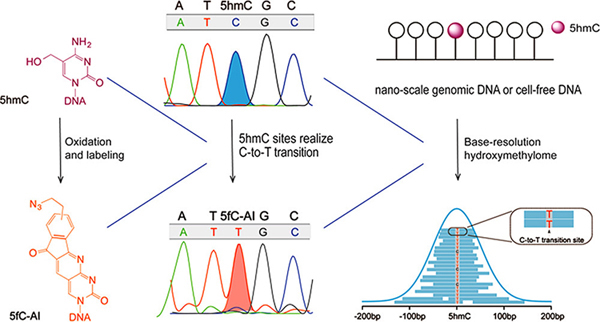
Abstract
High-resolution detection of genome-wide 5-hydroxymethylcytosine (5hmC) sites of small-scale samples remains challenging. Here, we present hmC-CATCH, a bisulfite-free, base-resolution method for the genome-wide detection of 5hmC. hmC-CATCH is based on selective 5hmC oxidation, chemical labeling and subsequent C-to-T transition during PCR. Requiring only nanoscale input genomic DNA samples, hmC-CATCH enabled us to detect genome-wide hydroxymethylome of human embryonic stem cells in a cost-effective manner. Further application of hmC-CATCH to cell-free DNA (cfDNA) of healthy donors and cancer patients revealed base-resolution hydroxymethylome in the human cfDNA for the first time. We anticipate that our chemical biology approach will find broad applications in hydroxymethylome analysis of limited biological and clinical samples.
Graphical Abstract
Active DNA demethylation in mammals is achieved via ten-eleven translocation (TET)-mediated oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), followed by thymine DNA glycosylase (TDG)-mediated excision of 5fC and 5caC.1–3 Emerging evidence reveals that 5hmC serves as not only an active DNA demethylation intermediate but also a stable DNA modification that plays distinct epigenetic roles and impacts a broad range of biological processes.4,5 Unlike 5mC, the total mass of 5hmC can vary significantly among different tissues.6,7 Furthermore, the reduction of 5hmC in DNA is a hallmark of cancer;8 recent studies also showed that 5hmC signature in cell-free DNA (cfDNA) could be a biomarker for human cancer.9–11
To detect genome-wide 5hmC distribution, different technologies have been developed.12,13 For instance, antibody-or biotin-based pull-down methods (namely 5hmC-DIP, hmC-Seal, CMS-seq and GLIB-seq) have been used to enrich 5hmC-containing genomic DNA for sequencing, although their resolution is limited. On the basis of the unique properties of endonucleases/exonucleases, several high-resolution 5hmC detection methods (namely Aba-seq, RRHP, Pvu-Seal-seq and SCL-exo) have also been reported. In particular, two methods, which are Tet-assisted bisulfite sequencing (TAB-seq) and oxidative bisulfite sequencing (oxBS-Seq), realize quantitative 5hmC detection at single-base resolution.14,15 However, the two approaches are bisulfite-dependent and hence cause significant DNA degradation, limiting their application in precious biological and clinical DNA samples. Moreover, they require a high sequencing depth for whole genome 5hmC detection and are thus very expensive.16 Previously, bisulfite-free chemistry for 5hmC has been demonstrated;17,18 however, their utility in detecting whole-genome hydroxymethylome has not been achieved.
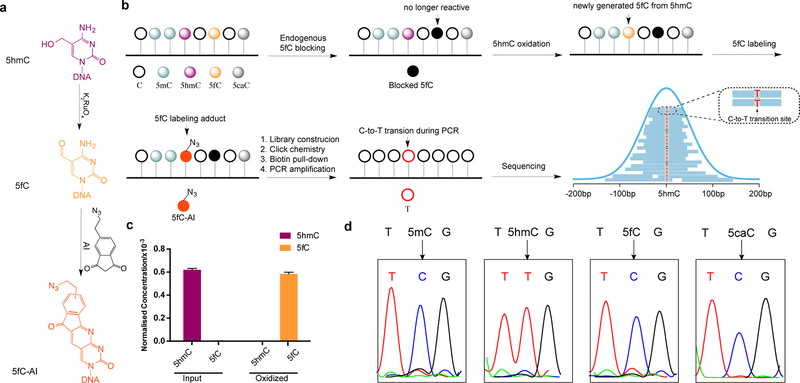
Here we present a bisulfite-free and cost-effective method that detects genome-wide 5hmC at single-base resolution. Our method is based on: (1) selective oxidation of 5hmC to 5fC (Figure 1a); (2) subsequent labeling of the newly generated 5fC (Figure 1a); and (3) 5fC labeling adduct caused C-to-T transition during DNA amplification. Because C, 5mC and 5caC remain inert during the chemical reactions and the endogenous 5fC is first blocked, our approach is specific for 5hmC (Figure 1b). We term such chemical-assisted C-to-T conversion of 5hmC sequencing “hmC-CATCH”.
Figure 1.
hmC-CATCH strategy and validation. (a) K2RuO4-mediated oxidation of 5hmC and the subsequent labeling reaction using an azido derivative of 1,3-indandione (“AI”). (b) Schematic diagram of hmC-CATCH. Endogenous 5fC is blocked with EtONH2. 5hmC is then oxidized to 5fC with K2RuO4. The newly generated 5fC can be specifically enriched and sequenced using the bisulfite-free, single-base 5fC method “fC-CET”.(c) Level of 5hmC and 5fC (normalized to C) in hESC genomic DNA before and after oxidation were quantified by LC-MS/MS. Data are represented as mean ± SEM, n=2. (d) hmC-CATCH of model DNA sequences with 5mC, 5hmC, 5fC or 5caC. Sanger sequencing results showed that 5hmC was completely converted to T, while 5mC, 5fC and 5caC were still read as C.
To achieve selective 5hmC oxidation, we utilized potassium ruthenate (K2RuO4), a ruthenium (+6) oxidant that is more oxidative than the reported oxidant potassium perruthenate (KRuO4, Ru7+). While KRuO4 caused severe DNA degradation15 (Figure S1a), K2RuO4-mediated oxidation is very mild but also complete (Figure S1b,c). We first tested K2RuO4-mediated oxidation on a 5hmC-containing, 9-mer synthetic model DNA, and found by MALDI-TOF that K2RuO4 is capable of efficiently converting 5hmC to 5fC (Figures S1d and S2). We then carried out the oxidation reaction on the genomic DNA of H1 human embryonic stem cells (hESCs), and demonstrated ~94% conversion rate of 5hmC to 5fC under the optimized conditions (Figure 1c). Importantly, 5mC content in the genomic DNA remains unchanged (Figure S3).
To ensure specific 5hmC detection, the endogenous 5fC in the genomic DNA was first blocked by O-ethylhydroxylamine (EtONH2),19 which renders it unreactive in the following reactions (Figure 1b and Figure S4). For the newly generated 5fC via 5hmC oxidation, we used an azido derivative of 1,3-indandione (or “AI”, which we reported previously in “fCCET”20) to achieve selective labeling. Because the adduct is read as a T instead of a C during PCR (Figure S5), such C-to-T transition is used as a direct readout of 5hmC. In addition, the azido group allows biotin conjugation and pull-down, thus enriching the 5hmC-containing DNA for detection (Figure 1b).
To test the specificity and efficiency of hmC-CATCH, we applied the method to multiple synthetic model DNAs containing either 5mC, 5hmC, 5fC or 5caC (Table S1). First, Sanger sequencing revealed that 5hmC is selectively converted to T, whereas 5mC, 5fC and 5caC stay as C (Figure 1d). Second, quantitative PCR analyses showed that only the 5hmC model DNA, but not the C or 5fC model sequences, was efficiently enriched (Figure S6).
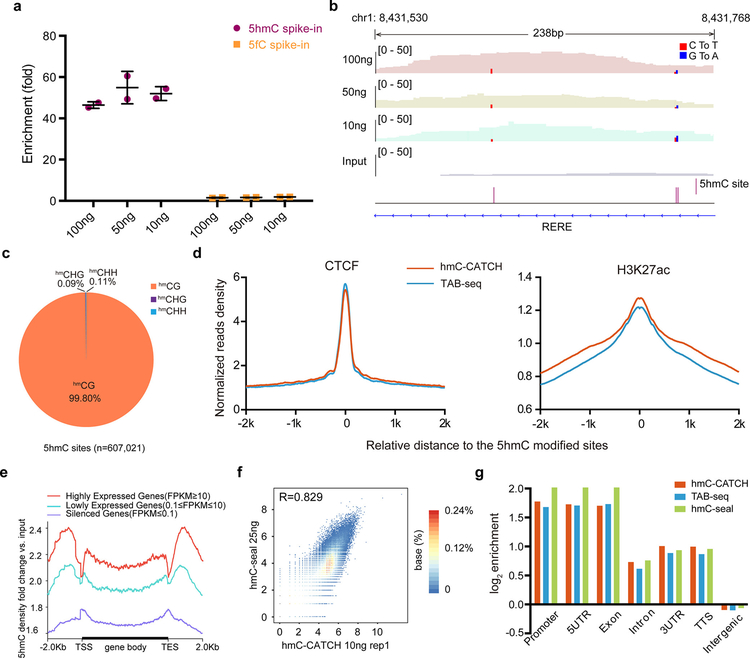
Because the chemistry of hmC-CATCH is very mild, we then applied it to the hydroxymethylome analysis using 100, 50 and 10 ng hESC genomic DNA, respectively (Table S2). For all samples, hmC-CATCH demonstrated efficient enrichment of 5hmC-containing spike-in DNAs during high-throughput sequencing (Figure 2a). Although slightly lower correlations were found for replicates using lower input material, the overall 5hmC profiles are highly reproducible between replicates (Figure S7a). At the single-base level, we observed high conversion rates for 5hmC (~80% and ~97% in the input and IP samples, respectively), contrasted to a low C-to-T rate for C (0.06%) and 5fC (3.3%) (Figure S8). A representative view of hmC-CATCH is shown in Figure 2b: within a 5hmC peak of ~200bp, three individual 5hmC sites (indicated by red and blue lines) were detected. In total, we detected 607, 021 5hmC sites in the 100 ng hESC samples using hmC-CATCH (Figure S9a,b). Among the 5hmC sites detected, 99.80% of them are in CG context and the remaining 0.20% in CH context (0.09% CHG and 0.11% CHH) (Figure 2c), which is comparable to the CH hydroxymethylation by TAB-seq (~0.11%). We found similar distribution of 5hmCG and 5hmCH sites in different genomic elements (Figure S9c), and 5hmC sites are enriched for H3K4me1, H3K27ac, open chromatin and binding regions of Pol II, CTCF and MYC (Figures 2d and S9d). We then examined the correlation between gene expression and 5hmC levels using hmC-CATCH results, and found an enrichment of 5hmC in gene bodies of more highly expressed genes (Figure 2e).
Figure 2.
Genome-wide, base-resolution maps of 5hmC in hESC genomic DNA. (a) The enrichment of spike-in probes by high throughput sequencing with nanoscale input materials (10, 50 and 100 ng). Data are represented as mean ± SEM, n=2. (b) Genome browser view of a representative 5hmC-enriched region, which contains three 5hmC sites (identified by C-to-T and G-to-A [for opposite strand] transition and indicated by red and blue bars). The entire view is approximately the length of a 5hmC peak. Results are shown from libraries generated with 10, 50 and 100 ng of starting hESC genomic DNA. (c) Distribution of 5hmCG and 5hmCH sites in hESCs. (d) Normalized read densities of 5hmC for CTCF and H3K27ac enriched regions in hESCs. CTCF(GSM822297) and H3K27ac(GSM466732) ChIP-Seq data are produced by the ENCODE Project Consortium. (e) Metagene profiles of fold change of 5hmC density in output versus input DNA. Genes are ranked according to their expression level in hESC RNA-Seq. (f) Scatterplots show high correlation between hmC-CATCH (using 10 ng genomic DNA, replicate 1) and hmC-Seal (using 25 ng genomic DNA), with Pearson correlation (r) displayed. Each dot represents a 5hmC-enriched peak and the number of points plotted is 82,893. The read counts were transformed to log2 base. (g) The relative enrichment of 5hmC sites detected by hmC-CATCH, TAB-seq and hmC-seal in different genomic elements.
Because hmC-CATCH simultaneously enriches 5hmC-containing DNA and preserves the base-resolution hydroxymethylome information, we compared the 5hmC profiles detected by hmC-CATCH with those by hmC-Seal and TAB-seq14,21 hmC-Seal is an enrichment-based method that detects 5hmC within tens to a few hundred nucleotides; we found that 5hmC profiles detected by the two methods showed ~87.3% overlap and correlated very well with each other (Figure 2f and Figure S7b–d). TAB-seq detects 5hmC at base resolution; we showed that using ~1/10 of the input material (100 ng in hmC-CATCH vs 1ug in TAB-seq (a recommended input amount)) and ~1/5 of the sequencing depth, hmC-CATCH detected very similar pattern of hydroxymethylome in the hESC genome (Figure 2g and Table S3). To further explore the cost-effective feature of hmC-CATCH, we down-sampled our sequencing data and found that using ~60 M sequencing reads (equivalent to a sequencing cost of approximately 200 USD), we can still recover ~84% 5hmC peaks (Figure S10a). In addition, base-resolution 5hmC sites can be realized at reasonable sensitivity and specificity as well (Figure S10b,c). Due to the pre-enrichment feature, hmC-CATCH is not absolute quantitative as TAB-seq; interestingly, using the normalized reads number, we found a good correlation between hmC-CATCH data and the quantitative data of TAB-seq for the gene body hydroxymethylation level (Figure S11).
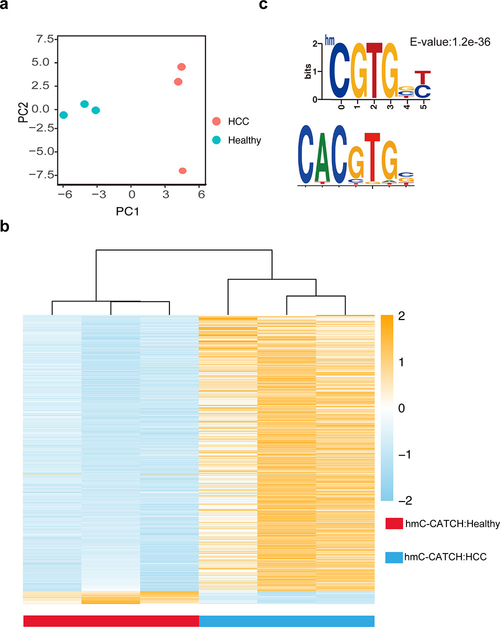
To further demonstrate the utility of hmC-CATCH in analyzing precious clinical samples, we next applied this approach to sequence the hydroxymethylome of cell-free DNA (cfDNA). Using cfDNA isolated from ~4 mL peripheral blood of three healthy individuals and three hepatocellular carcinoma (HCC) patients (Table S4 and S5), we identified 523,895–1,533,074 and 319,983–409,940 5hmC sites in healthy and cancer samples, respectively. We found very similar 5hmC enrichment profiles in cfDNA by hmC-CATCH and the previous data obtained from hmC-Seal9 (Figure S12a,b). Unbiased analysis using principal component analysis (PCA) revealed that the HCC patients can be readily separated from the healthy individuals, based on the hydroxymethylome obtained by hmC-CATCH (Figure 3a). Moreover, we identified 675 differential genes (Table S6), which could separate the HCC samples from the healthy samples (Figure 3b). These genes also showed differential hydroxymethylation in the previously reported cfDNA data by hmC-Seal9 (Figure S12c). Furthermore, using the single-base resolution 5hmC signals, we calculated the 5hmC motifs in cfDNA. We observed different sequence motifs in healthy and cancer samples (Figure S13); intriguingly, the top 5hmC motif specific for healthy individuals partially overlaps with the binding motif of the well-characterized oncogene MYC (Figure 3c). While further work is needed to determine if the observed hypohydroxymethylation at the MYC binding sites may contribute to carcinogenesis, our results demonstrate the robustness and sensitivity of hmC-CATCH in analyzing the base-resolution hydroxymethylome of limited samples.
Figure 3.
Genome-wide, base-resolution maps of 5hmC in human cell-free DNA. (a) PCA plot of normalized 5hmC reads number from healthy individuals and HCC (hepatocellular carcinoma) patients. (b) Heatmap of 675 differential genes detected by hmC-CATCH in healthy and HCC samples. Hierarchical clustering was performed across genes. (c) One representative promoter regions of 5hmC motif found only in healthy individuals (the upper panel) demonstrates partial sequence overlap with the known binding motif of MYC from the MotifMap Web site (the lower panel).
hmC-CATCH combines a known, selective 5hmC oxidation reaction15 with an established 5fC labeling and detection method,20 and hence is straightforward and enables convenient detection of hydroxymethylome at single base resolution. Because hmC-CATCH builds upon selective chemical reactions and is fully independent of enzymatic labeling and the usage of antibody, it avoids potential enzymatic preference and misinterpretation of genome-wide profiling data in the antibody-based techniques.22 More importantly, hmC-CATCH is bisulfite-free and causes no noticeable DNA degradation, thus allowing 5hmC detection in limited amount of biological and clinical samples. We applied this technique to generate 5hmC maps in nanoscale hESC genomic DNA and cfDNA, and showed that the results are in good agreement with those by previous methods. Additionally, it generated base-resolution hydroxymethylome at significantly reduced cost, and could be used to estimate 5hmC level as we did for the gene body hydroxymethylation. Such features may enable hmC-CATCH to have broad applications in obtaining refined 5hmC signatures for cancer biomarker identification.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank, Dr. Wen Zhou, Yifan Liu, Menghao Liu and Xiaoting Shu for technical assistance; Dr Dali Han and Xushen Xiong for bioinformatics assistance; and the Protein Core at the School of Life Sciences. Part of the analysis was performed on the High Performance Computing Platform of the Center for Life Science. This work was supported by the National Basic Research Foundation of China (nos. 2016YFC0900301 and 2014CB964900 to C.Y.), the National Natural Science Foundation of China (nos. 21522201 to C.Y.), SLS-Qidong Innovation Fund (to C.Y.) and National Institutes of Health (K01 HG006699 to Q.D. and R01 HG006827 to C.H.). C.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b08297.
Experimental details and supporting figures (PDF)
Model sequences and primers (XLSX)
Summary of hESC genomic DNA sequencing results (XLSX)
Clinical information for healthy and HCC samples (XLSX)
Summary of cfDNA sequencing results (XLSX)
Differential gene list (XLSX)
The authors declare the following competing financial interest(s): B.X., A.Z., C.Z. and C.Y. are co-inventors on filed patents (PCT/CN2014/087479 and 201710111600.9) for the labeling strategies of 5fC reported herein.
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number: GSE112048.
REFERENCES
- (1).Tahiliani M; Koh KP; Shen Y; Pastor WA; Bandukwala H; Brudno Y; Agarwal S; Iyer LM; Liu DR; Aravind L; Rao A Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324 (5929), 930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ito S; Shen L; Dai Q; Wu SC; Collins LB; Swenberg JA; He C; Zhang Y Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333 (6047), 1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).He YF; Li BZ; Li Z; Liu P; Wang Y; Tang Q; Ding J; Jia Y; Chen Z; Li L; Sun Y; Li X; Dai Q; Song CX; Zhang K; He C; Xu GL Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333 (6047), 1303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bachman M; Uribe-Lewis S; Yang X; Williams M; Murrell A; Balasubramanian S 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem 2014, 6 (12), 1049–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Shen L; Song CX; He C; Zhang Y Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem 2014, 83, 585–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kriaucionis S; Heintz N The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324 (5929), 929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Globisch D; Munzel M; Muller M; Michalakis S; Wagner M; Koch S; Bruckl T; Biel M; Carell T Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010, 5 (12), No e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Vasanthakumar A; Godley LA 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet 2015, 208(5), 167–77. [DOI] [PubMed] [Google Scholar]
- (9).Song CX; Yin S; Ma L; Wheeler A; Chen Y; Zhang Y; Liu B; Xiong J; Zhang W; Hu J; Zhou Z; Dong B; Tian Z; Jeffrey SS; Chua MS; So S; Li W; Wei Y; Diao J; Xie D; Quake SR 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 2017, 27(10), 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li W; Zhang X; Lu X; You L; Song Y; Luo Z; Zhang J; Nie J; Zheng W; Xu D; Wang Y; Dong Y; Yu S; Hong J; Shi J; Hao H; Luo F; Hua L; Wang P; Qian X; Yuan F; Wei L; Cui M; Zhang T; Liao Q; Dai M; Liu Z; Chen G; Meckel K; Adhikari S; Jia G; Bissonnette MB; Zhang X; Zhao Y; Zhang W; He C; Liu J 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 2017, 27 (10), 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tian X; Sun BF; Chen CY; Gao CC; Zhang J; Lu XY; Wang LC; Li XN; Xing YR; Liu RJ; Han X; Qi Z; Zhang XJ; He C; Han DL; Yang YG; Kan QC Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res 2018, 28 (5), 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Booth MJ; Raiber EA; Balasubramanian S Chemical methods for decoding cytosine modifications in DNA. Chem. Rev 2015, 115 (6), 2240–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Plongthongkum N; Diep DH; Zhang K Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet 2014, 15 (10), 647–61. [DOI] [PubMed] [Google Scholar]
- (14).Yu M; Hon GC; Szulwach KE; Song CX; Zhang L; Kim A; Li X; Dai Q; Shen Y; Park B; Min JH; Jin P; Ren B; He C Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012, 149 (6), 1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Booth MJ; Branco MR; Ficz G; Oxley D; Krueger F; Reik W; Balasubramanian S Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012, 336 (6083), 934–7. [DOI] [PubMed] [Google Scholar]
- (16).Rivera CM; Ren B Mapping human epigenomes. Cell 2013, 155 (1), 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hayashi G; Koyama K; Shiota H; Kamio A; Umeda T; Nagae G; Aburatani H; Okamoto A Base-Resolution Analysis of 5-Hydroxymethylcytosine by One-Pot Bisulfite-Free Chemical Conversion with Peroxotungstate. J. Am. Chem. Soc 2016, 138 (43), 14178–14181. [DOI] [PubMed] [Google Scholar]
- (18).Okamoto A; Sugizaki K; Nakamura A; Yanagisawa H; Ikeda S 5-Hydroxymethylcytosine-selective oxidation with peroxotungstate. Chem. Commun 2011, 47 (40), 11231–11233. [DOI] [PubMed] [Google Scholar]
- (19).Song CX; Szulwach KE; Dai Q; Fu Y; Mao SQ; Lin L; Street C; Li Y; Poidevin M; Wu H; Gao J; Liu P; Li L; Xu GL; Jin P; He C Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 2013, 153 (3), 678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xia B; Han D; Lu X; Sun Z; Zhou A; Yin Q; Zeng H; Liu M; Jiang X; Xie W; He C; Yi C Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Methods 2015, 12 (11), 1047–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Szulwach KE; Li XK; Li YJ; Song CX; Han JW; Kim S; Namburi S; Hermetz K; Kim JJ; Rudd MK; Yoon YS; Ren B; He C; Jin P Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells. PLoS Genet 2011, 7 (6), No e1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lentini A; Lagerwall C; Vikingsson S; Mjoseng HK; Douvlataniotis K; Vogt H; Green H; Meehan RR; Benson M; Nestor CE A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 2018, 15 (7), 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.