Abstract
Background
High-grade anal intraepithelial neoplasia (AIN2/3; HGAIN) is highly prevalent in human immunodeficiency virus positive (HIV+) men who have sex with men (MSM), but only a minority will eventually progress to cancer. Currently, the cancer risk cannot be established, and therefore all HGAIN is treated, resulting in overtreatment. We assessed host cell deoxyribonucleic acid (DNA) methylation markers for detecting HGAIN and anal cancer.
Methods
Tissue samples of HIV+ men with anal cancer (n = 26), AIN3 (n = 24), AIN2 (n = 42), AIN1 (n = 22) and HIV+ male controls (n = 34) were analyzed for methylation of 9 genes using quantitative methylation-specific polymerase chain reaction. Univariable and least absolute shrinkage and selection operator logistic regression, followed by leave-one-out cross-validation, were used to determine the performance for AIN3 and cancer detection.
Results
Methylation of all genes increased significantly with increasing severity of disease (P < 2 × 10-6). HGAIN samples revealed heterogeneous methylation patterns, with a subset resembling cancer. Four genes (ASCL1, SST, ZIC1,ZNF582) showed remarkable performance for AIN3 and anal cancer detection (area under the curve [AUC] > 0.85). ZNF582 (AUC = 0.89), detected all cancers and 54% of AIN3 at 93% specificity. Slightly better performance (AUC = 0.90) was obtained using a 5-marker panel.
Conclusions
DNA methylation is associated with anal carcinogenesis. A marker panel that includes ZNF582 identifies anal cancer and HGAIN with a cancer-like methylation pattern, warrantingvalidation studies to verify its potential for screening and management of HIV+ MSM at risk for anal cancer.
Keywords: anal intraepithelial neoplasia, anal cancer, human immunodeficiency virus, human papillomavirus, DNA methylation markers
Deoxyribonucleic acid methylation is associated with anal carcinogenesis and has potential to detect high-grade anal intraepithelial neoplasia. This is a novel, more effective anal cancer screening procedure for human immunodeficicency virus positive men who have sex with men.
Anal cancer is relatively uncommon in the general population (1–2 cases per 100 000 person-years) [1]. However, incidence rates are increasing and the highest incidence rates are seen in human immunodeficiency virus positive (HIV+) men who have sex with men (MSM), with rates up to 109 per 100000 person-years since the introduction of combination antiretroviral therapy [2].
Anal squamous cell carcinoma (SCC) accounts for the vast majority of anal cancers and, similar to cervical SCC, is caused by a persistent high-risk human papillomavirus (hrHPV) infection, most notably HPV16 [3]. Anal cancer develops through several precancerous stages, called anal intraepithelial neoplasia (AIN; graded 1–3) [3, 4]. AIN2 and AIN3, or high-grade AIN (HGAIN), can progress to invasive anal SCC, whereas AIN1, or low-grade AIN, is not seen as a real cancer precursor [4, 5]. Only limited data on the malignant progression of HGAIN exists, showing progression to anal cancer in only a minority of HGAIN cases (1 per 377 per year in HIV+ MSM in the combination antiretroviral therapy era) [2]. However, this results in a lifetime risk of approximately 10% of developing anal cancer [5].
HGAIN is found in 20–40% of all HIV+ MSM [2]. High-resolution anoscopy–guided biopsies with histopathological evaluation of lesions is the gold standard for identifying AIN, and ablative treatment of HGAIN to prevent cancer is highly recommended [4, 6]. The current practice of screening and treatment of HGAIN in HIV+ MSM is, however, far from ideal, because high-resolution anoscopy is difficult, costly, burdensome to the patient, and recurrence rates after treatment are high [6, 7]. Alternative screening methods have been evaluated. Anal cytology has, depending on the cytological cut-off point, either too low sensitivity or too low specificity for HGAIN detection [4]. HPV testing provides poor specificity, since multiple hrHPV are frequently found in HIV+ MSM [4]. HPV messenger ribonucleic acid (mRNA) analysis, HPV16/18 genotyping, and p16/Ki-67 dual staining also yielded suboptimal sensitivity and specificity in detecting HGAIN [8]. In addition, considering that only a small proportion of HGAIN cases will eventually progress to cancer, preferably only lesions with high malignant potential should be treated. Unfortunately, a marker that predicts the cancer risk is currently lacking. Therefore, in current clinical practice, all HGAIN lesions are treated, regardless of their malignant potential, resulting in significant overtreatment. Hence, there is an urgent need for biomarkers to accurately discriminate HGAIN cases that are at high risk of progression to cancer and in need of treatment, as opposed to HGAIN cases with low cancer risks.
Progression of HPV-induced precancerous lesions to invasive carcinomas is driven by an accumulation of genetic and epigenetic aberrations affecting host cell genes, resulting in the activation of oncogenes and/or inactivation of tumor suppressor genes [9]. Deoxyribonucleic acid (DNA) methylation is a type of epigenetic changes characterized by the addition of a methyl group to a cytosine located 5′ of a guanine in gene promoters. When promoter regions of tumor suppressor genes are hypermethylated, this generally leads to their inactivation. Therefore, DNA methylation, as an important epigenetic hallmark in HPV-induced carcinogenesis, could provide valuable biomarkers for the detection of anal (pre-)cancer [9].
Our group and others have shown that, in the cervix, DNA methylation analysis of host cell genes allows the detection of high-grade cervical intraepithelial neoplasia (HGCIN) with high cancer risks and carcinomas [9–11]. Following the analysis of genome-wide methylation profiles in HPV-positive cell lines and clinical specimens, we have identified various methylated genes that are associated with HPV-induced carcinogenesis, including ASCL1, GHSR, SST, ST6GALNAC3, ST6GALNAC5, WDR17, ZIC1, ZNF582, and ZNF583 [11, 12]. Given the similar pathogenesis of cervical and anal cancers, we considered these methylation markers as potential candidates for the detection of HGAIN and anal cancer.
Limited research on host cell DNA methylation in anal carcinogenesis has been performed [13–15]. In the present study, we evaluated DNA methylation of the aforementioned 9 genes at the different stages of anal carcinogenesis and assessed their diagnostic potential for the detection of HGAIN and anal cancer in HIV+ men.
MATERIALS AND METHODS
Clinical Specimens and Ethics
This study involved a cross-sectional series of 148 formalin-fixed paraffin-embedded anal (pre-)cancer and control tissue samples, obtained in 133 HIV+ men. Anal SCC specimens of HIV+ men, obtained between 1986 and 2011 (n = 26, including 1 highly suspicious for infiltration, 1 basosquamous carcinoma, and 1 verrucous carcinoma; median age 48.5 years), were retrieved from the pathology archives of the Amsterdam University Medical Centers, location Academic Medical Center (AMC) and Onze Lieve Vrouwe Gasthuis (OLVG), both in Amsterdam. A total number of 88 archival AIN biopsy specimens (AIN1: n = 22, median age 48; AIN2: n = 42, median age 45; AIN3: n = 24, median age 47) of HIV+ men were obtained in the course of a triple-arm trial on AIN treatment (2008–2011), as published previously [7]. Normal anal epithelium (including reactive epithelium) served as normal control samples (n = 34; median age 52), after histopathological confirmation. These biopsies were taken from non-suspected anal epithelium in HIV+ men who had biopsies taken from suspected lesions during routine screening for anal (pre-)cancer in HIV+ MSM at the AMC in 2016. For some patients, multiple biopsies taken from different sites were included. The median ages of the patients at time of biopsies did not significantly differ between histological groups (Kruskall-Wallis omnibus test: P = .35).
This study followed the ethical guidelines of the Institutional Review Board of the AMC and was approved under reference numbers 17/151 (SCC), 07/318 (AIN biopsies), and 05/031 (normal control samples).
Human Papillomavirus and DNA Methylation Analysis
For sample processing, HPV testing, and details on DNA methylation analysis, see Supplementary Data. We analyzed 9 methylation markers using 3 multiplex quantitative methylation-specific polymerase chain reaction (qMSP) assays, each targeting 3 genes and the reference gene, β-actin (ACTB). A quantification cycle (Cq) threshold of <32 for ACTB indicated sufficient DNA and adequate bisulfite conversion [16].
The ACTB quality control also showed that test performance was not related to sample age and all samples met the DNA quality criteria. Cq and ΔΔCq ratios were computed using the comparative Cq method. For GHSR, SST, and ZIC1, the target Cq values were normalized with the Cq value of reference gene ACTB (2-ΔCq x 100) to obtain Cq ratios. For the remaining genes, ΔΔCq ratios were calculated by comparing the target Cq values to the Cq values of ACTB and of the calibrator (2-ΔΔCq x 100) [17].
Statistical Analysis
To visualize methylation levels per disease category, boxplots were computed from the log2 transformed Cq and ΔΔCq ratios. Differences in levels of DNA methylation between the disease categories were assessed using the Kruskal-Wallis omnibus test, followed by post hoc testing using the Mann-Whitney U test and by Bonferroni multiple testing correction in cases with significant results.
To assess the diagnostic performance of each individual methylation marker, measured by its ability to distinguish between cases (AIN3 and anal SCC; [AIN3+]) and controls (normal control samples and AIN1; [≤AIN1]), we applied univariable logistic regression analyses on square root–transformed Cq and ΔΔCq ratios [11]. Due to the relatively small sample size, we combined the AIN3 and SCC samples as 1 group (referred to as cases) in the logistic regression analysis. To define a marker panel with optimal performance and to account for the multicollinearity between the markers (evaluated by the variance inflation factor), we performed least absolute shrinkage and selection operator (LASSO) logistic regression. We evaluated the models by leave-one-out cross-validation (LOOCV), whereby for each sample a predicted probability is obtained by the model that was trained on the remaining samples. Predicted probabilities (values ranging from 0 to 1), representing the risk for an underlying AIN3 and SCC, were calculated per sample. The performance of the models was visualized using receiver operating characteristic (ROC) curves. Subsequently, the area under the curve (AUC) was calculated, and sensitivity and specificity were determined from an optimum threshold.
Statistical analyses were performed in R Statistical Software using packages ggplot2 and pROC (version 3.2.5; Foundation for Statistical Computing, Austria) and IBM SPSS Statistics software (version 22; IBM Corporation, Armonk, NY). Reported P values are 2-sided and a value ≤0.05 was considered statistically significant.
RESULTS
Human Papillomavirus Genotyping of Tissue Samples
Overall HPV positivity was high (93.2%), with all (100%) of the AIN (AIN1-3) biopsies showing HPV positivity. Moreover, 96.2% (25 of 26) of SCC and 76.5% (26 of 34) of normal control samples were HPV positive (see Table 1 for hrHPV types and Supplementary Table 1 for all HPV types). Multiple HPV types were found in 38.1% of HPV-positive samples. Positivity for hrHPV types [18, 19] varied, with 27.3% in AIN1, 85.7% in AIN2, 95.8% in AIN3, and 88.0% in HPV-positive SCC. Interestingly, hrHPV was also found in 57.7% of HPV-positive normal control samples. The most predominant HPV type was HPV16, accounting for 42.9% of AIN2 and 79.2% of AIN3 lesions, and for 56.0% of all HPV-positive anal SCC. HPV16 was most often seen as a single infection. HPV genotyping data per sample are shown in Figure 3.
Table 1.
HPV Prevalence and High-risk HPV Genotype Distribution Per Histological Category in WTS of Anal Samples Obtained from HIV+ Men
| HPV Genotyping Results per Histological Category | |||||
|---|---|---|---|---|---|
| HPV Type | Normal (n = 34) | AIN1 (n = 22) | AIN2 (n = 42) | AIN3 (n = 24) | SCC (n = 26) |
| Overall HPV positivity | 26/34 (76.5%) | 22/22 (100%) | 42/42 (100%) | 24/24 (100%) | 25/26 (96.2%) |
| Multiple HPV types in WTS | 11/26 (42.3%) | 7/22 (31.8%) | 13/42 (31.0%) | 10/24 (41.7%) | 12/25 (48.0%) |
| High-risk HPV | |||||
| Overall high-risk HPV positivity | 15/26 (57.7%) | 6/22 (27.3%) | 36/42 (85.7%) | 23/24 (95.8%) | 22/25 (88.0%) |
| HPV16 | 4/26 (15.4%) | 2/22 (9.1%) | 18/42 (42.9%) | 19/24 (79.2%) | 14/25 (56.0%) |
| HPV18 | 3/26 (11.5%) | 2/42 (4.8%) | 1/24 (4.2%) | 1/25 (4.0%) | |
| HPV31 | 5/26 (19.2%) | 2/22 (9.1%) | 3/42 (7.1%) | 1/24 (4.2%) | 3/25 (12.0%) |
| HPV33 | 1/26 (3.8%) | 2/42 (4.8%) | 3/24 (12.5%) | 3/25 (12.0%) | |
| HPV35 | 1/26 (3.8%) | 3/42 (7.1%) | 1/24 (4.2%) | 1/25 (4.0%) | |
| HPV39 | 2/26 (7.7%) | 1/42 (2.4%) | 2/24 (8.3%) | ||
| HPV45 | 1/26 (3.8%) | 1/42 (2.4%) | 4/25 (16.0%) | ||
| HPV51 | 5/26 (19.2%) | 3/22 (13.6%) | 8/42 (19.0%) | 1/24 (4.2%) | 3/25 (12.0%) |
| HPV52 | 2/26 (7.7%) | 2/22 (9.1%) | 1/42 (2.4%) | 2/24 (8.3%) | 2/25 (8.0%) |
| HPV56 | 1/26 (3.8%) | 1/42 (2.4%) | 2/24 (8.3%) | ||
| HPV58 | 1/22 (4.5%) | 2/42 (4.8%) | |||
HPV risk classification based on International Agency for Research on Cancer [18, 19]. Type-specific positivity includes those contributed by multiple infections. N of tissue samples tested = 148 (including 9 HPV negative). Prevalence of probable/possible high-risk HPV types and low-risk HPV types is shown in Supplementary Table 1.
Abbreviations: AIN, anal intraepithelial neoplasia (grades 1–3); HIV+, human immunodeficiency virus positive; HPV, human papillomavirus; Normal, normal control samples; MSM, men who have sex with ; SCC, anal squamous cell carcinoma; WTS, whole tissue sections.
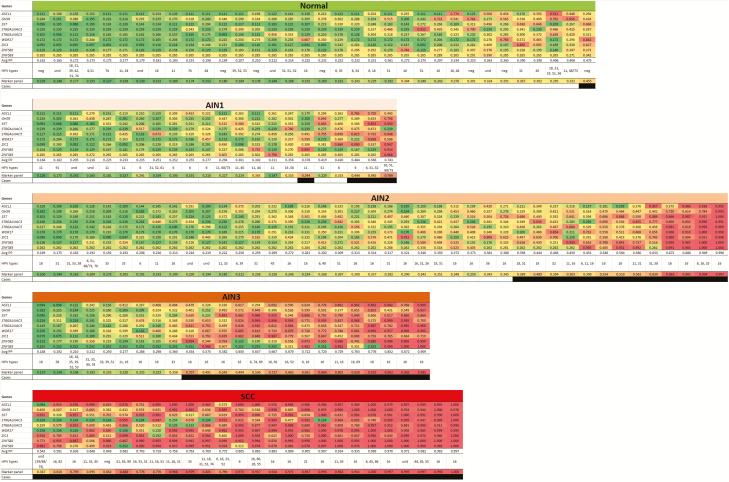
Figure 3.
DNA methylation pattern of the 9 methylation markers and the optimal marker panel (ASCL1, ST6GALNAC3, WDR17, ZIC1, ZNF582; based on LASSO; rows) per histological category. Predicted probabilities are shown per sample (column) in the different histological subgroups and colored from green (predicted probability of 0) to red (predicted probability of 1) based on LOOCV. In each group, samples are ordered based on their average predicted probability (Avg PP). HPV genotypes were found per sample in whole tissue sections. Black boxes represent samples detected using the optimal marker panel (classified as case by LASSO using predicted probability threshold ≥0.38). Abbreviations: AIN, anal intraepithelial neoplasia (grades 1–3); HPV, human papillomavirus; LASSO, least absolute shrinkage and selection operator; LOOCV, leave-one-out cross-validation; Normal, normal control samples; SCC, anal squamous cell carcinoma; und, undetermined.
Methylation Levels in Various Histological Categories of Anal Disease
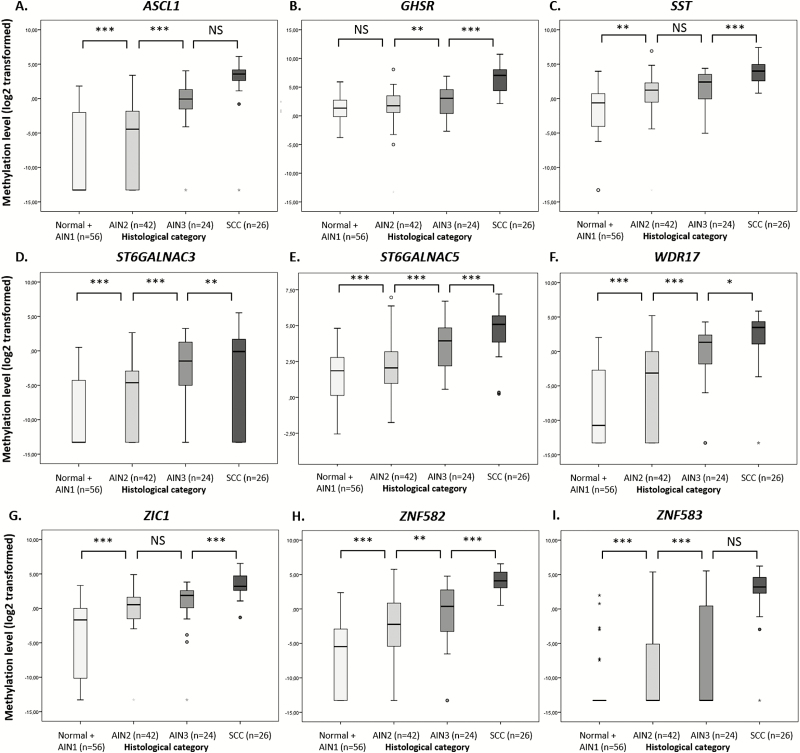
Methylation levels of all 9 markers increased significantly with the severity of disease (Figure 1): that is, with progression from normal control samples/AIN1 through AIN2 and AIN3 towards anal SCC (Kruskal-Wallis omnibus test: P < 2 × 10-6). In the majority of cases, a significant increase in DNA methylation was also observed between individual consecutive disease categories.
Figure 1.
Methylation levels increase with severity of anal disease. DNA methylation levels are shown relative to a reference gene β-actin (log2-transformed Cq and ΔΔCq ratios; Y-axis) in the different histological categories of anal tissue samples of human immunodeficiency virus positive men (X-axis) for 9 markers: (A) ASCL1, (B) GHSR, (C) SST, (D) ST6GALNAC3, (E) ST6GALNAC5, (F) WDR17, (G) ZIC1, (H) ZNF582, and (I) ZNF583. * and ◦ indicate extreme and mild outlier samples, respectively. Abbreviations: AIN, anal intraepithelial neoplasia (grades 1–3); Normal, normal control samples; SCC, anal squamous cell carcinoma. Differences between histological categories upon Kruskal-Wallis omnibus test, followed by post hoc testing using the Mann-Whitney U test and Bonferroni multiple testing correction: *P < .05, **P < .01, ***P < .001, NS: not significant.
Fold changes in methylation levels (expressed as Cq/ΔΔCq ratio) for all genes increased with the severity of disease, compared to the reference (median of normal control samples and AIN1; Supplementary Table 2).
Diagnostic Performance of the Individual Methylation Markers
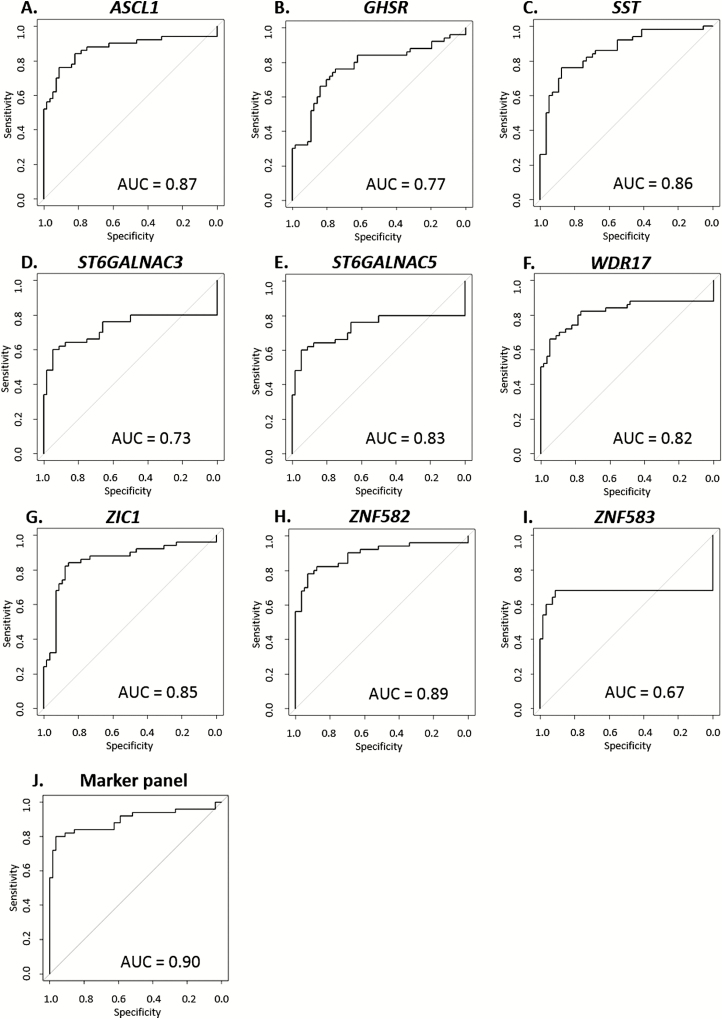
Using univariable logistic regression analyses, we assessed the diagnostic performance of the 9 markers, measured by their ability to distinguish cases (AIN3 and SCC; [AIN3+]; n = 50) from controls (normal control samples and AIN1; [≤AIN1]; n = 56). All markers were able to significantly distinguish cases from controls (P < .001). The performance of the models, before and after LOOCV, is summarized in Table 2 and the corresponding ROC curves of the LOOCV are depicted in Figure 2. Although all markers generally had good performance, the markers ASCL1, SST, ZIC1, and ZNF582 had an AUC > 0.85. ZNF582 performed best, with an AUC of 0.89. At the optimum predicted probability threshold (≥0.40), this translates to a sensitivity of 78% and a specificity of 93% while detecting all (26 of 26) anal cancers. Using this threshold, ZNF582 classified 54.2% of AIN3 (13 of 24), 42.9% of AIN2 (18 of 42), 13.6% of AIN1 (3 of 22), and 2.9% of normal control samples (1 of 34) as cases.
Table 2.
Results of Logistic Regression Analysis on Diagnostic Performance of the 9 Individual Markers and the Optimal Marker Panel
| Methylation Marker | ASCL1 | GHSR | SST | ST6GALNAC3 | ST6GALNAC5 | WDR17 | ZIC1 | ZNF582 | ZNF583 | Marker Panel |
|---|---|---|---|---|---|---|---|---|---|---|
| nonCV AUC | 0.89 | 0.79 | 0.87 | 0.79 | 0.84 | 0.85 | 0.87 | 0.91 | 0.80 | 0.93 |
| LOOCV AUC | 0.87 | 0.77 | 0.86 | 0.73 | 0.83 | 0.82 | 0.85 | 0.89 | 0.67 | 0.90 |
| Sensitivity | 0.76 | 0.76 | 0.76 | 0.60 | 0.78 | 0.66 | 0.84 | 0.78 | 0.68 | 0.82 |
| Specificity | 0.91 | 0.75 | 0.88 | 0.95 | 0.85 | 0.95 | 0.86 | 0.93 | 0.91 | 0.91 |
Reported are nonCV and LOOCV AUC. Sensitivity and specificity from LOOCV for the optimum threshold per marker and marker panel (ASCL1, ST6GALNAC3, WDR17, ZIC1, and ZNF582) are based on LASSO. Endpoint: AIN3 and anal SCC (AIN3+) in anal tissue samples of HIV+ men.
Abbreviations: AIN, anal intraepithelial neoplasia (grades 1–3); AUC, area under the curve; HIV+, human immunodeficiency virus positive; LOOCV, leave-one-out cross-validation; nonCV, non–cross-validated; SCC, anal squamous cell carcinoma.
Figure 2.
Diagnostic performance visualized with receiver operating characteristic curves of the leave-one-out cross-validation for [AIN3+] detection, for 9 individual methylation markers: (A) ASCL1, (B) GHSR, (C) SST, (D) ST6GALNAC3, (E) ST6GALNAC5, (F) WDR17, (G) ZIC1, (H) ZNF582, (I) ZNF583, and (J) the optimal marker panel (ASCL1, ST6GALNAC3, WDR17, ZIC1, and ZNF582) based on least absolute shrinkage and selection operator. The endpoint is AIN3 and anal squamous cell carcinoma [AIN3+]. Abbreviations: AIN, anal intraepithelial neoplasia (grades 1–3); AUC, area under the curve.
The DNA methylation pattern of the 9 genes, depicted by predicted probabilities, is shown in Figure 3. What can be noted is a heterogeneous pattern in AIN2 and AIN3, subsets of which have low methylation levels (green) and high methylation levels (red). Normal control samples have predominantly low methylation levels, whereas cancers have high methylation levels, both showing less heterogeneity.
Definition and Diagnostic Performance of a Marker Panel
To define a marker panel with optimal performance for [AIN3+] detection, we performed LASSO logistic regression. The LASSO model resulted in a panel of 5 markers (ASCL1, ST6GALNAC3, WDR17, ZIC1, and ZNF582) that yielded an AUC of 0.90 following LOOCV (Figure 2). A predicted probability threshold of ≥0.38 to classify a sample as a case generated a sensitivity of 82% and a specificity of 91% for [AIN3+] (Table 2; marker panel column). Using this marker panel and threshold, 100% of cancers (26 of 26), 62.5% of AIN3 (15 of 24), 19.0% of AIN2 (8 of 42), and 9.1% AIN1 (2 of 22) were classified as cases (see Figure 3’s black boxes).
DISCUSSION
We investigated HPV’s presence and host cell DNA methylation in the multistep pathway of anal carcinogenesis using a unique cross-sectional series of anal tissue samples of HIV+ men. HPV genotyping of all anal tissue samples revealed a high overall HPV positivity. The HPV16 prevalence increased towards AIN3, and was detected in the majority of SCC, though at a lower prevalence than published in a recent meta-analysis [20]. Using qMSP, 9 methylation markers were evaluated for their diagnostic performance in the detection of AIN3 and anal cancer. All markers showed a significant correlation between methylation level and the severity of disease, reaching the highest methylation levels in SCC. This indicates that DNA methylation of these genes is associated with disease progression during anal carcinogenesis. Investigating their diagnostic performance (ie, to detect AIN3 and SCC), we identified 4 genes that, after LOOCV, had an AUC > 0.85 (ASCL1, SST, ZIC1, and ZNF582). Of these genes, ZNF582 had the highest AUC (0.89), showing good sensitivity (78%) and specificity (93%) and detecting all cancers. Our effort to compose an optimal marker panel resulted in a panel of 5 markers (ASCL1, ST6GALNAC3, WDR17, ZIC1, and ZNF582) that showed similar diagnostic performance as ZNF582, albeit with higher sensitivity, as the panel classified several AIN3 samples as cases, which were missed by ZNF582 alone. Given the preferred use of a maximum of 3 markers that can be tested in a single multiplex qMSP assay, the AUC values and predicted probabilities (Figures 2 and 3) indicate that a marker panel consisting of ZNF582 combined with ASCL1 and/or ZIC1 is most likely the optimal choice for further validation studies [16].
Natural history data suggest that HGAIN cases, similar to HGCIN cases, encompasses a heterogeneous group of lesions, of which only a small proportion will eventually progress to anal cancer [2, 5, 11, 22, 23]. Ideally, we should be able to identify, and subsequently treat, only those HGAIN cases with high malignant potential. We are among the first to show that histopathologically similar AIN2 and AIN3 lesions display heterogeneous methylation patterns. Part of the AIN2 (19%) and AIN3 (63%) samples showed high methylation levels and a so-called cancer-like methylation profile, while the remaining HGAIN samples demonstrated low or intermediate methylation levels. Similar results have been seen in HGCIN, where methylation markers were found to have an extremely high sensitivity for detecting HGCIN with a longstanding preceding HPV infection (>5 years), indicating a high short-term cancer risk [9, 10, 21, 23]. These methylation markers have now been clinically validated for the detection of so-called “advanced HGCIN” and cervical cancer in women with hrHPV-positive cervical scrapes and self-collected cervico-vaginal specimens [24–26].
Since we analyzed a cross-sectional series of HIV+ anal samples, and longitudinal follow-up is missing, we cannot prove that HGAIN samples with a cancer-like methylation profile do, indeed, have a higher risk of progression to cancer compared to HGAIN samples characterized by low methylation levels. Hence, further research on the role of host cell methylation during the longitudinal course of anal carcinogenesis is warranted. Restriction of treatment to HGAIN cases with a high risk of progression to cancer could resolve the current problem of overtreatment of HGAIN lesions with low carcinogenic potential.
Until now, remarkably limited research on DNA methylation in anal carcinogenesis has been performed. Only 2 studies have demonstrated the value of increased methylation of several other host cell genes in a limited set of anal cancer and HGAIN samples, compared to controls [13, 14]. Another 2 additional studies on viral DNA methylation showed a heterogeneous pattern of HPV16 upstream regulatory region methylation in AIN, although its application for HGAIN detection remains to be established [27, 28]. In contrast, our study was based on DNA methylation of host cell genes, making it HPV-genotype independent and, therefore, also accounting for non-HPV16–related HGAIN and SCC cases. A recent study combined methylation analyses of several hrHPV types and a single host cell gene (EPB41L3), presenting a reasonable performance for detecting HGAIN and SCC, yet only analyzing 5 cancer samples [15]. A prominent strength of our study is the relatively large sample size, with 148 anal tissue specimens that represent the complete spectrum of anal carcinogenesis.
We recognize that, by focusing on HIV+ men in this study, our data cannot be directly extrapolated to the general population or to other risk groups, such as HIV-negative MSM, HIV+ women, women with a history of HPV-induced (pre-)cancer, or otherwise immunocompromised patients.
We believe another future application of methylation marker testing in anal (pre-)cancer screening would be on less-invasive anal swabs instead of biopsies, to triage men at an even earlier stage. As a result, the burdensome high-resolution anoscopy procedure and biopsies could then be withheld in men with a negligible risk of anal cancer development based on methylation analysis from the swab. In this manner, anal cancer screening can be patient-friendlier and likely also more (cost-)effective, improving the quality of life of HIV+ MSM.
In conclusion, we established that host cell DNA methylation is significantly associated with anal carcinogenesis. HGAIN samples revealed a heterogeneous methylation pattern, with histopathologically similar AIN2 and AIN3 lesions showing either low methylation levels or a cancer-like methylation pattern. We identified several methylation markers, such as ASCL1, ZIC1, and ZNF582, with good diagnostic performance for the detection of SCC and AIN3 with a cancer-like methylation pattern, suggestive of a higher cancer risk. Further validation studies are warranted to verify their potential as biomarkers for the screening and management of HIV+ MSM at risk for anal cancer.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank W. Verlaat, S. Mongera, and H. van den Munckhof for excellent technical advice and assistance and H. E. Nobel, Dr K. C. M. Gosens, and Dr M. L. Siegenbeek van Heukelom for taking biopsies.
R. D. M. S., J. M. P., and H. J. C. dV. were the principal investigators of the study. R. P. vdZ., O. R., H. J. C. dV., J. M. P., and R. D. M. S. designed the study. R. P. vdZ., O. R., C. J. M. vN., W. G. V. Q., H. J. C. dV., J. M. P., and R. D. M. S. were involved in providing and collecting clinical material and data collection. R. P. vdZ., A. P. vS. and S. D. performed the laboratory work. C. J. L. M. M. and R. D. M. S. developed the quantitative methylation-specific polymerase chain reaction assays used in this study. G. E. L. vdB. provided some of the clinical material. R. P. vdZ. and R. D. M. S. performed the data analysis. R. P. vdZ., I. C.-T. and P. W. N. performed the statistical analysis. R. P. vdZ. managed the database. R. P. vdZ., O. R., H. J. C. dV., J. M. P., and R. D. M. S. drafted the manuscript. All authors critically reviewed the manuscript and approved the final version. All authors had full access to all of the data in the study and can take responsibility for the integrity of the data and the accuracy of the data analysis, and believe that the manuscript represents honest work. R. D. M. S. affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained.
Financial support. This work was supported by the Aidsfonds (Dutch Aids foundation; High-Risk High-Gain call 2016; grant number P-26606) and the KWF Kankerbestrijding (Dutch Cancer Society; grant number 2016–10781).
Potential conflicts of interest. R. D. M. S. and C. J. L. M. M. are minority stock holders of Self-screen B.V., a spin-off company of Amsterdam University Medical Centers, location Vrije Universiteit Medical Center (VUmc) that owns patents on methylation markers and human papillomavirus detection. C. J. L. M. M. is part-time director of Self-screen B.V. since September 2017; has received a speakers’ fee from GSK, Qiagen, and SPMSD/Merck; served occasionally on the scientific advisory board (expert meeting) of GSK, Qiagen, and SPMSD/Merck; has been co-investigator on a SPMSD sponsored trial, of which his institute received research funding; has a very small number of Qiagen shares; and was minority shareholder of Diassay B.V. until April 2016. H. J. C. dV. received money or goods for research from Medigene, Gilead, and MSD; money for presentations from Abott and Janssen; and money for advice to Medigene and Novartis. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. van der Zee RP, Richel O, de Vries HJ, Prins JM. The increasing incidence of anal cancer: can it be explained by trends in risk groups?Neth J Med 2013; 71: 401–11. [PubMed] [Google Scholar]
- 2. Machalek DA, Poynten M, Jin F, et al. . Anal human papillomavirus infection and associated neoplastic lesions in men who have sex with men: a systematic review and meta-analysis. Lancet Oncol 2012; 13:487–500. [DOI] [PubMed] [Google Scholar]
- 3. Hoots BE, Palefsky JM, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer 2009; 124:2375–83. [DOI] [PubMed] [Google Scholar]
- 4. Darragh TM, Winkler B. Anal cancer and cervical cancer screening: key differences. Cancer Cytopathol 2011; 119:5–19. [DOI] [PubMed] [Google Scholar]
- 5. Berry JM, Jay N, Cranston RD, et al. . Progression of anal high-grade squamous intraepithelial lesions to invasive anal cancer among HIV-infected men who have sex with men. Int J Cancer 2014; 134:1147–55. [DOI] [PubMed] [Google Scholar]
- 6. Palefsky JM. Anal cancer prevention in HIV-positive men and women. Curr Opin Oncol 2009; 21:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richel O, de Vries HJ, van Noesel CJ, Dijkgraaf MG, Prins JM. Comparison of imiquimod, topical fluorouracil, and electrocautery for the treatment of anal intraepithelial neoplasia in HIV-positive men who have sex with men: an open-label, randomised controlled trial. Lancet Oncol 2013; 14:346–53. [DOI] [PubMed] [Google Scholar]
- 8. Wentzensen N, Follansbee S, Borgonovo S, et al. . Human papillomavirus genotyping, human papillomavirus mRNA expression, and p16/Ki-67 cytology to detect anal cancer precursors in HIV-infected MSM. AIDS 2012; 26:2185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014; 14:395–405. [DOI] [PubMed] [Google Scholar]
- 10. De Strooper LM, Meijer CJ, Berkhof J, et al. . Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014; 7:1251–7. [DOI] [PubMed] [Google Scholar]
- 11. Verlaat W, Snijders PJF, Novianti PW, et al. . Genome-wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical precancer and cancer. Clin Cancer Res 2017; 23:3813–22. [DOI] [PubMed] [Google Scholar]
- 12. Verlaat W, Snoek BC, Heideman DAM, et al. . Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin Cancer Res 2018; 24:3456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Martins CR, Fansler ZB, et al. . DNA methylation in anal intraepithelial lesions and anal squamous cell carcinoma. Clin Cancer Res 2005; 11:6544–9. [DOI] [PubMed] [Google Scholar]
- 14. Hernandez JM, Siegel EM, Riggs B, et al. . DNA methylation profiling across the spectrum of HPV-associated anal squamous neoplasia. PLoS One 2012; 7:e50533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorincz AT, Nathan M, Reuter C, et al. . Methylation of HPV and a tumor suppressor gene reveals anal cancer and precursor lesions. Oncotarget 2017; 8:50510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Snellenberg S, De Strooper LM, Hesselink AT, et al. . Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape. BMC Cancer 2012; 12:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101–8. [DOI] [PubMed] [Google Scholar]
- 18. Muñoz N, Bosch FX, de Sanjosé S, et al. ; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003; 348:518–27. [DOI] [PubMed] [Google Scholar]
- 19. Schiffman M, Clifford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer 2009; 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin C, Franceschi S, Clifford GM. Human papillomavirus types from infection to cancer in the anus, according to sex and HIV status: a systematic review and meta-analysis. Lancet Infect Dis 2018; 18:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bierkens M, Hesselink AT, Meijer CJ, et al. . CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013; 133:1293–9. [DOI] [PubMed] [Google Scholar]
- 22. De Strooper LM, van Zummeren M, Steenbergen RD, et al. . CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol 2014; 67:1067–71. [DOI] [PubMed] [Google Scholar]
- 23. Wilting SM, Steenbergen RD, Tijssen M, et al. . Chromosomal signatures of a subset of high-grade premalignant cervical lesions closely resemble invasive carcinomas. Cancer Res 2009; 69:647–55. [DOI] [PubMed] [Google Scholar]
- 24. De Strooper LMA, Verhoef VMJ, Berkhof J, et al. . Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol Oncol 2016; 141:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hesselink AT, Heideman DA, Steenbergen RD, et al. . Methylation marker analysis of self-sampled cervico-vaginal lavage specimens to triage high-risk HPV-positive women for colposcopy. Int J Cancer 2014; 135:880–6. [DOI] [PubMed] [Google Scholar]
- 26. Verhoef VM, Heideman DA, van Kemenade FJ, et al. . Methylation marker analysis and HPV16/18 genotyping in high-risk HPV positive self-sampled specimens to identify women with high grade CIN or cervical cancer. Gynecol Oncol 2014; 135:58–63. [DOI] [PubMed] [Google Scholar]
- 27. Wiley DJ, Huh J, Rao JY, et al. . Methylation of human papillomavirus genomes in cells of anal epithelia of HIV-infected men. J Acquir Immune Defic Syndr 2005; 39:143–51. [PubMed] [Google Scholar]
- 28. Molano M, Tabrizi SN, Garland SM, et al. ; SPANC Study Team. CpG methylation analysis of HPV16 in laser capture microdissected archival tissue and whole tissue sections from high grade anal squamous intraepithelial lesions: a potential disease biomarker. PLoS One 2016; 11:e0160673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.