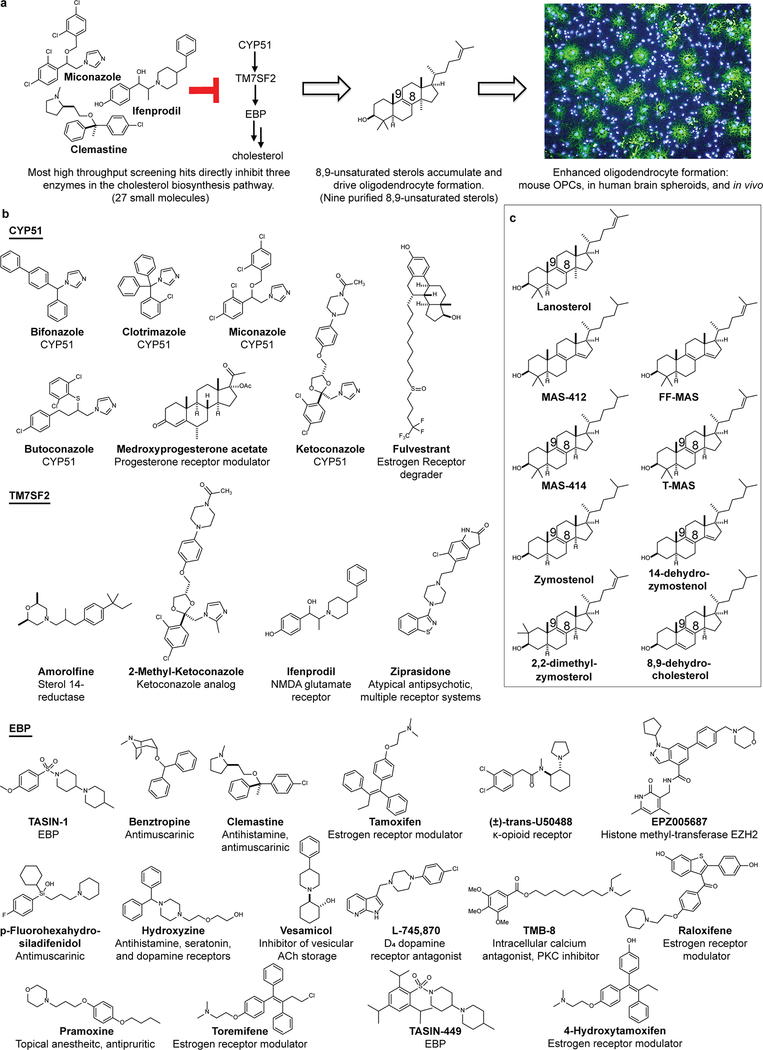
Abstract
Regeneration of myelin is mediated by oligodendrocyte progenitor cells (OPCs), an abundant stem cell population in the CNS and the principal source of new myelinating oligodendrocytes. Loss of myelin-producing oligodendrocytes in the central nervous system (CNS) underlies a number of neurological diseases, including multiple sclerosis (MS) and diverse genetic diseases1–3. Using high throughput chemical screening approaches, we and others have identified small molecules that stimulate oligodendrocyte formation from OPCs and functionally enhance remyelination in vivo4–10. Here we show a broad range of these pro-myelinating small molecules function not through their canonical targets but by directly inhibiting CYP51 (cytochrome P450, family 51), TM7SF2, or EBP (emopamil binding protein), a narrow range of enzymes within the cholesterol biosynthesis pathway. Subsequent accumulation of the 8,9-unsaturated sterol substrates of these enzymes is a key mechanistic node that promotes oligodendrocyte formation, as 8,9-unsaturated sterols are effective when supplied to OPCs in purified form while analogous sterols lacking this structural feature have no effect. Collectively, our results define a unifying sterol-based mechanism-of-action for most known small-molecule enhancers of oligodendrocyte formation and highlight specific targets to propel the development of optimal remyelinating therapeutics.
Results
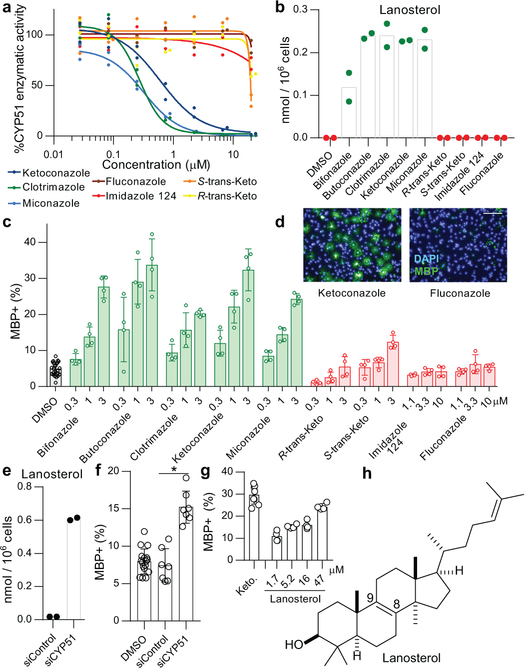
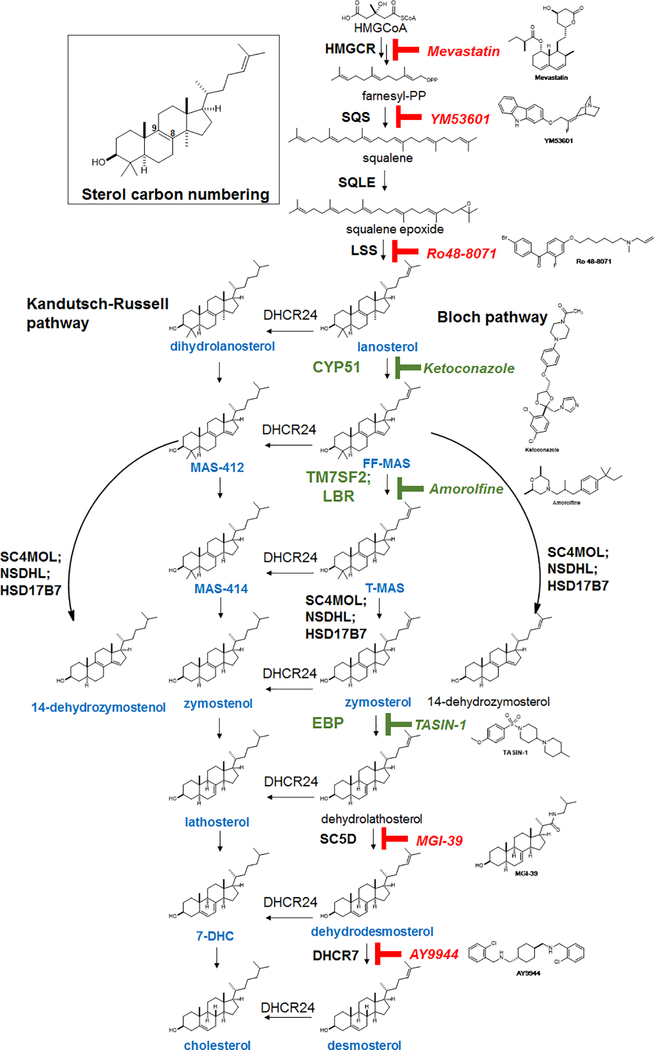
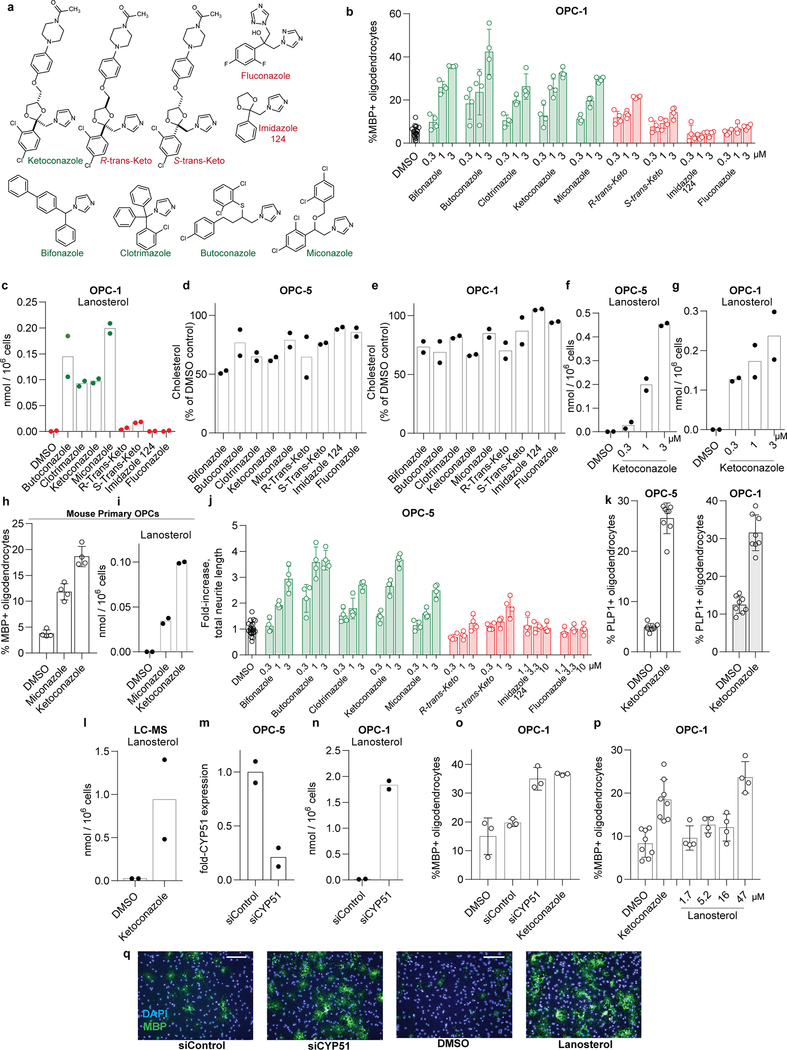
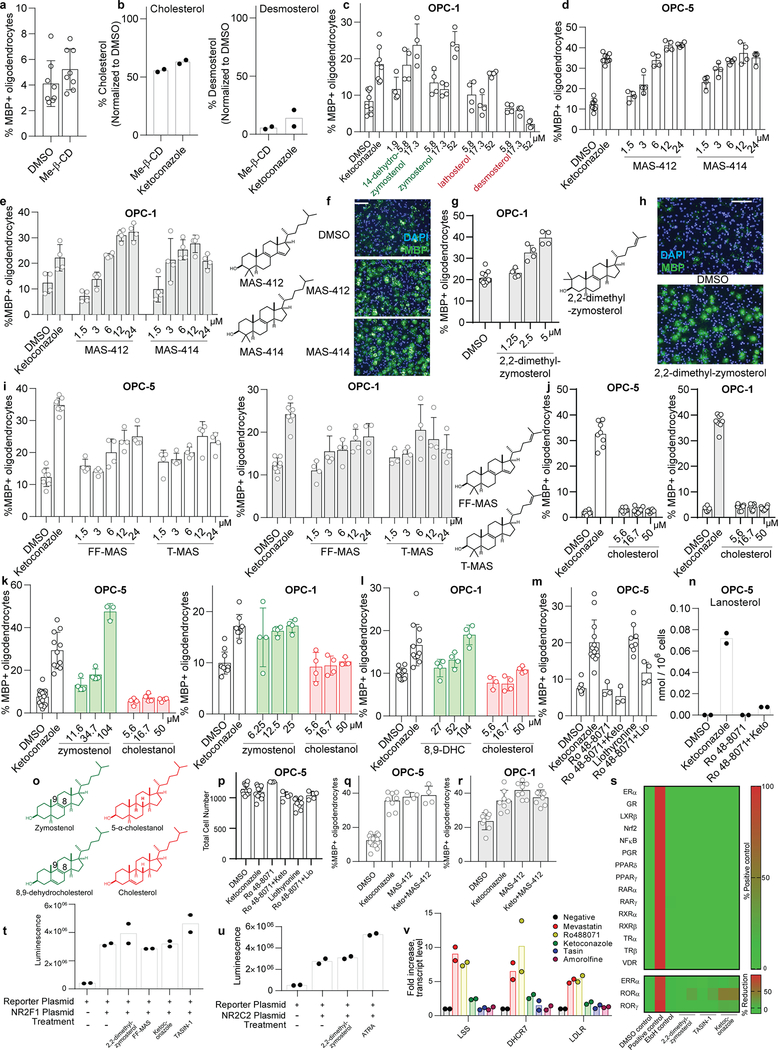
Previously we identified structurally-diverse imidazole antifungal drugs as a robust class of small molecules that stimulate the generation of new mouse and human oligodendrocytes and enhance remyelination in mouse disease models 4. Imidazole antifungals mediate their effects in yeast by inhibiting CYP51, an essential enzyme for sterol biosynthesis in both fungal and mammalian cells (for a detailed cholesterol biosynthesis diagram, see Extended Data Figure 1). We noted that across a panel of nine azole-containing molecules, the ability to inhibit CYP51 in vitro and in OPCs predicted enhanced formation of myelin basic protein-positive (MBP+) oligodendrocytes from mouse epiblast stem cell-derived OPCs (Fig. 1a-d, Extended Data Fig. 2a-c). To measure CYP51 inhibition in OPCs, we used gas chromatography / mass spectrometry (GCMS) to quantitate increased levels of CYP51’s substrate (lanosterol) and decreased cholesterol levels (Fig. 1b, Extended Data Fig. 2c-e).11,12,13 For ketoconazole, the dose-response for accumulation of lanosterol closely resembled the dose-response for enhanced oligodendrocyte formation (Extended Data Fig. 2f,g). Notably, we confirmed all effects of small molecules on oligodendrocyte formation and sterol levels using a second, independently isolated batch of OPCs, and key results were also validated using mouse primary OPCs (Extended Data Fig. 2b-i; see Methods for details of OPC derivations). Additionally, the effects of azole molecules were confirmed using an orthogonal image quantitation approach, a second oligodendrocyte marker, and an LC/MS method for detecting cellular sterols (Extended Data Fig. 2j-l).
Figure 1. Imidazoles inhibit CYP51 to enhance oligodendrocyte formation.
a) Rat CYP51 enzymatic activity following treatment with azoles. n = 2 independent enzymatic assays. b) GC/MS-based quantitation of lanosterol levels in OPCs treated with the indicated azoles at 2.5 μM. n = 2 wells per condition. c, f, g) Percentage of MBP+ oligodendrocytes generated from OPCs following treatment with azoles (c), cell permeable siRNA reagents (f), and lanosterol (g). n ≥ 4 wells per condition; for exact well counts in all figures, see Statistics and Reproducibility. In f, *, P = 0.0005, two-tailed Student’s t-test. d) Representative images of OPCs treated with the indicated azoles. Nuclei are labeled with DAPI (blue), and oligodendrocytes are indicated by immunostaining for myelin basic protein (green). Scale bar, 100 μm. e) GC/MS-based quantitation of lanosterol levels in OPCs treated with the indicated reagents. n = 2 wells per condition. h) Structure of lanosterol. All bar graphs indicate mean ± standard deviation. Experiments in c, d, and g are representative of three independent experiments, while b, e, and f are representative of two independent experiments using OPC-5 cells; for validation in an independent derivation of OPCs, see Extended Data Fig. 2.
We next used RNA interference and metabolite supplementation to independently confirm the role of CYP51 in oligodendrocyte formation. Cell-permeable siRNA reagents depleted CYP51 transcript levels in OPCs by 80% 14, led to significant accumulation of lanosterol, and enhanced formation of MBP+ oligodendrocytes (Fig. 1e-f, Extended Data Fig. 2m-o). Additionally, we treated OPCs directly with purified lanosterol and observed enhanced formation of MBP+ oligodendrocytes in a dose-responsive fashion (Fig. 1g-h, Extended Data Fig. 2p-q). These findings support CYP51 as the functional target of imidazole antifungals in OPCs and suggest that accumulation of sterol intermediates may play a direct role in enhancing oligodendrocyte formation.
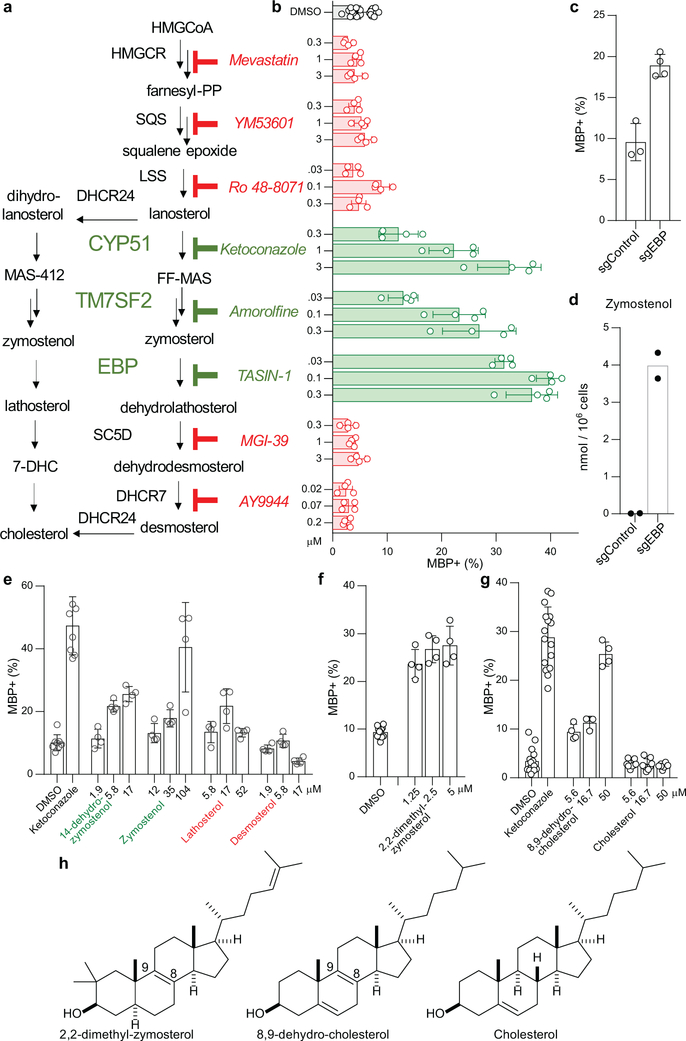
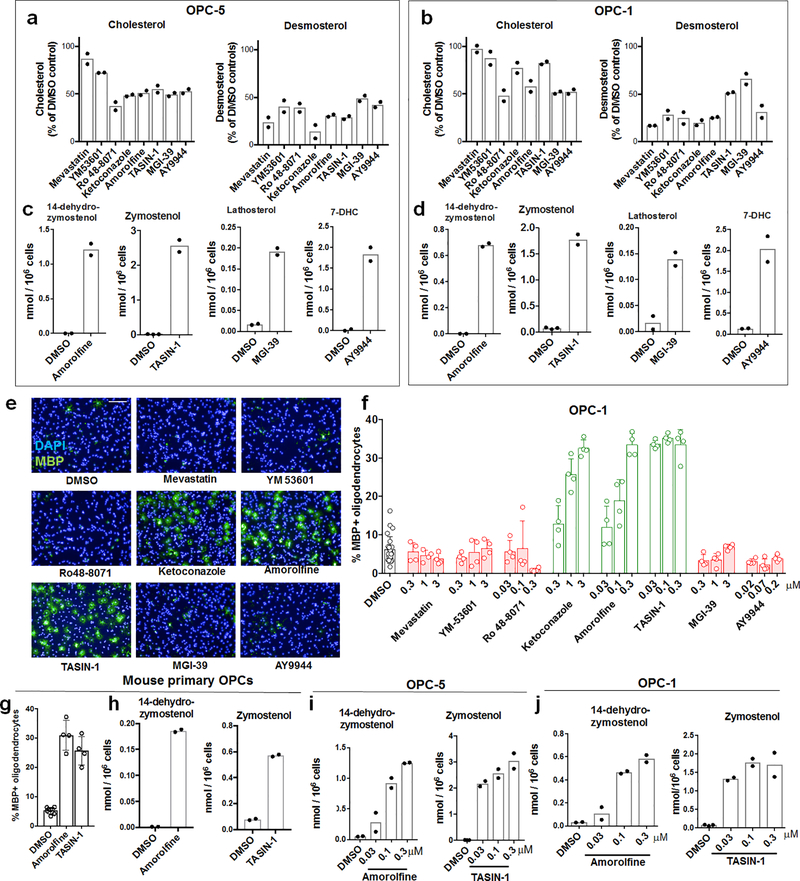
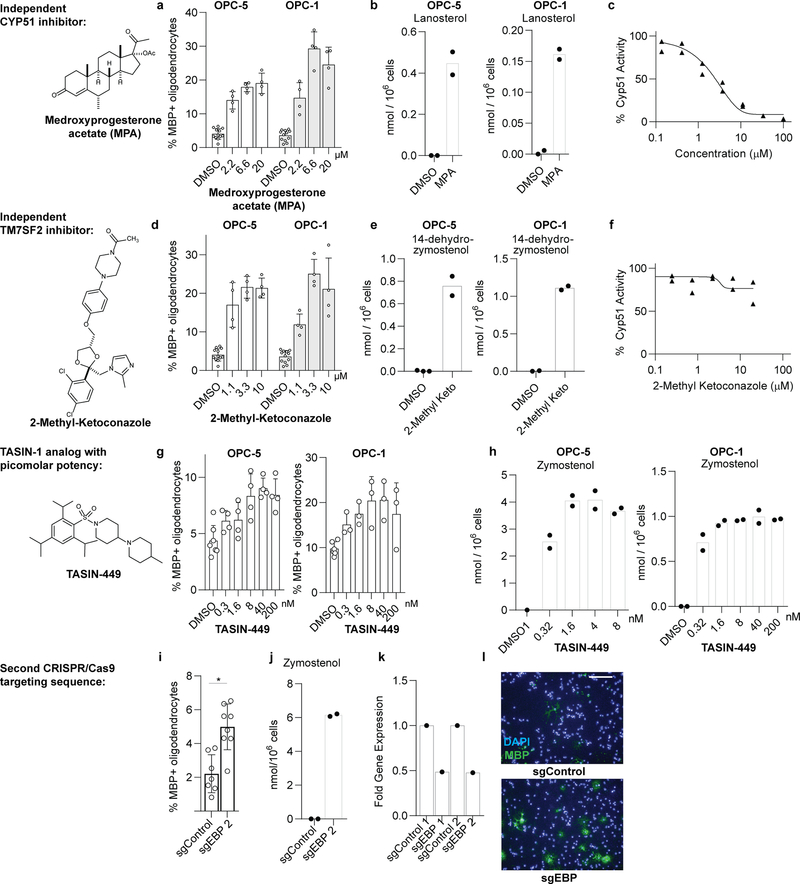
Since CYP51 inhibition was sufficient to induce the formation of oligodendrocytes, we used a chemical genetics approach to test whether modulation of other steps in cholesterol biosynthesis has a similar effect. (Fig. 2a, Extended Data Fig. 1). We used GC/MS-based sterol profiling in OPCs to validate a panel of eight small molecules as selectively inhibiting their known enzyme targets within the cholesterol biosynthesis pathway. (Extended Data Fig. 3a-d; see Source Data for abundance of all quantitated metabolites in all GCMS-based sterol profiling experiments). Only molecules targeting CYP51 (ketoconazole), TM7SF2 (amorolfine15), and EBP (TASIN-116) enhanced formation of MBP+ oligodendrocytes, whereas inhibitors of the five other pathway enzymes were ineffective (Fig. 2b, Extended Data Fig. 3e-h). Treatments had little effect on cell number (Extended Data Fig. 3e). Concentrations of amorolfine and TASIN-1 that enhanced oligodendrocyte formation also led to accumulation of 14-dehydrozymostenol and zymostenol, (Extended Data Fig. 3i-j). Moreover, distinct structural classes of inhibitors of CYP51, TM7SF2, and EBP comparably enhanced oligodendrocyte formation, including at picomolar doses (Extended Data Fig. 4a-h)17.
Figure 2. Small-molecule inhibition of CYP51, TM7SF2, or EBP enhances oligodendrocyte formation via accumulation of 8,9-unsaturated sterols.
a) Abbreviated cholesterol biosynthesis pathway. For greater detail, see Extended Data Figure 1. b) Percentage of MBP+ oligodendrocytes generated from OPCs treated with the indicated pathway inhibitors. n ≥ 4 wells per condition. c) Percentage of MBP+ oligodendrocytes generated from OPCs expressing Cas9 and guide RNA targeting EBP. n ≥ 3 wells per condition. d) Functional validation of Cas9-based targeting of EBP using GC/MS-based quantitation of zymostenol levels. n = 2 wells per condition. e, f, g) Percentage of MBP+ oligodendrocytes generated from OPCs with the indicated purified sterols. n ≥ 4 wells per condition. h) Structures of various sterols. All bar graphs indicate mean ± standard deviation. See Statistics and Reproducibility for exact well counts. Experiments in b-g are representative of two or more independent experiments using OPC-5 cells; for validation in an independent derivation of OPCs, see Extended Data Fig. 3–5.
We also used CRISPR/Cas9 targeting to evaluate the effects of genetic suppression of EBP. OPCs expressing Cas9 and guide RNA targeting EBP demonstrated reduced EBP transcript levels, robust accumulation of the expected intermediate zymostenol, and enhanced formation of oligodendrocytes under differentiation-permissive conditions (Fig. 2c,d, Extended Data Fig. 4k). Two independent guide RNA sequences provided comparable results (Extended Data Fig. 4i-l). In total, this genetic and chemical genetic analysis suggests that inhibition of the cholesterol biosynthesis pathway within a limited window of enzymes between CYP51 and EBP is sufficient for enhancing the formation of oligodendrocytes.
The efficacy of these small molecules and genetic perturbations is not mediated by simple reduction of sterol levels, as statin drugs and methyl β-cyclodextrin treatment depleted cholesterol levels comparably without enhancing oligodendrocyte formation (Fig. 2b, Extended Data Fig. 3a,b, 5a,b). Because treatment of OPCs with CYP51’s substrate lanosterol enhanced oligodendrocyte formation, we examined the effects of other purified sterols. Treatment of OPCs with 8,9-unsaturated sterols, including 14-dehydrozymostenol (accumulates following TM7SF2 inhibition) and zymostenol (accumulates following EBP inhibition), enhanced the formation of MBP+ oligodendrocytes. By contrast, sterols lacking the 8,9 unsaturation, including cholesterol itself,18 were ineffective (Fig. 2e,h, Extended Data Fig. 5c). A total of nine natural and unnatural 8,9-unsaturated sterols enhanced oligodendrocyte formation from OPCs, with 2,2-Dimethyl-zymosterol optimally potent among the sterols evaluated to date (Fig. 2f; Extended Data Fig. 5d-l, o). Conversely, preventing the accumulation of 8,9-unsaturated sterols in OPCs by inhibition of lanosterol synthase with Ro 48–8071 abrogated the enhanced oligodendrocyte formation observed from the CYP51 inhibitor ketoconazole. (Extended Data Fig. 5m,n,p). Additionally, analogs of either zymostenol or 8-dehydrocholesterol that lack the 8,9-unsaturation were inactive, highlighting the 8,9-unsaturation as a crucial structural feature for activity in OPCs (Fig. 2g; Extended Data Fig. 5k,l). Finally, co-treating OPCs with ketoconazole and MAS-412 provided no further benefit over ketoconazole alone, confirming a redundant mechanism (Extended Data Fig. 5q-r). Together these findings indicate that the accumulation of 8,9-unsaturated sterols in OPCs is a central mechanism for enhancing oligodendrocyte formation whether these sterols arise from small-molecule inhibition of cholesterol biosynthesis enzymes or are supplied to OPCs in purified form.
Most of the 8,9-unsaturated sterols shown here to enhance oligodendrocyte formation have previously been shown to function as signaling lipids in oocytes by inducing the resumption of meiosis19,20. While the direct cellular targets of 8,9-unsaturated ‘meiosis-activating sterols’ remain poorly understood, past work suggested nuclear hormone receptors (NHRs) may play a role19. We evaluated 2,2-dimethylzymosterol and the pathway inhibitors ketoconazole and TASIN-1 in cell-based reporter assays for 20 NHRs, but no molecule showed significant activity in any assay (Extended Data Fig. 5s-u). Additional experiments discounted a role for SREBP2, which transcriptionally regulates cholesterol homeostasis, suggesting these sterols act by mechanisms beyond NHRs or SREBP2 (Extended Data Fig. 5v). Together these studies suggest a novel role for the ‘meiosis-activating sterols’ in promoting oligodendrocyte formation.
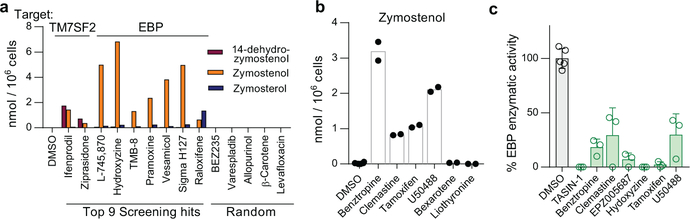
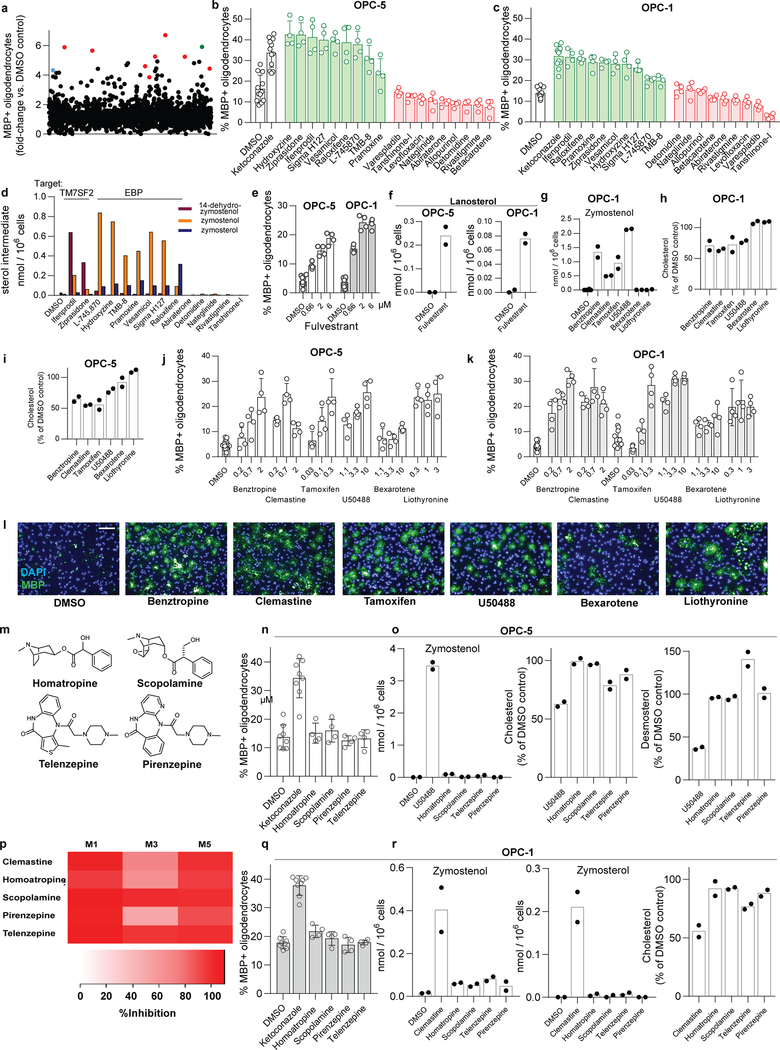
In parallel, we executed a screen of over 3,000 bioactive small molecules and approved drugs at a uniform dose of 2 μM (Extended Data Fig. 6a). In addition to molecules previously annotated as enhancing OPC differentiation 5,6,9,we also identified many confirmed hits whose known targets did not cluster into easily discernible categories (Supporting Table 1). Among the top 10 novel enhancers of oligodendrocyte formation, we noted four molecules had previously been shown to inhibit TM7SF2 or EBP in CNS-derived cells11,21. In fact, GCMS-based sterol profiling revealed that all 10 top hits led to 8,9-unsaturated sterol accumulation at the screening dose, while randomly selected library members had no effect on sterol levels or oligodendrocyte formation. (Fig. 3a, Extended Data Fig. 6b-f).
Figure 3. Inhibition of TM7SF2 and EBP is a unifying mechanism for many small-molecule enhancers of oligodendrocyte formation.
a) Quantitation of sterol levels in OPCs treated with the indicated molecules at 2 μM (one well per condition; for validation in a second derivation of OPCs, see Extended Data Fig. 6). (b) Quantitation of sterol levels in OPCs treated with the indicated previously-reported enhancers of oligodendrocyte formation (n = 2 wells per condition except DMSO, n = 6). Representative of two independent experiments. c) Quantitation of EBP enzymatic activity in a biochemical assay. All treatments 10 μM. n = 3 independent enzymatic assays, except DMSO, n = 5. Bars indicate mean; error bars indicate standard deviation. p-Fluorohexahydro-sila-difenidol abbreviated as Sigma H127.
Given the frequency of cholesterol pathway modulators within our top screening hits, we assessed whether any previously-reported enhancers of remyelination identified by drug screening might also induce sterol intermediate accumulation. At concentrations that promoted oligodendrocyte formation, benztropine, clemastine, tamoxifen, and U50488 induced accumulation of zymostenol and zymosterol and decreased basal sterol levels, indicative of inhibition of EBP in OPCs (Fig. 3b, Extended Data Fig. 6g-l). Tamoxifen has been shown previously to directly inhibit EBP enzymatic activity 22,11,23, and we confirmed that benztropine, clemastine, tamoxifen, U50488, and several HTS hits all inhibited EBP directly in a biochemical assay (Fig. 3c). 22 By contrast, liothyronine and bexarotene showed minimal effects on sterol levels in OPCs (Fig. 3b, Extended Data 6g), consistent with their known functions as modulators of transcription factor function and confirming that many, but not all, treatments that enhance oligodendrocyte formation cause 8,9-unsaturated sterol accumulation.
While each of these bioactive small molecules has a previously annotated ‘canonical’ target, extensive structure-activity relationship data reveal that the ability to inhibit EBP, rather than the canonical target, predicts enhanced oligodendrocyte formation. For example, we validated a panel of six muscarinic receptor antagonists that all showed near-completed inhibition of the M1, M3, and M5 muscarinic receptor subtypes at the HTS dose of 2 μM (Extended Data Fig. 6m,p). Among these molecules, only clemastine and benztropine inhibited EBP in OPCs, and only clemastine and benztropine enhanced oligodendrocyte formation (Extended Data Fig. 6j,k,m-r). Likewise, among selective estrogen receptor modulators, toremifine and ospemifine are structurally near-identical and show comparable cellular antiestrogen activity. However, only toremifine inhibited EBP in OPCs, and only toremifine enhanced oligodendrocyte formation (Extended Data Fig. 7a-g). Conversely, while 4-hydroxy-tamoxifen as expected showed 100-fold enhanced cellular antiestrogen activity relative to tamoxifen, both molecules have comparable potency for inhibition of EBP and comparable potency for enhancing oligodendrocyte formation (Extended Data Fig. 7h-j). Finally, the leading novel hit from our HTS, EPZ005687, was annotated as an inhibitor of the histone methyltransferase EZH2. However, analysis of three additional structurally-related EZH2 inhibitors revealed that EPZ005687 uniquely inhibited EBP and enhanced oligodendrocyte formation (Extended Data Fig. 7k-r). Across various antimuscarinics, SERMs, and EZH2 inhibitors, the ability to inhibit EBP, rather than each molecule’s canonical activity, predicts enhanced oligodendrocyte formation.
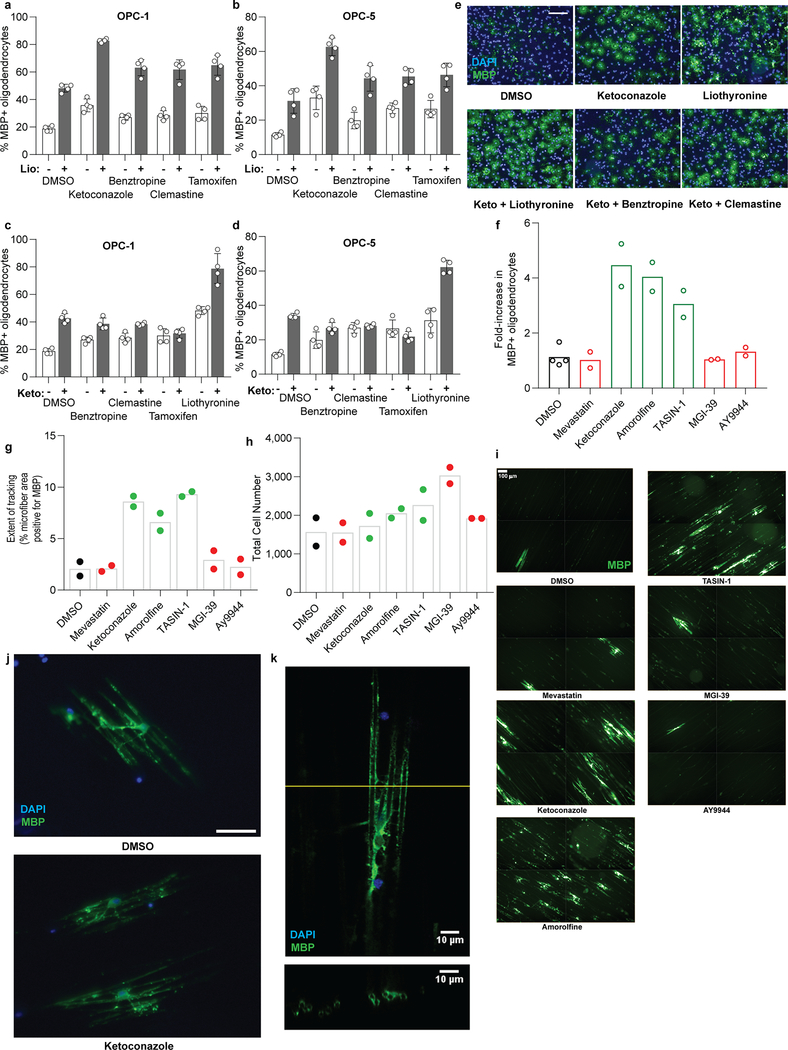
We next tested the potential for combinations of small molecules to show additive or non-additive effects. Combining the thyroid hormone agonist liothyronine with a range of sterol-modulating OPC differentiation-inducing treatments produced additive effects on oligodendrocyte formation, indicating that these molecules likely function by mechanisms other than thyroid hormone receptor signaling to enhance oligodendrocyte generation (Extended Data Fig. 8a-b). In contrast, combinations of ketoconazole at a maximally effective dose with benztropine, clemastine, tamoxifen, or U50488 did not enhance differentiation above levels seen for ketoconazole alone (Extended Data Fig. 8c-e), consistent with these molecules sharing 8,9-unsaturated sterol accumulation as a common mechanism for induction of oligodendrocyte formation.
Since our in vitro OPC assays only model the initial differentiation event into oligodendrocytes, we next tested whether sterol pathway modulation also enhanced subsequent oligodendrocyte maturation and myelination in vitro and in vivo. First, we cultured OPCs on electrospun microfibers to assess the effects of sterol pathway modulators on oligodendrocytes’ ability to track and wrap along axon-like substrates 24,25. Ketoconazole (CYP51), amorolfine (TM7SF2), and TASIN-1 (EBP) all significantly enhanced MBP+ oligodendrocyte tracking along and wrapping of the microfibers. By contrast, inhibition of other enzymes up- or downstream in the pathway had little effect on oligodendrocyte maturation and ensheathment of microfibers (Extended Data Fig. 8f-k).
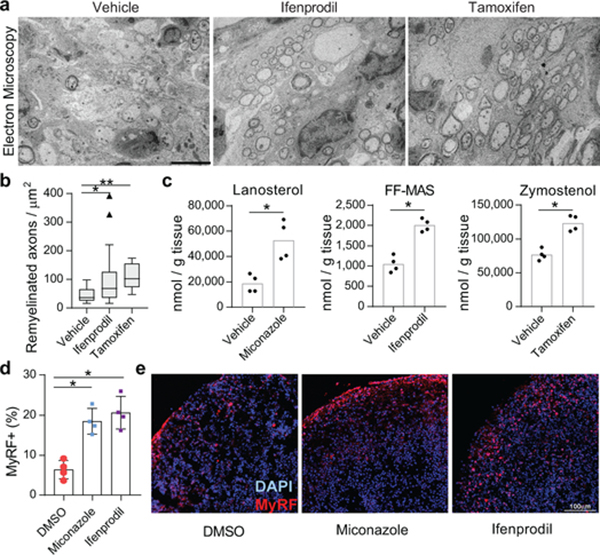
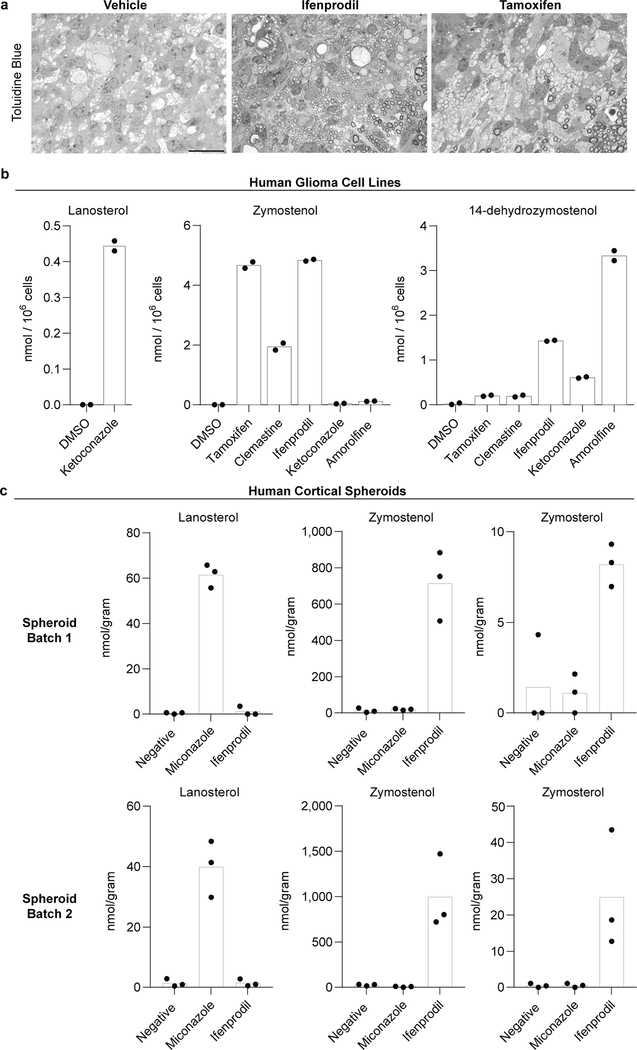
Previously we established that the imidazole antifungal miconazole, which targets CYP51, penetrates the mouse blood brain barrier and enhances remyelination in mouse models of demyelination 4. Here we evaluated brain-penetrant molecules with affinity for TM7SF2 (ifenprodil) and EBP (tamoxifen) using a well-established mouse model where injection of lysolecithin is used to create focal lesions of demyelination in the dorsal column white matter of the adult spinal cord 26. In vehicle treated animals, profiles of sparsely distributed remyelinating axons characterized by thin myelin sheaths were detected mainly at the periphery of the lesion, while ultrastructural analyses revealed unmyelinated axons or axons with a single wrap of myelin (Fig. 4a,b). By contrast, following 8 days of treatment with ifenprodil or tamoxifen, remyelination was widespread throughout the lesion (Fig. 4a,b, Extended Data Fig. 9a), consistent with a recent report regarding tamoxifen9. Critically, we used GCMS-based sterol profiling of brain tissue from mice treated with miconazole, ifenprodil, and tamoxifen to demonstrate that these therapeutic dosing regimens all led to significant accumulation of 8,9-unsaturated sterols within the mouse brain, indicating inhibition of CYP51, TM7SF2, and EBP (Fig. 4c). Collectively, these data show that small molecule inhibitors of CYP51, TM7SF2, and EBP can engage their sterol pathway targets and significantly enhance remyelination in mice.
Figure 4. Accumulation of 8,9-unsaturated sterols enhances remyelination in vivo and in human brain spheroids.
a) Representative electron microscopy images of LPC-lesioned dorsal spinal cord from mice treated with ifenprodil or tamoxifen. Scale bar, 5 μm. b) Tukey plot showing quantitation of remyelinated axons in LPC-lesioned spinal cord from mice in panel a. n = 6 animals per group except vehicle, n = 4. **, P = 0.0004; *, P = 0.048, two-tailed Mann-Whitney test. c) Quantitation of brain sterol levels in mice treated with miconazole, ifenprodil, or tamoxifen. n = 4 animals per group. P = 0.0007 for miconazole, P = 0.0003 for ifenprodil, P = 0.0006 for tamoxifen, two-tailed Student’s t-test. d) Quantitation of MyRF+ oligodendrocytes within human myelinating cortical spheroids following treatment with miconazole (2 μM) or ifenprodil (2 μM). n = 4 spheroids per treatment condition. P = 0.0009 for miconazole and P = 0.0009 for ifenprodil, two-tailed Student’s t-test. e) Representative images of spheroids. DAPI+ nucelei (blue) and MyRF+ oligodendrocytes (red) are labeled. Scale bar, 100 μm. In c and d, bar graphs indicate mean and error bars indicate standard deviation.
Finally, the oligodendrocyte-enhancing and sterol-modulating activities of leading pathway inhibitors extend to human cells and tissue. Various small molecules caused accumulation of the expected 8,9-unsaturated sterol intermediates both in a human glioma cell line and in human pluripotent stem cell-derived cortical spheroids27, confirming that these molecules similarly engage the sterol synthesis pathway in mouse and human cells and CNS tissue (Extended Data Fig. 9b-c). Critically, miconazole and ifenprodil also significantly enhanced the generation human oligodendrocytes in a three-dimensional human pluripotent stem cell-derived cortical spheroid model, indicating conservation of function across species (Fig. 4d-e).
Here we have defined a dominant mechanism shared by many small molecule enhancers of remyelination: elevation of 8,9-unsaturated sterol intermediate levels by inhibition of a narrow range of cholesterol biosynthesis enzymes between CYP51 and EBP. In all we have characterized twenty-seven small molecules as both enhancing myelination and elevating 8,9-unsaturated sterol intermediate levels11,21,23. Mechanistically, several lines of evidence support a central signaling role for 8,9-unsaturated sterols in the observed enhanced oligodendrocyte formation, including the ability of nine independent 8,9-unsaturated sterols to enhance the formation of oligodendrocytes when supplied to OPCs (Extended Data Fig. 10).
Myelin is cholesterol-enriched, and past work has established that genetic or pharmacological treatments that inhibit early enzymes in cholesterol biosynthesis lead to hypomyelination in vivo 28–30. Our work supports these observations, as inhibition of HMGCoA reductase and squalene synthesis have neutral-to-negative effects on oligodendrocyte formation in our assays (Fig. 2b, Extended Data Fig. 3). These enzymes catalyze steps prior to the synthesis of the first sterol intermediate, so their inhibition prevents the synthesis of all cellular sterols. Our findings establish an alternate paradigm in which the cholesterol biosynthesis pathway can be leveraged to enhance the formation of new oligodendrocytes by targeting later steps whose inhibition does not cause net depletion of cellular sterols. Instead, acute inhibition of CYP51, TM7SF2, or EBP during OPC differentiation induces a ‘sterol shift’ in which a minority of cellular cholesterol is diverted to 8,9-unsaturated sterol intermediates that signal to enhance oligodendrocyte formation. Importantly, we and others have independently shown that multiple molecules now annotated by us as enhancing 8,9-unsaturated sterol intermediate levels can regenerate functional myelin in vivo, as evidenced by reversal of paralysis in mice with MS-like disease 4,6,5. Ultimately, our work demonstrates that modulating the sterol landscape in OPCs can enhance the formation of oligodendrocytes and points to new therapeutic targets, potent inhibitors for these targets, and metabolite-based biomarkers to accelerate the development of optimal remyelinating therapeutics.
METHODS
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper [and its supplementary information files] or are available from the corresponding author upon request. Source data for all GCMS-based sterol profiling experiments and animal experiments are provided with the paper.
Statistics and reproducibility:
No statistical methods were used to predetermine sample size. Data were expressed as mean ± standard deviation and P values were calculated using an unpaired two-tailed Student’s t-test for pairwise comparison of variables with a 95% confidence interval and n-2 degrees of freedom, where n is the total number of samples, in all figures except Figure 4b. In Figure 4b, P values were calculated using an unpaired two-tailed Mann-Whitney test with at 95% confidence interval and the data plotted as a Tukey box and whisker plot. Boxes indicate the interquartile range, and the horizontal line represents the median. Biological replicates: Figure 1: c) n = 4 wells per condition, except DMSO, n = 24; f) n = 17 wells for DMSO, n = 7 for siControl & siCYP51; g) n = 8 wells for DMSO and n = 4 for lanosterol; Figure 2: b) n = 4 wells per condition, except DMSO, n = 24. c) n = 3 wells for sgControl and n = 4 for sgEBP. e, f, g) n = 4 wells per condition, except n = 8 for DMSO and n = 7 ketoconazole in e, n = 12 for DMSO in f, n = 16 for DMSO and ketoconazole, n = 8 for cholesterol in g. Independent experiments: Figures 2b and 2f are representative of three and 2c, e, g are two independent experiments using OPC-5 cells; for validation in an independent derivation of OPCs, see Extended Data Fig. 3, 4 and 5.
Small molecules:
The identity and purity of small molecules were authenticated by LC/MS before use (Supporting Table 2). The following compounds were purchased from Sigma-Aldrich as a solid: Ketoconazole, Miconazole, Clotrimazole, Fluconazole, Fulvestrant, Ifenprodil, Benztropine, Bexarotene, Tamoxifen, 4-Hydroxytamoxifen, Medroxyprogesterone acetate, Ospemifene, GSK343, Trans-U50488, Methyl-β-cyclodextrin, 5α−Cholestan-3β-ol, and Cholesterol. The following compounds were purchased from Cayman Chemicals as a solid: Liothyronine, Clemastine, AY9944, YM53601 and Ro-48–8071. The following compounds were obtained from Janssen Pharmaceuticals as a solid: 2-Methyl-ketoconazole, R-trans-Ketoconazole, and S-trans-Ketoconazole. Mevastatin was purchased as a solid from Selleck Chemicals. The following compounds were purchased from Selleck Chemicals as a 10 mM DMSO solution: Bifonazole, Butoconazole, Amorolfine, Toremifene, EPZ005687, EPZ6438, UNC1999, Hydroxyzine, Ziprasidone, p-Fluorohexahydro-sila-difenidol (abbreviated in figures as Sigma H127), Vesamicol, Raloxifene, L-745,870, TMB-8, Pramoxine, Varespladib, Tanshinone-I, Levofloxacin, Nateglinide, Abiraterone, Allopurinol, Detomidine, Rivastigmine, Beta carotene, BEZ-235, Scopolamine, and Homatropine. Pirenzepine and Telenzepine were purchased from Sigma-Aldrich as a 10 mM DMSO solution. Cholesterol biosynthetic intermediates were purchased from Avanti Polar Lipids as a solid: Lanosterol, Zymosterol, Zymostenol, Lathosterol, Desmosterol, 7-dehydrodesmosterol, FF-MAS (4,4-dimethyl-5α-cholesta-8,14,24-trien-3β-ol), 8,9-dehydrocholesterol, and 2,2-dimethylzymosterol (2,2-dimethyl-5α-cholesta-8,24-dien-3β-ol). 14-dehydrozymostenol (5α-cholesta-8,14-dien-3β-ol), MAS-412 (4,4-dimethyl-5α-cholesta-8,14-dien-3β-ol), and MAS-414 (4,4-dimethyl-5α-cholesta-8-en-3β-ol) were provided by Franz Bracher, Ludwig-Maximilians University of Munich. Imidazole 124 31, TASIN-1 16, TASIN-449 17, and MGI39 32 were synthesized as reported. T-MAS (4,4-dimethyl-5α-cholesta-8,24-dien-3β-ol) from HPLC purification of yeast extracts was provided by Jing Jin and William K. Wilson, Rice University.
Mouse OPC preparation:
To rigorously assess effects of small molecule and genetic treatments on OPCs, all treatments were assayed in two batches of epiblast stem cell-derived OPCs, and key results were confirmed using mouse primary OPCs. OPCs were generated from two separate EpiSC lines, EpiSC5 (giving rise to OPC-5 OPCs) and 129O1 (giving rise to OPC-1 OPCs). Unless otherwise noted, results in OPC-5 cells are presented in Figures 1–4 while results in OPC-1 are presented in the Extended Data.
EpiSC-derived OPCs were obtained using in vitro differentiation protocols and culture conditions described previously 33. To ensure uniformity throughout all in vitro screening experiments, EpiSC-derived OPCs were sorted to purity by fluorescence activated cell sorting at passage five with conjugated CD 140a-APC (eBioscience, 17–1401; 1:80) and NG2-AF488 (Millipore, AB5320A4; 1:100) antibodies. Sorted batches of OPCs were expanded and frozen down in aliquots. OPCs were thawed into growth conditions for one passage before use in further assays. Cultures were regularly tested and shown to be mycoplasma free.
To obtain mouse primary OPCs, whole brain was removed from post-natal day 2 pups anesthetized on ice. Brains were placed in cold DMEM/F12, and the cortices were isolated and the meninges were removed. The cortices were manually chopped and processed with the Tumor Dissociation Kit (Miltenyi) and incubated at 37 °C for 10 minutes. The cell suspension was filtered through a 70 μM filter and centrifuged at 200×g for 4 minutes at room temperature. The cells were washed in DMEM/F12, re-centrifuged and plated in poly-Ornithine and Laminin-treated flasks containing DMEM/F12 supplemented with N2 Max, B27 (ThermoFisher), 20ng/mL FGF, and 20ng/mL PDGF. OPCs were passaged once prior to treatment. Media was changed every 48 hours.
In vitro phenotypic screening of OPCs:
EpiSC-derived OPCs were grown and expanded in poly-ornithine (PO) and laminin-coated flasks with growth medium (DMEM/F12 supplemented with N2-MAX (R&D Systems), B-27 (ThermoFisher), GlutaMax (Gibco), FGF2 (10 μg/mL, R&D systems, 233-FB-025) and PDGF-AA (10 μg/mL, R&D systems, 233-AA-050) before harvesting for plating. The cells were seeded onto poly-D-lysine 96-well CellCarrier or CellCarrierUltra plates (PerkinElmer) coated with laminin (Sigma, L2020; 15 μg/ml) using multi-channel pipet. For the experiment, 800,000 cells/mL stock in differentiation medium (DMEM/F12 supplemented with N2-MAX and B-27) was prepared and stored on ice for 2 h. Then, 40,000 cells were seeded per well in differentiation medium and allowed to attach for 30 min before addition of drug. For dose–response testing of all molecules except sterols, a 1000x compound stock in dimethyl sulphoxide (DMSO) was added to assay plates with 0.1 μL solid pin multi-blot replicators (V & P Scientific; VP 409), resulting in a final primary screening concentration of 1x. Sterols were added to cells as an ethanol solution (0.2% final ethanol concentration). Positive control wells (ketoconazole, 2.5 μM) and DMSO vehicle controls were included in each assay plate. Cells were incubated under standard conditions (37 °C, 5% CO2) for 3 days and fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 20 min. Fixed plates were washed with PBS (200 μL per well) twice, permeabilized with 0.1% Triton X-100 and blocked with 10% donkey serum (v/v) in PBS for 40 min. Then, cells were labelled with antibodies recognizing MBP (Abcam, ab7349; 1:200) or PLP1 (1:1,000, clone AA3, generously provided by B. Trapp, Cleveland Clinic) for 16 h at 4° C followed by detection with Alexa Fluor conjugated secondary antibodies (1:500) for 45 min. Nuclei were visualized by DAPI staining (Sigma; 1 μg/ml). During washing steps, PBS was added using a multi-channel pipet and aspiration was performed using Biotek EL406 washer dispenser (Biotek) equipped with a 96-well aspiration manifold.
High-content imaging and analysis:
Plates were imaged on the Operetta High Content Imaging and Analysis system (PerkinElmer) and a set of 6 fields captured from each well resulting in an average of 1200 cells being scored per well. Analysis (PerkinElmer Harmony and Columbus software) began by identifying intact nuclei stained by DAPI; that is, those traced nuclei that were larger than 300 μm2 in surface area. Each traced nucleus region was then expanded by 50% and cross-referenced with the mature myelin protein (MBP) stain to identify oligodendrocyte nuclei, and from this the percentage of oligodendrocytes was calculated. In some experiments, PLP1 staining was performed instead of MBP, or the total process length of MBP+ oligodendrocytes was calculated as previously described. 4
OPCs differentiation and sterol profiling after Methyl-β-cyclodextrin treatment:
EpiSCs derived OPCs harvested from culture flasks were resuspended in 10 mL of differentiation medium to a final cell density of 500,000 cells/mL, to this cell-culture grade water or Methyl-β-cyclodextrin (1 mM) was added and incubated at 37 C. After 30 min the cells were washed twice with differentiation medium (5 mL), and spilt into two portions for differentiation and sterol profiling. The 1000,000 cells/condition were directly processed as described in GC/MS-based sterol profiling to measure the endogenous in sterol levels. For differentiation, the cells were resuspended in differentiation medium to a final cell density of 800,000 cells/mL and plated in a PDL/laminin coated 96-well CellCarrierUltra plate. After 72 h, the cells were fixed, stained, imaged and quantified as described above.
High-throughput screening of 3,000 bioactive small molecules:
EpiSC-derived OPCs were grown and expanded in poly-ornithine and laminin-coated flasks before harvesting for plating. Cells were dispensed in differentiation media supplemented with Noggin (R&D Systems; 100 ng/ml), Neurotrophin 3 (R&D Systems; 10 ng/ml), cAMP (Sigma; 50 μM), and IGF-1 (R&D Systems; 100 ng/ml)) using a Biotek EL406 Microplate Washer Dispenser (Biotek) equipped with 5 μL dispense cassette (Biotek), into poly-D-lysine/ laminin (Sigma, L2020; 4 μg/ml)-coated sterile, 384-well, CellCarrier ultra plates (PerkinElmer), to a final density of 12,500 cells per well and allowed to attach for 45 min before addition of drug. A 3 mM stock of bioactive compound library in dimethylsulphoxide (DMSO) were prepared in an Abgene storage 384-well plate (ThermoFisher Scientific; AB1055). These were added to assay plates using a 50 nL solid pin tool attached to Janus automated workstation (Perkin Elmer), resulting in a final screening concentration of 2 μM. Cells were incubated at 37 °C for 1 hour and then T3 (Sigma; 40 ng/ml) was added to all wells except negative controls, to which FGF (20 ng/ml) was added instead. Negative controls and T3-alone were included in each assay plate. After incubation at 37° C for 72 h, cells were fixed, washed and stained similar to 96-well OPC assay protocol, although all the washing steps were performed using a Biotek EL406 Microplate Washer Dispenser (Biotek) equipped with a 96-well aspiration manifold. Cells were stained with DAPI (Sigma; 1 μg/ml) and MBP antibody (Abcam, ab7349; 1:100). Plates were imaged on the Operetta High Content Imaging and Analysis system (PerkinElmer) and a set of 4 fields captured from each well resulting in an average of 700 cells being scored per well. Analysis was performed as in High-Content Imaging and Analysis, above. All plates for the primary screen were processed and analyzed simultaneously to minimize variability. Molecules causing more than 20% reduction in nuclear count relative to DMSO control wells were removed from consideration, and hits were called on the basis of largest fold-increase in percentage of MBP+ oligodendrocytes relative to DMSO controls within the same plate. When selecting the leading hits for further experiments, molecules obtained in previous screens were omitted, including imidazole antifungals and clemastine.
GC/MS-based sterol profiling:
EpiSC-derived OPCs were plated at 0.5 million cells per ml in PDL- and laminin-coated six or twelve well plate with differentiation media. After 24 hours, cells were dissociated with Accutase, rinsed with saline, and cell pellets were frozen. For sterol analyses, cells were lysed in methanol (Sigma-Aldrich) with agitation for 30 minutes and cell debris removed by centrifugation at 10,000 rpm for 15 min. Cholesterol-d7 standard (25,26,26,26,27,27,27-2H7-cholesterol, Cambridge Isotope Laboratories) was added before drying under nitrogen stream and derivatization with 55 μl of bis(trimethylsilyl)trifluoroacetamide/trimethylchlorosilane to form trimethylsilyl derivatives. Following derivatization at 60 °C for 20 minutes, 1 μl was analyzed by gas chromatography/mass spectrometry using an Agilent 5973 Network Mass Selective Detector equipped with a 6890 gas chromatograph system and a HP-5MS capillary column (60 m × 0.25 mm × 0.25 μm). Samples were injected in splitless mode and analyzed using electron impact ionization. Ion fragment peaks were integrated to calculate sterol abundance, and quantitation was relative to cholesterol-d7. The following m/z ion fragments were used to quantitate each metabolite: cholesterol-d7 (465), FF-Mas (482), cholesterol (368), zymostenol (458), zymosterol (456), desmosterol (456, 343), 7-dehydrocholesterol (456, 325), lanosterol (393), lathosterol (458), 14-dehydrozymostenol (456). Calibration curves were generated by injecting varying concentrations of sterol standards and maintaining a fixed amount of cholesterol-D7. The human glioma cell line GBM528 was a gift of Jeremy Rich (Cleveland Clinic). These cells were validated as unique by STR profiling (Miller et al., Nature, 2017 547 355–359).
LC/MS-based sterol profiling:
Sterols were extracted after treatment of OPC-5 OPCs with ketoconazole as described in GC/MS-based sterol profiling above. Picolinate derivatization, chromatographic separation, and mass spectrometric detection were performed as reported previously.34 Peaks from selective reaction monitoring were integrated to calculate sterol abundance, and quantitation was relative to cholesterol-d7.
Human cortical spheroids:
Human cortical spheroids were generated as described previously 27 with modifications to enable the inclusion and differentiation of OPCs. In brief, spheroids were treated with miconazole or ifenprodil (2 μM) from days 62–72 and assayed on day 93 for MyRF+ oligodendrocytes (Rabbit anti-MyRF antibody was generously provided by M. Wegner and used at 1:1000). [Reference to be updated when spheroid manuscript publishes].
CYP51 enzymatic assay:
CYP51 enzymatic activity was measured using a reported method with slight modifications 35: rat CYP51 (Cypex, Inc.) was used as enzyme; reaction volume was 500 μl; reaction time was 30 minutes; lanosterol concentration was 50 μM; and reactions were quenched with 500 μl isopropanol. Finally, 15 μl of each reaction/isopropanol mixture was injected onto a SCIEX Triple Quad 6500 LC-MS/MS system using an APCI ion source in positive ion mode with a Shimadzu UFLC-20AD HPLC and a Phenomenex Kinetix C18XB 50×2.1×2.6 column at 40 °C.
EBP enzymatic assay:
EBP enzymatic activity was measured using a reported method with slight modifications22: active EBP was obtained from mouse microsomes, inhibitors were added, zymostenol was added at a final concentration of 25 μM in a final reaction volume of 500 μl, and the reaction incubated at 37 °C for 2 h. Sterols were extracted using 3 × 1 ml hexanes, cholesterol-d7 was added to enable quantitation, and the pooled organics were dried (Na2SO4) and evaporated under nitrogen gas. Samples were then silylated and analyzed using GC/MS as described above.
siRNA treatments:
Cell-permeable siRNAs were obtained as pools of 4 individual siRNAs targeting mouse CYP51, or a non-targeting control (Accell siRNAs, Dharmacon. Pooled CYP51 siRNA sequence: GUCUGUUUUGAGAUUAGU; CGACUAUGCUUCGUUUAUA; CGCUGCUCUUCAAUAGUAA; CUAUUAAGUUAUUGUGAAC. Non-targeting control siRNA: UGGUUUACAUGUCGACUAA). For differentiation analysis, cells were plated in 96-well plate (as detailed above) and treated with 1 μM pooled siRNA suspended in RNAse free water diluted in differentiation media (as detailed above). For sterol analysis cells were plated in a six-well plate at 300,000 cells per well in standard differentiation media supplemented with PDGF (R&D Systems, 20 ng/ml), neurotrophin 3 (R&D Systems; 10 ng/ml), cAMP (Sigma; 50 μM), IGF-1 (R&D Systems; 100 ng/ml), noggin (R&D Systems; 100 ng/ml). At 24 hours, 1 μM siRNA was added to the media. Cells were grown for three more days in siRNA containing media, with growth factor supplementation every 48h, before harvesting and processing for GC/MS analysis as detailed above.
CRISPR/Cas9-mediated targeting of EBP:
Guide RNA sequences were obtained using the Broad Brie library and manufactured by IDT. Nucleotide sequences (sgRNA sequence: GAAACGCAATCACTACCCAT (sgEBP); GGGGCCTAATTGTGATCACG (sgEBP2)) were prepared and inserted into the LentiCRISPRv2 plasmid (Addgene, 52961) using the instructions from GeckoLibrary preparation: briefly, Fastdigest BsbmB1 (fermentas) was used for plasmid digestion, T4 PNK (NEB M0201S) for nucleotide annealing, and Quick Ligase (NEB M2200S) for sgRNA insertion. Insertion was confirmed by sanger sequencing. Hek293T cells were transfected using Lenti-x shots as per manufacturer’s protocol (Clontech). After 24 hours the media was changed to OPC media for collection of virus. 48 hours later the media was collected, supplemented with FGF, PDGF, and protamine sulfate (Sigma, 8ug/mL), and used to transduce OPCs. 24 hours later the media was changed to non-virus containing media for 48 hours. Cells underwent two 48 hour stretches of puromycin selection (Invitrogen). After 24 hours of recovery in non-selection media, cells were plated for differentiation, GCMS, and qPCR as described above.
Focal demyelination, drug treatment and histological analysis:
Focal demyelination in the dorsal column of the spinal cord was induced by the injection of 1% LPC solution. 12 week old C57BL/6 female mice were anesthetized using isoflurane and T10 laminectomies were performed. 1μl of 1% LPC was infused into the dorsal column at a rate of 15 μl/hour. At day 4, animals were randomized into treatment groups prior to treatment (2 animals were excluded due to surgical complications). Between days 4 and 11 post laminectomy, animals received daily injections of either vehicle or drug intraperitoneally. Drugs were dissolved in DMSO or corn oil and then diluted with sterile saline for injections such that final doses were 2 mg/kg for Tamoxifen and 10 mg/kg for Ifenprodil. This experiment was done in a blinded manner: compounds were coded to ensure the researchers performing the experiments were unaware of the treatment being administered to each animal. All animals were euthanized 12 days post laminectomy (n= 4–6 per group). Mice were anesthetized using ketamine/xylazine rodent cocktail and then euthanized by transcardial perfusion with 4% PFA, 2% glutaraldehyde, and 0.1 M sodium cacodylate. Samples were osmicated, stained en bloc with uranyl acetate and embedded in EMbed 812, an Epon-812 substitute (EMS). 1 μm sections were cut and stained with toluidine blue and visualized on a light microscope (Leica DM5500B). The number of myelinated axons per unit area was counted from sections obtained from the middle of each lesion and then averaged over each treatment group. All sections within the lesion area were scored (Vehicle, 10 sections; Tamoxifen, 11 sections; Ifenprodil, 28 sections). A Mann-Whitney statistical analysis was performed to assess statistical significance.
Analysis of mouse brain sterol levels:
Ten to twelve week old male C57BL/6 mice were injected with 2mg/kg tamoxifen, 10 mg/kg ifenprodil, or 10 mg/kg miconazole dissolved in corn oil (tamoxifen) or DMSO (ifenprodil, miconazole) in sterile saline daily for three days. Mice were anaesthetized with isoflurane and perfused with phosphate buffered saline to remove blood from the brain. Brains were collected and flash frozen using liquid nitrogen. The samples were pulverized and 50–100 milligrams of tissue were collected for further processing. A modified Folch protocol was used for extraction of sterols.36 Briefly, samples were resuspended in a 2:1 chloroform/methanol mixture and homogenized. Cell debris was removed by centrifugation at 4000 g for 10 min. The solution was dried under air and resuspended in hexane with a cholesterol-D7 standard and dried again. Lipids were derivatized with 70 μl of bis(trimethylsilyl)trifluoroacetamide; 2 μls were injected and analyzed by GC/MS as described above.
Estrogen-dependent cell proliferation assay:
Estrogen-dependent cell proliferation was measured as previously described with minor modifications 37. After growth in estrogen-free media (Phenol red-free RPMI supplemented with 10% charcoal stripped fetal bovine serum) for 5 days, cells were seeded at 2,500 cells/well into 96 well plates. The following day 3x drug containing media was added to triplicate wells and cells were allowed to grow for an additional 5 days at 37 ˚C in standard a 5% CO2 humidified incubator. Total DNA per well was measured using an adaptation of the method of Labarca and Paigen 38. At this time media was removed, cells were washed one time with 0.25x PBS and 100 μl of distilled water was added. Plates were frozen and thawed to enhance cell lysis and 200 μl of 10 μg/ml Hoechst 33258 (Sigma-Aldrich, St. Louis, MO.) in 2M NaCl, 1mM EDTA, 10mM Tris-HCl pH 7.4 was added. After incubation at room temperature for 2 hours, plates were read in a SpectraMax i3 fluorescent plate reader (Molecular Devices, Sunnyvale, CA) with excitation at 360nm and emission at 460nm. All values were converted to microgram DNA per well using a standard curve derived from purified salmon testes DNA. T47D cells were provided by the Translational Research Shared Resource of the Case Comprehensive Cancer Center and used without further authentication beyond the observed estrogen-dependent cell proliferation.
Oligodendrocyte formation and imaging on electrospun microfibers:
A 12-well plate containing Mimetex aligned scaffold (microfiber plate, AMSBIO, AMS.TECL-006–1X, Electrospun poly-L-lactide Scaffold, 2 μM fibre diameter cell crown inserts) was prepared as previously described 24. Briefly, fiber inserts were sterilized with 70% ethanol and washed with PBS before being coated with polyornithine and laminin. After laminin coating, 100,000 cells/mL of EpiSC-derived OPCs (1.5 mL/well) were plated in differentiation medium. After 24 h the media was replaced with fresh media containing small molecule treatments. Every 48 h the media was replaced with fresh compound containing media for a total of 14 days. Plates were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and blocked with 10% donkey serum (v/v) in PBS for 60 min. Plates were stained for MBP (Abcam, ab7349; 1:100) and DAPI staining (Sigma; 5 μg/ml). After staining, the inserts were moved into new 12-well plate and covered with 2 mL of PBS before imaging in Operetta high content Imaging and analysis system. Plates were imaged on the Operetta High Content Imaging and Analysis system (PerkinElmer) and a set of 8 fields captured from each well resulting in an average of 45,000 cells being scored per well. Analysis (PerkinElmer Harmony and Columbus software) identified intact nuclei stained by DAPI and calculated the MBP signal intensity per cell per well. Microfiber insert tracking images were taken using a Leica DMi8 with a 20x Dry/NA 0.40 objective. Microfiber plate inserts were mounted using Flouromount-G (SouthernBiotech) and allowed to partially harden before coverslips were added and the insert ring was removed. Confocal images were obtained on a Leica SP8 confocal scanning microscope, with 40x oil/NA 1.30 objective. Confocal stacks of 0.336 μm z-steps were taken at 1024×1024. Each fluorophore was excited sequentially and all contrast and brightness changes were applied consistently between images.
A separate analysis approach was performed on an independent experiment performed as above except the small-molecule treatment was limited to the first 4 days of the 14 day culture period. After staining, the fiber inserts were mounted on a glass slide (Fisherbrand Superfrost Plus Microscope Slides) using Fluormount-G (Southern Biotech) with a cover glass (Fisherbrand Microscope Cover Glass) and dried at RT in dark for 36 h. The mounted inserts were imaged on the Operetta High Content Imaging and Analysis system (PerkinElmer) and a set of 22 fields captured from each condition resulting in an average of 2000 cells being scored per well. The total microfiber area was calculated using bright filed imaging and a spot-finding function (area larger than 2 px2). The MBP+ pixel area within the defined microfiber area was then defined and the percentage of the total microfiber area calculated.
CYP51 qPCR:
Cells were plated at 500,000 cells per well in a six-well plate and were grown in standard differentiation media supplemented with PDGF, neurotrophin 3, cAMP, IGF-1, and noggin for four days as described above. At 24 hours, cells were treated with 1 μM siRNA. Growth factors were added every 48 hours. After three days of siRNA treatment, RNA was isolated with the RNeasy Mini Kit (Qiagen), and cDNA was made using High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems). Exon spanning primers for ActinB (Thermo-Fisher, Taqman, Mm02619580_g1) and CYP51 (Thermo-Fisher, Taqman, Mm00490968_m1) were used for detection of relative RNA levels by quantitative real time PCR (Applied Biosystems, 7300 Realtime PCR system). Cycle time and outliers were calculated using Applied Biosystems’ 7300 System Sequence Detection Software version 1.4.
EBP qPCR:
OPCs were accutased and 1 million cells per cell line was spun down and RNA was isolated with the RNeasy Mini Kit (Qiagen), DNA was removed using the DNA-free RNA removal kit (Invitrogen), and cDNA was made using High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems). Primers for exon 5 of EBP (Forward primer: TGTGCGAGGAGGAAGAAGAT, Reverse Primer: GATAGGCCACCCCGTTTATT) and GAPDH (Forward primer: AGGTCGGTGTGAACGGATTTG; Reverse primer: GGGGTCGTTGATGGCAACA) were manufactured by IDT and gene expression was asses using Power SYBR® Green Master Mix (Applied Biosystems) were used for detection of relative RNA levels by quantitative real time PCR (Quantstudio 7 flex system). Cycle time and outliers were calculated using QuantStudio™ Software V1.3.
Muscarinic Receptor Antagonism Assay:
GeneBLAzer M1-NFAT-bla CHO-K1 cells (or M3- or M5-NFAT-bla CHO-K1 cells)(ThermoFisher) were thawed into Assay Media (DMEM, 10% dialyzed FBS, 25 mM HEPES pH 7.3, 0.1 mM NEAA). 10,000 cells/well were added to a 384-well TC treated assay plate and incubated 16–24 h at 37 ˚C. 4 μl of a 10x stock of antimuscarinic molecules was added to the plate and incubated 30 min. 4 μl of 10x control agonist Carbachol at the pre-determined EC80 concentration was added to wells containing antimuscarinic molecules. The plate was incubated 5 h and 8 μl of 1 μM Substrate + Solution D Loading Solution was added to each well and the plate was incubated 2 h at room temperature before reading on a fluorescence plate reader.
SREBP qPCR:
Cells were plated at 1 million cells per well in a six-well plate and were grown in standard differentiation media supplemented with with DMSO, Mevastatin (2.5 μM), Ro 48–8071 (500 nM), Ketoconazole (2.5 μM), Tasin-1 (100 nM), or Amorolfine (100 nM). At 24 hours, RNA was isolated with the RNeasy Mini Kit (Qiagen), and cDNA was made using High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems). Exon spanning primers ActinB (Thermo-Fisher, Taqman, Mm02619580_g1), LSS (Thermo-Fisher, Taqman, Mm00461312_m1), LDLR (Thermo-Fisher, Taqman, Mm01177349_m1), and DHCR7 (Thermo-Fisher, Taqman, Mm00514571_m1) were used for detection of relative RNA levels by quantitative real time PCR (Applied Biosystems, 7300 Realtime PCR system). Cycle time and outliers were calculated using Applied Biosystems’ 7300 System Sequence Detection Software version 1.4.
NR2C2 and NR2F1 Luciferase assays:
Forty-eight hours prior to transfection, 100,000 Hek293T cells were plated per well in a 24 well plate. HEK293T cells were chosen because they were used previously in this assay.39 NR2C2 (Origene, MR221079) or NR2F1 (gift from Dr. Christian Schaaf) and NGFI promoter reporter plasmid (generous gift of C. Schaaf) were transfected using Lipofectamine 2000 (Thermo-Fisher, 11668027) as per manufacturer’s protocol. After 16 hours, Hek293 cells were treated with the compounds (2,2-dimethyl-zymosterol 5 μM, FF-MAS 10 μM, Ketoconazole 2.5 μM, Tasin-1 100 nM, Mevastatin 2.5 μM, Liothyronine 3 μM, and all-trans retinoic acid 5 μM). 32 hours later cells were lysed using a firefly Luciferase assay system (Promega, E1500) and readout using Synergy Neo2 High Performance plate reader.
Nuclear receptor profiling:
Luciferase reporter assays performed by Indigo Biosciences were used to assess interaction of 2,2-dimethylzymosterol (5 μM), Ketoconazole (2.5 μM), and Tasin-1 (250 nM) with human ERα, GR, LXRβ, NFκB, NRF2, PGR, PPARα, PPARγ, RARα, RARγ, RXRα, RXRβ, TRα, TRβ and VDR in agonist mode and ERRα, RORα and RORγ in inverse-agonist mode. The reporter for these assays is firefly luciferase linked with either the genetic response elements (GRE) or the Gal4 upstream activation sequence (UAS). These cells also express either the native receptor or a receptor in which the native N-terminal DNA binding domain (DBD) has been replaced with that of the yeast Gal4 DBD. The specifics of each assay are shown in the table below. In brief, a suspension of Reporter Cells was prepared in Cell Recovery Medium (CRM; containing 5% (RORγ) or 10% charcoal stripped FBS for others). 100 μl of the Reporter Cell suspension was dispensed into wells of a white 96-well assay plate. Test compound, reference compounds, and the respective vehicle were diluted into INDIGO’s Compound Screening Medium (CSM; containing 5% (RORγ) or 10% charcoal stripped FBS for others). 100 μl of each treatment medium was dispensed into duplicate assay wells pre-dispensed with Reporter Cells. Assay plates were incubated at 37◦C for 24 hr. Following the incubation period, for agonist and inverse-agonist assays, treatment media were discarded and 100 μl/well of Luciferase Detection Reagent was added. RLUs were quantified from each assay well to determine agonist or inverse-agonist activity using the following assay designs:
ERα (NR3A1);Native Receptor;ER GRE-Luciferase
ERRα (NR3B1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
GR (NR3C1);Native Receptor;GR GRE-Luciferase
LXRβ (NR1H2);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
PGR (NR3C3);Native Receptor;PGR GRE-Luciferase
PPARδ (NR1C2);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
PPARγ (NR1C3);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RARα (NR1B1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RARγ (NR1B3);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RORα (NR1F1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RORγ (NR1F3);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RXRα (NR2B1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
RXRβ (NR2B2);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
TRα (NR1A1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
TRβ (NR1A2);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
VDR (NR1I1);Gal4 DBD hybrid Receptor;Gal4 UAS-Luciferase
NF-κB;Native NF-κB;NF-κB GRE-Luciferase
NRF2;Native Receptor;ARE-Luciferase
Animal Welfare:
All animal experiments were performed in accordance with protocols approved by the Case Western Reserve University and George Washington University Institutional Animal Care and Use Committees.
Extended Data
Extended Data Figure 1. Expanded cholesterol synthesis pathway diagram.
The cascade cyclization of squalene epoxide, catalyzed by lanosterol synthase (LSS), provides the first sterol, lanosterol. Processing of lanosterol to cholesterol can proceed via the Kandutsch-Russell and/or Bloch pathways, which use the same enzymes and process substrates that vary only in the presence or absence of the C24 double bond. Intermediates in blue have been confirmed in our GC/MS-based sterol profiling assay using authentic standards. Sterol 14-reductase activity in mouse is shared by two genes, TM7SF2 and LBR. Consistent with past reports (ref. 21), inhibition of sterol 14-reductase activity can lead to accumulation of the expected upstream intermediate (FF-MAS) or 14-dehydrozymostenol, also known as cholesta-8,14-dien-3-β-ol. Green indicates enzyme targets and small molecules whose inhibition promotes oligodendrocyte formation.
Extended Data Figure 2. CYP51 is the functional target by which imidazole antifungals enhance oligodendrocyte formation.
a) Azole molecules with varying degrees of potency for mammalian CYP51 inhibition. Throughout, green labels indicate molecules considered active, while red labels indicate inactive molecules. b) Percentage of MBP+ oligodendrocytes generated from a second, independent derivation of OPCs (OPC-1) at 72 h following treatment with the indicated concentrations of azoles. n = 4 wells per condition except DMSO (n = 24), with > 1,000 cells analyzed per well. c) GC/MS-based quantitation of lanosterol levels in a second derivation of OPCs (OPC-1) treated 24 h with the indicated azoles at 2.5 μM. n = 2 wells per condition. d, e) GC/MS-based quantitation of cholesterol levels in OPCs (OPC-5 and OPC-1) treated 24 h with the indicated azoles at 2.5 μM. n = 2 wells per condition. f, g) GC/MS-based quantitation of lanosterol levels in OPCs (OPC-5, OPC-1) treated 24 h with the indicated doses of ketoconazole. n = 2 wells per condition. Concentrations shown in panels f and g mirror those shown in Fig. 1c and panel b. h) Percentage of MBP+ oligodendrocytes generated from mouse primary OPCs at 72 h following treatment with the indicated imidazole antifungals at 3 μM. n = 4 wells per condition, with > 1,000 cells analyzed per well. i) GC/MS-based quantitation of lanosterol levels in mouse primary OPCs treated 24 h with the indicated imidazole antifungals at 3 μM. n = 2 wells per condition. j) Assessment of oligodendrocyte formation using an alternative image quantitation metric, fold increase in total neurite length. Panel is a re-analysis of data shown in Fig. 1c. n = 4 wells per condition except DMSO (n = 24), with > 1,000 cells analyzed per well. k) Percentage of oligodendrocytes generated from OPCs at 72 h following treatment with ketoconazole (2.5 μM) as measured by PLP1 immunostaining. Left, OPC-5; right, OPC-1. n = 8 wells per condition, with > 1,000 cells analyzed per well. l) LC/MS-based quantitation of lanosterol levels in OPC-5 treated 24 h with ketoconazole at 2.5 μM. n = 2 wells per condition. m) CYP51 mRNA levels measured by RT-qPCR following 96-h treatment with non-targeting or CYP51-targeting pools of cell-permeable siRNAs. n = 2 wells per condition. n) GC/MS-based quantitation of lanosterol levels in OPC-1 treated 96 h with the indicated pooled siRNA reagents. n = 2 wells per condition. o) Percentage of MBP+ oligodendrocytes generated from a second, independent batch of OPCs (OPC-1) at 72 h following treatment with the indicated reagents. n = 3 wells per condition, with > 1,000 cells analyzed per well. p) Percentage of MBP+ oligodendrocytes generated from an independent derivation of OPCs at 72 h following treatment with exogenous lanosterol. n = 4 wells per condition except DMSO and Ketoconazole (n = 8), with > 1,000 cells analyzed per well. q) Representative images of OPC-5 cells treated 72 h with the indicated siRNA reagents and lanosterol. Nuclei are labeled with DAPI (blue), and oligodendrocytes are indicated by immunostaining for myelin basic protein (green). Scale bar, 100 μm. All bar graphs indicate mean ± standard deviation, panels b, d, h, i, k, l, o and p are representative of two independent experiments, and all findings have been confirmed in a second independent derivation of OPCs (Fig. 1).
Extended Data Figure 3. Effect of small-molecule inhibition of the cholesterol biosynthesis pathway on enhancing oligodendrocyte formation.
a) GC/MS-based quantitation of sterol levels in OPCs (OPC-5) treated 24 h with the indicated inhibitors of cholesterol biosynthesis. Left, cholesterol; right, desmosterol. n = 2 wells per condition. Inhibitors were used at the following doses unless otherwise noted: mevastatin, ketoconazole, MGI-39, 2.5 μM; YM53601, 2 μM; Ro 48–8071, amorolfine, TASIN-1, 100 nM; AY9944, 200 nM. b) GC/MS-based quantitation of sterol levels in a second derivation of OPCs (OPC-1). Left, cholesterol; right, desmosterol. n = 2 wells per condition. c) GC/MS-based quantitation of the sterol intermediates expected to accumulate following treatment of OPCs with the indicated inhibitors of cholesterol biosynthesis for 24 h. n = 2 wells per condition. d) GC/MS-based quantitation of the sterol intermediates expected to accumulate following treatment of a second derivation of OPCs (OPC-1) with the indicated inhibitors of cholesterol biosynthesis for 24 h. n = 2 wells per condition. In c and d, no accumulation of other sterol intermediates indicative of off-target effects within the cholesterol pathway were observed (see Source Data). e) Representative images of OPC-5 cells treated 72 h with the indicated small molecules. All treatments are at the highest concentration shown in Fig. 2b. Scale bar, 100 μm. f) Percentage of MBP+ oligodendrocytes generated from a second batch of OPCs (OPC-1) at 72 h following treatment with the indicated cholesterol pathway inhibitors. n = 4 wells per condition, except DMSO, n = 24, with > 1,000 cells analyzed per well. g) Percentage of MBP+ oligodendrocytes generated from mouse primary OPCs at 72 h following treatment with the indicated cholesterol pathway inhibitors at 300 nM. n = 4 wells per condition, except DMSO, n = 8, with > 1,000 cells analyzed per well. h) GC/MS-based quantitation of sterol intermediate levels in mouse primary OPCs treated 24 h with the indicated inhibitors of cholesterol biosynthesis at 300 nM. Left, 14-dehydrozymostenol levels following treatment with amorolfine; Right, zymostenol levels following treatment with TASIN-1. n = 2 wells per condition. i, j) GC/MS-based quantitation of sterol intermediate levels in OPC-5 (i) and OPC-1 (j) treated 24 h with the indicated doses of inhibitors of cholesterol biosynthesis. Left, 14-dehydrozymostenol levels following treatment with amorolfine; Right, zymostenol levels following treatment with TASIN-1. n = 2 wells per condition. Concentrations shown in panel i mirror those shown in panel f. All bar graphs indicate mean +/− standard deviation, and panels a, c, e-h are representative of two independent experiments.
Extended Data Figure 4. Effect of independent chemical-genetic and genetic modulators of CYP51, sterol 14 reductase and EBP on oligodendrocyte formation and cholesterol biosynthesis.
a, d, g) Percentage of MBP+ oligodendrocytes generated from two independent derivation of OPCs at 72 h following treatment with the indicated concentrations of medroxyprogesterone acetate (a), 2-methyl ketoconazole (d) or TASIN-449 (g). n = 4 wells per condition, except DMSO, n = 12 in a and d. In g, for OPC-5, n = 4 except DMSO, n = 7; for OPC-1, n = 3 except DMSO, n = 6. b, e, h) GC/MS-based quantitation of sterol levels in two independent derivations of OPCs treated 24 h with medroxyprogesterone acetate at 10 μM (a), 2-methyl ketoconazole at 2.5 μM (e) and TASIN-449 at the indicated concentrations (h). n = 2 wells per condition. c, f) Rat CYP51 enzymatic activity following treatment with varying concentrations of medroxyprogesterone acetate (c) and 2-methyl ketoconazole (f) as measured by LC/MS-based quantitation of the CYP51 product FF-MAS. n = 2 independent enzymatic assays. i) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-5) infected with lentivirus expressing Cas9 and an independent guide RNA targeting EBP (cf. Fig. 2c). 8 wells per condition, with > 1,000 cells analyzed per well. Two-tailed Student’s t-test, * P = 0.0009. j) Functional validation of CRISPR-based targeting of EBP with a second sgRNA using GC/MS-based quantitation of zymostenol levels. n = 2 wells per condition. k) EBP mRNA levels measured by RT-qPCR in OPCs (OPC-5) infected with lentivirus expressing Cas9 and either of two guide RNAs targeting EBP. One well per condition, with results validated in an independent experiment. l) Representative images of the oligodendrocyte formation assay shown in Main Figure 2c. Nuclei are labeled with DAPI (blue), and oligodendrocytes are indicated by immunostaining for myelin basic protein (green). Scale bar, 100 μm. All bar graphs indicate mean +/− standard deviation, and panels a, d, g, i and k are representative of two independent experiments.
Extended Data Figure 5. Effect of 8,9-unsaturated sterols on oligodendrocyte formation.
a) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-5) at 72 h following treatment with methyl β-cyclodextrin (1 mM) for 30 min at 37 °C. n = 8 wells per condition, with > 1,000 cells analyzed per well. b) GC/MS-based quantitation of cholesterol (left) and desmosterol (right) levels in OPCs (OPC-5) treated with methyl β-cyclodextrin (Me-β-CD) at 1 mM or ketoconazole at 2.5 μM. n = 2 wells per condition. c, d) Percentage of MBP+ oligodendrocytes generated from OPC-1 (c) and OPC-5 cells (d) at 72 h following treatment with the indicated purified sterol intermediates. n = 4 wells per condition, except n = 8 for DMSO and ketoconazole, with > 1,000 cells analyzed per well. Green text highlights metabolites that accumulate after treatments that enhance oligodendrocyte formation (Fig. 2e, Extended Data Fig. 3c). e) Percentage of MBP+ oligodendrocytes generated from OPC1 at 72 h following treatment with MAS-412 and MAS-414. n = 4 wells per condition, with > 1,000 cells analyzed per well. f) Representative images of OPC5 cells treated 72 h with DMSO, MAS-412, or MAS-414 (3 μM). Nuclei are labeled with DAPI (blue), and oligodendrocytes are indicated by immunostaining for myelin basic protein (green). Scale bar, 100 μm. g) Percentage of MBP+ oligodendrocytes generated from OPC-1 at 72 h following treatment with 2,2-dimethyl-zymosterol. n = 4 wells per condition except DMSO (n = 12), with > 1,000 cells analyzed per well. h) Representative images of OPC-5 cells treated 72 h with vehicle and 2,2-dimethyl-zymosterol (2.5 μM). Nuclei are labeled with DAPI (blue), and oligodendrocytes are indicated by immunostaining for myelin basic protein (green). Scale bar, 100 μm. i) Percentage of MBP+ oligodendrocytes generated from OPC-5 (left) and OPC-1 (right) at 72 h following treatment with FF-MAS or T-MAS. n = 4 wells per condition except DMSO and Ketoconazole (n = 8), with > 1,000 cells analyzed per well. j) Percentage of MBP+ oligodendrocytes generated from OPC-5 and OPC-1 OPCs at 72 h following treatment with the indicated concentrations of cholesterol. n = 8 wells per condition, with > 1,000 cells analyzed per well. k, l) Percentage of MBP+ oligodendrocytes generated from OPC-5 and OPC-1 at 72 h following treatment with the indicated concentrations of sterols that are structurally identical aside from the presence or absence of the 8,9 double bond (structures in panel o). n ≥ 3 wells per condition (see dot plots as replicate values vary by condition), with > 1,000 cells analyzed per well. m) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-5) at 72 h following treatment with the indicated small molecules or combinations of small molecules (ketoconazole, 2.5 μM; Ro 48–8071, 11 nM; liothyronine, 3 μM). n = 3 wells per condition, except DMSO n = 11, ketoconazole n = 13, liothyronine n = 8 & liothyronine + Ro 48–8071 n = 4, with > 1,000 cells analyzed per well. n) GC/MS-based quantitation of lanosterol levels in OPCs (OPC-5) treated 24 h with the indicated small molecules or combinations of small molecules at concentrations stated in m. n = 2 wells per condition. o) Structures of zymostenol, 8,9-dehydrocholesterol, 5α-cholestanol, and cholesterol. p) Total cell number as measured by counting of DAPI+ nuclei in the experiment presented in panel m. q, r) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC5 and OPC-1) at 72 h following treatment with the indicated small molecules or combinations of small molecules in two independent batches of OPCs (ketoconazole, 2.5 μM; MAS412, 5 μM). In q, n = 16 for DMSO, 8 for Ketoconazole, and 4 for remaining bars. In r, n = 8 wells per condition. s) Luciferase reporter assays were used to assess if 2,2-Dimethylzymosterol (5 μM), Ketoconazole (2.5 μM), and TASIN-1 (250 nM) modulate human ERα, GR, LXRβ, NFkB, NRF2, PGR, PPARδ, PPARγ, RARα, RARγ, RXRα, RXRβ, TRα, TRβ and VDR transcriptional activity in agonist mode and ERRα, RORα and RORγ in inverse-agonist mode. n = 2 wells per condition and n = 3 wells per positive control conditions. t) Effects of sterols (2,2-dimethylzymosterol 5 μM, FF-MAS 10 μM) and small molecules (Ketoconazole 2.5 μM, TASIN-1 100 nM) on the NR2F1-mediated activation of a NGFI-A promoter driven luciferase reporter. n = 2 wells per condition. u) Effects of 2,2-dimethylzymosterol (5 μM) on NR2C2-mediated activation of a NGFI-A promoter driven luciferase reporter in comparison to cells transfected with reporter only, untreated, or treated with a previously reported positive control (all-trans retinoic acid, ATRA, 5 μM). n = 2 wells per condition. v) LSS, DHCR7, LDLR mRNA levels measured by RT-qPCR following 24 h treatment with DMSO, Mevastatin (2.5 μM), Ro 48–8071 (500 nM), Ketoconazole (2.5 μM), TASIN-1 (100 nM), or Amorolfine (100 nM). n = 2 wells. All bar graphs indicate mean +/− standard deviation, and panels a-n, and t-v are representative of two independent experiments.
Extended Data Figure 6. Inhibiting CYP51, TM7SF2 and EBP is a unifying mechanism for many small-molecule enhancers of oligodendrocyte formation identified by high-throughput screening.
a) Percentage of MBP+ oligodendrocytes (relative to DMSO control wells) generated from OPCs (OPC-1 derivation) at 72 h following treatment with a library of 3,000 bioactive small molecules, each at 2 μM. Each dot represents the result for one small molecule in the library. Red, imidazole antifungals; blue, clemastine; green, EPZ005687, the top novel hit molecule which is discussed in Extended Data Fig. 7 below. b, c) Percentage of MBP+ oligodendrocytes generated from OPCs (Left: OPC-5; Right: OPC-1) at 72 h following treatment with ketoconazole, nine top molecules identified by bioactives screening (green), and nine randomly chosen library members (red) at a uniform dose of 5 μM. n = 4 wells per condition except DMSO and Ketoconazole, n = 12 wells, with > 1,000 cells analyzed per well. d) GC/MS-based quantitation of zymosterol, zymostenol, and 14-dehydrozymostenol levels in a second batch of OPCs treated 24 h with the indicated screening hits and randomly chosen library members at 2 μM. n = 1; for validation in a second derivation of OPCs, see Fig. 3a. Molecules are clustered by enzyme targeted (top labels). e) Percentage of MBP+ oligodendrocytes generated from OPCs at 72 h following treatment with the indicated doses of fulvestrant, one of the top 10 HTS hits. n = 4 wells per condition except DMSO, n = 12), with > 1,000 cells analyzed per well. f) GC/MS-based quantitation of lanosterol levels in OPCs treated 24 h with fulvestrant at 2 μM. n = 2 wells per condition. g, h, i) GC/MS-based quantitation of metabolite levels in OPCs treated 24 h with the indicated previously-reported enhancers of oligodendrocyte formation at the following doses: Benztropine, 2 μM; Clemastine, 1 μM; Tamoxifen, 100 nM; U50488, 5 μM; bexarotene, 1 μM; liothyronine, 3 μM. n = 2 wells per condition. j, k) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-5 left, OPC-1 right) at 72 h following treatment with the indicated previously-reported enhancers of oligodendrocyte formation. n = 4 wells per condition, except DMSO n = 20 for OPC-5 and n = 12 for OPC-1, with > 1,000 cells analyzed per well. All doses are μM. l) Representative images of OPCs treated 72 h with the indicated small molecules. All treatments in l are at the highest concentration shown in panel j. Scale bar, 100 μm. m) Structures of muscarinic receptor antagonists used in this study. n, q) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-5: top, OPC-1: bottom) at 72 h following treatment with ketoconazole or the indicated muscarinic receptor modulators at 2 μM, the concentration used during screening. n = 4 wells per condition except DMSO and Ketoconazole, n = 8, with > 1,000 cells analyzed per well. o) GC/MS-based quantitation of three metabolite levels in OPC-5 OPCs treated 24 h with U50488 (5 μM) or the indicated muscarinic receptor modulators (2 μM). Left, zymostenol; center, cholesterol; right, desmosterol. n = 2 wells per condition. p) Heatmap indicating inhibition of muscarinic receptor isoforms M1, M3, and M5 by the indicated small molecules (2 μM) assayed using GeneBLAzer NFAT-bla CHO-K1 cells. n = 2 wells per condition. r) GC/MS-based quantitation of three metabolite levels in OPC-1 OPCs treated 24 h with clemastine (1 μM) or the indicated muscarinic receptor modulators at 2 μM. n = 2 wells per condition. Left, zymostenol; center, zymosterol; right, cholesterol. p-Fluorohexahydro-sila-difenidol is abbreviated as Sigma H127. All bar graphs indicate mean +/− standard deviation, and panels b, c, e, i, j, k, n and q are representative of two independent experiments.
Extended Data Figure 7. Effect of selective estrogen receptor modulators and EZH2 inhibitors on cellular EBP function and oligodendrocyte formation.
a) Structures of selective estrogen receptor modulators used in this study. b) Effects of ospemifene and toremifene on the estrogen-dependent growth of T47D cells. n = 3 wells per condition. c, d) Percentage of MBP+ oligodendrocytes generated from two independent batches of OPCs at 72 h following treatment with ospemifene and toremifene. n = 4 wells per condition except DMSO and Ketoconazole, n = 8, with > 1,000 cells analyzed per well. e) Representative images of OPCs treated 72 h with the indicated small molecules. All molecules were treated at 300 nM. Scale bar, 100 μm. f, g) GC/MS-based quantitation of two metabolite levels in OPCs treated 24 h with ospemifene and toremifene at 300 nM. Left, zymostenol; right, cholesterol. n = 2 wells per condition. h) Percentage of MBP+ oligodendrocytes generated from two independent batches of OPCs at 72 h following treatment with tamoxifen and 4-hydroxytamoxifen. Left, OPC-5; right, OPC-1. n = 4 wells per condition, except DMSO, n = 6 for OPC-1 (right). i) Effects of tamoxifen and 4-hydroxytamoxifen on the estrogen-dependent growth of T47D cells. n = 3 wells per condition. j) GC/MS-based quantitation of zymostenol (left axis) and zymosterol levels (right axis) in OPC-5 and OPC-1 treated 24 h with tamoxifen and 4-hydroxytamoxifen at the indicated concentrations. n = 2 wells per condition. Keto = Ketoconazole. k) Percentage of MBP+ oligodendrocytes generated from OPCs at 72 h following treatment with the indicated structurally analogous EZH2 inhibitors. n = 4 wells per condition, except DMSO, n = 12, with > 1,000 cells analyzed per well. l) Percentage of MBP+ oligodendrocytes generated from a second batch of OPCs at 72 h following treatment with the indicated structurally analogous EZH2 inhibitors. n = 4 wells per condition, except DMSO, n = 12, with > 1,000 cells analyzed per well. m) Percentage of MBP+ oligodendrocytes generated from mouse primary OPCs at 72 h following treatment with EPZ005687. n = 4 wells per condition, except DMSO, n = 12, with > 1,000 cells analyzed per well. n) Structure of EPZ005687 and structurally analogous EZH2 inhibitors. o) Representative images of OPCs treated 72 h with the indicated EZH2 inhibitors. All treatments are at 2 μM. Scale bar, 100 μm. p) GC/MS-based quantitation of two sterol intermediates following treatment of OPCs with the indicated EZH2 inhibitors at 1 μM for 24 h. Left, zymostenol; right, zymosterol. n = 2 wells per condition. q) GC/MS-based quantitation of two sterol intermediates following treatment of a second derivation of OPCs with the indicated EZH2 inhibitors at 1 μM for 24 h. Left, zymostenol; right, zymosterol. n = 2 wells per condition. r) GC/MS-based quantitation of two sterol intermediates following treatment of mouse primary OPCs with EPZ005687 at 2 μM for 24 h. Left, zymostenol; right, zymosterol. n = 2 wells per condition. All bar graphs indicate mean +/− standard deviation, and panels c, d, h, k-o, and r are representative of two independent experiments.
Extended Data Figure 8. Effect of combinations of small-molecule treatments on oligodendrocyte formation, and ability of oligodendrocytes to track along and wrap electrospun microfibers after single small molecule treatments.
a, b) Percentage of MBP+ oligodendrocytes generated from OPCs (OPC-1: Left, OPC-5: Right) at 72 h following treatment with the indicated combinations of liothyronine and enhancers of oligodendrocyte formation. Unless noted, the following concentrations were used: ketoconazole, 2.5 μM; benztropine, 2 μM; clemastine 2 μM; tamoxifen 200 nM; liothyronine, 3 μM. n = 4 wells per treatment condition, with > 1,000 cells analyzed per well. Lio = liothyronine. c, d) Percentage of MBP+ oligodendrocytes generated from OPCs at 72 h following treatment with the indicated combinations of ketoconazole and enhancers of oligodendrocyte formation. n = 4 wells per treatment condition, with > 1,000 cells analyzed per well. Keto = ketoconazole.e) Representative images of OPCs treated 72 h with the indicated small molecules. Small-molecule concentrations are as in panel a. Scale bar, 100 μm. f) Fold-increase of MBP+ oligodendrocytes following plating of OPCs (OPC-5) onto microfibers and treatment for 14 days with the indicated pathway modulators. n = 2 wells per condition, except DMSO, n = 4. g) In an independent experiment, OPCs (OPC-5) were plated onto microfibers, treated with small molecules for 4 days, and fixed and stained after 14 days. The extent to which MBP+ oligodendrocytes tracked along the microfiber substrate was measured. n = 2 wells per condition. h) Total DAPI+ cell number in the experiment described in g. i) Representative images highlighting tracking along the microfiber substrate. Each image is a montage of 4 separate images within the same well. Green, MBP. Scale bar, 100 μm. j) High-resolution images of MBP+ oligodendrocytes tracking along microfibers. Green, MBP; Blue, DAPI. Ketoconazole, 2.5 μM. Scale bar, 50 μm. k) Confocal imaging of OPCs seeded onto aligned microfibers and treated 14 days with ketoconazole (2.5 μM). The plane of the cross-section is highlighted in yellow and the cross-section, in which green fluorescence appears to encircle several microfibers, is shown in the bottom panel. Green, MBP; Blue, DAPI. All bar graphs indicate mean +/− standard deviation, and panels a-d are representative of two independent experiments.
Extended Data Figure 9. Effect of oligodendrocyte-enhancing small molecules on sterol levels in human cells and human cortical spheroids.
a) Representative images of toluidine blue stained sections of LPC-lesioned dorsal spinal cord from mice treated 8 days with ifenprodil (10 mg per kg) or tamoxifen (2 mg per kg). Scale bar, 20 μm. b) GC/MS-based quantitation of three metabolite levels in human glioma cells (GBM528) treated 24 h with the indicated small molecules at the following concentrations: Tamoxifen, 100 nM; Clemastine, 2 μM; Ifenprodil, 2 μM; Ketoconazole, 2.5 μM; Amorolfine, 100 nM. Left, lanosterol; center, zymostenol; right, 14-dehydrozymostenol. n = 2 wells per condition. c) GC/MS-based quantitation of three metabolite levels in two independent batches of human cortical spheroids treated 24 h with the indicated small molecules at 2 μM. Left, lanosterol; center, zymostenol; right, zymosterol. n = 3 spheroids per condition; representative of two independent experiments.
Extended Data Figure 10. 27 small molecules and nine purified 8,9-unsaturated sterols shown here to enhance the formation of oligodendrocytes.
a) Schematic showing the proposed mechanism of action for enhancing oligodendrocyte formation by diverse small molecules which enhance oligodendrocyte formation. b) Molecules that enhance oligodendrocyte formation are grouped by enzyme inhibited (GCMS analysis in OPCs): CYP51, top; sterol 14-reductase, center; EBP, bottom. c) Purified 8,9-unsaturated sterols that enhance oligodendrocyte formation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant NS095280 (R.H.M., P.J.T), Conrad N. Hilton Foundation Pilot Innovator in MS Award (D.J.A.), Mallinckrodt Foundation Grant Award (D.J.A), philanthropic support from the Peterson, Fakhouri, Long, Goodman, Geller, Judge, and Weidenthal families, and unrestricted support from the CWRU School of Medicine. Additional support was provided by the Small-Molecule Drug Development, Proteomics, and Translational Research Shared Resources of the Case Comprehensive Cancer Center (P30 CA043703). We gratefully acknowledge use of the Leica SP8 confocal microscope in the Light Microscopy Imaging Facility at CWRU made available through the Office of Research Infrastructure (NIH-ORIP) Shared Instrumentation Grant (S10 OD016164). The authors thank M. Drumm, T. Miller, B. Karl, O. Iyoha-Bello, J. Pink., P. Conrad, R. Lee, X. Li, D. Schlatzer, K. Polak, Janssen Pharmaceuticals, CXR Biosciences, ThermoFisher, Avanti Polar Lipids, and the P. Scacheri Lab for technical assistance and discussion.
Footnotes
Competing Interests. The authors acknowledge competing interests. See online edition for details (currently in Methods).
Competing Interests: The authors declare the following competing interests: D.J.A, P.J.T, Z.H., D.A., M.S.E. and R.H.M. are inventors on patents and patent applications that relate to this work and have been licensed to Convelo Therapeutics, Inc., which seeks to develop remyelinating therapeutics. D.J.A. and P.J.T. hold equity in Convelo Therapeutics, Inc. and receive consulting income from Convelo Therapeutics, Inc. After resubmission of this work, D.C.F. became an employee of Convelo Therapeutics, Inc.
MAIN REFERENCES
- 1.Goldman SA, Nedergaard M & Windrem MS Glial progenitor cell-based treatment and modeling of neurological disease. Science 338, 491–495, doi: 10.1126/science.1218071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fancy SP et al. Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol 225, 18–23, doi: 10.1016/j.expneurol.2009.12.020 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Franklin RJ & Ffrench-Constant C Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci 9, 839–855, doi: 10.1038/nrn2480 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Najm FJ et al. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature 522, 216–220, doi: 10.1038/nature14335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshmukh VA et al. A regenerative approach to the treatment of multiple sclerosis. Nature 502, 327–332, doi: 10.1038/nature12647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mei F et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med 20, 954–960, doi: 10.1038/nm.3618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mei F et al. Identification of the Kappa-Opioid Receptor as a Therapeutic Target for Oligodendrocyte Remyelination. J Neurosci 36, 7925–7935, doi: 10.1523/JNEUROSCI.1493-16.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang JK et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat Neurosci 14, 45–53, doi: 10.1038/nn.2702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez GA et al. Tamoxifen accelerates the repair of demyelinated lesions in the central nervous system. Sci Rep 6, 31599, doi: 10.1038/srep31599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lariosa-Willingham KD et al. A high throughput drug screening assay to identify compounds that promote oligodendrocyte differentiation using acutely dissociated and purified oligodendrocyte precursor cells. BMC Res Notes 9, 419, doi: 10.1186/s13104-016-2220-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korade Z et al. The Effect of Small Molecules on Sterol Homeostasis: Measuring 7-Dehydrocholesterol in Dhcr7-Deficient Neuro2a Cells and Human Fibroblasts. J Med Chem 59, 1102–1115, doi: 10.1021/acs.jmedchem.5b01696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giera MM, C.; Bracher F Analysis and Experimental Inhibition of Distal Cholesterol Biosynthesis. Chromatographia 78, 343, doi: 10.1007/s10337-014-2796-4 (2015). [DOI] [Google Scholar]
- 13.Giera M, Plossl F & Bracher F Fast and easy in vitro screening assay for cholesterol biosynthesis inhibitors in the post-squalene pathway. Steroids 72, 633–642, doi: 10.1016/j.steroids.2007.04.005 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Mir F & Le Breton GC A novel nuclear signaling pathway for thromboxane A2 receptors in oligodendrocytes: evidence for signaling compartmentalization during differentiation. Mol Cell Biol 28, 6329–6341, doi: 10.1128/MCB.00482-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jachak GR et al. Silicon Incorporated Morpholine Antifungals: Design, Synthesis, and Biological Evaluation. ACS Med Chem Lett 6, 1111–1116, doi: 10.1021/acsmedchemlett.5b00245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L et al. Selective targeting of mutant adenomatous polyposis coli (APC) in colorectal cancer. Sci Transl Med 8, 361ra140, doi: 10.1126/scitranslmed.aaf8127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jef DeBrabander JWS, Wang Wentian, Nijhawan Deepak, Theodoropoulos Pano. Targeting Emopamil Binding Protein with Small Molecules that Induce an Abnormal Feedbak Response by Lowering Endogenous Cholesterol Biosynthesis. (2016).
- 18.Saher G et al. Therapy of Pelizaeus-Merzbacher disease in mice by feeding a cholesterol-enriched diet. Nat Med 18, 1130–1135, doi: 10.1038/nm.2833 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Byskov AG, Andersen CY & Leonardsen L Role of meiosis activating sterols, MAS, in induced oocyte maturation. Mol Cell Endocrinol 187, 189–196 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Grondahl C Oocyte maturation. Basic and clinical aspects of in vitro maturation (IVM) with special emphasis of the role of FF-MAS. Dan Med Bull 55, 1–16 (2008). [PubMed] [Google Scholar]
- 21.Canfran-Duque A et al. Atypical antipsychotics alter cholesterol and fatty acid metabolism in vitro. J Lipid Res 54, 310–324, doi: 10.1194/jlr.M026948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moebius FF et al. Pharmacological analysis of sterol delta8-delta7 isomerase proteins with [3H]ifenprodil. Mol Pharmacol 54, 591–598 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Gylling H et al. Tamoxifen and toremifene lower serum cholesterol by inhibition of delta 8-cholesterol conversion to lathosterol in women with breast cancer. J Clin Oncol 13, 2900–2905, doi: 10.1200/JCO.1995.13.12.2900 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Bechler ME, Byrne L & Ffrench-Constant C CNS Myelin Sheath Lengths Are an Intrinsic Property of Oligodendrocytes. Curr Biol 25, 2411–2416, doi: 10.1016/j.cub.2015.07.056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S et al. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat Methods 9, 917–922, doi: 10.1038/nmeth.2105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi S et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann Neurol 65, 304–315, doi: 10.1002/ana.21581 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–678, doi: 10.1038/nmeth.3415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miron VE et al. Statin therapy inhibits remyelination in the central nervous system. Am J Pathol 174, 1880–1890, doi: 10.2353/ajpath.2009.080947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klopfleisch S et al. Negative impact of statins on oligodendrocytes and myelin formation in vitro and in vivo. J Neurosci 28, 13609–13614, doi: 10.1523/JNEUROSCI.2765-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saher G et al. High cholesterol level is essential for myelin membrane growth. Nat Neurosci 8, 468–475, doi: 10.1038/nn1426 (2005). [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 31.Godefroi EF, Heeres J, Van Cutsem J & Janssen PA The preparation and antimycotic properties of derivatives of 1-phenethylimidazole. J Med Chem 12, 784–791 (1969). [DOI] [PubMed] [Google Scholar]
- 32.Giera M, Renard D, Plossl F & Bracher F Lathosterol side chain amides: a new class of human lathosterol oxidase inhibitors. Steroids 73, 299–308, doi: 10.1016/j.steroids.2007.10.015 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Najm FJ et al. Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nat Methods 8, 957–962, doi: 10.1038/nmeth.1712 [nmeth.1712 pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honda A et al. Highly sensitive quantification of key regulatory oxysterols in biological samples by LC-ESI-MS/MS. J Lipid Res 50, 350–357, doi: 10.1194/jlr.D800040-JLR200 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Warrilow AG, Parker JE, Kelly DE & Kelly SL Azole affinity of sterol 14alpha-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob Agents Chemother 57, 1352–1360, doi: 10.1128/AAC.02067-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folch J, Lees M & Sloane Stanley GH A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226, 497–509 (1957). [PubMed] [Google Scholar]
- 37.Pink JJ & Jordan VC Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res 56, 2321–2330 (1996). [PubMed] [Google Scholar]
- 38.Labarca C & Paigen K A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 102, 344–352 (1980). [DOI] [PubMed] [Google Scholar]
- 39.Bosch DG et al. NR2F1 mutations cause optic atrophy with intellectual disability. Am J Hum Genet 94, 303–309, doi: 10.1016/j.ajhg.2014.01.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper [and its supplementary information files] or are available from the corresponding author upon request. Source data for all GCMS-based sterol profiling experiments and animal experiments are provided with the paper.