Abstract
An antibody fragment that recognizes the tertiary structure of a target protein with high affinity can be utilized as a crystallization chaperone. Difficulties in establishing conformation‐specific antibodies, however, limit the applicability of antibody fragment‐assisted crystallization. Here, we attempted to establish an alternative method to promote the crystallization of target proteins using an already established anti‐tag antibody. The monoclonal antibody NZ‐1 recognizes the PA tag with an extremely high affinity. It was also established that the PA tag is accommodated in the antigen‐binding pocket in a bent conformation, compatible with an insertion into loop regions on the target. We, therefore, explored the application of NZ‐1 Fab as a crystallization chaperone that complexes with a target protein displaying a PA tag. Specifically, we inserted the PA tag into the β‐hairpins of the PDZ tandem fragment of a bacterial Site‐2 protease. We crystallized the PA‐inserted PDZ tandem mutants with the NZ‐1 Fab and solved the co‐crystal structure to analyze their interaction modes. Although the initial insertion designs produced only moderate‐resolution structures, eliminating the solvent‐accessible space between the NZ‐1 Fab and target PDZ tandem improved the diffraction qualities remarkably. Our results demonstrate that the NZ‐1‐PA system efficiently promotes crystallization of the target protein. The present work also suggests that β‐hairpins are suitable sites for the PA insertion because the PA tag contains a Pro‐Gly sequence with a propensity for a β‐turn conformation.
Keywords: crystallization chaperone, protein crystallography, monoclonal antibody, antigen‐binding fragment, epitope tag insertion
Short abstract
Introduction
Antibody fragments such as antigen‐binding fragments (Fab) and variable fragments (Fv) can serve as crystallization chaperones in protein crystallography.1, 2, 3, 4 Antibody fragments increase the probability of forming inter‐molecular contacts required for lattice formation. This strategy is particularly important for difficult‐to‐crystallize proteins such as membrane proteins covered with fluidic detergent micelles and glycoproteins containing flexible sugar moieties. Although various immunization and screening techniques have been developed,5, 6, 7 the establishment of antibodies that promote co‐crystallization is still a difficult task. We, therefore, attempted to explore an alternative strategy to promote crystallization by inserting the epitope sequence of an established antibody into target proteins and subsequently preparing complexes with antibody fragments. Specifically, we utilized a monoclonal antibody NZ‐1 and its epitope tag. NZ‐1 was originally generated by immunizing rats with the platelet‐aggregation‐stimulating domain of human podoplainin and the epitope was identified as the tetradecapeptide PA14 (EGGVAMPGAEDDVV).8 Although the central portion, PA10 (GVAMPGAEDD), is indispensable and sufficient for specific binding with NZ‐1,9 PA14 and PA12 (GVAMPGAEDDVV) both show exceptionally high affinity and slow dissociation from NZ‐1.10 Due to these properties, the PA12 epitope has been developed as the PA tag for protein purification. In addition, a crystallographic analysis of the NZ‐1 Fab–PA14 peptide complex has shown that the epitope binds to NZ‐1 in a bent conformation with a Type II β‐turn between Pro‐5 and Gly‐6, making it possible to insert a PA tag into the loop regions of target proteins.11 In fact, a PA‐insertion has already been used as a probe to detect conformational states of a modular cell‐surface receptor by comparing the accessibilities of the PA tag to NZ‐1.11 NZ‐1 labeling of an inserted PA tag is also useful for locating subunits or domains within macromolecular complexes by electron microscopy (EM) imaging.12, 13 In this study, we further examined the applicability of the inserted PA tag‐NZ‐1 Fab pair in generating well‐diffracting crystals of target proteins.
As a target for PA‐insertion, we selected the periplasmic soluble fragment from a Site‐2 protease (S2P) homolog from the hyper‐thermophilic bacterium Aquifex (A.) aeolicus.14 This soluble fragment contains two tandemly arranged PSD95/Dlg/ZO‐1 (PDZ) domains (hereafter, the soluble fragment is referred to as the PDZ tandem). In our previous work, we obtained crystals of the PDZ tandem fragment, which diffracted X‐rays up to 2.8 Å resolution.15 We found that the PDZ tandem was loosely packed in the crystal with a relatively high solvent content (74%), which is probably due to the abundance of flexible lysine residues on the molecular surface. In that study, we generated a monoclonal antibody against the PDZ tandem to utilize the Fab fragment as the crystallization chaperone. As a result, we improved the crystallographic resolution to 2.2 Å and determined the crystal structure more precisely.15 The successful previous application of the standard approach for chaperone‐assisted crystallography makes the PDZ tandem a good system to test and evaluate a novel approach.
Here, we tested if we could insert the PA tag into the PDZ tandem without disrupting the tertiary structure and improve the resolution by using the NZ‐1 Fab as the crystallization chaperone.
Results
Insertion of the PA tag into β‐hairpins and structure determination of the Fab‐bound form
The canonical PDZ domain consists of six β‐strands, βA‐F, and two α‐helices, αA–B. Both of the two PDZ domains (PDZ‐N and ‐C) forming the PDZ tandem are circular permutants of the canonical PDZ‐fold (Fig. 1). As a result, the non‐canonical PDZ domains of the PDZ tandem possess the N‐terminus before βC and the C‐terminus after βB while βF and βA are connected by a loop. We selected these two βF‐βA loops of PDZ‐N and ‐C as the PA‐insertion sites (Fig. 1(A)) because both of the two loops protrude outside of the PDZ tandem fragment. In the present work, we specify the respective PDZ tandem mutants based on the residues flanking the PA tag sequence and the length of the insertion. For instance, the mutant with the PA12 sequence inserted between Arg‐181 and Glu‐184 in the PDZ‐N is termed as PDZ tandem (181‐PA12‐184). Similarly, the PDZ‐C mutant with the PA‐insertion is termed as PDZ tandem (263‐PA12‐266).
Figure 1.
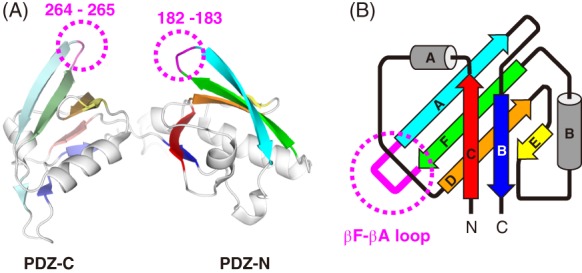
3D structure of the PDZ tandem from the A. aeolicus Site‐2 protease homolog. (A) Ribbon model of PDZ tandem. The six β‐strands of the respective PDZ domains are colored differently where PDZ‐N and PDZ‐C are shown in bright and pale colors, respectively. The deleted loop residues for the PA‐insertion in the β‐hairpins are indicated in magenta with dotted circles. (B) Topology diagram of the circular‐permutant PDZ domain. The loop connecting the βF and βA strands is colored in magenta and indicated with a dotted circle. In the present work, the PA tag was inserted here.
The two PDZ tandem mutants were produced as glutathione‐S‐transferase (GST)‐fusion proteins. The mutant fragments with the PA‐insertion were monodisperse after removal of the GST portion. In size‐exclusion chromatography, both of the mutants eluted at elution volumes comparable to that of the wild type (Supporting Information Fig. S1(A–C)), indicating that the PA insertion did not affect the folding of the PDZ domains. As a control, we introduced an L259R mutation to PDZ tandem (263‐PA12‐266). Leu‐259 participates in the hydrophobic core and contributes to the structural integrity of the PDZ‐C domain. The mutation was therefore expected to cause partial unfolding of the PDZ tandem fragment. As expected, we observed a significant peak shift to a smaller elution volume than that of wild type in the size‐exclusion chromatography (Supporting Information Fig. S1(D)). The peak shifts observed in the PDZ tandem (181‐PA12‐184) and (263‐PA12‐266) were negligible compared with that observed in the L259R mutant, which is supportive of the correct folding for both of the PDZ tandem (181‐PA12‐184) and (263‐PA12‐266). Subsequently, we prepared complexes with the NZ‐1 Fab. Both mutants bound stably to the NZ‐1 Fab, and almost no dissociation was observed during fractionation by size‐exclusion chromatography. After purification, we attempted to crystallize the mutants alone and in complex with the NZ‐1 Fab. However, we obtained no crystals of the solitary mutants at least within the search space of our screening. As mentioned above, the wild‐type PDZ tandem tends to crystallize through loose lattice contacts. Both of the PA‐insertion sites were involved in lattice contacts for the wild‐type crystal, so there is a possibility that the insertions disrupt that form.
Although the mutants seemed to become less crystallizable, we discovered several crystallization conditions for both of the two mutants in complex with the NZ‐1 Fab. Many poorly diffracting co‐crystals were screened before we selected crystals that allowed us to determine crystal structures to resolutions of 3.2 Å and 4.0 Å for PDZ tandems (181‐PA12‐184) and (263‐PA12‐266) in complex with the NZ‐1 Fab, respectively (Figs. 2(A–C) and 3(A–C)). Inspection of the crystal packing indicated that not only the NZ‐1 Fab but also the PDZ tandem substantially contributed to the lattice formation in both cases. In particular, the moderate resolution of the co‐crystal with PDZ tandem (181‐PA12‐184) was probably due to loose lattice contacts. In the course of model building, we observed weak and disordered electron densities for the PDZ‐C portion. PDZ‐C is connected with the PA‐inserted PDZ‐N domain through a short linker, and the PDZ tandem, therefore, showed conformational flexibility. The weak electron densities suggested that PDZ‐C was involved in the lattice contacts to some extent, but we could not assign a reliable model for this portion in the end [Fig. 2(A)].
Figure 2.
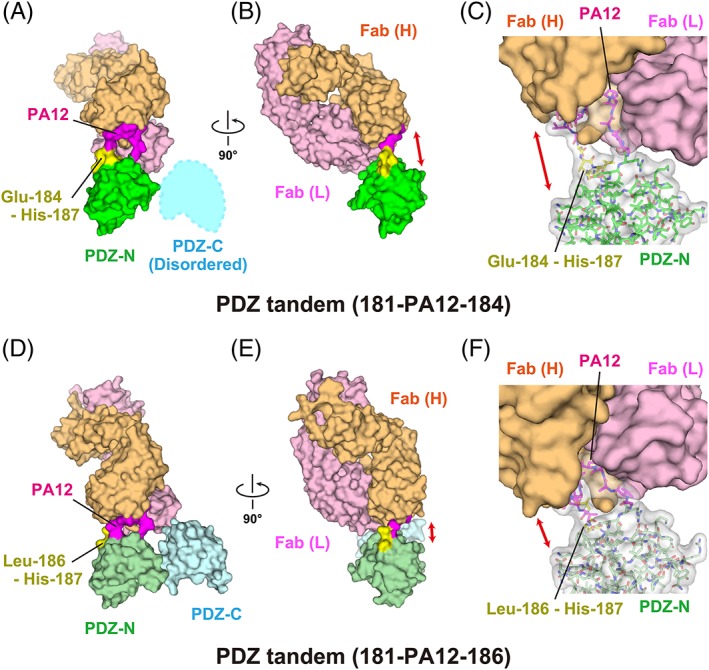
Complex formation of the PDZ tandem with the NZ‐1 Fab through a PA‐inserted PDZ‐N domain. (A, B) Surface model of PDZ tandem (181‐PA12‐184) in complex with the NZ‐1 Fab in two different views. The inserted PA tag is shown in magenta. The residues undergoing significant structural changes compared with the wild type, as shown in Figure 5, are highlighted in yellow. (C) Close‐up view of the binding site. The PDZ‐N domain and the inserted PA tag are shown as stick models with a transparent surface. The solvent‐accessible space between the rigidly folded part of the PDZ‐N domain and the NZ‐1 Fab is indicated with a double‐headed arrow. (D, E) Surface model of PDZ tandem (181‐PA12‐186) in complex with the NZ‐1 Fab in two different views. (F) Close‐up view of the binding site. The models are colored as in (A, B, and C) while the PDZ domains are shown in pale colors.
Figure 3.
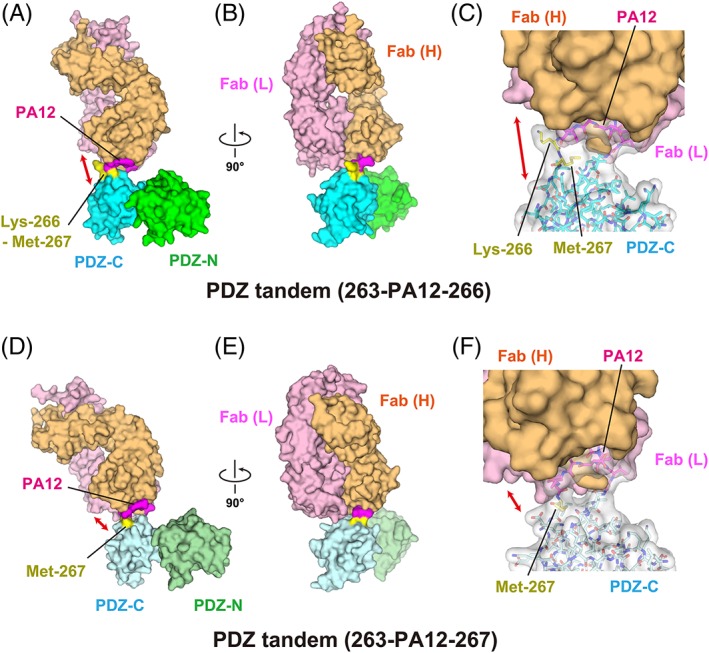
Complex formation of the PDZ tandem with the NZ‐1 Fab through a PA‐inserted PDZ‐C domain. (A, B) Surface model of PDZ tandem (263‐PA12‐266) in complex with the NZ‐1 Fab in two different views. The inserted PA tag is shown in magenta. The residues undergoing significant structural changes compared with the wild type, as shown in Figure 6, are highlighted in yellow. (C) Close‐up view of the binding site. The PDZ‐C domain and the inserted PA tag are shown in stick models with a transparent surface. The solvent‐accessible space between the rigidly‐folded part of the PDZ‐C domain and the NZ‐1 Fab is indicated with a double‐headed arrow. (D, E) Surface model of PDZ tandem (263‐PA12‐267) in complex with NZ‐1 Fab in two different views. (F) Close‐up view of the binding site. The models are colored as in (A, B, and C) but the PDZ domains are shown in pale colors.
Structural details of PDZ tandem (181‐PA12‐184) in the Fab‐bound form
In the NZ‐1 Fab‐PDZ tandem (181‐PA12‐184) complex, the interaction mode of the complementarity determining regions with the inserted PA tag was essentially consistent with that observed in the NZ‐1 Fab‐PA14 co‐crystal, excepting the involvement of a metal ion [Fig. 4(A, B)]. The electron density indicated that a metal ion is co‐ordinated by Glu‐8 and Asp‐9 from the PA tag together with Asp‐75 from the NZ‐1 heavy chain, although these two PA residues were recognized by Lys‐76 of the heavy chain in the NZ‐1 Fab‐PA14 complex [Supporting Information Fig. S2(A)]. The metal ion is presumed to be a cadmium ion present in the crystallization buffer. However, we could not determine the identities of the residual electron densities due to the absence of anomalous data near the absorption edge. Likewise, coordination geometry, such as monodentate or bidentate co‐ordination by carboxyl groups, could not be unambiguously assigned due to the poor quality of electron density map.
Figure 4.
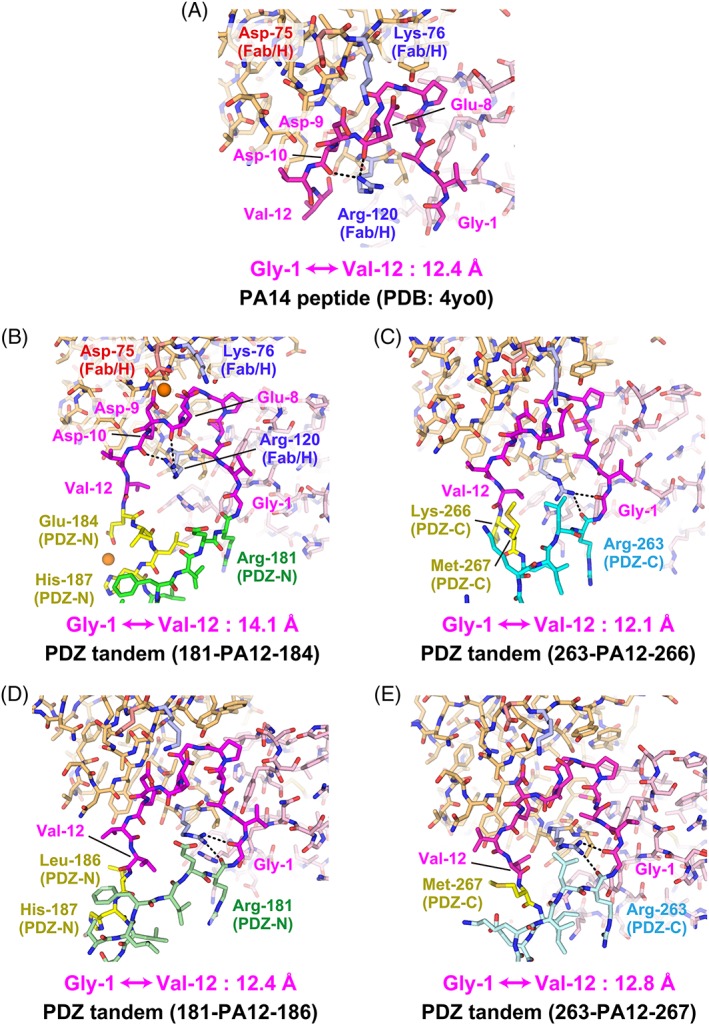
Conformation of the PA tag in the antigen‐binding pocket. (A) The PA14 peptide in complex with the NZ‐1 Fab (PDB code: 4YO0). The PA peptide is shown in magenta while the heavy and light chains of the NZ‐1 Fab are colored with light orange and light pink, respectively. Lys‐76 and Arg‐120 of the heavy chain are highlighted in blue where the hydrogen bonds formed by the side chain of Arg‐120 are indicated with dotted lines. Asp‐75 of the heavy chain is also highlighted in red. (B) PDZ tandem (181‐PA12‐184) in complex with the NZ‐1 Fab. The PDZ‐N domain is shown in green where the residues showing significant structural changes compared with the wild type are highlighted in yellow as shown in Figure 5. Metal ions presumed to be cadmium ions are shown with sphere models. (C) PDZ tandem (263‐PA12‐266) in complex with the NZ‐1 Fab. The PDZ‐C domain is shown in cyan where the residues showing significant structural changes compared to the wild type are highlighted in yellow as shown in Figure 6. (D) PDZ tandem (181‐PA12‐186) in complex with the NZ‐1 Fab. (E) PDZ tandem (263‐PA12‐267) in complex with the NZ‐1 Fab. The models are colored as in previous figures but the PDZ‐N and ‐C domains are shown in pale colors. The distances between the Cα atoms of Gly‐1 and Val‐12 are indicated under the respective models.
As the NZ‐1 Fab rigidifies the main chain conformation of the PA tag when it is accommodated into the antigen‐binding pocket, the Cα atoms of Gly‐1 and Val‐12 were separated by 14.1 Å, which is comparable to the 12.4 Å distance in the NZ‐1 Fab‐PA14 co‐crystal [Fig. 4(A,B)]. The separation of these two residues caused significant conformational changes in the target PDZ‐N domain at the junction with the PA tag. For instance, Glu‐184 and Val‐185 that originally belonged to the βA strand were separated from their hydrogen‐bonding partners of the βF strand. In addition, the inter‐strand hydrogen bonds were also broken at Leu‐186 and His‐187 due to a main chain distortion [Fig. 5(A–C)]. These conformational changes at the junction are expected to increase the flexibility of the relative positioning between the PDZ tandem and the NZ‐1 Fab. Glu‐184 and His‐187 were also co‐ordinated to a metal ion with a fixed conformation in the co‐crystal, but these side chains will likely be solvent exposed and disordered under metal‐free conditions. In contrast, we observed that the overall fold of the target PDZ‐N domain was hardly affected by PA‐insertion and Fab‐binding. The 76 Cα atoms of PDZ‐N excluding the four junction residues, Glu‐184 to His‐187, could be superposed onto to those of the wild type with a root‐mean‐square deviation (RMSD) of 0.697 Å.
Figure 5.
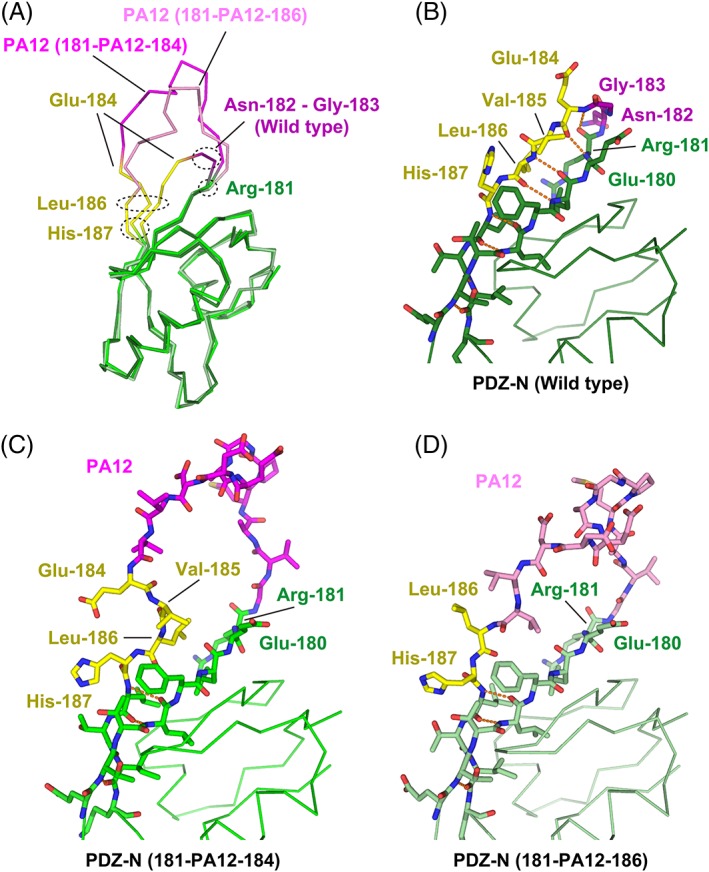
Structural change of the PDZ‐N domain around the PA insertion site. (A) Superposition of the Cα traces. The wild‐type PDZ‐N domain and the two PA‐inserted mutants are displayed as varying shades of the same color scheme. The rigidly‐folded part of the PDZ‐N domain is shown in green where the residues undergoing significant structural changes due to the PA insertion (Glu‐184 to His‐187) are highlighted in yellow. The two residues (Asn‐182 and Gly‐183) deleted to construct the PDZ tandem (181‐PA12‐184) are shown in dark magenta. The PA12 residues of PDZ tandem (181‐PA12‐184) and (181‐PA12‐186) are shown in bright magenta and light magenta, respectively. Close‐up view of the PA insertion site of the wild type PDZ‐N domain (B), PDZ tandem (181‐PA12‐184) (C), and PDZ tandem (181‐PA12‐186) (D). Inter‐strand hydrogen bonds are shown as orange dotted lines.
Structural details of PDZ tandem (263‐PA12‐266) in the Fab‐bound form
The recognition mode of the PA tag was also conserved in the NZ‐1 Fab‐PDZ tandem (263‐PA12‐266) complex where the Cα atoms of Gly‐1 and Val‐12 were separated by 12.1 Å [Fig. 4(C) and Supporting Information Fig. S2(B)]. However, a slight difference was found in the side chain conformation of Arg‐120 on the NZ‐1 heavy chain. It had been shown in the NZ‐1 Fab‐PA14 complex that this residue penetrated into the loop formed by the PA tag and seemed to fix the loop in the antigen‐binding pocket where the guanidinium group of Arg‐120 interacted with the main chain carbonyl groups of Glu‐8 and Asp‐10 of PA14 [Fig. 4(A)]. In contrast, Arg‐120 pointed to Arg‐263 of PDZ‐C and Gly‐1 of PA12 in the NZ‐1 Fab‐PDZ tandem (263‐PA12‐266) complex [Fig. 4(C)]. As in the above case of PDZ tandem (181‐PA12‐184), some junction residues of the βA strand, such as Lys‐266 and Met‐267, were displaced by the PA‐insertion [Fig. 6(A–C)]. In particular, Lys‐266 made no direct contacts with the NZ‐1 Fab nor the remainder of PDZ‐C [Figs. 3(C) and Supporting Information Fig. S2(B)]. Both Lys‐266 and Met‐267 appeared to be highly mobile although they are located relatively close to one of the symmetry neighbors in the crystal packing.
Figure 6.
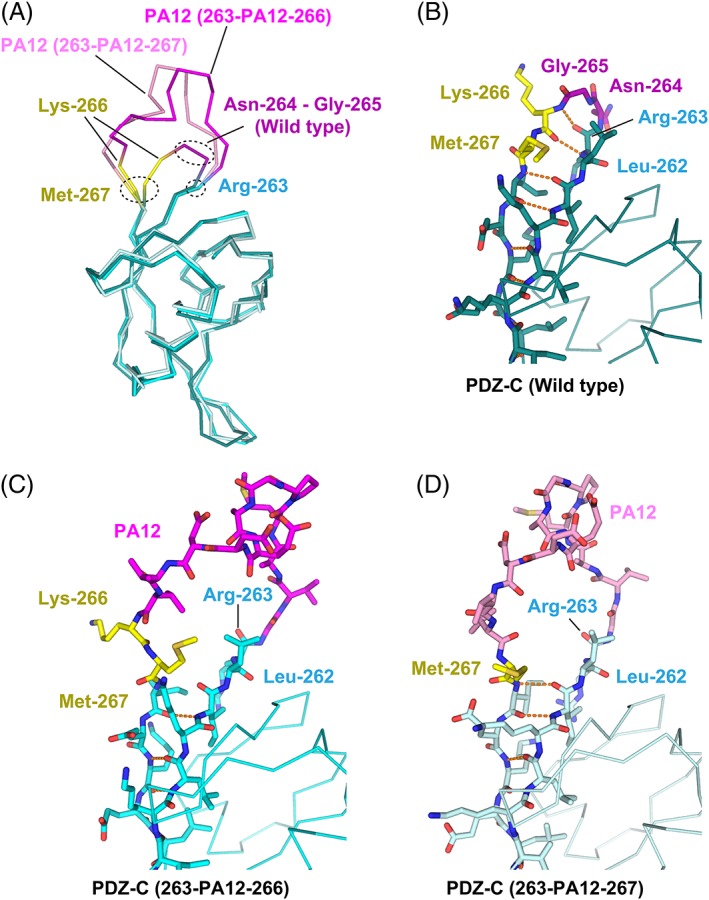
Structural change of the PDZ‐C domain around the PA insertion site. (A) Superposition of the Cα traces. The wild‐type PDZ‐N domain and the two PA‐inserted mutants are displayed as varying shades of the same color scheme. The rigidly folded part of the PDZ‐C domain is shown in cyan where the residues undergoing significant structural changes due to the PA insertion (Lys‐266 and Met‐267) are highlighted in yellow. The two residues (Asn‐264 and Gly‐265) deleted to construct the PDZ tandem (263‐PA12‐266) are shown in dark magenta. The PA12 residues of PDZ tandem (263‐PA12‐266) and (263‐PA12‐267) are shown in bright magenta and light magenta, respectively. Close‐up view of the PA insertion site of the wild‐type PDZ‐C domain (B), PDZ tandem (263‐PA12‐266) (C), and PDZ tandem (263‐PA12‐267) (D). Inter‐strand hydrogen bonds are shown as orange dotted lines.
Compared with the PDZ‐N domain of PDZ tandem (181‐PA12‐184), the structure of the PA‐inserted PDZ‐C domain deviated from that of the wild type to a greater extent. The 77 Cα atoms of PDZ‐C excluding the PA tag were superposed on those of the wild type with an RMSD of 0.980 Å. Unexpectedly, the largest deviation was due to a conformational change of the βA‐βB loop, rather than one near the PA‐insertion site. It had been found that the βA–βB loop was relatively flexible in the crystallographic analysis of the wild‐type PDZ tandem.15 The conformation of this loop region is easily affected by lattice contacts because it largely projects away from the domain. Accordingly, the RMSD decreased to 0.696 Å when the six residues belonging to the βA–βB loop were excluded from the superposition.
Optimization of PA‐insertion sites to improve diffraction qualities of co‐crystals
The above structural analysis does not support the conclusion that fold destabilization from insertion of the PA tag caused the reduced resolution for the co‐crystal structure. Apparently, the inserted PA tag did not disrupt the fold topology of the PDZ domains in either mutant. Another possibility was that the flexibility between the PA‐inserted PDZ domain and NZ‐1 Fab increased the thermal vibration of the entire complex in the crystal lattice and decreased the resolutions of the co‐crystals. In both cases, the NZ‐1 Fab was tethered to the PDZ tandem mutants only through the inserted PA tag, without direct contacts with the remainder of the target PDZ tandem [Figs. 2(C) and 3(C)], while it formed lattice contacts with both the NZ‐1 Fab and the PDZ tandem from the symmetry neighbors. Furthermore, some residues located at the junction with the PA tag appeared to be highly flexible in both mutants, as mentioned above. These observations suggested that the junction residues permit some extent of a swinging motion of the NZ‐1 Fab with respect to the PDZ domain. Hence, we constructed two new mutants, PDZ tandem (181‐PA12‐186) and (263‐PA12‐267), to make more rigid complexes by deleting the respective solvent‐exposed residues at the junction sites.
The two modified mutants were stable and monodisperse after purification, as shown by the elution profiles of the size‐exclusion chromatography [Supporting Information Fig. S1(E, F)]. We successfully obtained co‐crystals with the NZ‐1 Fab for both of them. As hypothesized, the resolutions of the X‐ray diffraction were improved remarkably as compared to those of the respective pre‐modified mutants. The co‐crystal structures with PDZ tandem (181‐PA12‐186) and (263‐PA12‐267) were finally refined to resolutions of 2.0 Å and 2.6 Å, respectively [Figs. 2(D–F) and 3(D–F)]. In the complex with PDZ tandem (181‐PA12‐186), the 12.4 Å separation of the Cα atoms in Gly‐1 and Val‐12 caused a flip‐out of Leu‐186 at the junction into the solvent, but the inter‐strand hydrogen bonds with the βF strand were maintained in the downstream of this residue [Figs. 4(D) and 5(D), and Supporting Information Fig. S2(C)]. In the complex with the PDZ tandem (263‐PA12‐267), it appeared as if removing Lys‐266 had no noticeable effect on the main chain conformation of the flanking residues and the inter‐strand hydrogen bonds [Fig. 6(A, D)]. The recognition mode of the PA tag as well as the distance between Gly‐1 and Val‐12 were essentially consistent with those from other complexes [Figs. 4(E) and Supporting Information Fig. S2(D)].
Although the removal of the junction residues noticeably changed the orientation of the NZ‐1 Fab with respect to the PDZ tandem, the NZ‐1 Fab again made no direct contacts with the PDZ tandem other than at the inserted PA tag in both complexes (Figs. 2 and 3). It is, therefore, presumed that some flexibility still remained in the relative arrangement between the NZ‐1 Fab and the PDZ tandem. Again, the PA‐insertion and Fab‐binding in these new mutants has little effect on the fold of the target PDZ domains. The 76 Cα atoms of PDZ‐N in PDZ tandem (181‐PA12‐186) were superposed on their counterparts from the wild‐type structure with an RMSD of 0.630 Å. The PDZ‐C in PDZ tandem (263‐PA12‐267) again showed a slightly larger structural change from that of the wild type, but the RMSD was reduced to 0.722 Å for 71 Cα atoms excluding the βA–βB loop region. In contrast, the relative arrangement of PDZ‐N and ‐C domains is flexible and varies between the individual models.
High‐resolution structure of PDZ tandem (263‐PA12‐267) in the Fab‐free form
In contrast to the pre‐modified mutant, PDZ tandem (263‐PA12‐267) produced crystals in the Fab‐free form as well, which resulted in a structure determined to 1.9 Å resolution [Fig. 7(A)]. The electron density of the inserted PA tag sequence was also sufficiently clear to build the model. Of note, the PA tag was partly incorporated into the β‐hairpin structure of PDZ‐C. Gly‐1 and Val‐2 participated in the βF strand, and formed hydrogen bonds with Asp‐10, Val‐11, and Val‐12 included in the βA strand [Fig. 7(B)]. In addition, the Type II β‐turn was constituted by Met‐4, Pro‐5, Gly‐6, and Ala‐7 in the typical conformation whereas the inter‐strand hydrogen bonds were disrupted at Ala‐3, Glu‐8, and Asp‐9. The extended β‐hairpin protrudes out of PDZ‐C to a length of about 16 Å, and it makes contacts with four neighboring molecules in the crystal [Fig. 7(C)]. It appears that the lattice contacts with the β‐hairpin generate a rigidly packed crystal with a solvent content of 44.5%, which is much lower than that of the Fab‐free wild type crystal.
Figure 7.
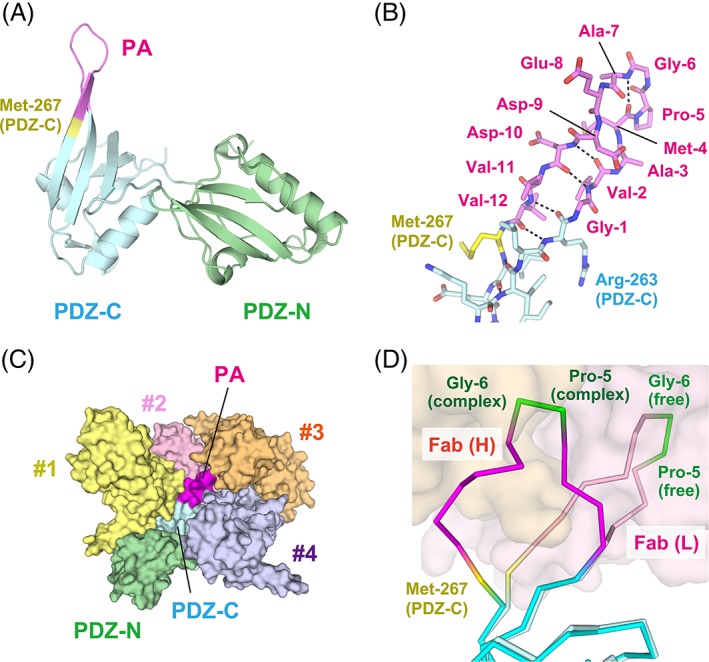
Crystal structure of PDZ tandem (263‐PA12‐267) in the Fab‐free form. (A) Ribbon model of the entire PDZ tandem. The inserted PA tag and Met‐267 are colored in magenta and yellow, respectively. The N‐ and C‐terminal residues of the PA tag are incorporated into the β‐strands of the target PDZ‐C domain. (B) Stick model of the inserted PA tag. Inter‐strand hydrogen bonds are indicated with dotted lines. (C) Lattice contacts for PDZ tandem (263‐PA12‐267). PDZ tandem (263‐PA12‐267) is shown in surface model with the same coloring as in (A). The inserted PA tag makes direct contacts with four neighboring molecules (#1–#4) in the crystal lattice. (D) Conformational change of the inserted PA tag upon the Fab‐binding. The Cα trace model in bright colors represents the conformation in the Fab‐bound form while the pale‐colored model represents the Fab‐free form. The NZ‐1 Fab in the complex is shown as a surface model. Met‐267 alone has undergone a conformational change in the target PDZ‐C domain. In contrast, Pro‐5 and Gly‐6 maintain a β‐turn conformation after Fab‐binding.
Comparison between the Fab‐free and Fab‐bound forms indicated that the structure of target PDZ‐C domain remarkably changed in Met‐267 at the PA tag junction as a result of Fab‐binding. The βA–βB loop of PDZ‐C also showed a larger conformational change with a 6 Å shift at the Cα atom of Lys‐279 in the β‐turn, which is presumably affected by lattice contacts. However, the main chain conformation was maintained between the Fab‐free and Fab‐bound forms with an RMSD of 0.823 Å for the 71 Cα atoms excluding the PA tag and the βA–βB loop region. These observations indicate that the inserted PA tag can adopt a conformation suitable for the interaction with NZ‐1 without affecting the folding of the target PDZ‐C domain [Fig. 7(D)].
Discussion
Application of antibody fragments has brought great success for structural biology. In particular, crystallographic analyses of membrane proteins have been accelerated dramatically by utilizing antibody fragments as crystallization chaperones.1, 2, 4 One benefit for crystallization from antibody fragments is expanded solvent‐exposed area in targets such as membrane proteins. Antibody fragments are also useful for crystallizing other difficult‐to‐crystallize proteins such as glycoproteins with conformational flexibilities.16 Most of the antibody fragment‐assisted crystallizations have been performed using an antibody that specifically recognizes the tertiary structure of the target protein. Our study further raised the possibility that an anti‐tag antibody can be applied to the same kind of crystallization strategy under the condition that the tag sequence can be inserted into the target protein without disrupting its tertiary structure. In the present work, we selected the β‐hairpins in the PDZ tandem fragment of a bacterial S2P homolog as the insertion sites because the Fab‐bound PA tag is known to adopt a loop‐like structure with a β‐turn between Pro‐5 and Gly‐6. Although the size of the PA tag is not negligible compared with the total size of the individual 80‐residue PDZ domains, the PA‐insertion did not disrupt the fold topology of the target PDZ domains for any of the mutants constructed in the present work (Supporting Information Fig. S1). Furthermore, our results have shown that the inserted PA tag can serve as a scaffold to generate a ready‐to‐crystallize complex between the NZ‐1 Fab and the target PDZ tandem. In fact, we obtained co‐crystals with the NZ‐1 Fab for all of the PA‐inserted mutants, even in cases where crystallization was not successful for the Fab‐free form.
In addition, our results strongly indicated that optimization of the insertion site should be an important process for obtaining a co‐crystal with a higher diffraction quality. We attempted to insert the PA tag into the two different sites and initially obtained co‐crystals with moderate diffraction qualities for both cases. Based on our inspection of the co‐crystal structures, we hypothesized that the conformational flexibilities between the NZ‐1 Fab and the target PDZ domains lowered the diffraction qualities. The NZ‐1 Fab did not form any specific interactions with the target PDZ domains, but was tethered to them only through the inserted PA tag within the complex [Figs. 2(C, F) and 3(C, F)]. Hence, the solvent‐accessible space present between the NZ‐1 Fab and the rigidly folded part of the PDZ domain is thought to increase thermal vibration throughout the entire complex. To test the hypothesis, we modified both of the two mutants by eliminating the flexible residues present at the insertion sites. Ultimately, the two modified mutants produced the co‐crystals with higher diffraction qualities than those of the respective pre‐modified mutants. The averaged temperature factors in the co‐crystals significantly decreased after the modification of the insertion sites (Table 2), although it is not a direct measurement of decreased conformational flexibilities in the complexes. Remarkably, the diffraction qualities were also higher than that of the Fab‐free wild‐type crystal. These results support the case that the NZ‐1 Fab plays a beneficial role in the crystallization and improved diffraction quality of the PDZ tandem mediated through the inserted PA tag.
Table 2.
Refinement Statistics
| Construct | PDZ tandem (181‐PA12‐184) | PDZ tandem (181‐PA12‐186) | PDZ tandem (263‐PA12‐266) | PDZ tandem (263‐PA12‐267) | PDZ tandem (263‐PA12‐267) |
|---|---|---|---|---|---|
| Form | Fab‐bound | Fab‐bound | Fab‐bound | Fab‐bound | Fab‐free |
| Resolution limits (Å) | 48.35–3.20 (3.28–3.20) | 48.23–2.00 (2.05–2.00) | 48.64–4.00 (4.10–4.00) | 50.09–2.60 (2.67–2.60) | 42.75–1.90 (1.95–1.90) |
| R work a | 0.278 (0.442) | 0.213 (0.312) | 0.216 (0.304) | 0.199 (0.294) | 0.200 (0.286) |
| R free a | 0.313 (0.457) | 0.260 (0.314) | 0.273 (0.347) | 0.251 (0.403) | 0.254 (0.392) |
| No. of atoms | 3,928 | 5,046 | 4,724 | 5,012 | 1,581 |
| PDZ tandem | 745 | 1,470 | 1,439 | 1,480 | 1,482 |
| PDZ‐N | 745 | 787 | 714 | 755 | 751 |
| PDZ‐C | 0 | 683 | 725 | 725 | 731 |
| NZ‐1 Fab | 3,183 | 3,325 | 3,285 | 3,350 | ‐ |
| Solvent | 0 | 251 | 0 | 182 | 99 |
| Average B‐factor (Å2) | 115.31 | 47.64 | 132.48 | 42.22 | 32.32 |
| PDZ tandem | 121.57 | 50.16 | 146.55 | 55.99 | 31.92 |
| PDZ‐N | 121.57 | 46.53 | 160.94 | 63.81 | 31.34 |
| PDZ‐C | 54.35 | 132.37 | 47.85 | 32.52 | |
| NZ‐1 Fab | 113.85 | 46.22 | 126.32 | 36.46 | ‐ |
| Solvent | ‐ | 51.55 | ‐ | 36.09 | 38.25 |
| RMSD from ideality | |||||
| Bond length (Å) | 0.002 | 0.004 | 0.008 | 0.004 | 0.008 |
| Bond angle (°) | 0.73 | 1.11 | 0.82 | 1.06 | 1.67 |
| Ramachandran plot | |||||
| Favored (%) | 92.45 | 98.36 | 92.76 | 96.36 | 98.90 |
| Outlier (%) | 0.60 | 0.16 | 0.82 | 0 | 0 |
| PDB code | 6AL1 | 6ICC | 6ICF | 6AL0 | 6AKQ |
Values in parentheses are for highest‐resolution shell.
R work is the crystallographic R‐factor (R cryst) for the working set used for the refinement. R cryst = Σh||F obs(h)|−|F calc(h)||/Σh| F obs(h)|, where F obs(h) and F calc(h) are the observed and calculated structure factors, respectively.
R free is R cryst calculated for the test set consisting of 5% of reflections excluded from the refinement.
The structure determination of PDZ tandem (263‐PA12‐267) has demonstrated that the inserted PA has potential to form a β‐hairpin structure. The formation of the β‐hairpin structure of the PA tag in the Fab‐free PDZ tandem (263‐PA12‐267) crystal is probably not only affected by the lattice contact but also dependent on the sequence context. Nevertheless, it is highly possible that β‐hairpins are generally competent as PA‐insertion sites. As the Pro‐Gly sequence generally has a higher β‐turn propensity,17, 18 it is presumed that Pro‐5 and Gly‐6 at the middle of the PA tag sequence are predisposed to form a turn at the insertion site. When incorporated as a β‐hairpin, the PA tag is expected to dock to the NZ‐1 Fab with an intact β‐turn conformation between Pro‐5 and Gly‐6. Furthermore, it is noteworthy that other than for Met‐267 the main chain conformations of the residues flanking the PA tag are almost identical in both the Fab‐free and Fab‐bound PDZ tandem (263‐PA12‐267) [Fig. 7(D)]. It seems that the inter‐strand hydrogen bonds maintained the folding of the target PDZ domain during the Fab‐binding, which also suggests that the β‐hairpins are suitable sites for PA insertion.
Although we here inserted the PA tag into a protein for which the 3D structure was initially available, it is presumed that the same co‐crystallization strategy can be applied to proteins whose 3D structures have not yet been determined experimentally. Accumulation of structural data in Protein Data Bank (PDB) and development of structure prediction algorithms19, 20 should enable us to predict the positions of loop regions that become candidates for PA‐insertion sites in proteins without an initial mid‐ to low‐resolution structure model. For instance, glycoproteins secreted to extracellular space or ectodomain fragments of membrane proteins should be ideal targets for the PA insertion as they are abundant in the β‐hairpin structures. Although multi‐pass membrane proteins are generally α‐helical in the transmembrane regions, the loop regions connecting the α‐helices can be candidates for the insertion sites. Based on our structural analysis, an additional criterion can be proposed to select candidate insertion sites. It has been shown in the complex structures that docking to the NZ‐1 Fab separates Gly‐1 and Val‐12 of the PA tag by at least 12 Å (Fig. 4). Therefore, PA tag insertion sites that conform to this distance constraint can be selected from a predicted structural model. In fact, some residues belonging to the βA strand were deleted along with the loop residues for PA insertion in the two modified mutants of the PDZ tandem. The additional deletions extended the end‐to‐end distance of the PA tag in the host proteins and thereby minimized the structural distortion upon the Fab‐binding in both two cases. We expect some difficulty in finding an optimal insertion site in the case of de novo structure determination. Nevertheless, once initial co‐crystal structures can be determined it should be possible in principle to improve the diffraction qualities of co‐crystals from low‐resolution starting points by optimizing the insertion sites, as we demonstrated in this work.
In addition to the merit for protein crystallography, it is also highly probable that complex formation with an antibody fragment through an inserted epitope will be advantageous for structural determination via cryo‐EM. In recent years, cryo‐EM analysis has undergone dramatic development and has produced a growing number of atomic models for difficult‐to‐crystallize targets.21, 22, 23 However, it is still difficult to resolve high‐resolution cryo‐EM structures for targets with smaller particle sizes and lower symmetries. Several characteristics of our approach should have benefit toward these technical challenges in cryo‐EM analysis. An antibody fragment bound to the target should increase the size of the particle and serve to identify the orientation. While the conformational flexibility between the target and the bound antibody is expected to disturb the averaging procedure, it could be addressed by optimizing the insertion site based on the preliminary structural analysis as discussed above.
Conclusion
The present work demonstrates that the NZ‐1‐PA system has potential to be applied to antibody fragment‐assisted crystallization provided that the insertion sites of the PA tag are properly selected and optimized. In particular, proteins possessing β‐hairpins are promising candidates for PA‐insertion targets. The next issue to be addressed is whether or not our strategy can be applied to structure determination of more difficult targets such as α‐helical integral membrane proteins and modular glycoproteins with larger molecular sizes.
Materials and Methods
Purification of the PA‐inserted PDZ tandem mutants in the Fab‐free and Fab‐bound forms
The PDZ tandem fragment (Residues 115–292) was produced as an N‐terminal GST (glutathione S transferase) ‐fusion protein, in which a TEV protease recognition site was incorporated between the GST and PDZ tandem sequences, as reported previously.15 Each PA‐inserted construct was prepared by amplifying the whole plasmid DNA by PCR using a primer containing the DNA sequence of the PA tag. Escherichia coli BL21(DE3) cells transformed with the expression plasmid were grown at 37°C to an OD600 of 0.7 in a medium containing 10 g of bactotryptone, 5 g of yeast extract and 10 g of NaCl per liter supplemented with 50 μg/mL ampicillin, followed by induction of overexpression with 0.1 mM isopropyl β‐d‐thiogalactopyranoside and incubation at 25°C for additional 12 h. Cells were harvested by centrifugation and lysed by sonication in Tris buffered saline (TBS) composed of 10 mM Tris‐Cl (pH 7.4) and 150 mM NaCl. The soluble fraction of the cell lysate was mixed with Glutathione‐Sepharose 4B resin (GE Healthcare UK Ltd, Buckinghamshire, England) and incubated at 4°C for 1 h. After washing out the unbound fraction with TBS, the PDZ tandem was cleaved off from the GST portion through on‐column digestion with TEV protease at 20°C for 12 h. The released PDZ tandem fragment was further purified using cation‐exchange chromatography HiTrap SP HP (GE Healthcare UK Ltd) and size‐exclusion chromatography Superdex 200 Increase 10/300 GL (GE Healthcare UK Ltd). Gel Filtration Standard (Bio‐Rad) was used as the molecular weight marker in size‐exclusion chromatography. The purified PDZ tandem was mixed with the NZ‐1 Fab at a molar ratio of 2:1 and incubated on ice for 30 min. NZ‐1 was obtained from the Antibody Bank (http://www.med-tohoku-antibody.com/topics/antibody.htm) at Tohoku University (Miyagi, Japan). Briefly, NZ‐1 was produced in Hybridoma‐SFM (Thermo Fisher Scientific Inc., Waltham, MA, USA), and was purified using Protein G (GE Healthcare UK Ltd). The NZ‐1 Fab was prepared by papain digestion according to the protocol as described previously.11 The mixture of PDZ tandem and NZ‐1 Fab was applied to size‐exclusion chromatography again to separate the PDZ tandem fragments into the Fab‐bound and Fab‐free forms. The final protein samples were concentrated by ultrafiltration.
Crystallization and data collection
The initial crystallization screens were conducted using Index™ (Hampton Research) crystallization reagents. About 0.2 μL of the protein solutions and reagents, respectively, were dispensed into 96‐well plates using a Gryphon robotic crystallization system (Art Robbins Instruments) and equilibrated against 60 μL of the reservoir by the sitting‐drop vapor‐diffusion method. The crystals subjected to X‐ray crystallographic analysis were generated from crystallization buffers as follows. The Fab‐bound PDZ tandem (181‐PA12‐184) was crystallized in buffer containing 5 mM CoCl2, 5 mM NiCl2, 5 mM CdCl2, 5 mM MgCl2, 100 mM HEPES‐Na (pH 7.5), and 12% (wt./vol.) polyethylene glycol (PEG) 3350. The crystals of the Fab‐bound PDZ tandem (263‐PA12‐266) were obtained in buffer containing 200 mM ammonium citrate (pH 7.0), and 20% (wt./vol.) PEG 3350. For Fab‐bound PDZ tandem (181‐PA12‐186), the crystallization buffer contained 200 mM sodium citrate, and 12–15% (wt./vol.) PEG 3350. For PDZ tandem (263‐PA12‐267), the crystal of the Fab‐bound form was obtained from buffer containing 200 mM ammonium citrate (pH 7.0), and 15–20% (wt./vol.) PEG 3350 while the Fab‐free form was crystallized in buffer containing 200 mM Bis‐Tris‐Cl (pH 6.5), and 23–25% (wt./vol.) PEG 3350. Cryoprotectant was prepared by mixing the crystallization buffer and ethylene glycol with a volume ratio of 4:1. However, the crystals of the Fab‐bound PDZ tandem (181‐PA12‐184) were labile in the cryoprotectant prepared according to this protocol. Therefore, we prepared cryoprotectant containing the same concentration of precipitant as the crystallization buffer to prevent the crystals from dissolving. All of the crystals were quickly soaked in the respective cryoprotectant and frozen in liquid nitrogen. X‐ray diffraction data were collected with a photon counting pixel array detector Eiger X4M (Dectris) at Photon Factory (PF) BL‐1A (Tsukuba, Japan) or with PILATUS3 S 6M (Dectris) at PF BL‐17A. Data were processed and scaled with XDS24 and aimless.25 Intensities were converted to structure factors with the CCP4 programs where 5% of the unique reflections were randomly selected as a test set.26, 27 Data collection statistics are summarized in Table 1.
Table 1.
Data Collection Statistics
| Construct | PDZ tandem (181‐PA12‐184) | PDZ tandem (181‐PA12‐186) | PDZ tandem (263‐PA12‐266) | PDZ tandem (263‐PA12‐267) | PDZ tandem (263‐PA12‐267) |
|---|---|---|---|---|---|
| Form | Fab‐bound | Fab‐bound | Fab‐bound | Fab‐bound | Fab‐free |
| Space group | P212121 | P212121 | I422 | P4212 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) | 62.67, 83.04, 188.88 | 49.94, 84.29, 185.91 | 171.93, 171.79, 118.02 | 141.65, 141.68, 78.62 | 49.23, 54.80, 68.32 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| No. of monomers or complexes/a.s.u. | 1 | 1 | 1 | 1 | 1 |
| X‐ray source | PF/BL‐1A | PF/BL‐17A | PF/BL‐1A | PF/BL‐1A | PF/BL‐1A |
| Wavelength (Å) | 1.1000 | 0.9800 | 1.1000 | 1.1000 | 1.1000 |
| Resolution limits (Å) | 48.35–3.20 (3.42–3.20) | 48.23–2.00 (2.05–2.00) | 48.64–4.00 (4.47–4.00) | 50.09–2.60 (2.72–2.60) | 47.25–1.90 (1.94–1.90) |
| No. of unique reflection | 16,978 (3,004) | 54,069 (3,874) | 7,742 (2,144) | 25,220 (3,024) | 15,147 (948) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 6.6 (6.8) | 6.7 (6.7) | 13.5 (12.5) | 17.9 (18.5) | 13.4 (13.2) |
| I/σ(I) | 12.6 (1.8) | 16.1 (2.3) | 12.2 (3.9) | 12.5 (4.1) | 10.6 (2.1) |
| R merge a | 0.124 (1.170) | 0.056 (0.874) | 0.186 (0.744) | 0.183 (0.808) | 0.176 (2.746) |
| CC (1/2) | 0.932 (0.916) | 0.940 (0.873) | 0.996 (0.911) | 0.993 (0.924) | 0.997 (0.790) |
Values in parentheses are for highest‐resolution shell.
R merge = ΣhΣi|I i(h)− < I(h) > |/ΣhΣi I(h), where I i (h) is the ith measurement.
Crystallographic analysis
For each crystal, initial phases were determined by the molecular replacement (MR) method by using Molrep28 in CCP4. The atomic coordinates of the A. aeolicus PDZ tandem15 (PDB code: 3WKL) and NZ‐1 Fab with the bound PA14 peptide11 (PDB code: 4YO0) were used as search models. As the PDZ tandem arrangement is conformationally flexible, the model was separated into two portions, the PDZ‐N and ‐C domains. Similarly, the Fab model was also separated into the Fv and constant regions. Accordingly, the four partial structures were used as the search models in the calculation of MR phasing. After assignment of the respective models, manual fitting was performed with COOT29 and the updated models were refined with Refmac530 iteratively. For the Fab‐bound PDZ tandem (181‐PA12‐186) and the Fab‐free PDZ tandem (263‐PA12‐267), the initial models were improved by automatic model building with ARP/wARP31 prior to manual fitting. For the Fab‐bound PDZ tandem (181‐PA12‐184), the electron densities indicated the presence of metal ions bound to the protein surface. These metal ions were presumed to be cadmium ions included in the crystallization buffer. We displayed models of two ions together with the co‐ordinated amino acid residues in Figure 4(B) and Figure S2(A). However, we deleted the ions from the final model deposited in PDB because the entities of the electron densities could not be unambiguously assigned as explained in the Results section.
Stereochemical parameters of the final models were assessed with MolProbity.32 Refinement statistics are summarized and PDB accession codes are listed in Table 2. Structural superposition and RMSD calculation were performed by the pair‐wise alignment protocol using LSQKAB.33 Figures for protein structures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
Competing interests
The corresponding author declares no financial and non‐financial competing interests on behalf of all authors.
Supporting information
Supplementary Table 1 Summary of mass spectrometric analysis.
Supplementary Figure 1 Elution profiles of the PDZ tandem fragments in size‐exclusion chromatography. (A) PDZ tandem wild type. The PDZ tandem fragment with a molecular mass of 19.9 kDa, as shown in Suppl. Table 1, eluted at a volume of 17.4 mL. The peak positions of the molecular weight markers are indicated above the chart. (B) PDZ tandem (181‐PA12‐184) (C) PDZ tandem (263‐PA12‐266). (D) PDZ tandem (263‐PA12‐266) with the additional L259R mutation. (E) PDZ tandem (181‐PA12‐186). (F) PDZ tandem (263‐PA12‐267). The respective elution volumes of the PDZ tandem mutants are indicated on the plots.
Supplementary Figure 2 Omit maps around the PA insertion sites. (A) PDZ tandem (181‐PA12‐184). (B) PDZ tandem (263‐PA12‐266). (C) PDZ tandem (263‐PA12‐266). (D) PDZ tandem (263‐PA12‐267). Omit maps with a contour level of 3 σ were calculated for the respective PA‐inserted mutants in complex with the NZ‐1 Fab. The residues shown in stick models were omitted from the model to calculate the mF o–DF c maps. In PDZ tandem (181‐PA12‐184), the metal ions (orange spheres) found around the PA insertion site were also omitted from the model for the map calculation.
Supplementary Figure 3 Succinimide formation at residue 150 of the PDZ‐N domain. (A) Simulated‐annealing omit map around the Snn residue. The mF o–DF c map with a contour level of 3 σ was calculated for PDZ tandem (181‐PA12‐186) in complex with the NZ‐1 Fab. The omitted residues were indicated with light magenta. (B) Two βD‐βE loops in the PDZ tandem. As shown in the yellow stick model, the βD‐βE loop of the PDZ‐C domain also possesses the same set of residues, that is, Asn‐235 and Gly‐236.
Acknowledgments
We are grateful to the beamline staff of Photon Factory (Tsukuba, Japan) for providing data collection facilities and support. We thank Makiko Neyazaki for technical support, Samuel Thomson for editing the manuscript, and Prof Junichi Takagi for useful discussion. This research is partially supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant nos. JP26291016 and JP17K19206 (to T.N.), by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from Japan Agency for Medical Research and Development (AMED) under Grant no. JP16am0101020 (to T.N.), and by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED under Grant no. JP17am0101078 (to Y.K.).
References
- 1. Ostermeier C, Iwata S, Ludwig B, Michel H (1995) Fv fragment‐mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Biol 2:842–846. [DOI] [PubMed] [Google Scholar]
- 2. Zhou Y, Morais‐Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel‐Fab complex at 2.0 a resolution. Nature 414:43–48. [DOI] [PubMed] [Google Scholar]
- 3. Hunte C, Michel H (2002) Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol 12:503–508. [DOI] [PubMed] [Google Scholar]
- 4. Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK (2007) Crystal structure of the human beta2 adrenergic G‐protein‐coupled receptor. Nature 450:383–387. [DOI] [PubMed] [Google Scholar]
- 5. Day PW, Rasmussen SG, Parnot C, Fung JJ, Masood A, Kobilka TS, Yao XJ, Choi HJ, Weis WI, Rohrer DK, Kobilka BK (2007) A monoclonal antibody for G protein‐coupled receptor crystallography. Nat Methods 4:927–929. [DOI] [PubMed] [Google Scholar]
- 6. Hino T, Iwata S, Murata T (2013) Generation of functional antibodies for mammalian membrane protein crystallography. Curr Opin Struct Biol 23:563–568. [DOI] [PubMed] [Google Scholar]
- 7. Bradbury AR, Sidhu S, Dubel S, McCafferty J (2011) Beyond natural antibodies: the power of in vitro display technologies. Nat Biotech 29:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kato Y, Kaneko MK, Kuno A, Uchiyama N, Amano K, Chiba Y, Hasegawa Y, Hirabayashi J, Narimatsu H, Mishima K, Osawa M (2006) Inhibition of tumor cell‐induced platelet aggregation using a novel anti‐podoplanin antibody reacting with its platelet‐aggregation‐stimulating domain. Biochem Biophys Res Commun 349:1301–1307. [DOI] [PubMed] [Google Scholar]
- 9. Ogasawara S, Kaneko MK, Price JE, Kato Y (2008) Characterization of anti‐podoplanin monoclonal antibodies: critical epitopes for neutralizing the interaction between podoplanin and CLEC‐2. Hybridoma (Larchmt) 27:259–267. [DOI] [PubMed] [Google Scholar]
- 10. Fujii Y, Kaneko M, Neyazaki M, Nogi T, Kato Y, Takagi J (2014) PA tag: a versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Prot Expr Purif 95:240–247. [DOI] [PubMed] [Google Scholar]
- 11. Fujii Y, Matsunaga Y, Arimori T, Kitago Y, Ogasawara S, Kaneko MK, Kato Y, Takagi J (2016) Tailored placement of a turn‐forming PA tag into the structured domain of a protein to probe its conformational state. J Cell Sci 129:1512–1522. [DOI] [PubMed] [Google Scholar]
- 12. Wang H, Han W, Takagi J, Cong Y (2018) Yeast inner‐subunit PA‐NZ‐1 labeling strategy for accurate subunit identification in a macromolecular complex through cryo‐EM analysis. J Mol Biol 430:1417–1425. [DOI] [PubMed] [Google Scholar]
- 13. Brown ZP, Arimori T, Iwasaki K, Takagi J (2018) Development of a new protein labeling system to map subunits and domains of macromolecular complexes for electron microscopy. J Struct Biol 201:247–251. [DOI] [PubMed] [Google Scholar]
- 14. Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353–358. [DOI] [PubMed] [Google Scholar]
- 15. Hizukuri Y, Oda T, Tabata S, Tamura‐Kawakami K, Oi R, Sato M, Takagi J, Akiyama Y, Nogi T (2014) A structure‐based model of substrate discrimination by a noncanonical PDZ tandem in the intramembrane‐cleaving protease RseP. Structure 22:326–336. [DOI] [PubMed] [Google Scholar]
- 16. Griffin L, Lawson A (2011) Antibody fragments as tools in crystallography. Clin Exp Immunol 165:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hutchinson EG, Thornton JM (1994) A revised set of potentials for beta‐turn formation in proteins. Protein Sci 3:2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu HJ, Chang HJ, Peng HP, Huang SS, Lin MY, Yang AS (2006) Assessing computational amino acid beta‐turn propensities with a phage‐displayed combinatorial library and directed evolution. Structure 14:1499–1510. [DOI] [PubMed] [Google Scholar]
- 19. Petersen B, Lundegaard C, Petersen TN (2010) NetTurnP‐‐neural network prediction of beta‐turns by use of evolutionary information and predicted protein sequence features. PLoS One 5:e15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng C, Kurgan L (2008) Prediction of beta‐turns at over 80% accuracy based on an ensemble of predicted secondary structures and multiple alignments. BMC Bioinformatics 9:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng Y (2018) Single‐particle cryo‐EM – How did it get here and where will it go. Science 361:876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, Subramaniam S (2016) Breaking cryo‐EM resolution barriers to facilitate drug discovery. Cell 165:1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhlbrandt W (2014) Biochemistry. The resolution revolution. Science 343:1443–1444. [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W (2010) Xds. Acta Crystallogr D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Cryst D69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D50:760–763. [DOI] [PubMed] [Google Scholar]
- 27. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30:1022–1025. [Google Scholar]
- 29. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Cryst D53:240–255. [DOI] [PubMed] [Google Scholar]
- 31. Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS (2001) ARP/wARP and molecular replacement. Acta Cryst D57:1445–1450. [DOI] [PubMed] [Google Scholar]
- 32. Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Cryst D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W (1976) A solution for the best rotation to relate two sets of vectors. Acta Cryst A32:922–923. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Summary of mass spectrometric analysis.
Supplementary Figure 1 Elution profiles of the PDZ tandem fragments in size‐exclusion chromatography. (A) PDZ tandem wild type. The PDZ tandem fragment with a molecular mass of 19.9 kDa, as shown in Suppl. Table 1, eluted at a volume of 17.4 mL. The peak positions of the molecular weight markers are indicated above the chart. (B) PDZ tandem (181‐PA12‐184) (C) PDZ tandem (263‐PA12‐266). (D) PDZ tandem (263‐PA12‐266) with the additional L259R mutation. (E) PDZ tandem (181‐PA12‐186). (F) PDZ tandem (263‐PA12‐267). The respective elution volumes of the PDZ tandem mutants are indicated on the plots.
Supplementary Figure 2 Omit maps around the PA insertion sites. (A) PDZ tandem (181‐PA12‐184). (B) PDZ tandem (263‐PA12‐266). (C) PDZ tandem (263‐PA12‐266). (D) PDZ tandem (263‐PA12‐267). Omit maps with a contour level of 3 σ were calculated for the respective PA‐inserted mutants in complex with the NZ‐1 Fab. The residues shown in stick models were omitted from the model to calculate the mF o–DF c maps. In PDZ tandem (181‐PA12‐184), the metal ions (orange spheres) found around the PA insertion site were also omitted from the model for the map calculation.
Supplementary Figure 3 Succinimide formation at residue 150 of the PDZ‐N domain. (A) Simulated‐annealing omit map around the Snn residue. The mF o–DF c map with a contour level of 3 σ was calculated for PDZ tandem (181‐PA12‐186) in complex with the NZ‐1 Fab. The omitted residues were indicated with light magenta. (B) Two βD‐βE loops in the PDZ tandem. As shown in the yellow stick model, the βD‐βE loop of the PDZ‐C domain also possesses the same set of residues, that is, Asn‐235 and Gly‐236.


