Abstract
Introduction
We sought to determine if a proteomic profile approach developed to detect Alzheimer's disease would distinguish patients with Lewy body disease from normal controls, and if it would distinguish dementia with Lewy bodies (DLB) from Parkinson's disease (PD).
Methods
Stored plasma samples were obtained from 145 patients (DLB n = 57, PD without dementia n = 32, normal controls n = 56) enrolled from patients seen in the Behavioral Neurology or Movement Disorders clinics at the Mayo Clinic, Florida. Proteomic assays were conducted and analyzed as per our previously published protocols.
Results
In the first step, the proteomic profile distinguished the DLB-PD group from controls with a diagnostic accuracy of 0.97, sensitivity of 0.91, and specificity of 0.86. In the second step, the proteomic profile distinguished the DLB from PD groups with a diagnostic accuracy of 0.92, sensitivity of 0.94, and specificity of 0.88.
Discussion
These data provide evidence of the potential utility of a multitiered blood-based proteomic screening method for detecting DLB and distinguishing DLB from PD.
Keywords: Dementia with Lewy bodies, Parkinson's disease, Proteomics, Blood biomarkers, Biomarker screening, Detection, Diagnostic accuracy
1. Background
Lewy body disease is the second most common neurodegenerative disease and clinically may present with dementia as dementia with Lewy bodies (DLB), or without dementia as Parkinson's disease (PD). DLB was first characterized as a dementia by Kosaka [1] and operationalized diagnostic criteria were initially put forth by McKeith [2] in 1992. Patients who meet consensus criteria for DLB commonly have Lewy-related pathology [3] at autopsy, and in a large dementia autopsy series [4], 25% were found to have Lewy-related pathology. The core clinical features of DLB include parkinsonism, fluctuating cognition, fully formed visual hallucinations, and a history of probable REM behavior disorder [5], [6], [7]. There is a subset of patients with Lewy-related pathology who are often not recognized clinically as having DLB [8], in large part because of concomitant Alzheimer's disease (AD)-related pathology. Furthermore, the more extensive the tau pathology the harder it is to recognize the DLB phenotype. Multimodality imaging helps to distinguish DLB from AD, but it is an expensive and less viable method for disease detection methods in community samples [9]. Therefore, a front-line, minimally invasive, and cost-effective screening method would be of tremendous value to the field.
A major impediment to the development of treatments and clinical trials for neurodegenerative diseases is the lack of sensitive and easily obtained diagnostic biomarkers [10], [11], [12], [13], [14]. The search for biomarkers with diagnostic and prognostic utility in neurodegenerative diseases has grown exponentially, with most work focusing on neuroimaging [15], [16], [17], [18] and cerebrospinal fluid (CSF) methodologies [11], [15], [17], [18], [19]. Some new promising evidence suggests that CSF may yield a potential biomarker for α-synuclein, but replication with a large sample will be needed [20]. While advanced imaging and CSF methods have tremendous potential as confirmatory diagnostic biomarkers of neurodegenerative diseases, accessibility and cost barriers preclude these from being utilized as the first step in this process [12], [13], [21]. Reliable biomarkers of DLB could have many uses, including early and preclinical diagnosis, tracking disease progression, and identifying disease endophenotypes [14], [21]. In addition, the advancement of biomarkers may serve to pave the road toward a precision medicine approach to identifying surrogates for therapeutic outcome measures and for the development of disease-modifying treatments [22].
There are no currently validated biomarkers for DLB [23]. It has been proposed that biological markers of the clinical conditions associated with DLB should be “cheap, reliable and reproducible, and make use of biological samples that are easy to obtain” (pg. 1) [13]. Blood-based biomarkers would fulfill these proposed criteria. In addition, it has been proposed that proteomic biomarker profiling is a promising method for discovering DLB biomarkers [21], [23] because a battery of markers covering a range of biological processes may be required to address the needs of such complex disorders [24]. In fact, profiling analytes associated with multiple diseases may highlight novel biological pathways for therapeutic interventions in the dementia syndromes [25]. Our work on blood-based biomarkers of AD and PD has consistently shown that a multimarker approach identifying biomarker profiles of disease presence can yield excellent results [26], [27], [28]. We hypothesize that our blood-based biomarker profile approach may serve to provide a cost- and time-effective means for establishing a rapidly scalable multitiered neurodiagnostic process [29], [30] for detecting neurodegenerative disease, including DLB. With this initial screening approach, appropriate referrals can be made for subsequent specialty examinations and confirmatory diagnostic biomarkers (imaging, CSF), following the multistage models used for diagnosing cancer [31]. For example, Groveman et al. [20] recently demonstrated the accuracy of a rapid and ultrasensitive seed amplification technique for detection of α-synuclein. In the present proposed context, a blood-based screening tool can be utilized to rule out the vast majority of patients who do not need to undergo lumbar puncture for biomarker confirmatory diagnostics. This approach can also be readily adopted to clinical trials thereby (1) increasing access to broader numbers of patients and (2) significantly reducing screening costs into such novel trials.
In our prior work, we have generated and cross-validated our AD proteomic profile across platforms [26], [32], cohorts [26], [28], [29], [33], [34], species (human, mouse) [32], tissue (brain, serum, plasma) [32], and ethnicities (non-Hispanic white, Mexican American) [26], [35], which is currently being prospectively tested in primary care settings. In our initial pilot work, this same approach was highly accurate in discriminating PD from AD [32]. Here we test the hypothesis that our previously published proteomic profile approach to detecting AD [29], [32] would be successful in (1) detecting neurodegenerative disease due to synucleinopathy (DLB and PD vs controls) and (2) discriminating among neurodegenerative disease due to synucleinopathy (i.e., DLB vs PD). This study was conducted by examination of plasma samples from the Mayo Clinic, Jacksonville. Following the methods from our prior work, we also examined the impact of demographic factors (age, gender, education) on the proteomic profile. Here we utilized the same approach as in our prior work beginning with the discovery phase by using a multistep approach to determine if our approach can detect neurodegenerative disease and discriminate DLB from PD.
2. Methods
2.1. Subjects
The study sample included 145 patients (DLB n = 57, PD n = 32, normal control n = 56) seen through the Alzheimer's Disease Research Center and the Movement Disorders Center at the Mayo Clinic, Florida. All participants underwent a neurologic examination, a Mini-Mental State Examination and diagnosis was based on recent criteria [5], [36]. The DLB patients also underwent neuropsychological testing, had pathologic confirmation of diffuse or transitional Lewy body disease, and were specifically selected for this study if they had a documented response to cholinesterase inhibitors based on our prior work showing that DLB cases who respond to these medications are less likely to have imaging-based AD comorbid pathology [18]. Normal controls were recruited through the Alzheimer's Disease Research Center and were all cognitively normal based on neuropsychological testing. All PD-dementia cases were not included in this study.
2.2. Proteomics
Blood samples were collected as per the NACC–Alzheimer's Center guidelines, which also align with the recent guidelines published by an international working group [37]. Briefly, nonfasting sample was collected in an EDTA tube from participants while seated using a 21g needle, gently inverted 5–10 times and centrifuged at 2000 × g for 10 min before being aliquoted into cryovial (polypropylene) tubes and stored at −80°C. All processing was completed within a two-hour time frame. Samples remained in storage until shipped to the O'Bryant laboratory for assay. Plasma samples were assayed via a multiplex biomarker assay platform using electrochemiluminescence laboratory using the QuickPlex from Meso Scale Discovery as per our previously published methods using commercially available kits [29], [32]. The Meso Scale Discovery platform has been used extensively to assay biomarkers associated with a range of human diseases including AD [38], [39], [40], [41]. Electrochemiluminescence technology uses labels that emit light when electronically stimulated, which improves the sensitivity of detection of many analytes at very low concentrations. Electrochemiluminescence measures have well-established properties of being more sensitive and requiring less volume than conventional ELISAs [40], the gold standard for most assays. We recently reported the analytic performance of each of these markers for >1300 samples across multiple cohorts and diagnoses (normal cognition, MCI, AD) [29]. The assays are reliable and our experience with these assays show excellent spiked recovery, dilution linearity, coefficients of variation (CV), as well as detection limits. Inter- and intra-assay variability has been excellent. Internal QC protocols are implemented in addition to manufacturing protocols, including assaying consistent controls across batches and assay of pooled standards across lots. To further improve assay performance, assay preparation was automated using a customized Hamilton Robotics STARplus system. A total of 500 μL of plasma was utilized to assay the following markers (including CV and lowest level of detection [LLOD]) with CVs and LLODs calculated from this automated system using the Meso Scale Discovery plates: fatty acid–binding protein (CV = 2.2 LLOD = 13.277 pg/mL), beta 2 microglobulin (CV = 7.4, LLOD = 32.5 pg/mL), pancreatic polypeptide (CV = 4.1, LLOD = 390 pg/mL), CRP (CV = 2.4; LLOD = 2.41 pg/mL), ICAM-1 (CV = 4.6; LLOD = 1.8 pg/mL), thrombopoietin (CV = 2.2; LLOD = 33.1 pg/mL), α2 macroglobulin (CV = 2.8; LLOD = 5886 pg/mL), exotoxin 3 (CV = 18.74 LLOD = 3.25 pg/mL), tumor necrosis factor α (CV = 3.5; LLOD = 0.077 pg/mL), tenascin C (CV = 3.7; LLOD = 17 pg/mL), interleukin (IL)-5 (CV = 12.1; LLOD = 0.108 pg/mL), IL6 (CV = 4.6; LLOD = 0.081 pg/mL), IL7 (CV = 12.3; LLOD = 0.206 pg/mL), IL10 (CV = 6.7; LLOD = 0.071 pg/mL), IL18 (CV = 3.1; LLOD = 6.07 pg/mL), I309 (CV = 6.9; LLOD = 1.22 pg/mL), factor VII (CV = 2.7; LLOD = 49.9 pg/mL), VCAM 1 (CV = 2.3; LLOD = 6.13 pg/mL), TARC (CV = 5.9; LLOD = 0.21 pg/mL), and SAA (CV = 4.4; LLOD = 19 pg/mL). As can be seen, analytic performance was excellent with the average CVs across all plates for each analyte being well below standard research use only assays; all CVs<10 and 62% were <5%.
2.3. Statistical analysis
Statistical analyses were conducted using the R (V 3.3.3) statistical software [42], SPSS 24 (IBM) and SAS. Support vector machine (SVM) analyses were conducted to create proteomic profiles specifically for control versus Lewy body disease and then DLB versus PD. SVM is based on the concept of decision planes that define decision boundaries and is primarily a classifier method that performs classification tasks by constructing hyperplanes in a multidimensional space that separates cases of different class labels. Diagnostic accuracy was calculated via receiver operating characteristic (ROC) curves. First, SVM analyses were used to discriminate controls from Lewy body disease (i.e., DLB/PD) with resulting diagnostic accuracy statistics generated (step 1). Next, SVM analysis was restricted only to those with Lewy body disease to discriminate DLB from PD (step 2) with resulting diagnostic accuracy statistics generated. This two-step process was used to allow for the overall algorithm to be more robust and avoid multilevel analyses simultaneously, which reduces risk for error and sample overidentification. In addition, in our prior work, we have demonstrated that the overall profile differs among neurodegenerative diseases [32] and, therefore, the multistep process capitalizes on these overall proteomic profile fluctuations. Finally, samples from n = 53 AD cases were analyzed to provide preliminary analyses on a three-step approach to (1) detect neurodegenerative disease (AD/DLB/PD) from controls, (2) discriminate dementia (AD/DLB) from PD and (3) discriminate AD from DLB. These AD cases were also evaluated and clinically diagnosed by the Mayo Alzheimer's Disease Research Center. Demographic characteristics of the AD cases are provided in Table 1.
Table 1.
Demographic characteristics of the cohort
| Descriptor | DLB mean (SD) | PD mean (SD) | Normal control mean (SD) | AD mean (SD) |
|---|---|---|---|---|
| N | 57 | 32 | 56 | 53 |
| Age; mean (SD) | 76.03 (6.23) | 67.06 (11.58) | 76.16 (6.07) | 76.12 (5.95) |
| Education mean (SD) | 14.73 (3.56) | 15.74 (2.49) | 14.47 (2.87) | 13.68 (3.25) |
| Gender (%M) | 76.0 | 68.8 | 74.5 | 74.2 |
| MMSE score mean (SD) | 21.13 (6.8) | -- | 28.04 (1.64) | 18.30 (5.97) |
Abbreviations: DLB, dementia with Lewy bodies; PD, Parkinson's disease; MMSE, Mini-Mental State Examination.
3. Results
Descriptive statistics of the sample are provided in Table 1. The PD group was younger and included more females than the other two groups. As expected, the DLB group had lower scores on the Mini-Mental State Examination.
For the SVM analyses, a two-step analytic approach was taken. First, the SVM profile was used to differentiate Lewy body disease (DLB and PD) from controls. Second, the SVM analysis was used to differentiate DLB from PD. This two-step approach was used given our prior work suggesting that our proteomic profile can be highly accurate in detecting “neurodegenerative disease” in general [29] and therefore, our analyses for discriminating among neurodegenerative diseases could be refined even further without contamination of normal controls in the analytics.
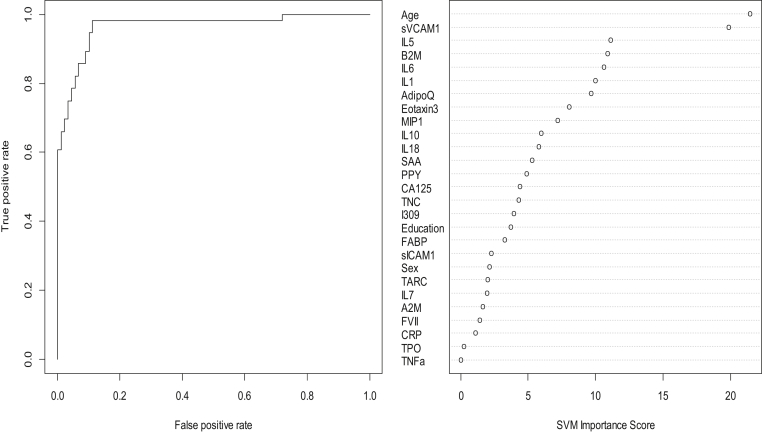
In step 1, our SVM-based proteomic profile was highly accurate in detecting Lewy body disease (DLB and PD) as compared with normal controls. The overall area under the ROC curve (AUC) of the proteomic profile was 0.94 with a sensitivity (SN) of 0.99 and specificity (SP) of 0.64. As with our prior work, inclusion of demographic variables (age, gender, education) increased the overall accuracy somewhat with an overall AUC of 0.97 with a SN decrease to 0.91 but SP increased to 0.86. Table 2 shows all of the correct and incorrect predictions, whereas the variable importance plot and ROC curve are presented in Fig. 1.
Table 2.
Diagnostic accuracy of blood test in step 1—discriminating control from Lewy body disease
| Confusion matrix for SVM classification for discriminating Lewy body disease from normal controls | ||
|---|---|---|
| Predicted | SVM model |
|
| DLB and PD | Normal control | |
| DLB and PD | 81 | 8 |
| NC | 8 | 48 |
| Sensitivity | 91.0% | |
| Specificity | 85.7% | |
| Area under the ROC curve | 0.9653 | |
Abbreviations: SVM, support vector machine; DLB, dementia with Lewy bodies; PD, Parkinson's disease.
Fig. 1.
ROC curve and variable importance plot for step 1—discriminating Lewy body disease from normal controls. Abbreviation: ROC, receiver operating characteristic.
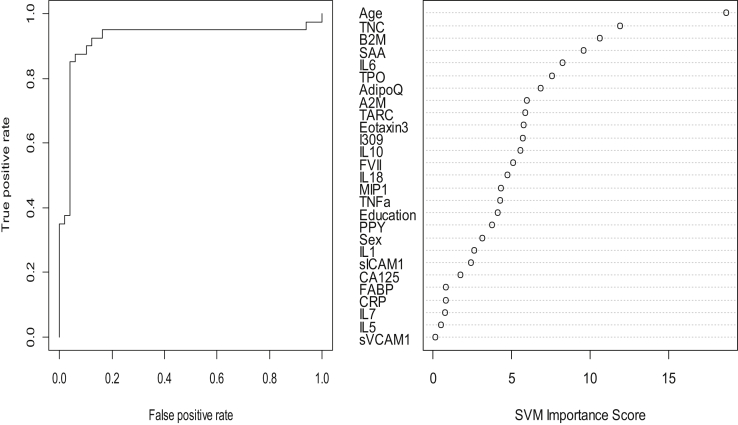
In the step 2, the overall SVM-proteomic profile also showed good accuracy at distinguishing DLB from PD. In this model, the AUC was 0.84 with SN = 0.95 and SN = 0.68. Inclusion of demographic variables improved the accuracy to AUC = 0.92, SN = 0.94 and SP = .88. Table 3 shows all the classifications (correct and incorrect), whereas the variable importance plot and ROC curve are presented in Fig. 2.
Table 3.
Diagnostic accuracy of blood test in step 2—discriminating between dementia with Lewy bodies and Parkinson's disease
| Confusion matrix for SVM classification for discriminating DLB from PD | ||
|---|---|---|
| Predicted | SVM model |
|
| DLB | PD | |
| DLB | 46 | 5 |
| PD | 3 | 35 |
| Sensitivity | 93.9% | |
| Specificity | 87.5% | |
| AUC | 0.9204 | |
Abbreviations: SVM, support vector machine; DLB, dementia with Lewy bodies; PD, Parkinson's disease; AUC, area under the ROC curve.
Fig. 2.
ROC curve and variable importance plot. Abbreviation: ROC, receiver operating characteristic.
Next we conducted preliminary analyses on a three-step algorithmic approach. Here, the full algorithm was applied (proteins + demographic variables). In the first step of the model, we sought to detect neurodegenerative disease (AD/DLB/PD) versus controls. With an optimized SVM risk threshold cutoff of −0.753, the AUC was 0.96 with an SN = 0.90 and SP = .89. In the second step, we sought to discriminate dementia (AD/DLB) from PD that yielded an AUC = 0.98, SN = 0.96, and SP = .97. In the third step, we sought to discriminate among dementias (DLB vs. AD) and found an AUC = 0.96, SN = 0.96, and SP = .97.
4. Discussion
The present study demonstrates, for the first time, that a multistep blood-based proteomic profile can accurately distinguish neurodegenerative disease due to synucleinopathy (DLB and PD) from normal controls (AUC = 0.97) and DLB from PD (AUC = 0.92). Recent work demonstrates that a CSF-based α-synuclein seeding technology can also achieve strong diagnostic accuracy in detecting neurodegenerative disease due to synucleinopathy (93% SN and 100% SP). Although that work requires cross-validation in larger studies, the advancement of the current work in tandem is promising for a sensitive and specific time- and cost-effective multistep approach for broad-based screening of DLB for prospective studies, clinical trials, and routine clinical practice.
The accuracy of the approach is directly due to the differing overall profiles, which is captured using advanced SVM analyses. Specifically, as can be seen from Fig. 1, Fig. 2, the variable importance plots are different in step 1 versus step 2. Therefore, by capitalizing on the complexity of the neurodegenerative disease due to synucleinopathy and the number of proteomics available, we can generate bioinformatics profiles. When reviewing the variable importance plots (Fig. 1, Fig. 2), the overall profiles for discriminating DLB/PD from controls was different compared with the profile for discriminating DLB from PD. The top 10 markers for discriminating DLB/PD from controls were as follows: age, sVCAM1, IL5, B2M, IL6, IL1, Adipo, Eotaxin, MIP1, and IL10. Not surprisingly, the top variable was age in both models. However, the top 2 proteins in this profile were the bottom 2 in the profile for discriminating DLB from PD. In fact, only age, B2M, IL6, adiponectin, and eotaxin overlapped in the top 10 markers in the algorithm (5 of top 10). Overall, the profile was a mix of inflammatory, metabolic, and vascular dysfunction, but at different levels between the categories. In our prior work, we have found that the AD profile is heavily inflammatory in nature as compared with PD and controls. In fact, the AD in adults with Down syndrome is also heavily inflammatory in nature. Therefore, while there are certainly disease-overlapping pathological processes depicted in this work, the profiles are different among categories. Prior work has demonstrated that there is a range of biological dysfunction across numerous neurodegenerative diseases. When tau and Aβ are present in DLB, they tend to occur at far less densities than what is typically seen in AD. A recent study showed that in DLB, α-synuclein is a key predictor of disease duration independently and synergistically with tau and amyloid β [Ferman et al., 2018]. It is possible that the proteomic profiles here are picking up on different levels of biological dysfunction due to differing levels of α-synuclein, amyloid, and tau pathology. Further work is needed to elucidate the pathological relevance of these overall proteomic profiles.
In our prior work, we have created and validated a proteomic signature for detecting AD across cohorts, species (humans, mice), and tissue (serum, plasma, brain) [26], [28], [29], [32]. Subsequently, we have proposed a multitiered neurodiagnostic process for detecting neurodegenerative disease beginning in primary care clinics using blood-based biomarkers [29], [30] which is now being prospectively studied in primary care settings (i.e., Alzheimer's Disease in Primary Care study). We have also demonstrated that our multiprotein algorithmic approach can discriminate AD from PD [32] as well as controls from “neurodegenerative disease” (i.e., AD, PD, DLB, Down syndrome) [29]. When the current work is compared with our prior work in AD [32], the synucleinopathy profile and DLB vs PD profile is different from the AD profile. Additional preliminary analyses were provided here to support the notion that the multimarker, multistep profile can also discriminate DLB and PD from AD. Given the sample size, these results are preliminary, but strongly supportive of further work. Therefore, the current work takes a significant step forward in the area of blood biomarkers for detecting neurodegenerative diseases as it sets the stage for a large-scale, multilevel proteomic-bioinformatic model that takes into account disease-specific profiles across numerous neurodegenerative diseases. The current team is currently assaying large numbers of samples across disease states to test this model.
There are weaknesses to the present study. First, the DLB cases included in this study were responders to cholinesterase inhibitors, who may be reflective of a specific subset of DLB patients. These cases were selected based on our prior work, suggesting that DLB cases that respond to cholinesterase inhibitors likely do not have imaging markers of AD. Therefore, it will be important to do additional work with larger, mixed DLB cases. Second, despite being a sizable proteomic study of Lewy body disease, the sample size is still relatively small and must be cross-validated in an independent sample. Therefore, the analyses were conducted with internal five-fold cross-validation rather than splitting the cohort into training and test samples. In addition, while the use of our previously validated proteomic profile is a significant strength, it is also possible that the inclusion of additional markers will aid in the detection of DLB as well as the discrimination of DLB from other neurodegenerative diseases. Given the recent resurgence of interest in blood-based biomarkers of amyloid using novel methodological advancements [43], [44], these markers should be considered in this work for additional refinement of discrimination among neurodegenerative diseases. In fact, the current team is currently including ultrasensitive markers of blood amyloid (Aβ40, Aβ42) total tau, neurofilament light chain, and α-synuclein in ongoing assays to determine how these markers enhance the accuracy of the models. Taken together, the current findings add substantially to a rapidly growing line of investigation suggesting that blood-based biomarkers can serve in a multitiered neurodiagnostic process for detecting a wide range of neurodegenerative diseases, including DLB.
Research in Context.
-
1.
Systematic review: Literature was identified and reviewed using PubMed. Several articles described the importance of rapid and cost-effective biomarkers for neurodegenerative diseases. However, no such blood-based biomarkers currently exist as a first step in a multitiered neurodiagnostic process.
-
2.
Interpretation: Our findings show that a blood-based biomarker profile can detect neurodegenerative disease (dementia with Lewy bodies [DLB]/Parkinson's disease) and distinguish DLB from Parkinson's disease cases.
-
3.
Future directions: This article provides support for the notion that a blood-based biomarker profiles can accurately detect DLB and even distinguish DLB from Parkinson's disease. Future work will be conducted to expand the sample size and also include other neurodegenerative disease categories such as Alzheimer's disease, Down syndrome, and traumatic brain injury. A blood-based biomarker profile for DLB would be of tremendous use for screening into novel therapeutic trials.
Acknowledgments
This grant was supported in part by a grant from the National Alzheimer's Coordinating Center (NACC-2016-04) and grants from the National Institute on Aging (R01AG058537, R01AG054073, R01AG058252, R01AG051848, P50AG016574). Z.K.W. is partially supported by the NIH/NIA (primary) and NIH/NINDS (secondary) 1U01AG045390-01A1, Mayo Clinic Center for Regenerative Medicine, the gift from Carl Edward Bolch, Jr., and Susan Bass Bolch, The Sol Goldman Charitable Trust, and Donald G. and Jodi P. Heeringa.
Footnotes
Conflict of Interests: S.E.O. has multiple pending and issued patents on blood biomarkers for detecting and precision medicine therapeutics in neurodegenerative diseases. He is a founding scientist and owns stock options in Cx Precision Medicine, Inc.
References
- 1.Kosaka K., Oyanagi S., Matsushita M., Hori A. Presenile dementia with Alzheimer-, Pick- and Lewy-body changes. Acta Neuropathol. 1976;36:221–233. doi: 10.1007/BF00685366. [DOI] [PubMed] [Google Scholar]
- 2.McKeith I.G., Perry R.H., Fairbairn A.F., Jabeen S., Perry E.K. Operational criteria for senile dementia of Lewy body type (SDLT) Psychol Med. 1992;22:911–922. doi: 10.1017/s0033291700038484. [DOI] [PubMed] [Google Scholar]
- 3.Fujishiro H., Ferman T.J., Boeve B.F., Smith G.E., Graff-Radford N.R., Uitti R.J. Validation of the neuropathologic criteria of the third consortium for dementia with lewy bodies for prospectively diagnosed cases. J Neuropathol Exp Neurol. 2008;67:649–656. doi: 10.1097/NEN.0b013e31817d7a1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker W.W., Luis C.A., Kashuba A., Luis M., Harwood D.G., Lowenstein D. Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis Assoc Disord. 2002;16:203–212. doi: 10.1097/00002093-200210000-00001. [DOI] [PubMed] [Google Scholar]
- 5.McKeith I.G., Boeve B.F., Dickson D.W., Halliday G., Taylor J.P., Weintraub D. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100. doi: 10.1212/WNL.0000000000004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferman T.J., Boeve B.F., Smith G.E., Lin S.C., Silber M.H., Pedraza O. Inclusion of RBD improves the diagnostic classification of dementia with Lewy bodies. Neurology. 2011;77:875–882. doi: 10.1212/WNL.0b013e31822c9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray M.E., Ferman T.J., Boeve B.F., Przybelski S.A., Lesnik T.G., Liesinger A.M. MRI and pathology of REM sleep behavior disorder in dementia with Lewy bodies. Neurology. 2013;81:1681–1689. doi: 10.1212/01.wnl.0000435299.57153.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday G.M., Holton J.L., Revesz T., Dickson D.W. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 2011;122:187–204. doi: 10.1007/s00401-011-0852-9. [DOI] [PubMed] [Google Scholar]
- 9.Kantarci K., Lowe V.J., Boeve B.F., Weigand S.D., Senjem M.L., Przybelski S.A. Multimodality imaging characteristics of dementia with Lewy bodies. Neurobiol Aging. 2012;33:2091–2105. doi: 10.1016/j.neurobiolaging.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen K., O'Bryant S.E., Hampel J.Q., Trojanowski T.J., Montine T.J., Jeromin A. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thal L.J., Kantarci K., Reiman E.M., Klunk W.E., Weiner M.W., Zetterberg H. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider P., Hampel H., Buerger K. Biological marker candidates of alzheimer's disease in blood, plasma, and serum. CNS Neurosci Ther. 2009;15:358–374. doi: 10.1111/j.1755-5949.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eller M., Williams D.R. α-Synuclein in Parkinson disease and other neurodegenerative disorders. Clin Chem Lab Med. 2011;49:403–408. doi: 10.1515/CCLM.2011.077. [DOI] [PubMed] [Google Scholar]
- 14.Sinha N., Firbank M., O'Brien J.T. Biomarkers in dementia with Lewy bodies: A review. Int J Geriatr Psychiatry. 2012;27:443–453. doi: 10.1002/gps.2749. [DOI] [PubMed] [Google Scholar]
- 15.Colloby S.J., Fairbank M.J., Pakrasi S., Lloyd J.J., Driver I., McKeith I.G. A comparison of 99mTc-exametazime and 123I-FP-CIT SPECT imaging in the differential diagnosis of Alzheimer's disease and dementia with Lewy bodies. Int Psychogeriatr. 2008;20:1124–1140. doi: 10.1017/S1041610208007709. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi M., Tashiro M., Arai H., Okamura N., Hara S., Higuchi S. Glucose hypometabolism and neuropathological correlates in brains of dementia with Lewy bodies. Exp Neurol. 2000;162:247–256. doi: 10.1006/exnr.2000.7342. [DOI] [PubMed] [Google Scholar]
- 17.McKeith I., O'Brien J., Walker Z., Tatsch K., Booij J., Darcourt J. Sensitivity and specificity of dopamine transporter imaging with 123I-FP-CIT SPECT in dementia with Lewy bodies: A phase III, multicentre study. Lancet Neurol. 2007;6:305–313. doi: 10.1016/S1474-4422(07)70057-1. [DOI] [PubMed] [Google Scholar]
- 18.Graff-Radford J., Boeve B.F., Pedraza O., Ferman T.J., Przybelski S., Lesnik T.G. Imaging and acetylcholinesterase inhibitor response in dementia with Lewy bodies. Brain. 2012;135:2470–2477. doi: 10.1093/brain/aws173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anonymous, Consensus report of the Working Group on: “Molecular and Biochemical Markers of Alzheimer's Disease” The Ronald and Nancy Reagan Research Institute of the Alzheimer's Association and the National Institute on Aging Working Group.[see comment][erratum appears in Neurobiol Aging 1998 May-Jun;19(3):285] Neurobiol Aging. 1998;19:109–116. [PubMed] [Google Scholar]
- 20.Groveman B.R., Orru C.D., Hughson A.G., Raymond L.D., Zanusso G., Ghetti B. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol Commun. 2018;6:7. doi: 10.1186/s40478-018-0508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henchcliffe C., Dodel R., Beal M.F. Biomarkers of Parkinson's disease and Dementia with Lewy bodies. Prog Neurobiol. 2011;95:601–613. doi: 10.1016/j.pneurobio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Hampel H., O'Bryant S.E., Castrillo J.I., Ritchie C., Rojkova K., Benda N. Precision Medicine: The Golden Gate for detection, treatment and prevention of Alzheimer's disease. J Prev Alzheimers Dis. 2016;3:243–259. doi: 10.14283/jpad.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho G.J., Liang W., Waragai M., Sekiyama K., Masliah E., Hashimoto M. Bridging molecular genetics and biomarkers in Lewy body and related disorders. Int J Alzheimer's Dis. 2011;2011:842475. doi: 10.4061/2011/842475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shtilbans A., Henchcliffe C. Biomarkers in Parkinson's disease: An update. Curr Opin Neurol. 2012;25:460–465. doi: 10.1097/WCO.0b013e3283550c0d. [DOI] [PubMed] [Google Scholar]
- 25.Hu W.T., Chen-Plotken A., Arnold S.E., Grossman M., Clark C.M., Shaw L.M. Biomarker discovery for Alzheimer's disease, frontotemporal lobar degeneration, and Parkinson's disease. Acta Neuropathol. 2010;120:385–399. doi: 10.1007/s00401-010-0723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Bryant S.E., Xiao G., Barber R., Reisch J., Doody R., Fairchild T. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67:1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Bryant S.E., Xiao G., Barber R., Reisch J., Hall J., Cullum C.M. A blood-based algorithm for the detection of Alzheimer's disease. Demen Geriatr Cogn Disord. 2011;32:55–62. doi: 10.1159/000330750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Bryant S.E., Xiao G., Barber R., Huebinger R., Wilhelmsen K., Edwards M. A Blood-Based Screening Tool for Alzheimer's Disease That Spans Serum and Plasma: Findings from TARC and ADNI. PLoS One. 2011;6:e28092. doi: 10.1371/journal.pone.0028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Bryant S.E., Edwards M., Johnson L., Hall J., Villarreal A.E., Britton G.B. A Blood Screening Test for Alzheimer's Disease. Alzheimers Dement. 2016;3:83–90. doi: 10.1016/j.dadm.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Bryant S.E., Mielke M.M., Rissman R.A., Lista S., Vanderstichele H., Zetterberg H. Blood-based biomarkers in Alzheimer disease: Current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2017;13:45–58. doi: 10.1016/j.jalz.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold L.S., Klein G., Carr L., Kessler L., Sullivan S.D. The emergence of diagnostic imaging technologies in breast cancer: Discovery, regulatory approval, reimbursement, and adoption in clinical guidelines. Cancer Imaging. 2012;12:13–24. doi: 10.1102/1470-7330.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Bryant S.E., Xiao G., Zhang F., Edwards M., German D.C., Yin X. Validation of a serum screen for alzheimer's disease across assay platforms, species, and tissues. J Alzheimers Dis. 2014;42:1325–1335. doi: 10.3233/JAD-141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villarreal A.E., O'Bryant S.E., Edwards M., Grajales S., Britton G.B., Panama Aging Research Initiative Serum-based protein profiles of Alzheimer's disease and mild cognitive impairmetn in elderly Hispanics. Neurodegener Dis Manag. 2016;6:203–213. doi: 10.2217/nmt-2015-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards M., Hall J., Williams B., Johnson L., O'Bryant S.E. Molecular markers of amnestic mild cognitive impairment among Mexican Americans. J Alzheimers Dis. 2016;49:221–228. doi: 10.3233/JAD-150553. [DOI] [PubMed] [Google Scholar]
- 35.O'Bryant S.E., Gupta V., Henriksen K., Edwards M., Jeromin A., Lista S. Biomarkers of Alzheimer's disease among Mexican Americans. J Alzheimer's Dis. 2013;34:841–849. doi: 10.3233/JAD-122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postuma R.B., Berg D., Stern M., Poewe W., Olanow C.W., Oertel W. MDS clinical diagnostic criteria for Parkinson's disease. Mov Disord. 2015;30:1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- 37.O'Bryant S.E., G V., Henriksen K., Edwards M., Jeromin A., Lista S. Guidelines for the standardization fo preanalytic variables for blood-based biomarker studies in Alzheimer's disease. Alzheimers Dement. 2015;11:549–560. doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alves G., Bronnick K., Aarsland D., Blennow K., Zetterberg H., Ballard C. CSF amyloid-β and tau proteins, and cognitive performance, in early and untreated Parkinson's Disease: The Norwegian ParkWest study. J Neurol Neurosurg Psychiatr. 2010;81:1080–1086. doi: 10.1136/jnnp.2009.199950. [DOI] [PubMed] [Google Scholar]
- 39.Bjerke M., Portelius E., Minthon L., Wallin A., Anckarsater H., Anckarsater R. Confounding factors influencing amyloid beta concentration in cerebrospinal fluid. Int J Alzheimer's Dis. 2010 doi: 10.4061/2010/986310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhle J., Regeniter A., Leppert D., Mehling M., Kappos L., Lindberg R.L. A highly sensitive electrochemiluminescence immunoassay for the neurofilament heavy chain protein. J Neuroimmunol. 2010;220:114–119. doi: 10.1016/j.jneuroim.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Oh E.S., Mielke M.M., Rosenberg P.B., Jain A., Fedarko N.S., Lyketsos C.G. Comparison of conventional ELISA with electrochemiluminescence technology for detection of amyloid-β in plasma. J Alzheimer's Dis. 2010;21:769–773. doi: 10.3233/JAD-2010-100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R_Development_Core_Team . 2009. R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org. Accessed January, 2019. [Google Scholar]
- 43.Nakamura A., Kaneko N., Villemagne V.L., Kato T., Doecke J., Dore V. High performance plasma amyloid-beta biomarkers for Alzheimer's disease. Nature. 2018;554:249–254. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 44.Wildburger N.C., Gyngard F., Guillermier C., Patterson B.W., Elbert D., Mawuenyega K.G. Amyloid-beta plaques in clinical Alzheimer's disease brain incorporate stable isotope tracer in vivo and exhibit nanoscale heterogeneity. Front Neurol. 2018;9:169. doi: 10.3389/fneur.2018.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]