Abstract
The use of Seleno‐methionine (SeMet) incorporated protein crystals for single or multi‐wavelength anomalous diffraction (SAD or MAD) to facilitate phasing has become almost synonymous with modern X‐ray crystallography. The anomalous signals from SeMets can be used for phasing as well as sequence markers for subsequent model building. The production of large quantities of SeMet incorporated recombinant proteins is relatively straightforward when expressed in Escherichia coli. In contrast, production of SeMet substituted recombinant proteins expressed in the insect cells is not as robust due to the toxicity of SeMet in eukaryotic systems. Previous protocols for SeMet‐incorporation in the insect cells are laborious, and more suited for secreted proteins. In addition, these protocols have generally not addressed the SeMet toxicity issue, and typically result in low recovery of the labeled proteins. Here we report that SeMet toxicity can be circumvented by fully infecting insect cells with baculovirus. Quantitatively controlling infection levels using our Titer Estimation of Quality Control (TEQC) method allow for the incorporation of substantial amounts of SeMet, resulting in an efficient and optimal production of labeled recombinant protein complexes. With the method described here, we were able to consistently reach incorporation levels of about 75% and protein yield of 60–90% compared with native protein expression.
Keywords: Seleno‐methionine (SeMet), baculovirus expression vector system, Sf9 and Hi5 insect cells, TEQC method, baculovirus infectivity, estimated multiplicity of infectivity (eMOI), the Mediator Head module, SeM‐TEQC method
Abbreviations
- AAA
amino acid analysis
- BEVS
baculovirus expression vector system
- E. coli
Escherichia coli
- FBS
fetal bovine serum
- hCG
human choriogonadotropin
- Hi5
Trichoplusia ni
- Leu
l‐leucine
- Met
l‐methionine
- Met+
l‐methionine‐containing
- Met−
l‐methionine‐free
- MAD
multi‐wavelength anomalous diffraction
- MOI
multiplicity of infection
- eMOI
estimated multiplicity of infection
- Phe
l‐phenylalanine
- SAD
single‐wavelength anomalous diffraction
- Sf9
Spodoptera frugiperda
- Se
selenium
- SeMet
Seleno‐l‐methionine
- SeAM
Se‐adenosylselenomethionine
- SeAH
Se‐adenosylselenohomocysteine
- SeCys
Selenocysteine
- S. cerevisiae
Saccharomyces cerevisiae
- TEQC method
Titer Estimation for Quality Control method
- Val
l‐valine
Introduction
Single or multi‐wavelength anomalous diffraction (SAD or MAD) phasing by utilizing Seleno‐methionine (SeMet)‐labeled proteins or protein complexes is almost synonymous with modern X‐ray crystallography.1, 2 This method has drastically reduced the need for the preparation of heavy atom derivatives, which is one of the most time‐consuming steps in X‐ray crystallography. The anomalous signal from SeMet can be used for phasing as well as for identifying methionine positions within the electron density map, thereby aiding model building in low‐resolution maps.3, 4
The production of SeMet‐labeled proteins in E. coli for X‐ray crystallography is well established and robust.1 Protocols exist for yeast Saccharomyces cerevisiae,3 yeast Pichia pastoris,5 the insect cells6, 7 as well as mammalian cells.8 While SeMet incorporation routinely achieves 100% for proteins expressed in E. coli, expression in eukaryotic cells is much more variable, with SeMet incorporation ranging from 50% to 90%, depending on the host cells and proteins being produced.5, 7, 8, 9, 10
The baculovirus expression system (BEVS) is an excellent tool for expressing recombinant proteins in insect cells. It is considered an attractive option for proteins as well as protein complexes and has been widely used for structural and functional studies.11, 12, 13 For the production of SeMet‐labeled proteins using the BEVS in the insect cells, the protocol by Bellizzi et al. has been widely cited.6 However, as pointed out by Cronin et al.,7 this protocol is suited for secreted proteins such as glycosylated lysosomal hydrolase6 and CD40,14 and is not necessarily applicable for intracellular expression. Moreover, this protocol requires several medium‐exchanges and dialyzed FBS added to the methionine‐free medium in order to maintain cell viability during methionine depletion. Since higher labor demand (more personnel cost) as well as requirement of additional reagents such as FBS will add to the overall cost of operation, this protocol could potentially become costly. To overcome some of these limitations, a protocol designed for secreted as well as intracellularly expressed proteins was published.7 However, this protocol still retains the cumbersome medium exchange in the midst of the operation, and yields were drastically reduced: 18–45% compared with the native protein. Low yield of SeMet‐labeled proteins is a serious problem as it limits crystallization conditions for screening, optimization, and production of crystals, and low incorporation rates reduce the available anomalous signal. Moreover, additional preparations are required to achieve desired protein yield leading to increased costs as well.
In our work on the yeast Mediator Head module, which is an essential sub‐complex of the Mediator complex, and a key component of transcription regulation in eukaryotes,15 we encountered a similar problem. The Mediator Head Module is composed of seven subunits with a total molecular mass of 223 kDa. We generated the recombinant complex in the insect cells using the MultiBac baculovirus expression vector system for structural and functional studies.4, 16 We wanted to use the SeMet‐labeled Mediator Head module for phasing as well as for model building purposes. Using a protocol similar to that from Cronin et al.7 or the protocol provided by the manufacturer (Expression Systems, Inc), we achieved only 10% or less recovery of the Mediator Head module compared to the native expression level. A substantial reduction of the SeMet‐labeled protein yield could likely be attributed to SeMet toxicity. Although this toxicity is wildly recognized in the field, there are no detailed experimental data available as to toxic dosages of SeMet to the insect cells. Chen and Bahl reported that 100 μg/mL of SeMet was toxic but they did not present dose‐dependent toxicity data, or experimental details.17 Further, there has been no investigation undertaken of potential ways to bypass or reduce the detrimental effects of SeMet on the insect cells, as it had been published for yeast S. cerevisiae.10, 18
Considering the limitations and intricacy of the current SeMet‐labeling methods in the insect cells,6, 7, 17 we decided to analyze critical aspects of SeMet incorporation in the insect cells in order to improve quality and quantity of SeMet‐labeled proteins. We report here that SeMet is indeed toxic to the insect cells, but such toxicity can be circumvented by high baculovirus infection levels. We developed a simple, quantitative, and easy‐to‐use protocol for the generation of SeMet‐labeled proteins or protein complexes in the insect cells.
Results
Sf9 and Hi5 insect cells are resilient to methionine depletion
We started off investigating the effect of methionine depletion on cell viability in Sf9 and Hi5 cells. We wanted to test if supplementing dialyzed FBS for cell maintenance under met‐depleted conditions6, 17 is necessary. Both cell lines were maintained in serum‐free ESF921 medium (Expression Systems Inc.). Cells, Hi5 or Sf9, were spun down and resuspended in either the Met‐containing ESF921 medium (control) or Met‐free ESF921 medium in a 50 mL culture with a cell density of 0.5 × 106 (Hi5) or 0.6 × 106 (Sf9) cells/mL. Cell viability (% of live cells) and cell density of each culture were monitored every 24 h for a total of 4 days (Fig. 1). Although growth of Hi5 cells in the methionine‐free medium began to be compromised on Day 3, Sf9 cells remained the same as the control culture during the 4‐day incubation period. More importantly, there is essentially no difference in cell viability with or without methionine in both cell lines [Fig. 1(A), 1(C)], suggesting strongly that both insect cell lines are resilient to methionine depletion for an extended period of time: Sf9 for at least 96 h and Hi5 for 48 h. Therefore, we concluded that (i) the addition of dialyzed FBS during methionine depletion is unnecessary and (ii) methionine depletion can be sustained for at least 48 h for Hi5 cells and 96 h for Sf9 cells.
Figure 1.
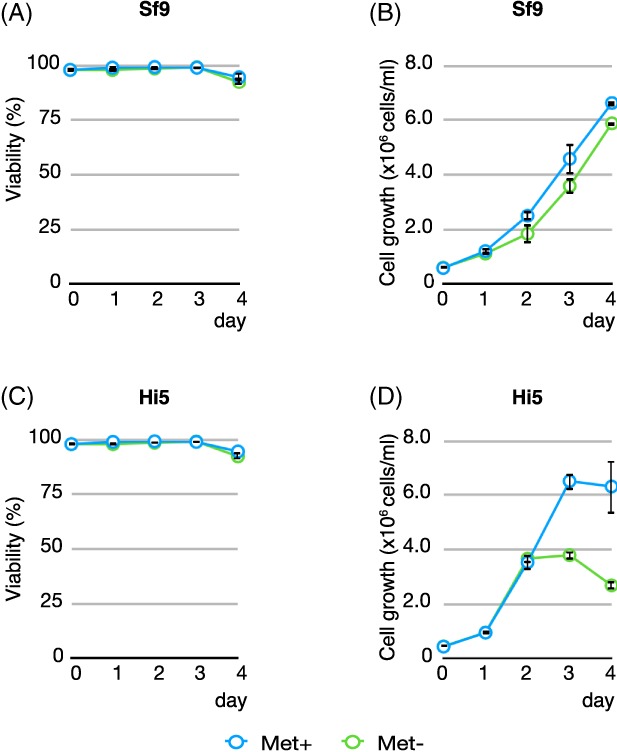
Effect of methionine depletion on Sf9 and Hi5 insect cells. Hi5 and Sf9 cells were grown in either Met‐free or Met‐containing medium as control. Cell viability (% of live cells) and cell density of each culture was measured every 24 h for a total of 4 days. (A) Cell viability of Sf9 cells with or without Met. (B) Cell density of Sf9 cells with or without Met. (C) Cell viability of Hi5 cells with or without Met. (D) Cell density of Hi5 cells with or without met. Measurements were performed in triplicates and averaged. Cyan: Met‐containing medium; green: Met‐free medium.
SeMet is toxic to the insect cells
Next, we examined SeMet toxicity in the insect cells in the presence or absence of Met by monitoring cell viability as well as overall cell growth. Sf9 or Hi5 cells were passaged with fresh ESF921 medium (Met+, control) or Met‐free ESF921 medium (Expression System, Inc.) to a cell density of 0.5 × 106 cells/mL in a 50 mL culture. Various amounts of SeMet, ranging from 0 mg/L to 200 mg/L, were added to both sets of cultures (Met+ or Met−). Cell viability (% of live cells) and cell density (total cell density) of each culture were monitored every 24 h for 4 days (Fig. 2). In the presence of Met, viability and growth of Sf9 cells were only slightly affected by SeMet at a concentration of 20–80 mg/L; at 120–200 mg/L, cell viability began to decrease [Fig. 2(A, B)]. Hi5 cells are more susceptible to the presence of SeMet, and their viability is dependent on the SeMet concentration in the medium [Fig. 2(E, F)]. In the Met‐free medium, the toxic effect of SeMet appears to manifest itself more prominently such that the cell viability drops to 40% in Sf9 cells [Fig. 2(C)] or 18% in Hi5 cells [Fig. 2(G)] at 80 mg/L SeMet. At 80 mg/L or higher, the cell viability of both cells declined to ∼20% [Fig. 2(C), 2(G)], and cell growth of both cell lines was severely compromised [Fig. 2(D), 2(H)]. These data clearly suggest that SeMet has detrimental effects on cell viability and cell growth per se, and impacts cell culture growth significantly at concentrations of 80 mg/L or higher in Met‐free medium. Based on the data presented here, the amounts of SeMet used in several publications6, 7, 14, 17, 19, 20 clearly fall in the toxic range.
Figure 2.
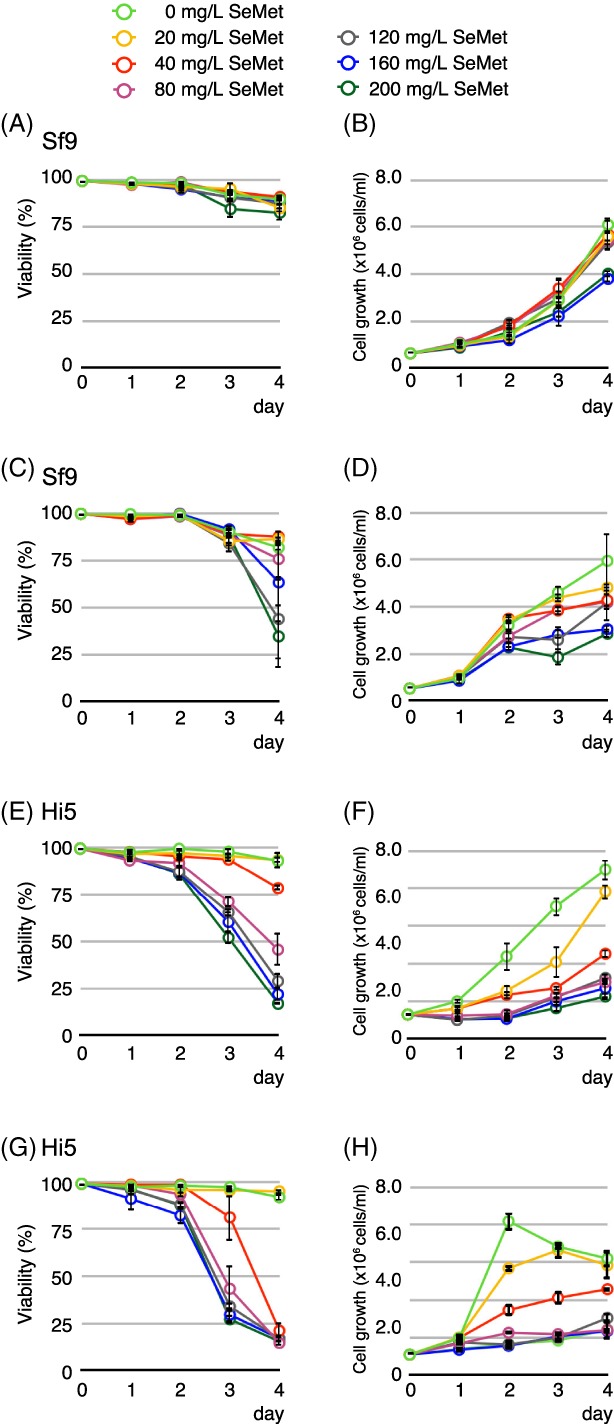
Effect of SeMet on Sf9 or Hi5 insect cells cultured in Met‐containing medium, or Met‐free medium. Hi5 or Sf9 cells were seeded in Met‐containing or Met‐free medium followed by the addition of SeMet to the final concentration of 0 (control), 20, 40, 80, 120, 160, and 200 mg/L. Cell viability (% of live cells) and cell density of each culture was measured every 24 h for a 4‐day span. (A) Cell viability of Sf9 cells in Met‐containing medium. (B) Cell growth of Sf9 cells in Met‐containing medium. (C) Cell viability of Sf9 cells in Met‐free medium. (D) Cell growth of Sf9 cells in Met‐free medium. (E) Cell viability of Hi5 cells in Met‐containing medium. (F) Cell growth of Hi5 cells in Met‐containing medium. (G) Cell viability of Hi5 cells in Met‐free medium. (H) Cell growth of Hi5 cells in Met‐free medium. Final concentration of SeMet: green: 0 mg/L; yellow: 20 mg/L; red: 40 mg/L; magenta: 80 mg/L; gray: 120 mg/L; blue: 160 mg/L; dark green: 200 mg/L.
SeMet toxicity can be circumvented by baculovirus infection
Having established that the insect cells are resilient to Met depletion for an extended period of time (Fig. 1) and that SeMet is indeed toxic to the insect cells at concentrations at 80 mg/L or higher under Met‐depleted conditions (Fig. 2), we investigated the relationship between SeMet incorporation and the yield of the SeMet‐labeled Mediator Head module. For this purpose, we set up cultures of Hi5 cells infected with the Mediator Head module virus at an estimated multiplicity of infection (eMOI) of 3.7, which gave consistently high yield in a native expression as we reported previously,21 and titrated increasing amounts of SeMet into the cultures (Fig. 3). In this experiment, Hi5 cells were chosen because the Mediator Head module expressed significantly better in Hi5 cells than Sf9 cells.21 The overall experimental scheme is described in [Fig. 3(A)]. For the ease of implementation, our labeling protocol was divided into 24 h (1 day) increments. On Day 0, Hi5 cells grown in log phase were spun down and resuspended in Met‐free medium. Cultures were incubated for 24 h to deplete Met, and after adjusting for cell density, the baculovirus expressing the Mediator Head module was added to each culture (Day 1) with an eMOI = 3.7.21 On Day 2, 0 (control)–200 mg/L SeMet was added to each culture and 100 mg/L Met was added to the native control culture. Cell density and viability were monitored every 24 h over a 4‐day period (Days 1–5). To our surprise, we saw no significant SeMet toxicity as assessed by cell density and viability over the measured time period [Fig. 3(B, C)]: Cell viability at 200 mg/L SeMet (52%) was similar to that of the native control with 100 mg/L Met (53%) added [Fig. 3(B)]. Our previous work showed that Hi5 cells were fully infected with an eMOI = 3.7.21 Thus, we hypothesize that high baculovirus infection levels could circumvent the SeMet toxicity in the insect cells.
Figure 3.
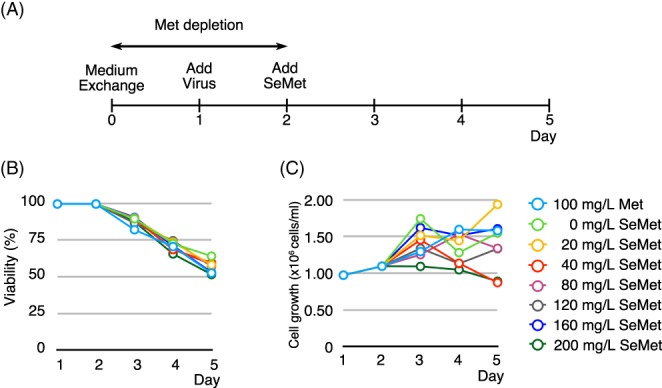
Effect of baculovirus infection on SeMet toxicity. (A) Overall experimental scheme. On Day 0, culture medium was exchanged from Met‐containing to Met‐free to deplete intracellular methionine. On Day 1, the baculovirus expressing the Mediator Head module with an eMOI = 3.7 was added to the culture. On Day 2, SeMet was added to each culture to the final concentration of 0 (control), 20, 40, 80, 120, 160, and 200 mg/L. As for the control, native expression was set up such that Met with a final concentration of 100 mg/L was added. Cell viability (% of live cells) (B) and cell density (C) of each culture was measured every 24 h for a 4‐day period (Days 1–5). Cyan: 100 mg/L of Met, for SeMet, green: 0 mg/L; yellow: 20 mg/L; red: 40 mg/L; magenta: 80 mg/L; gray: 120 mg/L; blue: 160 mg/L; dark green: 200 mg/L.
Cell viability and recovery of the SeMet‐labeled Mediator Head module in the presence of a toxic amount of SeMet depends on infectivity (or eMOI)
If a high baculovirus infection level could evade the SeMet toxicity, thereby maintaining cell viability, it should lead to higher recovery of SeMet‐labeled proteins or protein complexes. Therefore, we further hypothesized that optimal baculovirus infection is as critical for optimal SeMet‐labeled protein yield as it is for cell viability in terms of evading SeMet toxicity: optimal baculovirus infection leads to optimal SeMet‐labeled protein yield.
To test this hypothesis, we set up a virus titration such that the infectivity of the insect cells at the time of SeMet addition varied from low (22%) to high (∼100% infected cells) in order to see if there is a correlation among infectivity, cell viability, and SeMet‐labeled protein yield in the presence of a toxic amount of SeMet. We previously developed the Titer Estimation of Quality Control (TEQC) method, which enables us to quantitatively control virus infection levels.21 Using our TEQC method, the titer value of our virus stock was estimated and cultures with a total of five different estimated initial infectivities (infectivity 24 h after addition of virus: eI24), ranging from 22%, 39%, 63%, 86% to 98%, were set up. These initial infectivities correspond to estimates of multiplicity of infection (eMOIs) of 0.25 (22%), 0.5 (39%), 1.0 (63%), 2.0 (86%), and 4.0 (98%), respectively. It should be noted that the mathematical relationship between an initial infectivity (I24) and MOI is non‐linear: I24 = 1−e−MOI.21 The corresponding eMOI, instead of infectivities, were used for the experiments in Fig. 4, since this experimental condition is more convenient to set up. For instance, an eMOI = 2 implies twice as much virus volume was added than for eMOI = 1. As for controls, native expressions were set up in parallel using identical eMOI conditions. As shown previously, Hi5 cells are better suited for the expression of the Mediator Head module,21 and thus, Hi5 cells were used for this experiment. Following the same experimental scheme [Fig. 3(A)], after Met depletion and adjustment of cell density, the baculovirus was added to each culture with eMOI from 0.25 to 4.0 (Day 1). On Day 2 (24 h after virus addition), SeMet was added at a final concentration of 160 mg/L, which was shown to be highly toxic in the absence of infection [Fig. 2(G)], or Met was added at a final concentration of 100 mg/L as a native expression. Cell viability of each culture was monitored every 24 h for a 4‐day period (Days 1–5) [native: Fig. 4(A); SeMet: Fig. 4(B)]. On Day 5, cells were harvested, and the Mediator Head module was purified and quantified. Comparison in cell viability between a native vs. SeMet containing expressions on Day 5 is displayed in Fig. 4(C).
Figure 4.
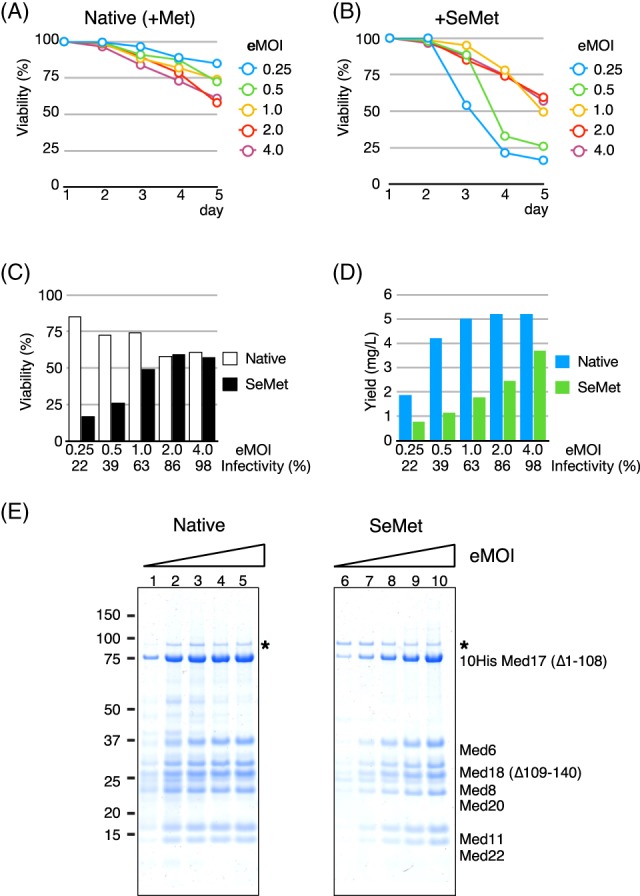
Cell viability and recovery of the SeMet‐labeled Mediator Head module in the presence of SeMet depends on infectivity (or MOI). (A) and (B) Cell viability as a function of estimated multiplicity of infection (eMOI), or initial infectivity. Following the experimental scheme described in Fig. 3(A), after Met depletion process, the recombinant baculovirus expressing the Mediator Head module at eMOI ranging from 0.25 (cyan), 0.5 (green), 1.0 (yellow), 2.0 (red), and 4.0 (purple) was added on Day 1 followed by addition of Met at a final concentration of 100 mg/L as native expression control, or SeMet at a final concentration of 160 mg/L on Day 2. Cell viability at each condition was monitored during a 4‐day period (Days 1–5), and cell viability (%) at each condition was plotted. (A) Native expression. (B) SeMet labeling expression. (C) Comparison of cell viability between native vs. SeMet containing expressions at Day 5 is displayed. White box: Native expression, black box: SeMet labeling expression. (D) Recovery of the Mediator Head module in native or SeMet‐labeling expression condition as a function of estimated multiplicity of infection (eMOI), or initial infectivity. Cells were harvested on Day 5. The Mediator Head module was affinity‐purified and quantified. The complex yield from each culture was measured and the data were plotted as a function of eMOI or initial infectivity. Yield was displayed as amount of the protein complex (mg) obtained from 1 L culture. Initial infectivities (%) correspond to eMOI were displayed below eMOI values. Cyan box: Native expression, green box: SeMet‐labeling expression. (E) SDS‐PAGE of the Mediator Head module obtained from the native expression on the left and SeMet‐labeling expression on the right. (*): Contaminant from the insect cells.
In the native expressions, cell viability went down to ∼60% at eMOI greater than 2.0, which corresponded to the infectivity >86% on Day 5, while at low eMOIs (0.25, 0.5, or 1.0) or infectivity = 22%, 39%, or 63%, cell viability was in a range of 72–85% [Fig. 4(A, C)]. In the presence of SeMet (160 mg/L), cell viability was severely compromised at low eMOIs (0.25, or 0.5) or infectivity <39% [Fig. 4(B, C)]. We speculate that this is due to a large percentage of cells being uninfected at the time of SeMet addition, and thus, far more prone to SeMet toxicity than infected cells. In contrast, at the higher eMOI range (2 or greater, or infectivity >86%), cell viability went down to 57% [Fig. 4(B, C)], which was almost the same cell viability of the culture infected with eMOI = 4.0 (infectivity 98% or higher) of the native expression (61%) [Fig. 4(C)], indicating that cell viability at higher eMOI was not significantly affected under SeMet‐labeling conditions: fully infected insect cells are far more resilient to SeMet toxicity than the uninfected.
Next, we looked into virus infectivity (or eMOI) and protein complex recovery. Protein yields were compared with those from native expression [Fig. 4(D)]. As for the native expression, protein complex yield appeared to reach a plateau at eMOI = 1.0 or higher [Fig. 4(D, E)]. In contrast, in the presence of SeMet, highly infected cells (eMOI = 4) produced more protein complex than lower eMOI [Fig. 4(D, E)]. For example, the condition of eMOI = 4 yielded 71% of the protein complex compared with that of the native expression, whereas only 27% of the native set up was recovered from cells infected with a low eMOI of 0.5. There is no significant difference in cell viability between cultures set up with eMOI =2.0, and 4.0. However, the protein complex yield recovered from the culture with eMOI = 4.0 was ∼34% better than that eMOI = 2.0.
Taken all together, the data strongly suggest that a recovery of SeMet‐labeled protein complex correlates with the infectivity (or eMOI) [Fig. 4(C, D)]: generating a condition of full infection (eMOI ≥4.0) at the time of SeMet addition should lead to optimal recovery of SeMet‐labeled protein complexes.
Optimal SeMet concentrations for SeMet labeling of the Mediator Head module expressed in Hi5 or Sf9 insect cells
After establishing the optimal conditions for evading SeMet toxicity without compromising recombinant protein recovery – adding the baculovirus with eMOI ≥4.0 – we investigated whether the higher tolerance for SeMet affects its incorporation into the recombinant protein complex. Ultimately, it is desirable to obtain high yield as well as a high SeMet incorporation. To study the effects of various SeMet concentrations on their incorporation into the Mediator Head module, Hi5 insect cells were cultured in Met‐free medium followed by addition of the baculovirus expressing the Mediator Head module with an eMOI of 4 in order to fully infect cells. SeMet was added in concentrations ranging from 20 to 200 mg/L. As a control, a native expression test was set up in parallel with Met added at a final concentration of 100 mg/L. For each condition, the protein complex was purified and the yield was determined. The percentage of SeMet incorporation was measured by amino acid analysis (AAA). Consistent with the previously reported results,7 high concentration of SeMet resulted in higher incorporation of SeMet [Fig. 5(A)]. The SeMet incorporation gradually increased and peaked at 160 mg/L with 75% incorporation [Fig. 5(A)]. Consistent with the data in Figure 4(C), the recovery of the protein complex was comparable with native levels up to a SeMet concentration of 160 mg/L [Fig. 5(B)]. The peak recovery of the complex at 160 mg/L SeMet is about 75% of that of native protein yield. No increase in incorporation level was observed at 200 mg/L SeMet [Fig. 5(A)], but Mediator Head Module recovery was severely compromised at this concentration [Fig. 5(B)]. We concluded that for Hi5 cells, the maximum concentration of SeMet is 160 mg/L without overly compromising the recovery of the protein complex and achieve 75% SeMet incorporation.
Figure 5.
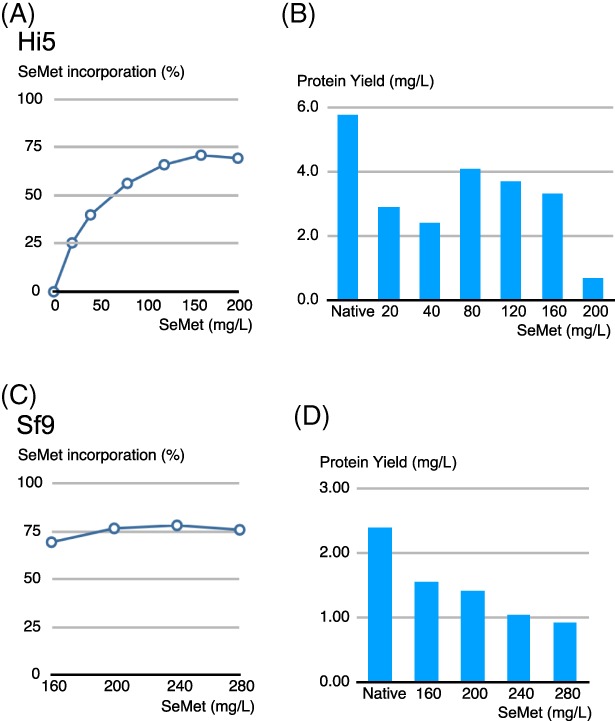
Effect of SeMet concentration on SeMet incorporation rate as well as on recombinant Mediator Head module recovery. Following the experimental scheme described in Fig. 3(A), Hi5 or Sf9 cells (200 mL in 1L shake flask) underwent the Met depletion process followed by the addition of the recombinant baculovirus expressing the Mediator Head module at eMOI of 4.0 on Day 1. On Day 2, SeMet was added to various final concentrations ranging from 20, 40, 80, 120, 160, and 200 mg/L. As for the control, a native expression was set up such that Met with a final concentration of 100 mg/L was added on Day 2. Cells were harvested on Day 5. The recombinant Mediator Head module was affinity‐purified21 and the protein complex yield at each condition was quantified. Plot of SeMet incorporation rate (%) into recombinant Mediator Head module expressed in Hi5 cells (A) or Sf9 cells (C) as a function of the concentration of SeMet added to the growth medium. Protein complex yield of the Mediator Head module expressed in Hi5 cells (B) or Sf9 cells (D) at each SeMet (or Met) concentration condition was plotted. Native: native expression where Met was added to a final concentration of 100 mg/L on Day 2; final concentrations of SeMet added on day 2 is indicated in the plot.
Next, we conducted the same experiment in order to find out the optimal SeMet concentration for expression of the Mediator Head module in Sf9 cells. Since Sf9 cells appear to accommodate more SeMet than Hi5 cells (Fig. 2),7 we started the SeMet titration from 160 mg/L all the way up to 280 mg/L [Fig. 5(C, D)]. In this range, 160–280 mg/L, SeMet incorporation reached 75% at 200 mg/L and there is no increase at higher concentration, suggesting that 75% is the upper limit for SeMet incorporation, consistent with the previous report.7 The recovery of SeMet‐labeled Mediator Head module decreased as the concentration of SeMet increases [Fig. 5(D)]. Considering SeMet incorporation rate as well as the complex recovery rate, we concluded that for Sf9 cells, the optimal concentration of SeMet is 200 mg/L without overly compromising the recovery of the protein complex and achieving 75% SeMet incorporation.
Application of our SeMet‐labeling method to multi‐protein complexes involved in RNA Polymerase II transcription
We further tested the generality of our labeling method by applying it to three other multi‐protein complexes involved in RNA Polymerase II transcription:22, 23 human Taf8–Taf10 heterodimer with a molecular mass of 81 kDa, yeast Mediator Middle module composed of seven subunits with a molecular mass of 190 kDa, and yeast TFIIF composed of three subunits with a molecular mass of 156 kDa, respectively. In our previous studies, the expression levels of human Taf8–Taf10, and yeast Mediator Middle module were fairly similar from low eMOI (0.5) to high eMOI (4.0) under native expression conditions. In fact, the optimal expression for the Mediator Middle module occurred with an eMOI = 0.5.21 Yeast TFIIF was chosen to test the SeMet labeling in Sf9 cells because it is one of only a few protein complexes we tested so far that expresses better in Sf9 cells than Hi5 cells.21 Following our protocol (supplemental protocol), Hi5 cells or Sf9 cells were cultured in Met‐free medium for 24 hours followed by addition of the baculoviruses expressing each of these complexes with eMOIs ranging from 0.5 to 4.0. SeMet was added to each culture at a final concentration of 160 mg/L SeMet for Hi5 cells, and 200 mg/L for Sf9 cells 24 hours after infection. In parallel, native expressions using the same eMOI were set up as controls. The protein complexes were purified, yields of protein complexes were determined, and the extents of SeMet incorporation were measured (Fig. 6).
Figure 6.
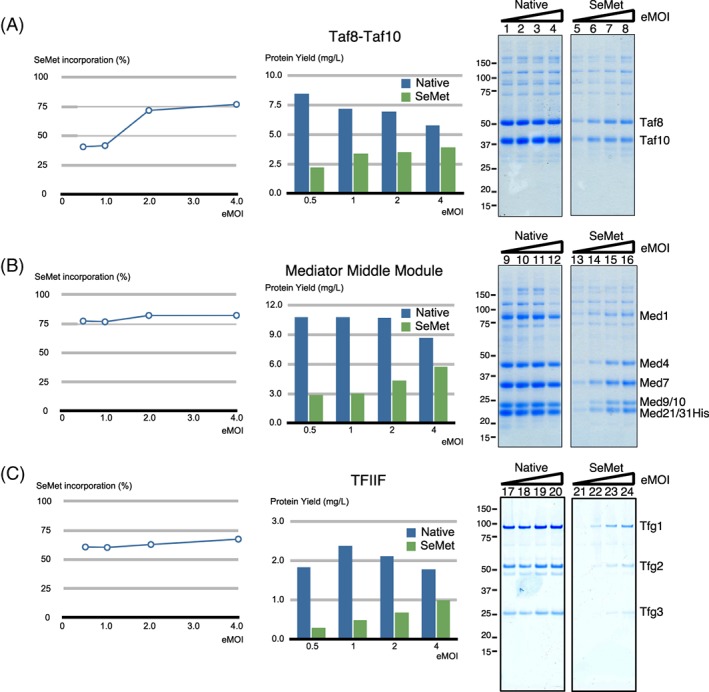
Application of the SeMet‐labeling method to human Taf8–Taf10, yeast Mediator Middle module, and yeast TFIIF complexes involved in RNA Polymerase II transcription. SeMet‐labeling method, “SeM‐TEQC,” used for the Mediator Head module was applied to human Taf8–Taf10, the yeast mediator middle module, and yeast TFIIF. Human Taf8–Taf10, and the yeast Mediator Middle module were expressed in Hi5 cells, and yeast TFIIF was expressed in Sf9 cells. Following the experimental scheme described in Fig. 3(A), Hi5 or Sf9 cells (200 mL in 1L shake flask) underwent Met depletion process. On Day 1, the recombinant baculovirus expressing human Taf8–Taf10, the yeast Mediator Middle module, or yeast TFIIF at an eMOI ranging from 0.5, 1.0, 2.0 and 4.0 was added followed by addition of Met at a final concentration of 100 mg/L as native expression control, or SeMet at a final concentration of 160 mg/L for Hi5 cells and 200 mg/L for Sf9 cells on Day 2. Cells were harvested on Day 5. For each condition, the three protein complexes were purified, the protein complex yields, as well as SeMet incorporation rates (%) were measured. Plot of SeMet incorporation rate (%) as a function of eMOI is displayed on the left; protein complex yields in native or SeMet‐labeling expression as a function of eMOI is displayed in the middle; and SDS‐PAGE of purified protein complex obtained from native or SeMet‐labeling expression are on the right. Lanes 1–4: eMOI = 0.5, 1.0, 2.0, 4.0; Lanes 5–8: eMOI = 0.5, 1.0, 2.0, 4.0. (A) Human Taf8–Taf10, (B) yeast mediator middle module, and (C) yeast TFIIF.
Consistent with our previous observation, the protein complex yields are fairly similar across different eMOI values for all three native complexes.21 Interestingly, the optimal eMOIs for all three complexes are at lower levels compared to the Mediator Head Module (Taf8–Taf10: 0.5; Mediator Middle module: 0.5; TFIIF: 1.0). However, in the presence of SeMet, the complex yields expressed at a low eMOI were substantially reduced and the highest recovery was achieved with an eMOI of 4.0 (> 98% infectivity), which is in line with our observations for the Mediator Head module. Compared with the native expression, yields of the complexes are ranging from 56% for TFIIF to almost 70% for Taf8–Taf10 and Mediator Middle module with an eMOI of 4.0 and SeMet incorporation of these three complexes reached 68–78%. Taken together, these results clearly indicate that our labeling method is applicable to multiple systems. Since the implementation of our TEQC protocol is the key for this SeMet‐labeling method, we name it SeM‐TEQC method.
Validation of our 24 h‐increment labeling protocol
One element of our experimental set up is a 24‐h infection period for the baculovirus prior to the addition of SeMet. If the Mediator Head module is expressed in this period, it would result in a production of non‐SeMet‐labeled protein complex, which may contribute to lowering the overall SeMet incorporation rate and may explain the maximal labeling of 75% we observed. Cronin et al.7 indicated that the addition of SeMet within the first 16 h following viral infection is critical for reducing a production of non‐SeMet‐labeled protein – a key point in their report. Therefore, we decided to examine how much of the protein complex is expressed in the first 24 h and thereafter. Following the experiment scheme illustrated in Fig. 3(A), we monitored expression of the Mediator Head module throughout the incubation period (Days 1–5) by western blotting using antibodies against 10His‐Med17 (Δ1‐108), Med18 (Δ109‐140), and Med11 subunits of the Mediator Head module. In this experiment, every 24 h, a small aliquot from the culture was taken at 24 h intervals, and expressions of the three representative subunits mentioned above were probed by western blotting. As for a control, we set up a native expression of the Mediator Head module to which Met was added on Day 2 instead of SeMet. In the first 24 h after addition of the virus, there was no detectable expression of any one of the three probed subunits in either the SeMet or native expression cultures (Fig. 7). These data argue against the idea of a substantial production of non‐SeMet‐labeled protein complex between Days 1 and 2, further substantiating our 24 h‐increment labeling protocol.
Figure 7.
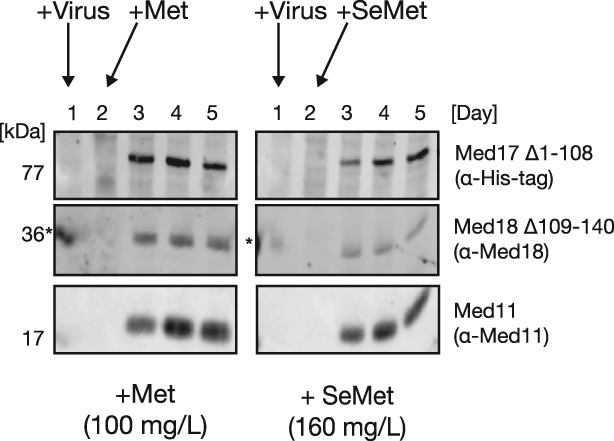
Expression profile of the Mediator Head module during SeMet labeling. Mediator Head Module production over the course of a standard SeMet‐labeling procedure [Fig. 3(A)] compared with native expression was monitored by Western Blot analysis. Hi5 cells were grown in a Met‐free medium for 24 h, followed by virus infection (Day 1). After a 24‐h incubation period, either Met or SeMet was added to a final concentration of 100 mg/L or 160 mg/L, respectively, and the cells cultured for another 72 h. Cell samples were collected every 24 h (Days 1–5), lysed, and subjected for SDS‐PAGE followed by Western blotting. Western Blot of three representative subunits, Med17, Med18, and Med11, of the Mediator Head Module probed with either anti‐His (Med17), anti‐Med18, and anti‐Med11, respectively. Time points of virus as well as Met or SeMet addition are indicated by arrows. Molecular mass (kDa) of each subunit is indicated on the left. (*) Signal bleed‐over from adjacent marker lane.
Use of SeMet‐labeled Mediator Head module for X‐ray crystallography
The SeMet‐labeled Mediator Head module was crystallized under similar conditions as the native Mediator Head module. The SeMet‐labeled crystals were isomophorous with native crystals with a similar size. SeMet‐containing crystals diffracted to 4.3 Å, and so did a representative native crystal. A scan of X‐ray‐induced fluorescence on the SeMet crystals demonstrated an absorption peak at 0.97948 A° (12,662 eV), consistent with the presence of SeMet. Diffraction data were collected at this wavelength, and processed with HKL200024 as described previously.4 The initial phases were determined by SIRAS using the Ta6Br14 derivative or SAD by using the K3Ir(NO3)6 derivative. The phasing results were quite similar for both derivatives. The initial phases were extended by density modification by program PARROT, using SeMet datasets descried previously.4 On the last step of the phasing, 98 SeMet peaks of a possible 141 sites (∼70%) in the unit cell were identified manually and were used to calculate the experimental phases by the program PHASER.25 The final experimentally phased map and the SeMet peaks were used for model building of the Mediator Head module as described previously.4
To evaluate anomalous differential Fourier map peak positions with SeMet positions, peaks were researched by CCP4 program FFT from anomalous difference Fourier peak height from more than 3.5σ to 8σ, and summarized in Table 1. A total of 97 SeMet peaks were higher than 4.0σ in 20–6 A°, suggesting high quality of anomalous signals from Selenium in the crystals. It should be noted that CCP4 program FFT26 identified 97 SeMet peaks within a unit cell while 98 were identified manually.
Table 1.
Peaks in 20–6 Å Anomalous
| Difference Fourier Map | ||
|---|---|---|
| σ Level | SeMet peaks | Noise peaks |
| <8 | 44 | 0 |
| <7 | 64 | 0 |
| <6 | 76 | 0 |
| <5 | 89 | 0 |
| <4 | 97 | 10 |
| <3.5 | 97 | 77 |
To further validate Selenium peaks, we compared the known positions of methionine residues in the high‐resolution structure of Med18–Med20, sub‐complex of the Mediator Head module previously determined by X‐ray analysis4 with the Selenium anomalous peaks by placing Med18–Med20 structure onto the 4.3 Å Mediator Head module map. Selenium anomalous difference Fourier map peak heights of Med18 and Med20 were summarized in Table 2. Peaks ranging from 3.0 σ to 9.6 σ were observed for 17 of possible 27 methionine sites for Med18, and five of possible nine methionine sites for Med20 were observed: 22 of a possible 36 sites (61%) (Table 2) – the methionine residues at N‐termini of both subunits were not included. Anomalous difference Fourier map peaks calculated from 20 to 6 Å at contoured at 3σ, nicely correlate with the location of known methionine sites in all three molecules found in the asymmetric unit [Fig. 8(A–C)].
Table 2.
Se Anomalous Difference Fourier Map Peak Height of Med18 (a) and Med20 (b)
| Met residue number | |||
|---|---|---|---|
| (a) Med18 | Mol1 | Mol2 | Mol3 |
| 1 | ND | ND | ND |
| 80 | ND | 7.0 | ND |
| 81 | 3.8 | 4.4 | 6.0 |
| 104 | ND | ND | ND |
| 150 | ND | ND | ND |
| 182 | 6.3 | 7.2 | 5.9 |
| 204 | 6.6 | 8.7 | ND |
| 224 | 6.6 | 8.7 | 6.9 |
| 276 | 4.9 | 4.7 | 3.9 |
| 296 | ND | 4.5 | 3.0 |
| (b) Med20 | |||
| 1 | ND | ND | ND |
| 57 | ND | 9.6 | 6.1 |
| 78 | ND | 6.8 | ND |
| 117 | ND | 4.8 | 3.5 |
Figure 8.
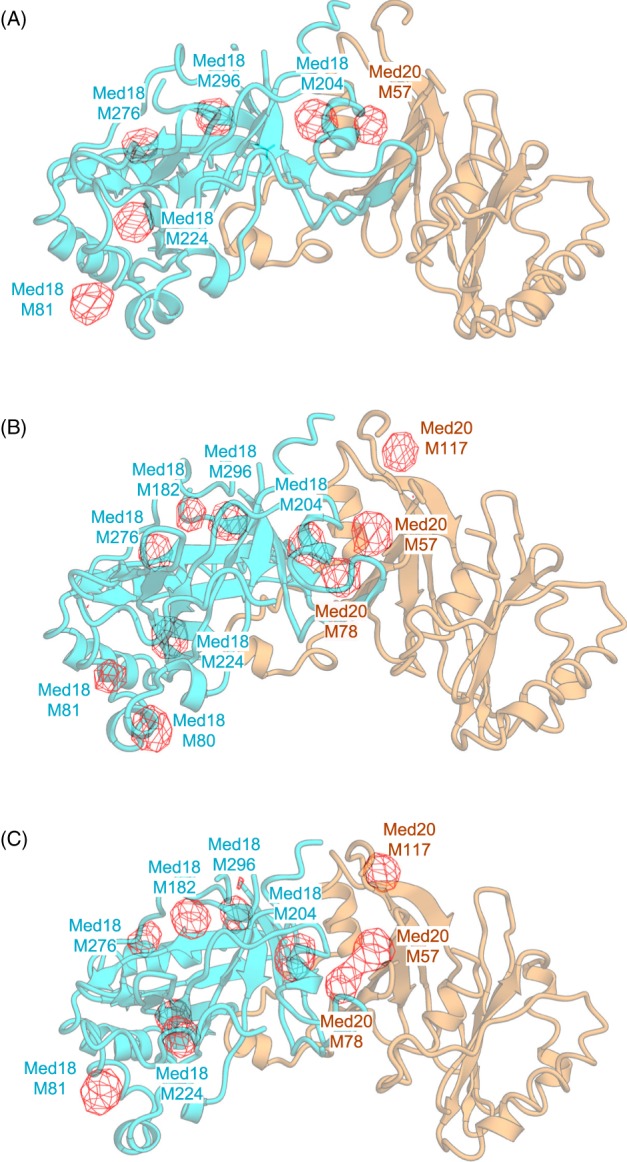
Comparison between Se anomalous peaks on the Med18–Med20 structure, and the known positions of methionine residues in the Med18–Med20 structure. Se Anomalous Difference Fourier Map was superimposed onto the cartoon view of the high‐resolution structure of the Med18–Med20 structure determined in the previous study.38 The selenium anomalous difference Fourier map calculated from 20 to 6 Å at contoured at 3 σ is shown in red and overlaid onto Med18–Med20. Three molecules in the asymmetric unit are displayed individually as (A)– (C). The height of the anomalous peaks is shown in Table 2. M: methionine residue (e.g., M57: methionine Residue 57).
Discussion
In this work, we developed a simple and robust protocol termed SeM‐TEQC method for productions of SeMet‐labeled protein complexes in the insect cells by addressing the issues related to (i) effect of Met depletion on the cells, (ii) timing of addition of SeMet, and most importantly, (iii) SeMet toxicity toward the insect cells. Our first finding was that the insect cells tolerate Met depletion for an extended period of time under otherwise normal growth conditions (Fig. 1), making medium exchanges with dialyzed FBS, or any other supplemental reagents during methionine starvation as reported previously6, 17 unnecessary. Second, based on our analysis, no detectable protein expression was observed 24 h following the virus infection (Fig. 7), eliminating the need for SeMet addition 16 h after viral infection7 and permitting a more feasible 24‐h schedule. Finally, and most importantly, we addressed the issue of the SeMet toxicity in the insect cells. This work outlines cell line‐dependent toxicity thresholds and establishes that the inherent SeMet toxicity can be evaded by optimizing baculovirus infection levels. Our previously developed TEQC method proved as an essential tool to limit SeMet toxicity as such it allows us to quantitatively control virus infection levels in a convenient and easy manner, and is crucial for optimal SeMet incorporation. In brief, to achieve full baculovirus infection, the eMOI needs to be 4.0 or higher, which corresponds to ∼98% infectivity.21 In retrospect, the condition used by Chen and Bahl corresponds to an MOI of 5–10 for SeMet‐labeling of human choriogonadotropin (hCG) in the insect cells, which turns out to be in the optimal range to evade SeMet toxicity, even though the authors failed to provide the reasoning to their MOI choice.17 Similarly, a high MOI was used in the works of Carfi et al.27 and Bellizzi et al.6 without any explanation as to why. On the contrary, Cronin et al. used a baculovirus MOI of 1 for infection, which is, according to our results, less optimal to evade SeMet toxicity and might explain the low recovery of SeMet‐labeled proteins they encountered.7
Possible mechanism of SeMet toxicity
Mechanism of SeMet toxicity toward eukaryotic cells has been studied mostly using the yeast Saccharomyces cerevisiae.10, 18 As these studies indicated, SeMet toxicity appears to come from two different mechanisms: SeMet likely generates reactive species, resulting in DNA damage,28 consistent with the observation that SeMet could effectively inhibit the insect cell growth.17 Alternatively, SeMet toxicity results from its metabolites, and a random incorporation of which promotes protein aggregation – SeMet causes proteotoxic stress resulting in cell death.29 How does baculovirus infection reduce SeMet toxicity? As reported, baculovirus infection results in cell cycle and DNA replication arrest.30, 31 Once infected, the insect cells are no longer able to divide. As a result, the effect of SeMet‐induced DNA damage could be neutralized, thereby maintaining overall cell viability as we observed (Figs. 3 and 4).
Considering the mechanism of SeMet toxicity being attributed to SeMet metabolites, we reasoned that SeMet toxicity could be minimized by reducing the amount of toxic SeMet metabolites (e.g., Selenocysteine) through impairing conversion of Se‐adenosylselenomethionine (SeAM) to Se‐adenosylselenohomocysteine (SeAH).32 In fact, this strategy worked well in the yeast S. cerevisiae by knocking out genes (SAM1, SAM2, or CYS3) encoding enzymes S‐adenosylmethionine synthetase, or Cystathionine gamma‐lyase in the pathway that produces toxic SeMet metabolites.10, 18 Accordingly, we attempted to apply a similar idea to disrupt conversion of SeMet to SeAM by the addition of SAM to the medium, which we hoped competes with SeAM as a substrate for methyltransferases, thereby minimizing the synthesis of SeMet metabolites. However, the addition of SAM to the growth medium did not reduce overall SeMet toxicity (data not shown). Thus, supplementing SAM to the insect cell culture may not effectively disrupt the metabolic pathway that produces SeMet metabolites as shown in yeast.10, 18 Thus, the main SeMet toxic effect may result from SeMet‐induced DNA damage and not via toxic SeMet metabolites. Alternately, it might be simply that a complete viral infection is forcing all excess SeMet toward incorporation into the recombinant protein, avoiding toxic cellular effects seen for uninfected cells. Regardless, deciphering the exact mechanisms is beyond the scope of this work and will have to be addressed at a later time.
Generality of the SeM‐TEQC method
The Mediator Head module was used as a model protein complex to develop the SeM‐TEQC method with subunits composition ranging from 15 kDa to 77 kDa. Moreover, we have shown applicability of our method by expanding our protocol to label human Taf8–Taf10 heterodimer with a molecular mass of 81 kDa, yeast Mediator Middle composed of seven subunits with a molecular mass of 190 kDa, and yeast TFIIF composed of three subunits with a molecular mass of 156 kDa. There was no significant difference among these complexes in terms of SeMet incorporation level, and protein yield compared to their corresponding native expressions, supporting the generality of our method. Having focused primarily on expression and SeMet incorporation of multi‐protein complex, application of our method on single‐subunits has been successfully achieved and led to the structure determination of the tight junction protein Claudin by SeMet SAD phasing,33 again proving the versatility of the SeM‐TEQC method.
As Cronin et al. pointed out,7 the advantage of the existing methods for secreted proteins, including engineered secrete expression,6, 17 lies in its ability to remove unlabeled proteins by medium exchange during the procedure, thereby increasing overall SeMet incorporation. For example, Chen and Bahl reported17 that SeMet incorporation rate of human choriogonadotropin (hCG) reached 84%, which is better than what our SeM‐TEQC method (∼75%) achieved. However, the report by Bellizzi et al.6 – this publication that has been widely cited for SeMet protocol for BEVS in the insect cells – indicated that their SeMet incorporation rate was 76%, which is comparable to that of the SeM‐TEQC method. However, our protocol was developed primarily for intracelluar multi‐protein complexes. It has not been applied to secreted proteins yet. It still remains to be seen how well SeM‐TEQC method works for those. Based on our current results, we would expect it to work equally good if not better than the existing methods.
Conclusions
Our discovery that SeMet toxicity can be circumvented by a high baculoviral infection led us to develop a simple and quantitative SeMet‐labeling method termed SeM‐TEQC. The overall scheme is illustrated in Fig. 9. This method does not require laborious procedures (medium exchange during the procedure) or additional reagents (e.g., dialyzed FBS). Thus, this method is more cost effective compared to the exiting protocols. The protocol steps are carried out in 24 h increments, making the procedure practical and easy to implement. Our SeM‐TEQC method enables an optimal production of SeMet‐labeled proteins or protein complexes with SeMet incorporation levels to about 75% and protein yield of 60–90% compared to the native protein expression.
Figure 9.
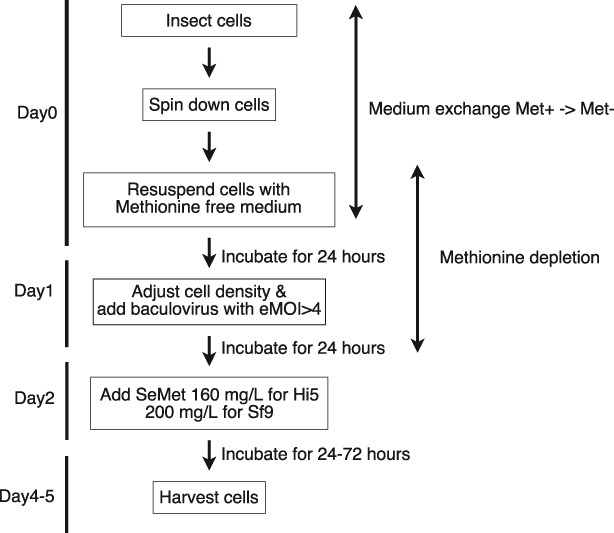
The overall experimental scheme for SeM‐TEQC method.
Materials and Methods
Maintenance of the insect cells, recombinant baculovirus production, virus storage, and virus titer estimation
Cells are maintained as described.21 The generation of recombinant baculoviruses was described.21, 34 The generation of frozen virus stocks was described.21, 35 The titer estimates of the recombinant baculoviruses (eTiters) were determined by TEQC method as described.21
Cell proliferation assay for methionine depleted conditions
Healthy dividing Hi5 or Sf9 insect cells were centrifuged in a 50 mL conical tube and methionine‐containing medium (ESF921) (Expression Systems, Davis, CA) was discarded. Cells were resuspended in Met‐free ESF921 medium (delta series, methionine deficient) (Expression Systems, Davis, CA), or as a control, methionine‐containing medium (ESF921) to set up a 50 mL culture with a cell density of 0.5 × 106 cells/mL for Hi5 cells or 0.6 × 106 cells/ml for Sf9 cells. Cell cultures were incubated on a shaker at 125 rpm at 27°C. Every 24 h for up to 96 h, the cell density as well as cell viability were measured with a TC20™ Automated Cell counter (Bio‐Rad).
Cell proliferation assay in presence of SeMet
Healthy dividing Hi5 or Sf9 insect cells were centrifuged in a 50 mL conical tube and methionine‐containing medium (ESF921) was discarded. Cells were resuspended in methionine‐free medium (ESF921), and as a control, methionine‐containing medium to set up a total of 7 flasks of 50 mL cultures with the cell density of 0.5 × 106 cells/ml for Hi5 cells or 0.6 × 106 cells/ml for Sf9 cells. Sets of cultures in methionine‐containing medium or in methionine‐depleted medium were set up in parallel. SeMet was added to each culture at the concentration indicated in the figure legends. Cell cultures were incubated at 27°C while shaking at 125 rpm. Every 24 h, the cell density and viability were measured with a TC20™ Automated Cell counter (Bio‐Rad).
Expressions of native and SeMet‐labeled protein complexes
Expressions of native multi‐protein complexes were performed in 200 mL cultures with 1.0 × 106 cells/ml of Hi5 cells or 1.5 × 106 cells/ml of Sf9. Cultures were infected with the recombinant baculoviruses at the indicated eMOI, which was calculated by our TEQC method.21 Each culture was incubated at 27°C for 96 h. The expression of SeMet‐labeled protein complexes is described in detail in the Supplemental protocol. Briefly, for medium exchange, Hi5 or Sf9 insect cells were centrifuged, methionine‐containing medium was discarded, and cells resuspended in 200 mL methionine‐depleted medium ESF921 to a cell density of 1.0 × 106 cells/mL for Hi5 cells or 1.5 × 106 cells/mL for Sf9 cells. Cell cultures were incubated on a shaker at 125 rpm for 24 h at 27°C in order to deplete endogenous methionine. After 24 h, the cell density was adjusted to 1.0 × 106 cells/ml in 200 mL for Hi5 cells or 1.5 × 106 cells/mL for Sf9 with methionine‐free medium. At this point, cells were infected with baculovirus at the various eMOI values indicated in the main text. After incubating for another 24 h at 27°C, SeMet was added to a final concentration of 20–200 mg/L and cells cultured for 72 h. Cells were harvested by centrifugation, cell pellets were frozen in liquid nitrogen, and stored at −80°C until use.
Purification of protein complexes
Purification procedures for the Mediator Head module, Taf8‐Taf10, TFIIF, and Mediator middle module were described previously.21
Determination of SeMet incorporation by amino acid analysis (AAA)
The incorporation of SeMet was determined by AAA carried out at The University of California Davis proteomics core facility. The details of AAA are described in (http://msf.ucdavis.edu/amino-acid-analysis/). Briefly, since Met is destroyed during hydrolysis with 6 N HCl, Met was determined by converting it to the acid stable form, methionine sulfone, with oxidation using performic acid, prior to the standard acid hydrolysis.36 Quantity of each amino acid was measured by AAA. Since valine (Val), leucine (Leu), and phenylalanine (Phe) are least affected by the oxidation process with performic acid, we used the average quantity of these three amino acids to compare the Met ratio in native and SeMet‐labeled proteins. The percentage of SeMet incorporation was calculated using the following equation:
SeMet incorporation rate = , where Met (n): quantity of Met under a native expression; Ave (n): average quantity of Val, Leu, and Phe combined under a native expression; Met (se): quantity of Met under SeMet‐labeling expression; Ave (se): average quantity of Val, Leu, and Phe combined under SeMet‐labeling expression. The detailed description of how this equation was derived is described in Supplemental materials. The ratio in quantity between Met and the average value of Val, Leu, and Phe under native as well as SeMet‐labeling expression condition was first calculated followed by determining the incorporation rate using the equation described above.
X‐ray crystallography for the SeMet‐labeled Mediator Head module
The SeMet‐labeled Mediator Head module crystals were obtained by the hanging‐drop vapor‐diffusion method as described previously.4 Diffraction data were collected at beamline 23ID at the Advanced Photon Source (APS) at Argonne National Laboratory. All diffraction data were processed with HKL2000. The structure was determined by SeMet single‐wavelength anomalous dispersion (SAD) after a sufficient number (98) of SeMet sites had been identified by a combination of the initial phases from Ta6Br14 and Iridium derivatives, and partial model SAD phases as described previously.4
Western blotting
Hi5 cells were infected with the recombinant baculovirus expressing the Mediator Head module with eMOIs = 4.0 after Met depletion. Twenty‐four hours after the addition of the virus, SeMet was added to the culture at a final concentration of 160 mg/L, and incubated for an additional 3 days. One milliliter of aliquot of cell culture was taken every 24 h, and cells were harvested in 1.5 mL tubes. Cell pellets were frozen in liquid nitrogen, and stored at −80°C until use. The preparation of cell lysate and the method for western blotting is described.21 The blot was probed for the Head module with anti–His tag monoclonal mouse antibody (Thermo Scientific Pierce) for 10×His‐tagged Med17, as well as with rabbit anti‐Med18 (anti‐Srb5) and rabbit anti‐Med11 antibodies.37 Detection was carried out with a ChemiDoc MP imaging system (BioRad) using Dylight 680 goat anti–rabbit IgG (Thermo Scientific Pierce) for Med18 and Med11, and Dylight 800 goat anti‐mouse IgG (Thermo Scientific Pierce) for anti‐His tag as secondary antibodies.
Supplemental material
Calculation of the SeMet incorporation rate using AAA data, and supplemental protocol for the SeM‐TEQC method.
Supporting information
Appendix S1: Supporting Information
Acknowledgments
We thank Dr T. Hurley for critical reading of the manuscript. We thank Dr John Schulze at The University of California Davis proteomics core facility for conducting amino acid analysis, and for providing the technical information. This research was supported by National Science Foundation grant MCB‐1157688, the National Institutes of Health (R01 GM111695), and Showalter Trust Fund to (Y.T.), Japan Science and Technology Agency (PRESTO, JPMJPR14L2) to (T.I.). X‐ray data were collected at the GM/CA‐CAT at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL. GM/CA‐CAT is funded by National Cancer Institute grant Y1‐CO‐1020 and National Institute of General Medical Sciences grant Y1‐GM‐1104. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract DE‐AC02‐06CH11357.
References
- 1. Hendrickson WA, Horton JR, LeMaster DM (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three‐dimensional structure. EMBO J 9:1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hendrickson WA, Ogata CM (1997) [28] phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol 276:494–523. [DOI] [PubMed] [Google Scholar]
- 3. Bushnell DA, Cramer P, Kornberg RD (2001) Selenomethionine incorporation in Saccharomyces cerevisiae RNA polymerase II. Structure 9:R11–R14. [DOI] [PubMed] [Google Scholar]
- 4. Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument‐Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y (2011) Architecture of the mediator head module. Nature 475:240–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kitajima T, Yagi E, Kubota T, Chiba Y, Nishikawa S, Jigami Y (2009) Use of novel selenomethionine‐resistant yeast to produce selenomethionyl protein suitable for structural analysis. FEMS Yeast Res 9:439–445. [DOI] [PubMed] [Google Scholar]
- 6. Bellizzi JJ, Widom J, Kemp CW, Clardy J (1999) Producing selenomethionine‐labeled proteins with a baculovirus expression vector system. Structure 7:R263–R267. [DOI] [PubMed] [Google Scholar]
- 7. Cronin CN, Lim KB, Rogers J (2007) Production of selenomethionyl‐derivatized proteins in baculovirus‐infected insect cells. Protein Sci 16:2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barton WA, Tzvetkova‐Robev D, Erdjument‐Bromage H, Tempst P, Nikolov DB (2006) Highly efficient selenomethionine labeling of recombinant proteins produced in mammalian cells. Protein Sci 15:2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carfi A, Gong H, Lou H, Willis SH, Cohen GH, Eisenberg RJ, Wiley DC (2002) Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Cryst D58:836–838. [DOI] [PubMed] [Google Scholar]
- 10. Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, Said M, Luft JR, Phizicky EM, DeTitta GT, Grayhack EJ (2007) Blocking S‐adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc Natl Acad Sci U S A 104:6678–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kost TA, Condreay JP, Jarvis DL (2005) Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol 23:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Massotte D (2003) G protein‐coupled receptor overexpression with the baculovirus‐insect cell system: a tool for structural and functional studies. Biochim Biophys Acta 1610:77–89. [DOI] [PubMed] [Google Scholar]
- 13. Assenberg R, Wan PT, Geisse S, Mayr LM (2013) Advances in recombinant protein expression for use in pharmaceutical research. Curr Opin Struct Biol 23:393–402. [DOI] [PubMed] [Google Scholar]
- 14. McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T (1999) Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci U S A 96:8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kornberg RD (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30:235–239. [DOI] [PubMed] [Google Scholar]
- 16. Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ (2010) Mediator head module structure and functional interactions. Nat Struct Mol Biol 17:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen WY, Bahl OP (1991) Selenomethionyl analog of recombinant human choriogonadotropin. J Biol Chem 266:9355–9358. [PubMed] [Google Scholar]
- 18. Bockhorn J, Balar B, He D, Seitomer E, Copeland PR, Kinzy TG (2008) Genome‐wide screen of Saccharomyces cerevisiae null allele strains identifies genes involved in selenomethionine resistance. Proc Natl Acad Sci U S A 105:17682–17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J (1998) Crystal structure of mouse H2‐M. Immunity 9:385–393. [DOI] [PubMed] [Google Scholar]
- 20. Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC (2002) Structure of the reovirus membrane‐penetration protein, Mu1, in a complex with is protector protein. Sigma3 Cell 108:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imasaki T, Wenzel S, Yamada K, Bryant ML, Takagi Y (2018) Titer estimation for quality control (TEQC) method: a practical approach for optimal production of protein complexes using the baculovirus expression vector system. PLoS One 13:e0195356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD (2005) Structural basis of eukaryotic gene transcription. FEBS Lett 579:899–903. [DOI] [PubMed] [Google Scholar]
- 23. Galbraith MD, Donner AJ, Espinosa JM (2010) CDK8: a positive regulator of transcription. Transcription 1:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 25. McCoy A, Grosse‐Kunstleve R, Adams P, Winn M, Storoni L, Read R (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Read RJ, Schierbeek AJ (1988) A phased translation function. J Appl Cryst 21:490–495. [Google Scholar]
- 27. Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC (2001) Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell 8:169–179. [DOI] [PubMed] [Google Scholar]
- 28. Seitomer E, Balar B, He D, Copeland PR, Kinzy TG (2008) Analysis of Saccharomyces cerevisiae null allele strains identifies a larger role for DNA damage versus oxidative stress pathways in growth inhibition by selenium. Mol Nutr Food Res 52:1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plateau P, Saveanu C, Lestini R, Dauplais M, Decourty L, Jacquier A, Blanquet S, Lazard M (2017) Exposure to selenomethionine causes selenocysteine misincorporation and protein aggregation in Saccharomyces cerevisiae . Sci Rep 7:44761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Braunagel SC, Parr R, Belyavskyi M, Summers MD (1998) Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology 244:195–211. [DOI] [PubMed] [Google Scholar]
- 31. Ikeda M, Kobayashi M (1999) Cell‐cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 258:176–188. [DOI] [PubMed] [Google Scholar]
- 32. Lazard M, Dauplais M, Blanquet S, Plateau P (2015) Trans‐sulfuration pathway seleno‐amino acids are mediators of selenomethionine toxicity in Saccharomyces cerevisiae . J Biol Chem 290:10741–10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y (2014) Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344:304–307. [DOI] [PubMed] [Google Scholar]
- 34. Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I (2006) Protein complex expression by using multigene baculoviral vectors. Nat Methods 3:1021–1032. [DOI] [PubMed] [Google Scholar]
- 35. Wasilko DJ, Lee SE, Stutzman‐Engwall KJ, Reitz BA, Emmons TL, Mathis KJ, Bienkowski MJ, Tomasselli AG, Fischer HD (2009) The titerless infected‐cells preservation and scale‐up (TIPS) method for large‐scale production of NO‐sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expr Purif 65:122–132. [DOI] [PubMed] [Google Scholar]
- 36. Hirs CHW. Determination of cystine as cysteic acid, 1967. Methods Enzymology. . London: Academic Press; p. 59–62. [Google Scholar]
- 37. Takagi Y, Kornberg RD (2006) Mediator as a general transcription factor. J Biol Chem 281:80–89. [DOI] [PubMed] [Google Scholar]
- 38. Lariviere L, Geiger S, Hoeppner S, Rother S, Strasser K, Cramer P (2006) Structure and TBP binding of the Mediator head subcomplex Med8–Med18–Med20. Nat Struct Mol Biol 13:895–901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


