Abstract
We previously showed that the first intron of genes exhibits several interesting characteristics not seen in other introns: 1) it is the longest intron on average in almost all eukaryotes, 2) it presents the highest number of conserved sites, and 3) it exhibits the highest density of regulatory chromatin marks. Here, we expand on our previous study by integrating various multiomics data, leading to further evidence supporting the functionality of sites in the first intron. We first show that trait-associated single-nucleotide polymorphisms (TASs) are significantly enriched in the first intron. We also show that within the first intron, the density of epigenetic chromatin signals is higher near TASs than in distant regions. Furthermore, the distribution of several chromatin regulatory marks is investigated in relation to gene expression specificity (i.e., housekeeping vs. tissue-specific expression), essentiality (essential genes vs. nonessential genes), and levels of gene expression; housekeeping genes or essential genes contain greater proportions of active chromatin marks than tissue-specific genes or nonessential genes, and highly expressed genes exhibit a greater density of chromatin regulatory marks than genes with low expression. Moreover, we observe that genes carrying multiple first-intron TASs interact with each other within a large protein–protein interaction network, ultimately connecting to the UBC protein, a well-established protein involved in ubiquitination. We believe that our results shed light on the functionality of first introns as a genomic entity involved in gene expression regulation.
Keywords: first intron, trait-associated SNPs, epigenetic regulatory marks
Introduction
Introns exist as components of gene structures in almost all eukaryotic genomes (Simpson et al. 2002; Koonin 2006). It has recently been revealed that they also contribute to organismal fitness, resulting in their maintenance within genomes. Interestingly, the density of intronic sequences in relatively complex mammalian genomes tends to be higher than that in primitive eukaryotes, such as yeast, Drosophila, and Caenorhabditis elegans; the size of introns is 4–5 times larger than the size of exons in the human genome, whereas a very small portion of the genome is allocated to introns in primitive eukaryotes (Nixon et al. 2002; Simpson et al. 2002; Wu et al. 2013). From an evolutionary point of view, it is also interesting that among different species, intron sizes vary more than exon sizes, indicating that introns may play important roles in determining species-specific characteristics and complexities (Gregory 2005; Taft et al. 2007). Studies demonstrating that introns are subject to considerable levels of evolutionary constraint resulting in sequence conservation have led to rejection of the classical understanding of introns (i.e., as nonfunctional, useless junk in the genome) (Gilbert 1978; Graur 1991; Li 1997; Bergman and Kreitman 2001; Halligan et al. 2004; Gazave et al. 2007). For instance, ∼23% of intronic sequences were found to be conserved in mouse–rat comparisons and 17% of intronic sites in a comparison of C. elegans and Caenorhabditisbriggsae (C. elegans Sequencing Consortium 1998; Jareborg et al. 1999; Shabalina and Kondrashov 1999; Vinogradov 2006). Related to these studies, it has been reported that there is a positive relationship between the length of the conserved intronic sequences in a gene and the number of functional domains in the protein expressed by that gene (Vinogradov 2006; Chorev et al. 2017).
First introns are particularly interesting compared with other downstream introns because first introns are the longest and the most selectively constrained and harbor more conserved blocks (Bradnam and Korf 2008; Park et al. 2014). In addition, first introns exhibit a higher density of regulatory elements or functional motifs than other downstream introns (Ladd and Cooper 2002; Majewski and Ott 2002; Halligan et al. 2004; Bradnam and Korf 2008; Park et al. 2014; Jo and Choi 2015). We have reported similar findings in the human genome (Park et al. 2014). We previously reviewed and summarized several independent studies demonstrating possible functional roles of introns (Jo and Choi 2015). Among the suggested functions of introns, our previous work focused on investigating the function of first introns as a possible location of gene expression regulation (Park et al. 2014).
In the present study, we expand on our previous work by integrating multiomics data with intronic site analyses, and we demonstrate the relationship between human diseases and functional clues reflected in gene expression and the enrichment of regulatory histone marks in first introns.
Materials and Methods
Obtaining Introns and Their Lengths in the Human Genome
We followed the same procedures employed in our previous study to extract information about exon–intron boundary positions (Park et al. 2014). Information about the positions of the exon–intron boundaries of genes was downloaded from the University of California Santa Cruz (UCSC) table browser (http://genome.ucsc.edu/cgi-bin/hgTables; last accessed January 2018, assembly: GRCh37/hg19, group: Genes and Gene Predictions, track: NCBI RefSeq, and output format: GTF), in which intron positions for a total of 16,439 genes are represented by unique gene symbols; when two or more RefSeq accession numbers were represented by a single gene symbol, the longest or most recent version of the transcript was chosen. Intronic sites in the present study were further refined through several filtering procedures (refer to Park et al. 2014). 1) The 300-bp regions around exon–intron boundaries were removed to avoid the inclusion of splicing regulatory signal sites. As described by Park et al. (2014), we excluded these regions in our analyses because our primary focus was the investigation of the functional roles of first introns as regions harboring transcriptional regulatory elements. According to Barash et al. (2010), splice site signals are located within 300 bp of a splice junction, which can represent a confounding effect for estimating evolutionary constraints in first introns purely based on sequences taking part in transcriptional regulation. 2) Genes that were too short (i.e., genes of <1 kb of a total length) or introns that were too long (genes > [third quartile + {interquartile range × 1.5}] of total length) were also removed from the analysis to avoid interference from extreme outliers. 3) Y chromosomes were removed, and 4) repeats were masked by RepeatMasker (http://repeatmasker.org/cgi-bin/WEBRepeatMasker). One can refer to our previous report for a more detailed explanation of our filtering procedures (Park et al. 2014).
Obtaining Information about Trait-Associated Single-Nucleotide Polymorphisms
We downloaded the “All associations v1.0” file from the GWAS Catalog database (https://www.ebi.ac.uk/gwas/docs/file-downloads) (MacArthur et al. 2016), containing information about trait-associated single-nucleotide polymorphisms (SNPs) (TASs) and their genomic locations. TAS positions determined based on the reference genome locations of the GRCh38/hg38 assembly were converted into TAS positions for the reference genome locations of GRCh37/hg19 by using the genomic information of GRCh38 assigned to GRCh37/hg19 provided in dbSNP151 (Sherry et al. 2001). As a result, a total of 59,382 TASs were retrieved. By mapping these TASs onto introns prepared through the aforementioned intron filtering procedure, 4,579 TASs were ultimately determined as intronic TASs, of which 1,631 TASs were located in the first intron.
Obtaining Regulatory Chromatin Marks and Mapping Them to Their Corresponding Genomic Positions
We also followed the same procedures applied in our previous study to obtain information about regulatory chromatin marks and to map them to genomic positions. The genomic positions of peaks for DNaseI-hypersensitive sites (DHSs); transcription factor binding sites (TFBSs); other regulatory chromatin marks, including H3K4me1, H3K4me3, H3K9me3, H3K27ac, and H3K27me3; and CCCTC-binding factor (CTCF) binding sites, produced for ENCODE Tier-1 cell lines GM12878, H1-hESC, and K562 by the ENCODE project were downloaded using the UCSC table browser (refer to the section on URLs). The genomic locations of peaks (i.e., regions of statistically significant signal enrichment) for each epigenetic mark were downloaded from the UCSC genome browser. We used the specific download links provided in table 1 of our previous article (Park et al. 2014), and we list the links in the URL section of the present article. After the peak regions for all of these chromatin signals were mapped onto the positions of filtered first intron sites, we estimated the proportion of the sites that overlapped with each chromatin signal.
Table 1.
Functional Annotation of 1,191 Genes Containing TASs within Their First Introns
| Term | Overlap (%) | P |
|---|---|---|
| Gene ontology biological process | ||
| Signal transduction | 6.04 | 1.94E-05 |
| Negative regulation of gene expression | 1.25 | 5.88E-05 |
| Adherens junction organization | 0.60 | 7.99E-05 |
| Negative regulation of cell growth | 0.11 | 8.31E-05 |
| Kidney development | 0.93 | 1.00E-04 |
| KEGG pathway | ||
| HTLV-I infection | 2.02 | 5.50E-05 |
| Inflammatory bowel disease (IBD) | 0.82 | 1.36E-04 |
| Cytokine–cytokine receptor interaction | 1.85 | 2.56E-04 |
| Intestinal immune network for IgA production | 0.65 | 3.80E-04 |
| Pathways in cancer | 2.56 | 5.67E-04 |
| Jensen disease analysis | ||
| Plasma membrane part | 9.57 | 1.03E-14 |
| Spanning component of plasma membrane | 9.15 | 2.41E-13 |
| Extracellular region part | 8.05 | 3.95E-09 |
| Extracellular region | 7.71 | 3.02E-08 |
| Membrane-bounded vesicle | 8.13 | 4.37E-09 |
| MGI mammalian phenotype analysis | ||
| Enlarged spleen | 15.75 | 4.75E-09 |
| Premature death | 9.95 | 2.77E-06 |
| No abnormal phenotype detected | 8.60 | 3.56E-06 |
| Decreased body weight | 9.00 | 1.07E-05 |
| Decreased body size | 9.82 | 1.18E-05 |
Note.—Overlap indicates the proportions of genes carrying TASs in the first intron that overlapped with the genes of each category; P values determined by Fisher’s exact test (i.e., a test of whether the proportion of genes assigned to each category is expected by random chance) were retrieved from DAVID and Enrichr (see Materials and Methods).
Obtaining Housekeeping Genes, Tissue-Specific Genes, Essential Genes, and Nonessential Genes
To obtain housekeeping genes, we compiled lists of housekeeping genes downloaded from two different studies (Eisenberg and Levanon 2013; Pan et al. 2013), including 3,804 genes from Eisenberg and Levanon (2013) and 2,516 genes from Pan et al. (2013), for a total of 4,864 genes (1,456 genes were listed in both studies). After overlapping the genes with the 16,439 genes that we selected after the indicated filtering procedure, a total of 4,159 housekeeping genes were obtained. A total of 2,011 tissue-specific genes were collected by reference to Chang et al. (2011), among which 1,886 genes were selected after overlapping them with the 16,439 genes. After removing genes assigned as both housekeeping genes and tissue-specific genes, a total of 4,082 housekeeping genes and 1,809 tissue-specific genes were ultimately selected. Genes that were neither housekeeping genes nor tissue-specific genes were grouped as “Others.” A total of 3,898 essential genes and 12,541 nonessential genes were retrieved from Chen et al. (2016) by overlapping the 4,420 essential genes and 16,345 nonessential genes from Chen et al. (2016) with the 16,439 genes. Several epigenetic chromatin marks indicated above were independently mapped to the first introns of these classes of genes, and the proportions of each chromatin mark allocated to the first introns were estimated for each class.
Obtaining Gene Expression Levels and Chromatin Marks from Normal Human Tissues
Levels of mRNA expression measured in reads per kilobase million (RPKM) from 11 human tissues were retrieved from the RNA-seq atlas (Krupp et al. 2012), as described in our previous report; a total of 32,384 transcripts were found to present expression values. A total of 14,759 genes from five different tissues (i.e., the tissue types that can be matched to ENCODE Chip-Seq data), including skeletal muscle, liver, lung, heart, and spleen, were selected from the atlas for our present study after overlapping them with our 16,439 genes. We used log2(RPKM + 1) values to report the expression levels of transcripts. Chromatin marks (including H3K4me1, H3K4me3, H3K9me3, and H3K27me3) in each of the five tissues were retrieved from the ENCODE database (supplementary table 1, Supplementary Material online). The relationship between the levels of gene expression and the enrichment of chromatin marks was then investigated for each tissue.
Gene Network Analysis and Functional Annotation
The GeneMANIA plug-in (version 3.5.0) (http://apps.cytoscape.org/apps/genemania; Warde-Farley et al. 2010) of Cytoscape (version 3.6.1) (http://www.cytoscape.org/; Shannon et al. 2003) was used to visualize the interactions of genes harboring TASs in their first introns; we selected “physical interaction” as the option for constructing interaction networks. Further functional annotations were conducted using DAVID (https://david.ncifcrf.gov/home.jsp; Dennis et al. 2003) and Enrichr (http://amp.pharm.mssm.edu/Enrichr/; Kuleshov et al. 2016), where information about gene ontology, KEGG pathways, Jensen diseases, and MGI mammalian phenotypes was obtained. Two types of estimates needed to evaluate the significance of functional annotations, overlap and P value, were retrieved from DAVID and Enrichr.
Statistical Tests
All statistical tests and preparation of graphical representations were performed using R (version 3.5.1; Ripley 2001) with R studio (version 1.1.463; Racine 2012), whereas the remaining analyses that required batching capabilities were conducted with in-house python scripts (version 3.6.0; http://www.python.org). To investigate the relationship between the proportion of each epigenomic mark and the numbers of introns in genes, Kendall’s rank correlation was performed, in which P values and τ were obtained, using “cor.test” with the “kendal” method of R package stats (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html). Linear regression lines were plotted using the “geom_smooth” function with the “lm” method of ggplot2 in the R package (https://cran.r-project.org/package=ggplot2). To investigate whether chromatin marks are distributed differently between different gene classes, such as housekeeping genes versus tissue-specific genes or essential genes versus nonessential genes, Wilcoxon tests were conducted using “stat_compare_means” in the R package ggpubr, in P values were obtained (https://rpkgs.datanovia.com/ggpubr/index.html). z-Score was estimated as follows: (, standard deviation of signal densities; , mean density of signal; and , bin density).
URLs
UCSC table browser:
http://genome.ucsc.edu/cgi-bin/hgTables
GWAS Catalog:
DNaseI-hypersensitive uniform peaks from ENCODE/Analysis:
http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeAwgDnaseUniform
Transcription factor ChIP-seq uniform peaks from ENCODE/Analysis:
http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeAwgTfbsUniform
Histone modifications by ChIP-seq from ENCODE/Broad Institute:
http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeBroadHistone
RNA-seq atlas:
http://medicalgenomics.org/rna_seq_atlas/download
Cytoscape:
GeneMANIA plug-in:
http://apps.cytoscape.org/apps/genemania
DAVID:
https://david.ncifcrf.gov/home.jsp
Enrichr:
http://amp.pharm.mssm.edu/Enrichr/
NCBI GEO database:
Results
TASs Are Enriched in First Introns More Than Expected by Random Chance
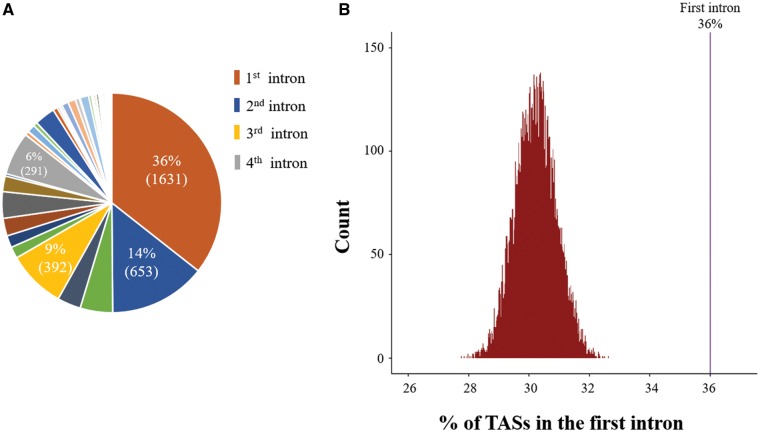
Enrichment of disease alleles in a certain genomic region can provide reasonable proof of the functionality of the region, as disease alleles in first introns should be rare if the first introns are mostly composed of nonfunctional sequences. We therefore tried to test how extensively first introns harbor disease alleles. For this purpose, we first collected TASs from the GWAS Catalog (see Materials and Methods). Up to 36% of the final 4,579 intronic TASs selected after extensive filtering procedures were found to be located in first introns, whereas only 14% and 9% were allocated in second and third introns, respectively (fig. 1A).
Fig. 1.
—Permutation experiment for TASs in the first intron. (A) Pie chart representing the proportions of trait-associated SNPs (TASs) at each ordinal position of introns within genes. Each proportion of TASs for each ordinal position of the intron indicated by each pie portion in the pie chart was estimated using a total of 4,579 TASs allocated to introns. The numbers in parentheses in each portion of the pie indicate the actual number of TASs found in each ordinal intron. Refer to the Materials and Methods for more detailed information on how we obtained intronic TASs. (B) Histograms of the proportions of TASs estimated during 10,000 random iterations. The observed proportion of TASs in the first intron (36%) is indicated by the purple line in the right panel, and the proportions of TASs estimated during the 10,000 permutations are represented by the histograms in the left panel.
It is not surprising that the largest proportion of TASs exist in first introns because first introns are longer than other downstream introns (Bradnam and Korf 2008; Park et al. 2014). Therefore, we asked whether the large proportion of TASs in first intron was due to functionality of the intron sites or if it was simply a by-product of their long length. To address this question, we tested whether the proportion of TASs in the first intron was significantly greater than expected by random chance by conducting a permutation analysis using the 4,579 intronic TASs and randomizing their positions across all intron regions. The proportions of TASs in first introns were estimated for each iteration during 10,000 random permutations and plotted in comparison with the observed proportion (36%), revealing how significantly the observed proportion is biased. As shown in figure 1B, none of the 10,000 permutations were able to produce the observed proportion, strongly excluding the possibility that the proportion of TAS in first introns is simply an artifact of their long length (P ≪ 0.000001). In other words, the fact that first introns exhibit the highest proportion TASs may reflect the functionality of first intron sites.
Higher Density of Epigenetic Chromatin Marks Near TASs
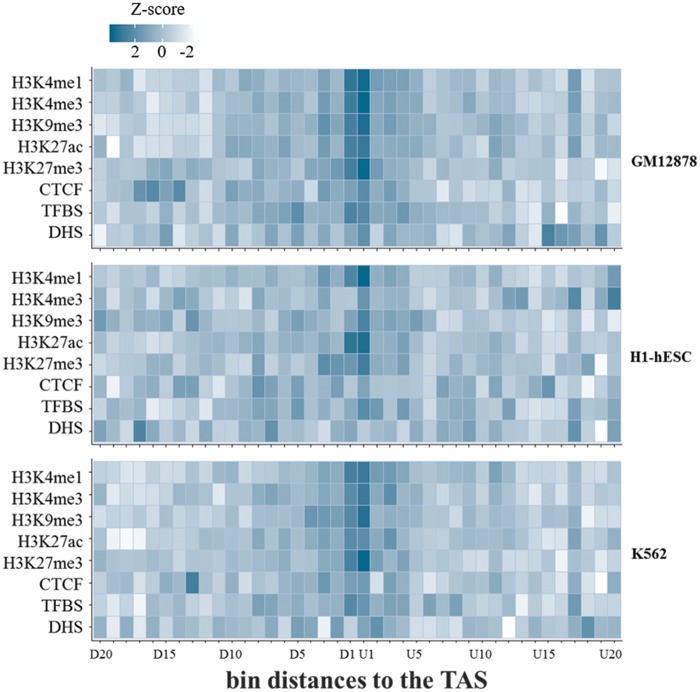
To confirm this conclusion, the relationship between the locations of TASs and epigenetic chromatin signals was subsequently investigated by comparing the density of epigenetic regulatory signals in the regions near TASs in the first intron with that in regions distant from TASs. To accomplish this, we mapped various epigenetic regulatory signals retrieved from ENCODE Tier-1 cell lines, including GM12878, H1-hESC and, K562 (see Materials and Methods), to the corresponding genomic sites and collected peaks located only in first introns. The first introns were then divided into 20 bins of 200 bp both upstream (from U1 to U20) and downstream (from D1 to D20) of the TASs, in which the z-scores of the proportions of sites in which each chromatin signal was located were calculated and are shown in the form of a heatmap (fig. 2). We found that the z-score, that is, the normalized difference of the proportion from the mean proportion of all bins, of most chromatin signals that we investigated was highest in the bin (D1 or U1) nearest the TAS position, indicating that the TAS positions tend to closely colocalize with the epigenetic regulatory signals (fig. 2). This conclusion was generally consistent in the three different Tier-1 cell lines (fig. 2). We next tested the statistical significance using a permutation experiment to determine how unlikely it would be for these densities or proportions of chromatin signals in the nearest bin, D1 or U1, to be generated at random, as shown in supplementary figure 1A–C, Supplementary Material online. As expected, this permutation experiment confirmed that the chromatin signals in the nearest bin to the TAS were significantly enriched. Interestingly, CTCF signals were found to be significantly decreased in the nearest TAS, which seems to be consistent with a previous finding that CTCF binding sites were depleted near H3K27me3 signals (Weth et al. 2014). The proportions of DHS marks did not significantly differ among different bin distances. We do not have a good explanation for this finding, but it seems that the DHS sites do not exactly correspond to the chromatin mark sites where TASs are located. Instead, they may reside sufficiently close to the sites of other chromatin marks to show the same trends (i.e., highest enrichment in the first intron and the higher conservation scores, as shown in our previous study; Park et al. 2014).
Fig. 2.
—Heatmaps of the proportions of epigenetic signals located at different bin distances to TASs. The locations of trait-associated SNPs (TASs) obtained from the GWAS Catalog (see Materials and Methods) were overlaid corresponding to reference genomic positions. Distances to the TASs within the first introns were then divided into 20 bins of 200 bp each. For example, D1 and U1 were 200-bp downstream and upstream of the TAS, respectively, and were plotted on the x axis. Genomic positions of several chromatin regulatory marks for GM12878 were derived from ENCODE (see Materials and Methods). The chromatin regulatory marks that we analyzed here included DNaseI-hypersensitive sites (DHSs), TFBSs, active chromatin marks (e.g., H3K4me1, H3K4me3, and H3K27ac), repressive chromatin marks (e.g., H3K9me3 and H3K27me3), and CTCF binding sites. After the peaks of each chromatin mark were mapped to the corresponding genomic position of each bin, heatmaps were generated with z-scores of the proportions of each chromatin peak for each bin (see Materials and Methods).
Higher Density of Active Chromatin Signals in Housekeeping Genes
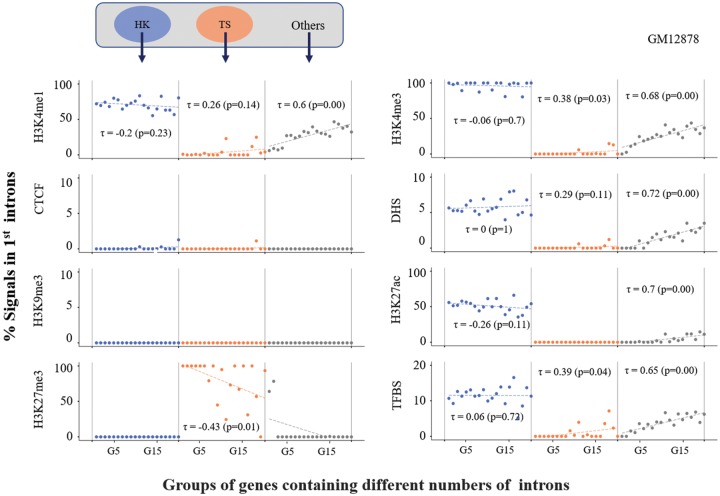
In our previous study, we showed that genes with greater numbers of introns tend to exhibit greater proportions of chromatin regulatory signals in their first introns (Park et al. 2014). Here, we further examined how this positive trend diverged within different classes of genes classified by expression specificity (i.e., housekeeping or tissue specific) or by gene essentiality (i.e., essential genes or nonessential genes) using chromatin data derived from the GM12878 cell line. In each class, the genes were subdivided into 20 different groups by the number of introns in the genes (from G1 to G20), similar to the procedure performed in our previous study (Park et al. 2014). Neither class of gene showed the positive trend in the GM12878 cell line; only the “Others” category (i.e., neither housekeeping nor tissue specific) showed the positive trend (fig. 3), whereas two other Tier-1 cell lines showed the ascending trend between the two measures (i.e., chromatin signals in first introns and the number of introns in genes) (supplementary fig. 2, Supplementary Material online).
Fig. 3.
—Proportions of epigenetic marks in the first introns of genes classified by the specificity of gene expression. Genes were classified by gene expression specificity as follows: housekeeping (HK) genes, tissue-specific (TS) genes, and “Others” (see Materials and Methods). For each class, we further grouped the genes by the number of introns within them. For instance, genes carrying one intron were grouped in G1 (i.e., genes carrying two exons within the gene structures), and genes with two introns were grouped in G2 (i.e., genes carrying three exons within the gene structures). Using the same epigenetic regulatory marks applied in figure 2, we estimated the proportion of each chromatin mark for each gene group and plotted the value in the graph; a total of up to 20 groups of genes (i.e., from G1 to G20) and the proportion of each epigenetic mark were plotted on the x axis and y axis, respectively. Linear regression analysis was applied for each group, from which Kendall’s τ coefficient was estimated, as shown in each graph.
A striking difference in the proportion of each chromatin signal was observed between housekeeping genes and tissue-specific genes in the GM12878 and K562 cell lines; housekeeping genes showed high proportions of active chromatin signals such as H3K4me1, H3K4me3, and H3K27ac, but few repressive chromatin marks, whereas tissue-specific genes showed high proportions of repressive chromatin marks such as H3K27me3, but few active chromatin marks, regardless of the number of introns (supplementary fig. 3, Supplementary Material online). No significant H3K9me3 marks (i.e., another type of repressive chromatin mark involved in maintaining heterochromatin) appeared in any class of genes. H1-hESC, a stem cell-derived cell line, presented few H3K27me3 signals in tissue-specific genes, unlike the other two cell lines (supplementary fig. 3, Supplementary Material online), which may indicate a cell-type specificity of each chromatin signal.
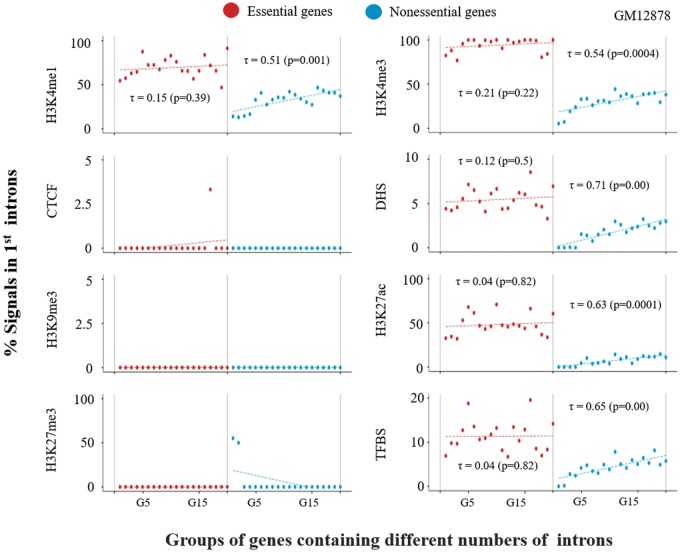
Next, we adapted the same analysis for essential and nonessential genes. Interestingly, nonessential genes showed an ascending trend between the number of introns in genes and the number of chromatin signals in the first introns (fig. 4), which was similar to what we observed for the “Others” category in GM12878, as shown in figure 3. Note that the Tier-1 cell lines K562 and H1-hESC produced a distribution pattern of chromatin signals that was similar, but not identical, to that observed for the GM12878 cell line (supplementary fig. 4, Supplementary Material online). Meanwhile, essential genes were found to exhibit significantly greater proportions of chromatin signals in their first introns than nonessential genes in all the three Tier-1 cell lines (supplementary fig. 5, Supplementary Material online). Notably, the proportions of chromatin signals in the first introns of nonessential genes were similar to those of the “Others” category rather, than those of tissue-specific genes, indicating that nonessential genes are not necessarily tissue-specific genes.
Fig. 4.
—Proportions of epigenetic marks in the first introns of genes classified by gene essentiality. Genes are classified into essential genes and nonessential genes by gene essentiality (see Materials and Methods). The same procedures used for figure 3 were applied in this analysis, where we grouped genes by the numbers of introns in the genes from G1 to G20 in the same way as for figure 3. For each group in each class, we estimated the proportions of chromatin marks as described in the figure 3 legend.
It can be reasonably concluded that the appearance of these interesting trends in the chromatin signal distribution in the first intron occurs because the first intron truly encapsulates functional sites that are tightly linked to gene expression regulation controlled by positioning chromatin marks.
Higher Density of Active Chromatin Signals in Genes with a High Expression Level
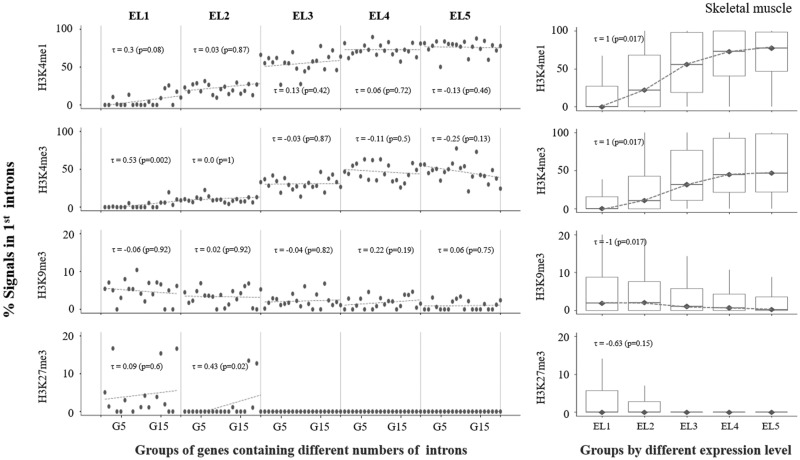
We then investigated how the distribution chromatin regulatory marks in the first intron differs in genes with different expression levels, for which genes were divided into five groups based on their expression levels, designated EL1 to EL5. For this analysis, we retrieved RNA-seq-based mRNA expression data measured in eleven normal human tissues (see Materials and Methods), from which RNA-seq data for five cell types for which ENCODE provides Chip-seq information, including skeletal muscle, heart, lung, liver, and spleen, were selected for analysis. We provide the results of the investigations performed for skeletal muscle here (fig. 5), and the remaining results from the other four cell types are provided in supplementary figure 6, Supplementary Material online.
Fig. 5.
—Proportions of epigenetic marks in the first introns of genes classified by levels of gene expression. Information about expression levels in skeletal muscle was obtained from the RNA-seq atlas (see Materials and Methods). Genes were classified into five groups, from EL1 to EL5, based on the level of gene expression; EL1 and EL5 represent the lowest and the highest levels of expression, respectively. Each class was further divided into 20 different groups by the number of introns within the genes (i.e., from G1 to G20); refer to figure 3 legend for this grouping procedure. We chose only four chromatin regulatory marks for this analysis, which included both active marks (H3K4me1 and H3K4me3) and repressive marks (H3K9me3 and H3K27me3). (A) The same procedures applied in figure 3 were used for this analysis. Refer to figure 3 legend. (B) The proportions of all genes within the same class were estimated for all classes, as represented by a box plot, showing the range of the proportion of each epigenetic mark in each class of genes.
In figures 3 and 4, the genes in each class were subdivided into 20 bins by the number of introns in the genes. As shown in figure 5A, in all classes from EL1 to EL5 in skeletal muscle cells, genes with a greater number of introns (G20) and genes with fewer introns (G1) tended to include greater proportions of active chromatin signals (H3K4me1 and H3K4me3) and repressive marks (H3K9me3 and H3K27me3), respectively, in their first introns. In addition, another interesting pattern was revealed in this analysis, in that genes with a higher expression level (EL5) and genes with a lower expression level (EL1) tended to exhibit greater proportions of active marks and repressive marks, respectively, in their first introns (fig. 5A), which we confirmed by estimating the proportions of chromatin marks after summing the genes from all 20 bins together in each class (fig. 5B). By combining these two observations, we reached another interesting conclusion that longer genes harboring a greater number of introns tend to exhibit higher expression levels, due to the greater numbers of active chromatin signals in their first introns, at least in skeletal muscle cells.
The detailed results for the other four cell types showed slight differences, partly due to the cell-type specificity of gene expression and the mechanical differences in its regulation, or to incompatibility between cell types in the gene expression data derived from the RNA-seq Atlas and the estimated chromatin signals provided by ENCODE (supplementary fig. 6, Supplementary Material online). Note that the RNA-seq data and chromatin regulatory signals analyzed here were not derived from the same cell or tissue sources, which caused some degree of inconsistency in the correlation patterns among different cell or tissue types.
Functional Implications of Genes with TASs in Their First Introns
Thus far, we have discussed observations that are only relevant when the assumption that portions of first introns are functional is true. Here, we present an additional observation that supports the functionality of first introns; we selected only genes that carry one or more TASs in their first introns, and functional annotations were mapped onto these genes. As a result, we found that a total of 1,191 genes included at least one TAS in their first introns, for which DAVID and Enrichr (see Materials and Methods) produced several interesting functional categories of genes, such as “signal transduction,” “negative regulation of gene expression,” and “adherens junction” (table 1). This result seems to be consistent with those of recent studies, demonstrating that introns are involved in maintaining a normal cellular state and DNA stability (Bonnet et al. 2017; Chorev et al. 2017).
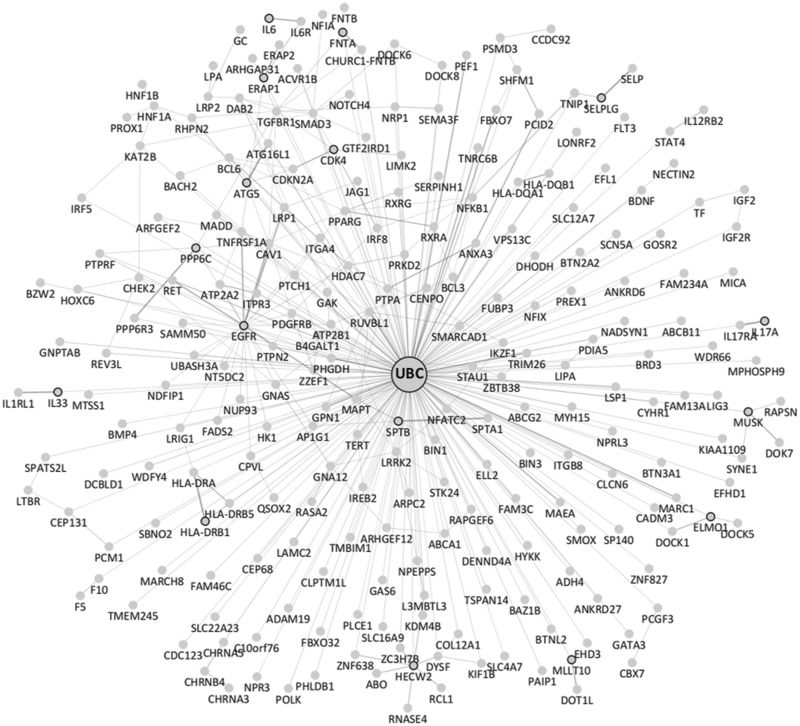
We then attempted to further classify the 1,191 genes by the number of TASs that they carry. Most of the genes carried only one TAS. However, some were found to carry more than 10 TASs within their first introns, such as RSPO3 and FADS2 (supplementary fig. 7, Supplementary Material online), and a total of 248 genes exhibited two or more first-intron TASs. Strikingly, when we analyzed protein–protein interactions for these genes harboring two or more first-intron TASs, a large interaction network connected to the UBC protein (i.e., a protein involved in protein ubiquitination for degradation) was generated (fig. 6). We investigated the extent of enrichment of the UBC-interacting proteins among the 248 genes with two or more TASs. A total of 51.3% (7,828 genes) of the 15,248 genes with no TASs were found to interact with UBC, whereas 64.9% (161 genes) of the 248 genes with two or more TASs interacted with UBC, showing that UBC-interacting genes were, in fact, significantly enriched among the 248 genes (P ≪ 0.0001; odds ratio = 1.75). The biological significance of the interactions of all of these ubiquitination-related genes harboring two or more first-intron TASs with each other within the large protein–protein interaction network is not clear. However, this finding may indicate that genes harboring several gene expression regulation sites within their first introns are likely to mainly function in cellular metabolism and ubiquitination.
Fig. 6.
—Genes carrying many TASs interacted with each other and with the UBC protein. GeneMANIA (a Cytoscape plug-in tool, see Materials and Methods) analysis performed by choosing the “physical interactions” option for genes with two or more first-intron trait-associated SNPs (TASs) showed that the genes interacted with each other as part of a large protein–protein interaction network that was ultimately connected to the UBC protein.
Discussion
We suggest here and in our previous work that the first intron is particularly interesting compared with all other downstream introns. An important question related to the maintenance of introns within the genome has been what selective advantage introns provide that can benefit species under the influence of natural selection over evolutionary time. A simple answer to this question would be that the functional benefits outweigh the costs to cells. In fact, many studies have investigated the functional benefits of introns, particularly the first introns of genes, and we previously summarized these benefits in two categories (Jo and Choi 2015; Bonnet et al. 2017; Chorev et al. 2017): 1) benefits provided by the direct functional roles of introns and 2) benefits provided by the indirect roles of introns.
The regulation of gene expression, mRNA splicing, or nonsense-mediated decay can typically be the result of the direct functional roles of first introns. For instance, a gene-targeting experiment in mice showed that the first intron of the Col1A1 gene plays a regulatory role in tissue-specific and developmental expression of the gene (Hormuzdi et al. 1998). Similarly, a transgenic transient expression experiment performed in Arabidopsisthaliana demonstrated that the first introns of PRF gene family members are functionally distinct in the regulation of gene expression; thus, the first introns of PRF1 and PRF2 affect high constitutive gene expression in vegetative tissues (Jeong et al. 2006). Several consistent findings have been in other monocot and dicot plants (Rose et al. 2008; Jeong et al. 2009; Morita et al. 2012). Additionally, Gallegos and Rose (2017) have suggested further importance of first introns (i.e., an important role in determining transcription start sites, TSSs), by showing that deletion of the promoter sequences of the UBC gene has little effect on the level of gene expression, as long as stimulatory sequences in the first intron are included. Moreover, it has been found that DNA methylation of the first intron is associated with gene expression, with an inverse relationship being observed between the levels of methylation and gene expression (Anastasiadi et al. 2018). The present study is consistent with these previous findings indicating that the first introns of genes are the genomic entity responsible for gene expression regulation. In the present study, we showed that chromatin regulatory marks were significantly enriched in the first intron and were associated with TASs, the specificity of expression, and gene essentiality.
It is somewhat surprising that only few studies have demonstrated the functional changes induced by intronic variations in relation to gene expression regulation, even though some intronic variants are known to be associated with diseases through perturbation of splicing regulation or microRNA binding (Xu and Taylor 2009; Chorev et al. 2017). In fact, there are some contradictory findings that support the idea of functionless introns as well. For example, introns evolve relatively freely due to a lack of selective constraints, and the rate of evolution of intronic sequences is more than 4-fold faster compared with degenerate sites (Parsch 2003; Halligan et al. 2004). In addition, no overt changes in the functions or levels of gene expression have been observed in many experiments designed for analyses in the presence or absence of introns for the same coding genes (Nott et al. 2003). Moreover, ∼3% of the human genome is known to be naturally intronless, yet introns somehow seem to maintain satisfactory expression levels (Grzybowska 2012).
On the other hand, the indirect benefits of introns can be explained by the negative relationship between intron length and the recombination rate. That is, natural selection favors longer introns in regions of low recombination to relax Hill–Robertson interference between two exons on both sides (Comeron and Kreitman 2000; Jo and Choi 2015).
It is well known that despite carrying the same genomic information within a given organism, different cells or tissues express different sets of genes. Epigenomic chromatin signals are key regulators in determining whether gene expression is turned on or off. The epigenomic chromatin signals in different positions in genomic regions vary greatly even among the cells or tissues of an individual organism, and complex combinatorial interactions among these signals and other expression regulatory machineries determine the shapes and patterns of gene expression in different cells or tissues within the same organism. In this context, it is interesting that various epigenomic chromatin signals are located in first introns more often than in all other introns and that the distribution of active chromatin marks and repressive chromatin marks differs significantly between housekeeping genes and tissue-specific genes and between essential genes and nonessential genes. However, in molecular biological experimental settings, it is very difficult to detect how alterations in chromatin signals that occur in first introns are reflected in changes in gene expression levels. This challenge may explain why many studies involving molecular biology-based experiments have failed to detect changes in gene expression between genes with and without introns for the same coding genes.
Although not all aspects of the functional importance of introns have yet been revealed, we no longer think that introns are junk regions in genomes. Further extensive investigation through integrating multiomics data sets with experimental validation may be necessary to gain a clear understanding of the selective advantages or functions that introns provide to organisms.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2016R1D1A1B03930411).
Literature Cited
- Anastasiadi D, Esteve-Codina A, Piferrer F.. 2018. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet Chromatin 11(1):37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, et al. 2010. Deciphering the splicing code. Nature 465(7294):53.. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Kreitman M.. 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11(8):1335–1345. [DOI] [PubMed] [Google Scholar]
- Bonnet A, et al. 2017. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol Cell 67(4):608–621.e6. [DOI] [PubMed] [Google Scholar]
- Bradnam KR, Korf I.. 2008. Longer first introns are a general property of eukaryotic gene structure. PLoS One 3(8):e3093.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282(5396):2012–2018. [DOI] [PubMed] [Google Scholar]
- Chang C, et al. 2011. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PLoS One 6(7):e22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lu G, Chen X, Zhao X, Bork P.. 2016. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 45(D1):D940–D944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorev M, Bekker AJ, Goldberger J, Carmel L.. 2017. Identification of introns harboring functional sequence elements through positional conservation. Sci Rep. 7:4201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M.. 2000. The correlation between intron length and recombination in Drosophila: dynamic equilibrium between mutational and selective forces. Genetics 156(3):1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, et al. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4(9):R60. [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY.. 2013. Human housekeeping genes, revisited. Trends Genet. 29(10):569–574. [DOI] [PubMed] [Google Scholar]
- Gallegos JE, Rose AB.. 2017. Intron DNA sequences can be more important than the proximal promoter in determining the site of transcript initiation. Plant Cell 29(4):843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Marqués-Bonet T, Fernando O, Charlesworth B, Navarro A.. 2007. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 8(2):R21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. 1978. Why genes in pieces? Nature 271(5645):501.. [DOI] [PubMed] [Google Scholar]
- Graur D. 1991. Fundamentals of molecular evolution. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Gregory TR. 2005. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 6(9):699.. [DOI] [PubMed] [Google Scholar]
- Grzybowska EA. 2012. Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem Biophys Res Commun. 424(1):1–6. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Eyre-Walker A, Andolfatto P, Keightley PD.. 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14(2):273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormuzdi SG, Penttinen R, Jaenisch R, Bornstein P.. 1998. A gene-targeting approach identifies a function for the first intron in expression of the alpha1(I) collagen gene. Mol Cell Biol. 18(6):3368–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jareborg N, Birney E, Durbin R.. 1999. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9(9):815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, et al. 2009. Roles of the first intron on the expression of Arabidopsis (Arabidopsis thaliana) genes for actin and actin-binding proteins. Plant Sci. 176(1):58–65. [Google Scholar]
- Jeong YM, et al. 2006. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 140(1):196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo B, Choi SS.. 2015. Introns: the functional benefits of introns in genomes. Genomics Inform. 13(4):112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2006. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 1:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp M, et al. 2012. RNA-seq atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28(8):1184–1185. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Cooper TA.. 2002. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 3(11):reviews0008. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. 1997. Molecular evolution. Sunderland (MA): Sinauer Associates, Inc. [Google Scholar]
- MacArthur J, et al. 2016. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Ott J.. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12(12):1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, et al. 2012. Differences in intron-mediated enhancement of gene expression by the first intron of cytosolic superoxide dismutase gene from rice in monocot and dicot plants. Plant Biotechnol. 29(1):115–119. [Google Scholar]
- Nixon JE, et al. 2002. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci U S A. 99(6):3701–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ.. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9(5):607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, et al. 2013. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS One 8(12):e80747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Hannenhalli S, Choi SS.. 2014. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genomics. 15:526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J. 2003. Selective constraints on intron evolution in Drosophila. Genetics 165(4):1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine JS. 2012. RStudio: a platform‐independent IDE for R and Sweave. J Appl Econom. 27(1):167–172. [Google Scholar]
- Ripley BD. 2001. The R project in statistical computing. MSOR Connect Newsl LTSN Maths Stats OR Netw. 1:23–25. [Google Scholar]
- Rose AB, Elfersi T, Parra G, Korf I.. 2008. Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20(3):543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Kondrashov AS.. 1999. Pattern of selective constraint in C. elegans and C. briggsae genomes. Genet Res. 74(1):23–30. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, MacQuarrie EK, Roger AJ.. 2002. Eukaryotic evolution: early origin of canonical introns. Nature 419(6904):270.. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS.. 2007. The relationship between non‐protein‐coding DNA and eukaryotic complexity. BioEssays 29(3):288–299. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 2006. “Genome design” model: evidence from conserved intronic sequence in human-mouse comparison. Genome Res. 16(3):347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Suppl 2):W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weth O, et al. 2014. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res. 42(19):11941–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. 2013. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci China Life Sci. 56(10):968–974. [DOI] [PubMed] [Google Scholar]
- Xu Z, Taylor JA.. 2009. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 37(Suppl 2):W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.