Abstract
Background
Whipple’s disease (WD) is a rare infection with Tropheryma whipplei that is fatal if untreated. Diagnosis is challenging and currently based on invasive sampling. In a case of WD diagnosed from a kidney biopsy, we observed morphologically-intact bacteria within the glomerular capsular space and tubular lumens. This raised the questions of whether renal filtration of bacteria is common in WD and whether polymerase chain reaction (PCR) testing of urine might serve as a diagnostic test for WD.
Methods
We prospectively investigated urine samples of 12 newly-diagnosed and 31 treated WD patients by PCR. As controls, we investigated samples from 110 healthy volunteers and patients with excluded WD or acute gastroenteritis.
Results
Out of 12 urine samples from independent, therapy-naive WD patients, 9 were positive for T. whipplei PCR. In 3 patients, fluorescence in situ hybridization visualized T. whipplei in urine. All control samples were negative, including those of 11 healthy carriers with T. whipplei–positive stool samples. In our study, the detection of T. whipplei in the urine of untreated patients correlated in all cases with WD.
Conclusions
T. whipplei is detectable by PCR in the urine of the majority of therapy-naive WD patients. With a low prevalence but far-reaching consequences upon diagnosis, invasive sampling for WD is mandatory and must be based on a strong suspicion. Urine testing could prevent patients from being undiagnosed for years. Urine may serve as a novel, easy-to-obtain specimen for guiding the initial diagnosis of WD, in particular in patients with extra-intestinal WD.
Keywords: Whipple’s disease, Tropheryma whipplei, real-time PCR, electron microscopy, fluorescence in situ hybridization
We found that in the majority of therapy-naive Whipple’s disease patients, Tropheryma whipplei is detectable in urine samples by polymerase chain reaction; therefore, urine may serve as a novel, easy-to-obtain diagnostic specimen for the initial diagnosis of Whipple’s disease.
(See the Editorial Commentary by Raoult on pages 1098–9.)
Whipple’s disease (WD) is a rare bacterial infection caused by Tropheryma whipplei that is usually fatal if untreated. Classical WD is a systemic infection associated with polyarthritis, chronic diarrhea, weight loss, and fever [1–3]. T. whipplei infection may, however, affect almost every organ or can be confined also to isolated organs (eg, joints, heart valves, skin) [4–7]. Consequently, symptoms may vary substantially depending on the location of infection and the immunologic host response, and may even present without any intestinal involvement. The key challenge for diagnosis is, therefore, rapid detection of the pathogen. To date, many patients receive diagnoses only years after onset of symptoms and, until then, are treated incorrectly for various other disorders [1, 8, 9].
Sample specimens for the diagnosis of WD are usually biopsies from the affected organs. Diagnosis is based on periodic acid–Schiff (PAS)-positive macrophages in biopsies and must be confirmed by an independent, specific method such as polymerase chain reaction (PCR) [1, 10–15]. In extra-intestinal WD, PCR can be more sensitive than histology [16]. Often, sampling of the affected organ is difficult or impossible: for example, in endocarditis. Stool and saliva have been suggested as diagnostic materials, since the prevalence and load of T. whipplei are far higher in samples from WD patients than from healthy subjects [5, 13, 17]. However, positive results are not a conclusive proof of infection—as opposed to direct detection of T. whipplei deoxyribonucleic acid (DNA) in affected organs—since asymptomatic carriage exists. The prevalence of healthy carriers with positive PCR results in their stool has been estimated to be >4% and even up to 12% in high-risk groups (ie, sewage workers or homeless people) for direct or indirect fecal-oral transmission [13, 18]. A very high prevalence (up to 75%) in stool samples of children under 5 years old has been reported for rural Senegal [19]. While diagnosis of WD is challenging, the disease usually can be successfully treated by antibiotic therapy if diagnosed early enough [20, 21]. Therefore, a non-invasive and safe approach to guide diagnosis of this deadly infection is highly desirable. Our aim was to assess whether PCR testing for T. whipplei DNA in urine could serve as diagnostic test in WD.
CASE REPORT
A 70-year-old man presented with weight loss, arthralgia, and a disturbance of short-term memory. He reported weight loss of 10 kg during the last 7 months, as well as listlessness, anhedonia, fatigue, and night sweats. During the past 6 years, he had been treated for seronegative rheumatoid arthritis with steroids, methotrexate, and leflunomide, without significant effect.
On physical examination, the patient was in a reduced general condition and cachectic (body mass index 18.5). He was afebrile and hemodynamically stable. Laboratory work-up demonstrated anemia (hemoglobin 9.4 g/dl), thrombocytopenia (platelets of 129 x 109/l), and an elevated C-reactive protein level (95 mg/l).
Bone scintigraphy and magnetic resonance imaging showed active, bilateral sacroiliitis. Re-assessment for an infectious cause included repeated blood cultures (extended incubation for 4 weeks), PCR for Chlamydia trachomatis from urine, an interferon gamma release assay, and serologic testing for Herpes simplex virus, Epstein Barr virus, cytomegalovirus, varizella-zoster virus, brucellosis, borreliosis, and syphilis, none of which provided evidence for an active infection.
Human leukocyte antigen (HLA) B27-negative spondyloarthritis was suspected and treatment with 50 mg weekly of Etanercept was started. Progressive thrombocytopenia (59 x 109/l) and persistent systemic inflammation developed. Rising creatinine from normal values to 2.1 mg/dl finally led to the performance of a renal biopsy.
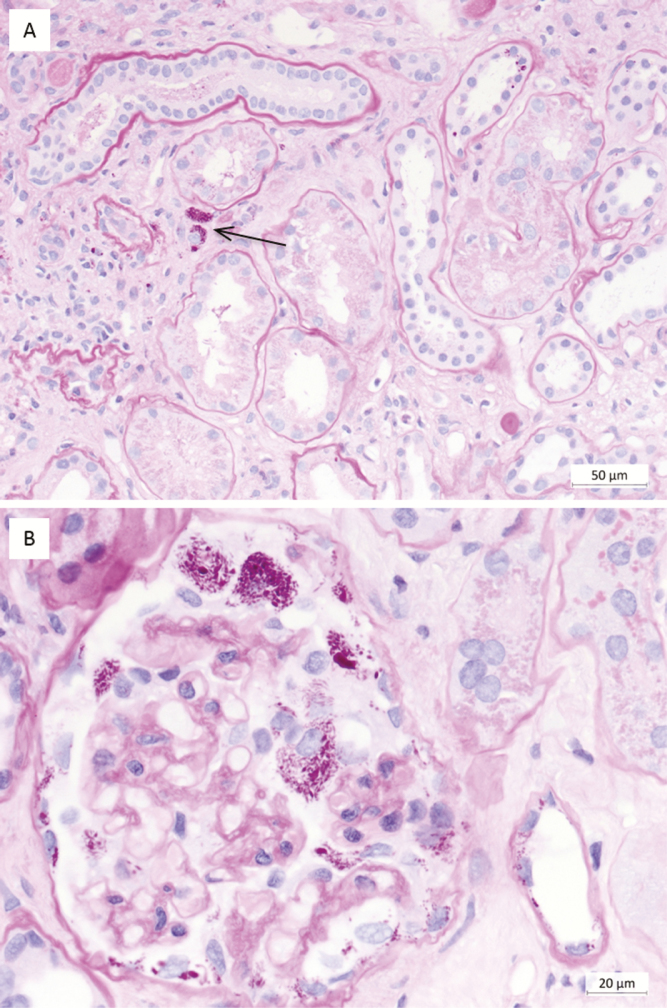
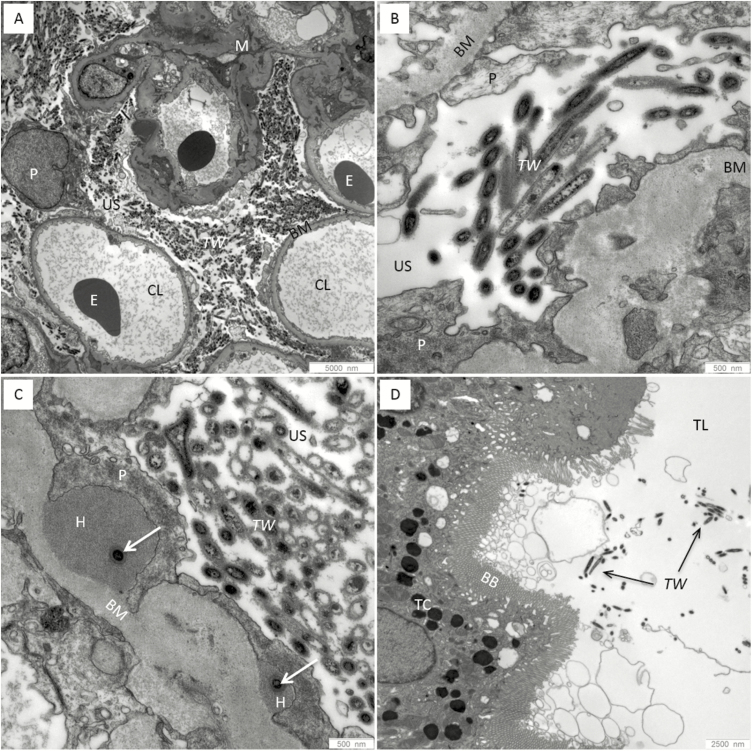
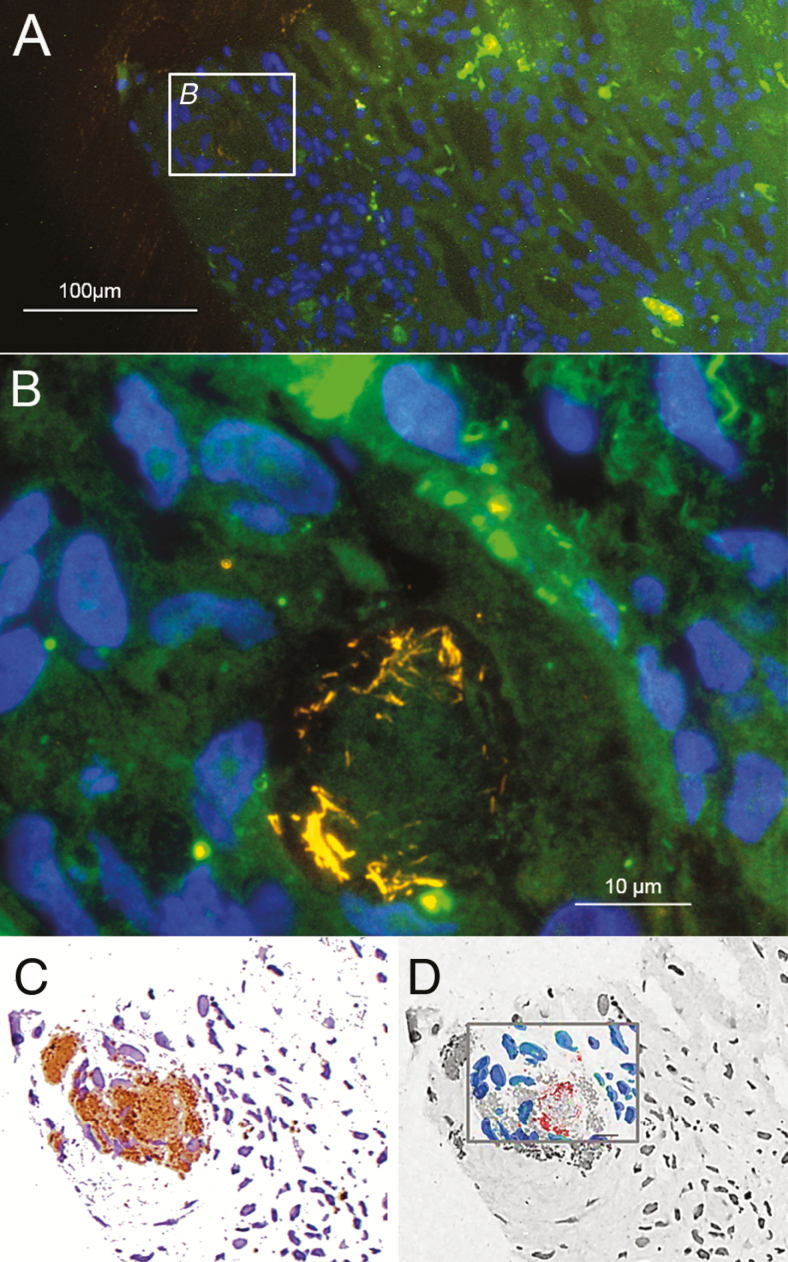
A histologic examination of the biopsy showed mild glomerulopathy of a post-/parainfectious type with additional, strongly PAS–positive particles within the glomerular capsular space, the cytoplasm of some glomerular and tubular epithelial cells, and a few interstitial cells (Figure 1). A PAS stain is routinely used as part of the workup of a renal biopsy. An electron microscopy revealed rod-shaped, 150 nm thick and up to 1.1 µm long structures bounded by a trilaminar wall, highly suggestive for bacteria, in the glomerular capsular space and in tubular lumens (Figure 2). A 16S ribosomal ribonucleic acid (rRNA)-gene PCR and sequence analysis, as well as specific quantitative real-time PCR, identified T. whipplei in high concentrations in the renal biopsy. This was confirmed by specific fluorescence in situ hybridization (FISH; Figure 3). Despite suspected central nervous system involvement, cardiac magnetic resonance imaging was normal and the cerebral spinal fluid was negative for T. whipplei DNA. Genotyping of the T. whipplei DNA isolated from both urine and renal biopsy tissue uniformly identified genotype 19, indicating that the patient was suffering from classical WD with a common T. whipplei strain [22].
Figure 1.
Histopathologic findings in the kidney biopsy of the index patient. (A) Periodic acid–Schiff (PAS): few macrophages in the interstitium with PAS-positive particles (Tropheryma whipplei) within the cytoplasm (arrow). (B) Periodic acid–Schiff: glomerulus with numerous, strongly PAS-positive, sickle-shaped particles (Tropheryma whipplei) within the urinary space and close to several visceral and parietal epithelial cells, as well as in the cytoplasm of some distal tubular epithelial cells (bottom right).
Figure 2.
Electron microscopic findings in the kidney biopsy of the index patient. (A and B) Numerous electron-dense rod-shaped structures (Tropheryma whipplei) in the urinary space outside the capillary lumens next to the podocytes, bounded by a trilaminar cell wall. (C) Several T. whipplei bacteria in the urinary space and occasionally (arrows) within electron-dense subepithelial humps, surrounded by the cytoplasm of a podocyte. (D) Few T. whipplei bacteria in the lumen of a proximal tubule.
Abbreviations: BB, brush border; BM, basement membrane; CL, capillary lumens; E, erythrocytes; H, electron-dense subepithelial humps; M, mesangium; P, podocytes; TC, tubular cell; TL, lumen; TW, Tropheryma whipplei; US, urinary space
Figure 3.
Tropheryma whipplei detected in the kidney biopsy of the index patient by FISH and immunohistochemistry. (A) Overview of the tissue sample showing the tissue background in green, T. whipplei (detected by REWHIPCy3 [26]) in orange, and nucleic acid stain DAPI in blue. (B) Inset B from panel A at a higher magnification, showing a cluster of T. whipplei cells in the tissue. (C) Immunohistochemistry of the identical microscopic view on the identical slide. Immunohistochemistry was performed with specific anti–T. whipplei antibodies. Antibody-stained regions shown in brown. (D) Overlay of the FISH signals from panel B in red and the DAPI-stained host cell nuclei in blue, with the immunohistochemistry image in panel C shown here in in black and white and the immunostain-positive region in gray. Note that only part of the antibody-stained region shows FISH signals. A FISH signal is only present in bacterial cells that contain ribosomes and are, therefore, presumably active at the time of sampling. A combination of FISH and immunohistochemistry, as shown here, may therefore provide therapy-relevant information that cannot be otherwise obtained. Abbreviations: DAPI, nucleic acid stain 4′,6- diamidino-2-phenylindole dihydrochloride; FISH, fluorescence in situ hybridization; REWHIPCy3, Tropheryma whipplei–specific FISH probe.
Upon diagnosis of WD, ceftriaxone was given intravenously for 14 days, followed by trimethoprim-sulfamethoxazole at 480 mg twice daily, with the dose adjusted to the current renal impairment. This led to a slow but dramatic clinical improvement with normalized C-reactive protein and blood parameters. The renal function recovered with a decrease of serum creatinine to 1.5 mg/dl, and the arthritis dissolved.
The observation of bacteria within the glomerular capsular space and in proximal tubular lumens (ie, in the primary urine) led to the hypothesis that T. whipplei could pass through the glomerular filter without destroying it. This raised the questions of whether (1) renal filtration of bacteria is a common event in WD and (2) PCR testing of urine might thus serve as a diagnostic test for the diagnosis of WD, even in patients without renal dysfunction.
MATERIALS AND METHODS
Study Design
Between August 2013 and June 2015, 70 potentially-eligible participants were included in this study (see Table 1 and Supplementary Figure S1). Out of these, WD was clinically considered a differential diagnosis in 27 participants, but no laboratory finding supported the diagnosis, and therefore they were grouped to the controls. Samples from all remaining 43 participants, including the index patient, were referred to the German Consiliary Laboratory (Deutsches Herzzentrum Berlin) and Charité–Universitätsmedizin Berlin for Tropheryma whipplei testing and were categorized into 3 groups according to their treatment status (Table 1): Group 1 consisted of newly-diagnosed WD patients who received no antibiotic treatment (n = 12), Group 2 consisted of newly-diagnosed WD patients who received a short-term antibiotic treatment (up to 14 days, n = 10), and Group 3 consisted of patients who had received treatment previously for 3–12 months (n = 21).
Table 1.
Patient and Control Group
| No | Group | State, Antibiotic Treatment | Male/Female Ratio | Age Range in Years, Median ± SD | Stool With Tropheryma whipplei DNAa | Urine With Tropheryma whipplei DNAa |
|---|---|---|---|---|---|---|
| 1 | WD group 1, initial diagnosis, n = 12 |
Untreated | 10/2 | 37–74, 61 ± 10 |
8/11, 72.7% | 9/12, 75.0% |
| 2 | WD group 2, n = 10 |
Short-term treatment for up to 14 days | 8/2 | 27–76, 54 ± 17 |
6/8, 75.0% | 2/10, 20.0% |
| 3 | WD group 3, n = 21 | Long-term treatment: 3–12 months; sampling 3–144 months after initiation of treatment |
11/10 | 51–79, 62 ± 8 |
5/21, 23.8% | 1/21, 4.7% |
| 4 | WD suspected, but not verified, n = 27 |
Untreated | 15/11 | 22–81, 53 ± 15 |
2/13, 15.4% | 0/27, 0.0% |
| 5 | Control group of patients with acute gastroenteritis, n = 43 |
Untreated | 27/16 | 21–83, 46 ± 19 |
4/43, 9.3% | 0/43, 0.0% |
| 6 | Control group of healthy volunteers, n = 40 |
Untreated | 14/26 | 20–73, 42 ± 14 |
5/40, 12.5% | 0/40, 0.0% |
Abbreviations: DNA, deoxyribonucleic acid; SD, standard deviation; T. whipplei, Tropheryma whipplei; WD, Whipple’s disease.
aAs detected by T. whipplei–specific real-time polymerase chain reaction.
All patients were asked as soon as possible after diagnosis to send their first-void urine and matched stool samples for an extended diagnostic work-up and molecular analysis. For most of the WD patients, creatinine and urea nitrogen levels in the blood were assessed (see Supplementary Table S1).
In total, 110 subjects were included as controls in the study and were divided into 3 further groups: Group 4 consisted of patients for whom a diagnosis of WD was excluded due to negative findings in diverse materials (n = 27 as mentioned above); Group 5 consisted of patients with acute gastroenteritis (n = 43); and Group 6 consisted of healthy volunteers (n = 40).
The study was approved by the Clinical Ethics Committee of the Charité. All authors vouch for the completeness and accuracy of the data and analyses.
Patients and Procedures
As defined previously [23], for diagnosis of WD we required at least 2 positive results from 1 relevant material, with positive results coming from either T. whipplei–specific PCR, PAS, or immunohistochemistry (from the best single molecular and morphological tests), in combination with clinical symptoms. For definitive identification of the pathogen in our hands, we pursued a positive PCR of the RNA polymerase beta subunit gene (rpoB) from at least 1 relevant material, plus partial 16S rRNA gene sequencing [12].
Clinical information, as well as results of the index test and reference standard results, were available to the performers of the index test within the routine diagnostic procedures (see Supplementary Table S1).
Tissue and fluid samples were investigated by PAS staining and T. whipplei–specific immunohistochemistry, as described previously [24]. For FISH, tissue was fixed and embedded in cold polymerizing methacrylate resin, sectioned, and hybridized with the pan-bacterial probe EUB338FITC, T. whipplei–specific FISH probe REWHIPCy3, nonsense probe NON338Cy5, and nucleic acid stain 4′,6- diamidino-2-phenylindole dihydrochloride (DAPI) [25–27]. Microscopic evaluations of the FISH signals in the histological tissue sections were carried out by epifluorescence microscopy.
In selected cases, FISH was applied to directly visualize and identify intact bacteria in situ and in urine sediments [26]. Here, up to 10 ml of first-void urine was centrifuged, fixed, and analyzed by FISH, as described before [28].
The presence of T. whipplei DNA in urine samples was determined for all patient and control specimens by real-time rpoB-PCR with hybridization probes, as reported previously [12]. The genotype of T. whipplei from the index patient was further characterized based on 4 highly-variable regions, as described previously [22].
A statistic analysis was performed with a prism graph pad per Fisher′s exact test.
RESULTS
Tropheryma whipplei in Kidney Tissue From the Index Patient
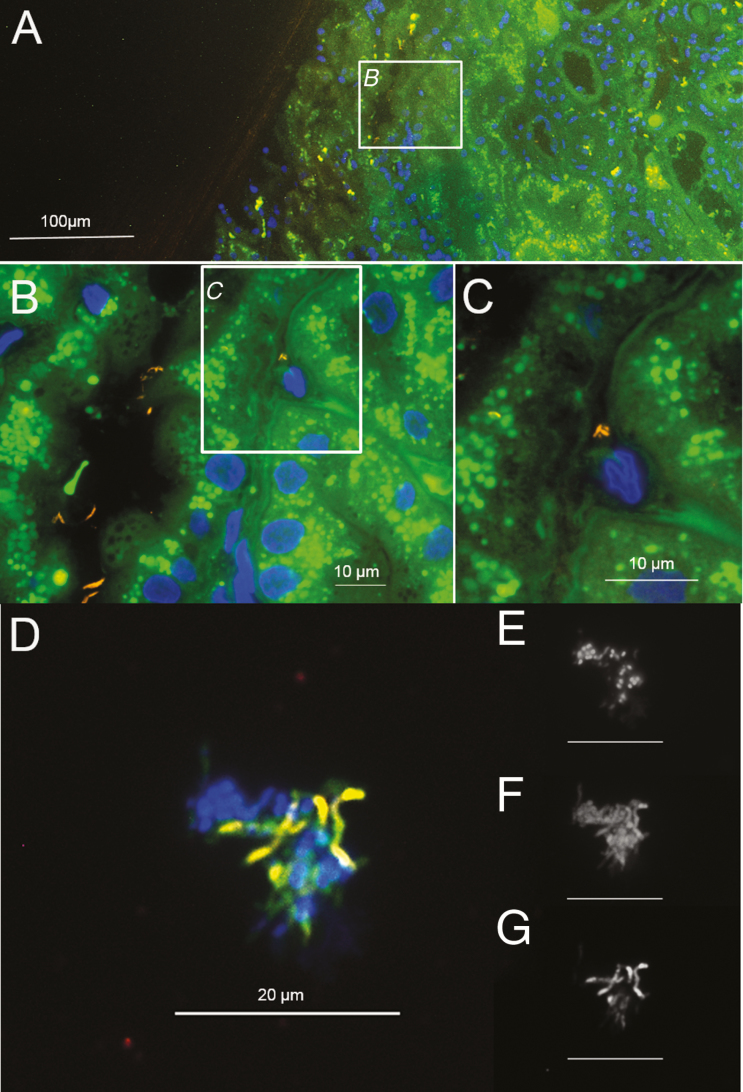
PAS staining showed few typical PAS-positive macrophages (Figure 1) and several PAS-positive particles in the glomerular urinary space, the cytoplasm of some glomerular and tubular epithelial cells, and in some tubuli (Figure 1). Electron microscopy revealed rod-shaped bacteria in the glomerular urinary space and in the tubular lumens as single cells or in clusters (Figure 2). Broad-range 16S rRNA-gene PCR and a subsequent sequence analysis unambiguously identified T. whipplei and T. whipplei–specific immunohistochemistry-confirmed infections with T. whipplei (Figure 3; immunohistochemistry antibody kindly provided by D. Raoult). Within immunohistochemistry-positive areas, single, morphologically-intact T. whipplei cells were visualized by the T. whipplei–specific FISH probe (Figure 3). The strong FISH signal indicated a high content of ribosomal RNA and, therefore, presumably, active cells (Figure 3) [29–31]. Single FISH-positive rods were also observed in the tubuli (Figure 4). All bacteria stained by the pan-bacterial probe EUB338FITC were also detected by the T. whipplei–specific probe, indicating no additional species, and no signal was obtained with the nonsense probe NON338Cy5 (data not shown).
Figure 4.
FISH of the kidney biopsy of the index patient and of a urine sample from a different patient showing morphologically-intact Tropheryma whipplei bacteria. (A) Overview of the kidney tissue of the index patient with the tissue background in green, T. whipplei (detected by REWHIPCy3 [26]) in orange, and the nucleic acid stain DAPI in blue. (B) At higher magnification, several T. whipplei bacteria are visible in tubules. (C) Inset from panel B at a higher magnification, showing single, morphologically-intact T. whipplei bacteria. (D) Urine sediment from another patient, showing morphologically-intact T. whipplei bacteria stained with REWHIPCy3 [26] (orange), among other bacteria with the pan-bacterial probe EUB338FITC [25] (green), and the nucleic acid stain DAPI (blue). (E) Identical microscopic view as D, showing only the nucleic acid stain DAPI in black and white. (F) Identical microscopic view as D, showing only pan-bacterial probe EUB338FITC [25] in black and white. (G) Identical microscopic view as D, showing only REWHIPCy3 [26] in black and white. Abbreviations: DAPI, 4′,6- diamidino-2-phenylindole dihydrochloride; FISH, fluorescence in situ hybridization; REWHIPCy3, Tropheryma whipplei–specific FISH probe.
Detection of Tropheryma whipplei in Urine by PCR and FISH
We detected T. whipplei DNA in 9 out of 12 urine samples from newly-diagnosed WD patients (Group 1) and 2 out of 10 patients after brief treatment (Group 2; Table 1). Upon antibiotic treatment for more than 3 months (Group 3), we found T. whipplei DNA in the urine of only 1 patient out of 31 (Table 2). Real-time PCR showed clearly positive results in duplicate in all positive cases, with crossing point values between 24.3 and 40.0 and typical melting curves.
Table 2.
Detection of Tropheryma whipplei in Urine by Polymerase Chain Reaction Under Antibiotic Treatment
| Group | Patient No. | Time of Diagnosis - Initial Specimen | Follow-up | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Before Treatment | Days After Start of Treatment | Months After Start of Treatment | |||||||||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 2 | 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | 27 | 30 | 33 | >36 | |||||
| 1 | 1 | + | Start of Treatment | End of treatment | |||||||||||||||||||||||||||
| 2 | + | + | − | − | − | − | |||||||||||||||||||||||||
| 3 | + | − | − | − | − | ||||||||||||||||||||||||||
| 4 | + | − | − | − | − | ||||||||||||||||||||||||||
| 5 | + | − | − | ||||||||||||||||||||||||||||
| 6 | + | − | − | − | − | ||||||||||||||||||||||||||
| 7 | + | − | − | − | − | ||||||||||||||||||||||||||
| 8 | − | ||||||||||||||||||||||||||||||
| 9 | + | − | − | − | |||||||||||||||||||||||||||
| 10 | − | − | − | − | − | ||||||||||||||||||||||||||
| 11 | − | − | − | − | − | ||||||||||||||||||||||||||
| 12 | + | − | − | − | − | ||||||||||||||||||||||||||
| 2 | 13 | − | |||||||||||||||||||||||||||||
| 14 | − | − | − | − | |||||||||||||||||||||||||||
| 15 | − | − | − | − | − | ||||||||||||||||||||||||||
| 16 | − | − | − | ||||||||||||||||||||||||||||
| 17 | − | − | |||||||||||||||||||||||||||||
| 18 | − | ||||||||||||||||||||||||||||||
| 19 | + | − | − | ||||||||||||||||||||||||||||
| 20 | − | − | − | − | − | ||||||||||||||||||||||||||
| 21 | + | − | − | − | − | − | − | ||||||||||||||||||||||||
| 22 | − | − | − | − | − | ||||||||||||||||||||||||||
| 3 | 23 | − | |||||||||||||||||||||||||||||
| 24 | − | ||||||||||||||||||||||||||||||
| 25 | − | − | − | − | − | ||||||||||||||||||||||||||
| 26 | − | − | |||||||||||||||||||||||||||||
| 27 | − | ||||||||||||||||||||||||||||||
| 28 | − | − | − | ||||||||||||||||||||||||||||
| 29 | − | ||||||||||||||||||||||||||||||
| 30 | − | ||||||||||||||||||||||||||||||
| 31 | − | ||||||||||||||||||||||||||||||
| 32 | − | ||||||||||||||||||||||||||||||
| 33 | − | − | |||||||||||||||||||||||||||||
| 34 | + | + | + | ||||||||||||||||||||||||||||
| 35 | − | ||||||||||||||||||||||||||||||
| 36 | − | ||||||||||||||||||||||||||||||
| 37 | − | ||||||||||||||||||||||||||||||
| 38 | − | ||||||||||||||||||||||||||||||
| 39 | − | − | |||||||||||||||||||||||||||||
| 40 | − | ||||||||||||||||||||||||||||||
| 41 | − | ||||||||||||||||||||||||||||||
| 42 | − | ||||||||||||||||||||||||||||||
| 43 | − | ||||||||||||||||||||||||||||||
In contrast, stool specimens in 6 out of 8 of the briefly-treated patients (Group 2) and 5 out of 21 of the long-term–treated patients (Group 3) still showed a positive PCR.
The urine of all patients in whom WD was excluded (Group 4) was negative for T. whipplei DNA. The same was true for patients with acute gastroenteritis (Group 5) and healthy volunteers (Group 6). Interestingly, these included 11 cases of healthy carriers whose stool samples were positive for T. whipplei at the same sampling time.
FISH visualized T. whipplei in the urine sediment of 3 WD patients (Figure 4). Of these, 1 patient had not received any previous antibiotic treatment at the time of sampling, while another patient had been treated for 7 days with intravenous ceftriaxone.
In Group 1, 1 patient with culture-negative endocarditis was diagnosed by pan-bacterial 16S rRNA-gene PCR and sequencing of the heart valve tissue. FISH visualized high numbers of T. whipplei cells within the tissue (Supplementary Figure S2). While in this case the stool samples were negative for T. whipplei and duodenal biopsies were not available, PCR detected T. whipplei from urine.
Statistical Analysis
The calculations of diagnostic accuracy measures of the urine testing in the initial diagnoses of WD results of patients from Group 1 versus Groups 4–6 resulted in a positive predictive value of 1 (95% confidence interval [CI] 0.70–1) and a negative predictive value of 0.97 (95% CI 0.92–0.99); however, these calculations are based on small patient numbers. Therefore, we calculated the relative risks for patients from Group 1 (n = 12) versus Groups 4–6 (n = 110) in Table 1. A positive T. whipplei–specific PCR from the urine before any antibiotic treatment was associated with a relative risk of WD of 37.67 (P value <.0001; 95% CI 12.33–115.1).
DISCUSSION
To our knowledge, the index patient represents the first case of T. whipplei infection of the kidney leading to renal failure that was diagnosed by histologic, ultrastructural, and molecular biological findings. Previous reports have shown possible renal involvement that could not finally be proven due to a lack of appropriate diagnostic techniques [32, 33]. Our index patient was misdiagnosed as having “seronegative rheumatoid arthritis” for years, and the immunosuppressive therapy probably led to massive bacterial growth. The impressive amount of T. whipplei bacteria within the glomerular capsular space (ie, in the primary urine) and the detection of active bacteria by FISH in the kidney tissue prompted the idea to test the urine for the presence of T. whipplei DNA, even in patients with normal renal functions.
Here, we describe the detection of T. whipplei DNA in 9 out of 12 urine samples from WD patients at initial diagnosis. In 3 cases, no T. whipplei DNA was found in the urine and, interestingly, the gastrointestinal tract samples of all 3 patients showed no granular PAS-positive macrophages. In these 3 cases, the diagnosis was established either by puncture of a hip prosthesis, by a lymph node biopsy, or in synovial fluid from a knee, raising the question of whether there was intestinal involvement as in “classical” WD or whether these were cases of localized infections.
T. whipplei was detected in the urine but not the stool sample from another patient with T. whipplei endocarditis. This 59-year-old male patient had been treated for rheumatoid arthritis for 6 years. WD had been missed by conventional endocarditis diagnostic tests (culture and serology). Since T. whipplei is among the most abundant infectious agents in culture-negative infective endocarditis, urine testing before open-heart surgery may prevent treatment with the wrong empirical antibiotics, guide diagnostic algorithms, and be lifesaving [4].
Since the molecular diagnosis of WD is possible, we learned that a large proportion of patients do not present with gastrointestinal symptoms, as in “classical” WD, but with a variety of symptoms, such as joint pain, weight loss, abdominal pain, lymphadenopathy, or fever. For these cases, it has been shown that gastrointestinal biopsies may miss WD, in particular in cases when the intestine is not the focus of symptoms and the biopsies have to be taken blindly [16]. In these extra-intestinal cases, a positive urine sample would have great impact on the initial diagnosis of WD, whereas stool testing alone would have led to a false suspicion for WD in 11.5% in our cohort (stool positive without WD, Table 1). We do not expect that the kidneys are always involved in WD to an extent that it is clinically obvious (eg, by elevated serum creatinine levels) and other symptoms usually predominate (see Supplementary Table S1). But it seems that T. whipplei is filtrated into urine if the cell numbers are high enough. In case of detection of PAS-positive macrophages and/or PAS-positive particles in the bowman’s space, we recommend a molecular work-up of the sample to exclude or to confirm T. whipplei.
Upon beginning antibiotic treatment, the detection rate of T. whipplei DNA in urine seems to drop quickly (Table 2). This is in clear contrast to the detection rate of T. whipplei DNA in stool samples that cannot discriminate untreated and short-term–treated patients. Only 1 patient of Group 3, who had a routine follow-up to exclude recurrence of the disease, had positive findings in the urine, without any clinical signs or hints from any other specimen for a relapse. This patient is currently being closely monitored. The rapid decrease of the detection of T. whipplei in urine under therapy possibly explains the results of other studies that tested only 3 of 8 urine samples as positive in PCR from confirmed WD patients [13] or 6 out of 890 urine samples from suspected WD patients [17].
All urine samples from the 110 control patients were negative. These controls included 11 healthy carriers with stool samples positive for T. whipplei at the identical sampling time. In contrast, a large proportion (9 of 12) of the urine samples from the initially-diagnosed WD patients were clearly positive for T. whipplei DNA. In our study, the detection of T. whipplei DNA in the urine of untreated patients correlated in all cases with WD.
To date, the early diagnosis of WD remains a challenge and invasive sampling is mandatory. Positive results from stools and saliva are difficult to interpret. Therefore, urine seems to be promising, since (1) it is easier to obtain than biopsies, (2) it is less loaded with the highly-diverse microbiota that are present in stools and hamper molecular detection of a specific pathogen, (3) urine samples are easier to standardize than stool samples since the T. whipplei load in the urine does not depend upon food intake, and (4) the detection of bacteria in urine indicates glomerular filtration from the blood, and thus might distinguish infected individuals from healthy carriers.
The potential of urine both for the exclusion of WD and for monitoring the effect of antibiotic therapy needs to be further investigated: for example, with regard to its relevance in the distinction of persistent or recurrent WD from immune reconstitution inflammatory syndrome [34]. Our results also raise questions about the role of the kidneys in WD. In addition to our index patient, who had impaired renal function that was likely caused by T. whipplei, some other WD patients also had elevated values for serum creatinine and blood urea nitrogen and 1 patient required kidney transplantation (patient 24, Supplementary Table S1). However, it was not possible to directly correlate a positive finding of T. whipplei DNA in the urine to the impaired renal function as seen by serum creatinine, estimated glomerular filtration rate (eGFR), and urea nitrogen levels. There were 8 patients from Group 1 with T. whipplei positive urine samples who had a normal renal function based on serum creatinine, and only 1 had a reduced eGFR. This implies that these 8 patients were not necessarily suffering from a real renal manifestation of WD, in terms of infection and the destruction of resident renal cells. Rather, it could be that active T. whipplei bacteria pass the glomerular filter without the relevant destruction of renal tissue. The exact mechanism of this phenomenon, of course, remains to be elucidated in future studies.
We would not recommend screening for T. whipplei by any sample type, because this would generate too many suspected cases and invasive sampling in a disease with an estimated incidence of 1:1 000 000. However, if a patient has a positive T. whipplei result in any sample, we recommend a staging, ideally before the start of therapy, to estimate the distribution and extent of the disease, including cerebrospinal fluid, duodenal biopsies, and urine tests. An initial staging helps to interpret any worsening of symptoms and may become important later for the detection of relapses and to differentiate between infection and immune reconstitution inflammatory syndrome.
WD has a very low prevalence, but is fatal if missed and has far-reaching impacts on the patient upon diagnosis, such as requiring the administration of antibiotic therapy for several months [35]. Therefore, invasive sampling for diagnostic material must be initiated, but it should be based on reasonable suspicion. Here, we provide evidence that urine testing for T. whipplei DNA can serve as a new, non-invasive test for the detection of WD. In particular, in extra-intestinal WD, urine is an important additional, easy-to-access material that can be tested by PCR for T. whipplei DNA before more invasive procedures need to be undertaken, which enables more patients to receive appropriate and life-saving therapies in time.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Michael J. Mihatsch for fruitful discussion of the morphologic findings, A. Stroux for revision of the statistics, S. Kasper and S. Reucher for technical assistance, Anne Gale for editorial assistance, and the Robert-Koch Institute for its continuing support of the German Consiliary Laboratory for Tropheryma whipplei.
Financial support. This work was supported by the Deutsche Forschungsgemeinschaft (grant number SFB1192 B6 to T. W.), the German Bundesministerium für Bildung und Forschung (grant number ZooGloW AP6 to T. S.), and by the Robert-Koch Institute (grant number 1369-373 Consiliary Laboratory for Tropheryma whipplei to A. M.).
Potential conflicts of interest. A. M. reports grants from the Robert-Koch Institute, Germany during the conduct of the study. T. S. reports grants from the German Federal Ministry of Education and Research during the conduct of the study. H. R. reports personal fees from Merck Sharp & Dohme (MSD), Pfizer, and Infectopharm outside the submitted work. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Schneider T, Moos V, Loddenkemper C, Marth T, Fenollar F, Raoult D. Whipple’s disease: new aspects of pathogenesis and treatment. Lancet Infect Dis 2008; 8:179–90. [DOI] [PubMed] [Google Scholar]
- 2. La Scola B, Fenollar F, Fournier PE, Altwegg M, Mallet MN, Raoult D. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple’s disease bacillus. Int J Syst Evol Microbiol 2001; 51:1471–9. [DOI] [PubMed] [Google Scholar]
- 3. Lagier JC, Cammilleri S, Raoult D. Classic Whipple’s disease diagnosed by (18)F-fluorodeoxyglucose PET. Lancet Infect Dis 2016; 16:130. [DOI] [PubMed] [Google Scholar]
- 4. Geissdörfer W, Moos V, Moter A, et al. High frequency of Tropheryma whipplei in culture-negative endocarditis. J Clin Microbiol 2012; 50:216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Angelakis E, Fenollar F, Lepidi H, Birg ML, Raoult D. Tropheryma whipplei in the skin of patients with classic Whipple’s disease. J Infect 2010; 61:266–9. [DOI] [PubMed] [Google Scholar]
- 6. Fenollar F, Célard M, Lagier JC, Lepidi H, Fournier PE, Raoult D. Tropheryma whipplei endocarditis. Emerg Infect Dis 2013; 19:1721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rickman LS, Freeman WR, Green WR, et al. Brief report: uveitis caused by Tropheryma whippelii (Whipple’s bacillus). N Engl J Med 1995; 332:363–6. [DOI] [PubMed] [Google Scholar]
- 8. Lagier JC, Lepidi H, Raoult D, Fenollar F. Systemic Tropheryma whipplei: clinical presentation of 142 patients with infections diagnosed or confirmed in a reference center. Medicine (Baltimore) 2010; 89:337–45. [DOI] [PubMed] [Google Scholar]
- 9. Glaser C, Rieg S, Wiech T, et al. Whipple’s disease mimicking rheumatoid arthritis can cause misdiagnosis and treatment failure. Orphanet J Rare Dis 2017; 12:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hinrikson HP, Dutly F, Nair S, Altwegg M. Detection of three different types of ‘Tropheryma whippelii’ directly from clinical specimens by sequencing, single-strand conformation polymorphism (SSCP) analysis and type-specific PCR of their 16S-23S ribosomal intergenic spacer region. Int J Syst Bacteriol 1999; 49(Pt 4):1701–6. [DOI] [PubMed] [Google Scholar]
- 11. Relman DA, Lepp PW, Sadler KN, Schmidt TM. Phylogenetic relationships among the agent of bacillary angiomatosis, Bartonella bacilliformis, and other alpha-proteobacteria. Mol Microbiol 1992; 6:1801–7. [DOI] [PubMed] [Google Scholar]
- 12. Moter A, Schmiedel D, Petrich A, et al. Validation of an rpoB gene PCR assay for detection of Tropheryma whipplei: 10 years’ experience in a National Reference Laboratory. J Clin Microbiol 2013; 51:3858–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenollar F, Laouira S, Lepidi H, Rolain JM, Raoult D. Value of Tropheryma whipplei quantitative polymerase chain reaction assay for the diagnosis of Whipple disease: usefulness of saliva and stool specimens for first-line screening. Clin Infect Dis 2008; 47:659–67. [DOI] [PubMed] [Google Scholar]
- 14. Moos V, Schneider T. Changing paradigms in Whipple’s disease and infection with Tropheryma whipplei. Eur J Clin Microbiol Infect Dis 2011; 30:1151–8. [DOI] [PubMed] [Google Scholar]
- 15. Relman DA, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med 1992; 327:293–301. [DOI] [PubMed] [Google Scholar]
- 16. Lehmann P, Ehrenstein B, Hartung W, Dragonas C, Reischl U, Fleck M. PCR analysis is superior to histology for diagnosis of Whipple’s disease mimicking seronegative rheumatic diseases. Scand J Rheumatol 2016: 46:1369–73. [DOI] [PubMed] [Google Scholar]
- 17. Edouard S, Fenollar F, Raoult D. The rise of Tropheryma whipplei: a 12-year retrospective study of PCR diagnoses in our reference center. J Clin Microbiol 2012; 50:3917–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keita AK, Brouqui P, Badiaga S, et al. Tropheryma whipplei prevalence strongly suggests human transmission in homeless shelters. Int J Infect Dis 2013; 17:e67–8. [DOI] [PubMed] [Google Scholar]
- 19. Keita AK, Bassene H, Tall A, et al. Tropheryma whipplei: a common bacterium in rural Senegal. PLoS Negl Trop Dis 2011; 5:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Feurle GE, Junga NS, Marth T. Efficacy of ceftriaxone or meropenem as initial therapies in Whipple’s disease. Gastroenterology 2010; 138:478–86; quiz 11–2. [DOI] [PubMed] [Google Scholar]
- 21. Lagier JC, Fenollar F, Lepidi H, Giorgi R, Million M, Raoult D. Treatment of classic Whipple’s disease: from in vitro results to clinical outcome. J Antimicrob Chemother 2013; 69:219–27 [DOI] [PubMed] [Google Scholar]
- 22. Li W, Fenollar F, Rolain JM, et al. Genotyping reveals a wide heterogeneity of Tropheryma whipplei. Microbiology 2008; 154:521–7. [DOI] [PubMed] [Google Scholar]
- 23. Günther U, Moos V, Offenmüller G, et al. Gastrointestinal diagnosis of classical Whipple disease: clinical, endoscopic, and histopathologic features in 191 patients. Medicine (Baltimore) 2015; 94:e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lepidi H, Costedoat N, Piette JC, Harlé JR, Raoult D. Immunohistological detection of Tropheryma whipplei (Whipple bacillus) in lymph nodes. Am J Med 2002; 113:334–6. [DOI] [PubMed] [Google Scholar]
- 25. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 1990; 56:1919–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mallmann C, Siemoneit S, Schmiedel D, et al. Fluorescence in situ hybridization to improve the diagnosis of endocarditis: a pilot study. Clin Microbiol Infect 2010; 16:767–73. [DOI] [PubMed] [Google Scholar]
- 27. Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993; 14:136–43. [DOI] [PubMed] [Google Scholar]
- 28. Gescher DM, Kovacevic D, Schmiedel D, et al. Fluorescence in situ hybridisation (FISH) accelerates identification of Gram-positive cocci in positive blood cultures. Int J Antimicrob Agents 2008; 32(Suppl 1):S51–9. [DOI] [PubMed] [Google Scholar]
- 29. Braubach P, Lippmann T, Raoult D, et al. Fluorescence in situ hybridization for diagnosis of Whipple’s disease in formalin-fixed paraffin-embedded tissue. Front Med (Lausanne) 2017; 4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferreira IS, Bettencourt AF, Gonçalves LM, et al. Activity of daptomycin- and vancomycin-loaded poly-epsilon-caprolactone microparticles against mature staphylococcal biofilms. Int J Nanomedicine 2015; 10:4351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bormann N, Koliszak A, Kasper S, et al. A short artificial antimicrobial peptide shows potential to prevent or treat bone infections. Sci Rep 2017; 7:1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schlumpf A, Marbet UA, Stöcklin E, et al. Chronic interstitial nephritis in Whipple’s disease. Klin Wochenschr 1983; 61:25–33. [DOI] [PubMed] [Google Scholar]
- 33. Dhib M, Heron F, Francois A, Bourreile J, Landrin I, Godin M. Kidney granuloma in Whipple’s disease. BMJ 1993; 307:1067–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moos V, Feurle GE, Schinnerling K, et al. Immunopathology of immune reconstitution inflammatory syndrome in Whipple’s disease. J Immunol 2013; 190:2354–61. [DOI] [PubMed] [Google Scholar]
- 35. Feurle GE, Moos V, Bläker H, et al. Intravenous ceftriaxone, followed by 12 or three months of oral treatment with trimethoprim-sulfamethoxazole in Whipple’s disease. J Infect 2013; 66:263–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.