R |
| 15 | 5602.57 | S. typhimurium | Blood culture | 09/14 | S | S | S | S | S | S | ++++ | ++ | R |
| 16 | 5905.07 | S. enteritidis | Stool | 08/15 | S | S | S | S | S | S | +++ | + | R |
| 17 | 5905.08 | S. enteritidis | Stool | 08/15 | S | S | S | S | S | S | ++++ | ++ | R |
| 18 | 5602.08 | S. enteritidis | Stool | 09/14 | S | S | S | S | S | S | +++ | ++ | R |
| 19 | 5512.03 | S. enteritidis | Blood culture | 08/14 | S | S | S | S | S | S | ++++ | + | R |
| 20 | 5603.72 | S. enteritidis | Blood culture | 09/14 | S | S | S | S | S | S | +++ | ++ | R |
| 21 | 4608.23 | S. paratyphi A | Stool | 12/10 | S | S | S | S | R | S | ++++ | R | R |
| 22 | 4504.56 | S. paratyphi A | Blood culture | 06/10 | S | S | S | S | R | I | ++++ | R | R |
| 23 | 6104.03 | S. paratyphi B | Blood culture | 02/16 | S | S | S | S | S | S | +++ | R | R |
| 24 | 6201.74 | S. paratyphi B | Stool | 05/16 | S | S | S | S | S | S | ++ | + | R |
| 25 | 5902.41 | S. typhimurium | Stool | 07/15 | S | S | S | S | S | S | ++++ | R | R |
| 26 | 5910.36 | S. typhimurium | Stool | 09/15 | S | S | S | S | S | S | ++++ | R | R |
| 27 | 4108.64 | S. oranienburg | Stool | 03/09 | S | S | S | S | S | S | + | R | R |
| 28 | 4310.33 | S. oranienburg | Stool | 12/09 | S | S | S | S | S | S | +++ | + | + |
| 29 | 1490.92 | S. choleraesuis | na | na | - | - | - | - | - | - | ++++ | ++ | ++++ |
| 30 | 6302.34 | S. enteritidis | Stool | 9/16 | S | S | S | S | S | S | ++ | + | R |
| 31 | 6101.40 | S. sonnei | Stool | 01/16 | S | S | R | S | S | S | +++ | + | +++ |
| 32 | 6105.15 | S. sonnei | Stool | 03/16 | S | S | R | S | S | S | +++ | + | +++ |
| 33 | 6108.73 | S. sonnei | Stool | 04/16 | - | - | - | - | - | - | +++ | ++++ | +++ |
| 34 | 6110.62 | S. sonnei | Stool | 04/16 | R | S | R | S | S | S | ++ | + | +++ |
| 35 | 6003.54 | S. flexneri | Stool | 10/15 | - | - | - | - | - | - | ++++ | R | +++ |
| 36 | 6004.50 | S. flexneri | Stool | 11/15 | S | S | R | S | R | S | ++ | R | ++ |
| 37 | 5906.08 | S. flexneri | Stool | 08/15 | S | S | S | S | S | S | ++ | R | ++ |
| 38 | 5509.52 | S. flexneri | Stool | 08/14 | R | S | R | R | S | S | R | R | R |
| 39 | 6306.26 | S. sonnei | Stool | 10/16 | S | S | R | S | S | S | +++ | ++++ | ++ |
| 40 | 5703.48 | S. sonnei | Stool | 11/14 | S | S | R | S | R | R | + | + | + |
| 41 | 5611.08 | S. sonnei | Stool | 11/14 | - | - | - | - | - | - | +++ | ++++ | +++ |
| 42 | 5605.11 | S. sonnei | Stool | 10/14 | S | S | R | S | S | S | ++ | ++ | ++ |
| 43 | 5402.22 | S. sonnei | Stool | 03/14 | S | S | R | S | R | R | ++ | ++ | ++ |
| 44 | 5312.31 | S. sonnei | Stool | 02/14 | R | S | S | S | S | S | ++ | ++ | ++ |
| 45 | 5203.63 | S. sonnei | Stool | 05/13 | S | S | R | S | S | S | ++ | + | ++ |
| 46 | 6209.65 | S. flexneri | Stool | 08/16 | - | - | - | - | - | - | +++ | R | ++ |
| 47 | 4907.58 | S. flexneri | Stool | 02/12 | S | S | R | R | R | R | +++ | R | ++++ |
| 48 | 4706.22 | S. flexneri | Stool | 04/11 | S | S | R | S | R | S | + | R | ++ |
| 49 | 4611.14 | S. flexneri | Stool | 01/11 | S | S | R | S | S | S | ++ | R | ++ |
| 50 | 4512.64 | S. flexneri | Stool | 09/10 | R | S | S | R | S | S | +++ | R | ++ |
Note. AMP, ampicillin; CRO, ceftriaxone; SXT, cotrimoxazole; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; R, resistant; I, intermediate; S, susceptible; na, not available; -, not tested.
a Strains were defined as susceptible to the bacteriophages when confluent lysis (ie, complete clearing: ++++), semi-confluent lysis (ie, clearing throughout but with faint hazy background: +++), opaque lysis (ie, turbidity throughout the cleared zone: ++), taches vierges (ie, a few individual plaques: +) were recorded. Strains showing no activity (ie, no clearing “R”) were defined as resistant.
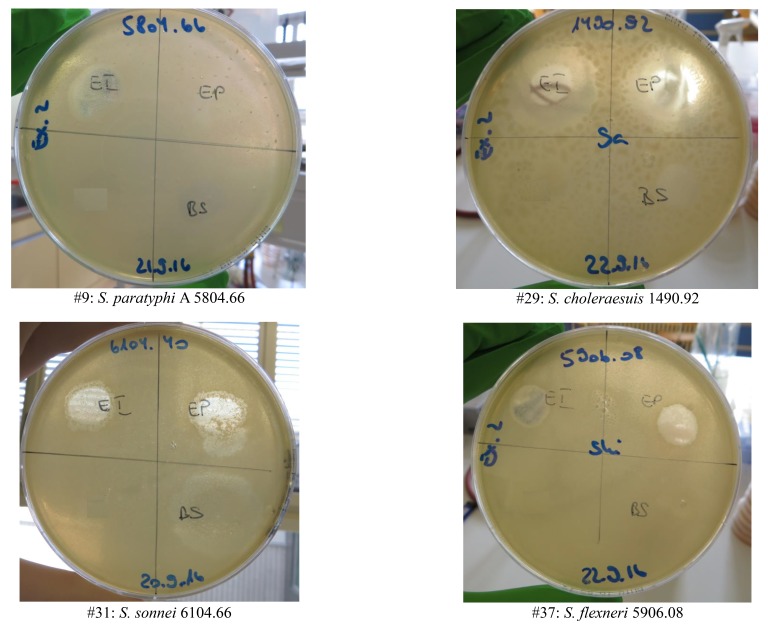
Supplementary Figure 1.
Examples of bacteriophage susceptibility results (see Supp. Table 1) for 2 Salmonella and 2 Shigella spp. strains. EI, Eliava INTESTI Bacteriophage cocktail; EP, Eliava PYO Bacteriophage (Eliava) cocktail; BS, Biochimpharm Septaphage Bacteriophage cocktail.
POTENTIAL CONFLICT OF INTERESTS
None
REFERENCES
- 1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17(1):7–15. PubMed PMID: 21192848. Pubmed Central PMCID: 3375761. doi: 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gu B, Cao Y, Pan S, Zhuang L, Yu R, Peng Z, Qian H, Wei Y, Zhao L, Liu G, Tong M. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents. 2012;40(1):9–17. PubMed PMID: 22483324. doi: 10.1016/j.ijantimicag.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 3. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–66. PubMed PMID: 10516787. Pubmed Central PMCID: 2557719 [PMC free article] [PubMed] [Google Scholar]
- 4. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–22. PubMed PMID: 23680352. doi: 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 5. Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol Rev. 2002;26(2):141–8. PubMed PMID: 12069879. [DOI] [PubMed] [Google Scholar]
- 6. Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. 2004;39(4):546–51. PubMed PMID: 15356819. doi: 10.1086/422726 [DOI] [PubMed] [Google Scholar]
- 7. Domingo-Calap P, Georgel P, Bahram S. Back to the future: bacteriophages as promising therapeutic tools. HLA. 2016;87(3):133–40. PubMed PMID: 26891965. doi: 10.1111/tan.12742 [DOI] [PubMed] [Google Scholar]
- 8. Vandenheuvel D, Lavigne R, Brussow H. Bacteriophage Therapy: Advances in Formulation Strategies and Human Clinical Trials. Annu Rev Virol. 2015;2(1):599–618. PubMed PMID: 26958930. doi: 10.1146/annurev-virology-100114-054915 [DOI] [PubMed] [Google Scholar]
- 9. Bao H, Zhang P, Zhang H, Zhou Y, Zhang L, Wang R. Bio-Control of Salmonella Enteritidis in Foods Using Bacteriophages. Viruses. 2015;7(8):4836–53. PubMed PMID: 26305252. Pubmed Central PMCID: 4576208. doi: 10.3390/v7082847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmadi M, Karimi Torshizi MA, Rahimi S, Dennehy JJ. Prophylactic Bacteriophage Administration More Effective than Post-infection Administration in Reducing Salmonella enterica serovar Enteritidis Shedding in Quail. Front Microbiol. 2016;7:1253 PubMed PMID: 27555842. Pubmed Central PMCID: 4977285. doi: 10.3389/fmicb.2016.01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mohammed M, Cormican M. Whole genome sequencing provides possible explanations for the difference in phage susceptibility among two Salmonella Typhimurium phage types (DT8 and DT30) associated with a single foodborne outbreak. BMC Res Notes. 2015;8:728 PubMed PMID: 26613761. Pubmed Central PMCID: 4661946. doi: 10.1186/s13104-015-1687-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karpe YA, Kanade GD, Pingale KD, Arankalle VA, Banerjee K. Genomic characterization of Salmonella bacteriophages isolated from India. Virus Genes. 2016;52(1):117–26. PubMed PMID: 26757942. doi: 10.1007/s11262-015-1269-7 [DOI] [PubMed] [Google Scholar]
- 13. Borie C, Albala I, Sanchez P, Sanchez ML, Ramirez S, Navarro C, Morales MA, Retamales AJ, Robeson J. Bacteriophage treatment reduces Salmonella colonization of infected chickens. Avian Dis. 2008;52(1):64–7. PubMed PMID: 18459298. doi: 10.1637/8091-082007-Reg [DOI] [PubMed] [Google Scholar]
- 14. Mai V, Ukhanova M, Reinhard MK, Li M, Sulakvelidze A. Bacteriophage administration significantly reduces Shigella colonization and shedding by Shigella-challenged mice without deleterious side effects and distortions in the gut microbiota. Bacteriophage. 2015;5(4):e1088124 PubMed PMID: 26909243. Pubmed Central PMCID: 4745833. doi: 10.1080/21597081.2015.1088124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kutateladze M, Adamia R. Phage therapy experience at the Eliava Institute. Med Mal Infect. 2008;38(8):426–30. PubMed PMID: 18687542. doi: 10.1016/j.medmal.2008.06.023 [DOI] [PubMed] [Google Scholar]
- 16. Zschach H, Joensen KG, Lindhard B, Lund O, Goderdzishvili M, Chkonia I, Jgenti G, Kvatadze N, Alavidze Z, Kutter EM, Hasman H, Larsen MV. What Can We Learn from a Metagenomic Analysis of a Georgian Bacteriophage Cocktail? Viruses. 2015;7(12):6570–89. PubMed PMID: 26703713. Pubmed Central PMCID: 4690881. doi: 10.3390/v7122958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; CLSI document M100-S26, 2016. Wayne, PA. [Google Scholar]
- 18. Martha R.J. Clokie, Kropiski AM. Bacteriophages. Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Book: Methods in Molecular Biology™ 2009;501. [Google Scholar]
- 19. Khan Mirzaei M, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One. 2015;10(3):e0118557 PubMed PMID: 25761060. Pubmed Central PMCID: 4356574. doi: 10.1371/journal.pone.0118557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fukushima M, Kakinuma K, Kawaguchi R. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J Clin Microbiol. 2002;40(8):2779–85. PubMed PMID: 12149329. Pubmed Central PMCID: 120687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11(10):675–87. PubMed PMID: 23979432. doi: 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 22. Bowen A, Hurd J, Hoover C, Khachadourian Y, Traphagen E, Harvey E, Libby T, Ehlers S, Ongpin M, Norton JC, Bicknese A, Kimura A, Centers for Disease C, Prevention. Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin - United States, May 2014-February 2015. MMWR Morb Mortal Wkly Rep. 2015;64(12):318–20. PubMed PMID: 25837241. [PMC free article] [PubMed] [Google Scholar]
- 23. Seiffert SN, Perreten V, Johannes S, Droz S, Bodmer T, Endimiani A. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob Agents Chemother. 2014;58(4):2446–9. PubMed PMID: 24468781. Pubmed Central PMCID: 4023741. doi: 10.1128/AAC.02417-13 [DOI] [PMC free article] [PubMed] [Google Scholar]