Abstract
In head and neck squamous cell cancer, the human epidermal growth factor receptor 1 (EGFR) is the dominant signaling molecule among all members of the family. So far, cetuximab is the only approved anti-EGFR monoclonal antibody used for the treatment of head and neck squamous cell cancer, but despite the benefits of adding it to standard treatment regimens, attempts to define a predictive biomarker to stratify patients for cetuximab treatment have been unsuccessful. We hypothesized that imaging with EGFR-specific radioligands may facilitate noninvasive measurement of EGFR expression across the entire tumor burden and allow for dynamic monitoring of cetuximab-mediated changes in receptor expression. Methods: EGFR-specific Affibody molecule (ZEGFR:03115) was radiolabeled with 89Zr and 18F. The radioligands were characterized in vitro and in mice bearing subcutaneous tumors with varying levels of EGFR expression. The protein dose for imaging studies was assessed by injecting 89Zr-deferoxamine-ZEGFR:03115 (2.4–3.6 MBq, 2 μg) either together with or 30 min after increasing amounts of unlabeled ZEGFR:03115 (1, 5, 10, 15, and 20 μg). PET images were acquired at 3, 24, and 48 h after injection, and the image quantification data were correlated with the biodistribution results. The EGFR expression and biodistribution of the tracer were assessed ex vivo by immunohistochemistry, Western blot, and autoradiography. To downregulate the EGFR level, treatment with cetuximab was performed, and 18F-aluminium fluoride-NOTA-ZEGFR:03115 (12 μg, 1.5–2 MBq/mouse) was used to monitor receptor changes. Results: In vivo studies demonstrated that coinjecting 10 μg of nonlabeled molecules with 89Zr-deferoxamine-ZEGFR:03115 allows for clear tumor visualization 3 h after injection. The radioconjugate tumor accumulation was EGFR-specific, and PET imaging data showed a clear differentiation between xenografts with varying EGFR expression levels. A strong correlation was observed between PET analysis, ex vivo estimates of tracer concentration, and receptor expression in tumor tissues. Additionally, 18F-aluminium fluoride-NOTA-ZEGFR:03115 could measure receptor downregulation in response to EGFR inhibition. Conclusion: ZEGFR:03115-based radioconjugates can assess different levels of EGFR level in vivo and measure receptor expression changes in response to cetuximab, indicating a potential for assessment of adequate treatment dosing with anti-EGFR antibodies.
Keywords: Affibody molecules, EGFR, 89Zr, 18F, cancer imaging
Globally, head and neck squamous cell cancer (HNSCC) is the sixth most common cancer; its treatment consists of surgery or radiotherapy, with or without concurrent chemotherapy or targeted therapy (1). Disappointingly, patient survival has not markedly improved in recent decades, and approximately 50% of patients with locally advanced disease will develop recurrence or metastases within 2 y. Such differences in outcome are largely driven by inter- and intrapatient heterogeneity in disease biology (2).
Of note, in HNSCC the human epidermal growth factor receptor 1 (EGFR) is the dominant signaling molecule among all members of the family (3). EGFR messenger RNA and receptor expression were found to be elevated in 92% and 38%–47% of cases, respectively (3,4). Furthermore, this aberrant receptor expression and activity were positively correlated with poor patient prognosis and resistance to radiation therapy (5). These findings led to the development and widespread implementation of specific anti-EGFR inhibitors, including monoclonal antibodies (mAbs) targeting the extracellular domain of EGFR (e.g., cetuximab, panitumumab, and zalutumumab) and small-molecule tyrosine kinase inhibitors targeting the intracellular domain (e.g., gefitinib and erlotinib) (6). So far, cetuximab is the only anti-EGFR therapy agent approved for the treatment of HNSCC. However, despite the clearly documented clinical benefits of cetuximab, attempts to define a predictive biomarker to stratify patients for treatment with the antibody have been unsuccessful. Remarkably, measurement of levels of EGFR protein expression or receptor activation (e.g., the presence of activating gene mutations) has not demonstrated any predictive value benefit to cetuximab treatment in HNSCC. In the context of radical, curative radiotherapy or chemoradiotherapy, dichotomizing EGFR expression into low versus high levels (defined as <50% or >50% positive cells in a clinical trial with radiotherapy and <80% or >80% positive cells in the RTOG0522 study with chemoradiotherapy) did not predict benefit from the addition of cetuximab (2,7,8). Similarly, in the EXTREME trial of first-line use of platin/5-fluorouracil/cetuximab in relapsed or metastatic disease, outcomes were essentially identical for patients with less than or more than 40% EGFR-positive tumor cells (9). As a consequence, at present the clinical use of cetuximab is not based on the measurement of intratumoral EGFR expression or, indeed, of any other biomarker. Against this background, it is important to consider the context in which EGFR has been assessed in previous clinical studies, in order to understand the limitations of the methodologies used. At this moment, no serum biomarker has been identified that can consistently classify an EGFR-positive subgroup of patients for targeted therapies; therefore, in most clinical trials, EGFR expression has been measured via immunohistochemical staining before the initiation of therapy at a single, static time point, and on a relatively small tissue sample (10). For patients with relapsed or metastatic disease, the assessment may have been performed on archival specimens and, furthermore, the reported immunohistochemistry data are subject to variability in the sensitivity and specificity of reagents, the use of different staining protocols, and concerns relating to the tissue sample that could have provided an unrepresentative view of heterogeneous receptor expression. Therefore, we hypothesized that incorporating imaging with EGFR-specific radioligands into routine clinical practice may not only facilitate a noninvasive, real-time measurement of EGFR expression across the patient’s entire tumor burden but also allow for dynamic monitoring of cetuximab-mediated receptor downregulation, providing a marker for adequate treatment dosing.
To date, the use of EGFR-specific molecular imaging for HNSCC patients has been limited to radiolabeled mAbs. Even though they have demonstrated high uptake in EGFR-positive tumors, their large molecular size (∼150 kDa) and consequential slow hepatobiliary clearance (e.g., cetuximab half-life in humans is about 95 h) resulted in poor contrast between tumor and normal tissues on images acquired at earlier time points (11,12). Accordingly, using smaller molecules, such as mAb fragments (e.g., F(ab′)2, Fab′, single-domain antibodies [∼15 kDa], or Affibody [Affibody AB] scaffolds [∼7 kDa]) that are characterized by rapid tumor penetration and fast blood clearance could facilitate high-contrast imaging as early as a few hours after injection (13). Indeed, these favorable pharmacokinetics have been demonstrated by PET imaging of EGFR-expressing xenografts using EGFR-specific Affibody molecules conjugated to several radioisotopes, such as 18F (14,15), 89Zr (16,17), and 11C (18). Of note, a study by Garousi et al. recently showed greater tumor-to-blood ratios 3 h after administration of 89Zr-deferoxamine (DFO)-ZEGFR:2377 than 48 h after injection of 89Zr-DFO-cetuximab, highlighting the advantage of imaging with smaller molecules (19).
Herein, we report the use of a radiolabeled Affibody molecule (ZEGFR:03115) to noninvasively measure differences in EGFR expression, both in vitro and in subcutaneous HNSCC xenograft models. An 89Zr-labeled conjugate was used to assess tumor-to-organ ratios at different time points and a 18F-labeled analog to measure the response to cetuximab treatment in vivo. These data support the hypothesis that a targeted PET agent can quantify EGFR expression level and may represent a powerful missing tool that will facilitate informed image-guided anti-EGFR therapeutic strategies in the clinic.
MATERIALS AND METHODS
Cell Lines
Human head and neck cancer cell line HN5 (EGFR++++) was provided by Prof. Kevin Harrington (Institute of Cancer Research); CAL27 (EGFR+++), Detroit562 (EGFR++), and human breast adenocarcinoma cell line MCF7 (EGFR+) were purchased from the American Type Culture Collection. Cells were cultured in Dulbecco modified Eagle medium (Gibco, Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Life Technologies) and grown as monolayers at 37°C in a humidified atmosphere containing 5% CO2.
Preparation of ZEGFR:03115-DyLight633, 89Zr-DFO- and 18F-Aluminium Fluoride (AlF)-NOTA Affibody Conjugates
The conjugation of DyLight633, DFO-maleimide, and maleimidoethylmonoamide NOTA to the Affibody molecules and the consequent 89Zr and 18F-AlF radiolabeling procedures are described in the Supplemental Data (supplemental materials are available at http://jnm.snmjournals.org).
Flow Cytometry Analysis of EGFR Expression
To assess the EGFR expression in the HN5, CAL27, Detroit562, and MCF7 cell lines, samples were incubated for 1 h at 4°C with a fluorescein isothiocyanate–conjugated EGFR-specific antibody (20 nM) (Santa Cruz Biotechnology). Afterward, the cells were washed in phosphate-buffered saline and analyzed on the BD LSRII flow cytometer (Becton Dickinson). The results were analyzed using FlowJo, version 10 (FlowJo, LLC).
Confocal Microscopy
The binding specificity of the Affibody molecule to EGFR was assessed by incubating HN5 cells with ZEGFR:03115-Dylight633 (1 μM) for 1 h at 37°C with and without preincubation with a 100-fold excess of the ZEGFR:03115. To study internalization of the conjugate, HN5 cells were incubated with ZEGFR:03115-Dylight633 (1 μM) for 1 h at 37°C and washed with phosphate-buffered saline, and confocal images were acquired at 3, 8, and 24 h after incubation. All cells were counterstained with Hoechst 33342 (nuclear stain; Thermo Fisher Scientific) and Lysotracker Green DND-26 (lysosome stain; Thermo Fisher Scientific) 1 h before the images were captured using the LSM700 confocal microscope (Carl Zeiss Inc.). Images (8 bit, 1,024 × 1,024) were analyzed using Zen software, version 2009 (Zeiss).
Immunoblotting
Western blotting was performed as previously described in the literature (20). Briefly, the proteins were immunoblotted with primary antibodies against EGFR, p-EGFR (Tyr1068), AKT (protein kinase B), p-AKT (S473), β-actin, or GAPDH (all from Cell Signaling Technology) overnight at 4°C. On the following day, membranes were rinsed and incubated with horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. The immunoblots were visualized after the addition of SuperSignal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific) and imaged with a ChemiDoc XRS+ System (Bio-Rad). The densitometry was performed using ImageJ (NIH).
In Vitro Binding Affinity and Specificity
The disassociation constants (Kd) of 89Zr- and 18F-AlF–based ZEGFR:03115 conjugates were assessed by a saturation binding assay. The CAL27 cells were incubated with increasing concentrations of radiolabeled ZEGFR:03115 (0.1–50 nM) for 2 h at 4°C. Nonspecific binding was determined by adding a 100-fold excess of unlabeled ZEGFR:03115. Cell-bound radioactivity was determined using the 2480WIZARD2 automatic γ-counter (PerkinElmer). To estimate the Kd, the data were plotted as the concentration of bound labeled Affibody (nM) versus concentration of the labeled Affibody added (nM). The specific binding was measured by subtracting the fraction of the nonspecific from the total binding and fitted using a nonlinear regression curve and a 1-site specific receptor-binding model using GraphPad Prism software, version 7.0.
To evaluate the binding specificity of ZEGFR:03115, we incubated HN5, CAL27, Detroit562, and MCF7 cells with either 89Zr-DFO-ZEGFR:03115 or 18F-AlF-NOTA-ZEGFR:03115 (20 nM) for 1 h at 4°C. To some of the cells, a 100-fold excess of either cetuximab (Merck), natural EGF ligand (Thermo Fisher Scientific), or unlabeled ZEGFR:03115 was added. Afterward, the cells were rinsed and trypsinized, and the radioactivity was measured using a γ-counter. Each experiment was normalized to the maximum cell-associated radioactivity and presented as the mean of 3 independent experiments (performed in triplicate) ± SEM.
Biodistribution
All experiments were performed in compliance with licenses issued under the U.K. Animals (Scientific Procedures) Act of 1986. Studies were compliant with the U.K. National Cancer Research Institute Guidelines for Animal Welfare in Cancer Research (21). Female NU(NCr)-Foxn1nu mice were purchased from Charles River Laboratories. To generate tumor xenografts, CAL27 (4 × 106), Detroit562 (6 × 106), or MCF7 (6.5 × 106) cells were inoculated in BD Matrigel matrix subcutaneously into the shoulder of the mouse. For the MCF7 xenograft model, a 17β-estradiol pellet (0.72 mg, 90-d release) was implanted in each mouse 48 h before cell inoculation. The tumors were allowed to grow until reaching an approximate volume of 100 mm3.
Dose-dependent 89Zr-DFO-ZEGFR:03115 uptake was assessed by injecting the 89Zr-DFO- ZEGFR:03115 into the tail vein (2 μg) either together with or 30 min after increasing amounts of unlabeled Affibody molecule (1, 5, 10, 15, and 20 μg). 89Zr-ZTaq (Taq DNA polymerase-specific Affibody molecule) (3–3.1 MBq, 2 μg) was used as a negative control. Animals were killed at 3, 24, and 48 h after injection. Blood was collected, the major organs were excised and weighed, and their associated radioactivity was measured using a γ-counter. Unless otherwise stated, the results for each tissue sample were calculated as percentage injected dose per gram of tissue (%ID/g) (n ≥ 3 mice ± SD).
Autoradiography
Dissected tumors were embedded in an optimal-cutting-temperature compound (Tissue-Tek; Sakura) and immediately snap-frozen on dry ice. The specimens were sectioned to a thickness of 6 μm using a cryomicrotome (Thermo Fisher Scientific) and mounted on slides. The slides were exposed to a photostimulable phosphor plate for 24 h and read using a Typhoon7000 phosphor imager (GE Healthcare Life Sciences). The recorded images were analyzed using ImageQuant TL, version 8.1 (GE Healthcare Life Sciences).
Immunohistochemistry
The tumors were fixed in formalin and were paraffin-embedded to be sectioned and further processed (Breast Cancer Now Histopathology Core Facility). Multiple sections were taken at regular intervals across each tumor, stained with hematoxylin and eosin (Leica) and an anti-EGFR mAb (Dako pharmDx) according to the manufacturer’s protocol, and imaged with the NanoZoomer-XR (Hamamatsu Photonics).
PET Imaging
The mice were anesthetized using isoflurane (1.5%–2% v/v in O2) before imaging. Using an Albira PET/SPECT/CT preclinical imaging system (Bruker), whole-body 15-min (89Zr) or 10-min (18F) static images were acquired with an energy window of 358–664 keV, followed by CT acquisition. PET data were reconstructed using a maximum-likelihood expectation-maximization algorithm (12 iterations), and scatter and attenuation corrections were applied using their respective CT scans. High-resolution CT scans were performed with the x-ray tube set up at a voltage of 45 kV, a current of 400 μA, 250 projections (1 s per projection), and a voxel size of 0.5 × 0.5 × 0.5 mm. The CT images were reconstructed using a filtered-backprojection algorithm. The PMOD software package (PMOD Technologies Ltd.) was used to analyze the images. The tumor volume was selected by first drawing volumes of interest around the tumor and selecting a 50% maximum pixel isocontour. The mean counts were extracted and converted into %ID/g using a calibration factor (MBq/g/counts) calculated by scanning a source (89Zr or 18F) of known activity and volume.
Cetuximab Treatment
Cetuximab treatment studies were conducted using HN5 xenografts, as this model is particularly sensitive to cetuximab treatment (22). Mice were randomized into either control (n = 6) or treatment (n = 7) groups and treated with a vehicle or cetuximab, respectively. The control mice were injected with 18F-AlF-NOTA-ZEGFR:03115 (12 μg, 1.5–2 MBq/mouse), imaged 1 h after conjugate administration, and killed for biodistribution study. Mice from the treatment group underwent cetuximab treatment (a 600-μg bolus injection every 3 d for a total of 4 doses delivered via intraperitoneal injection). Allowing 13 d for posttreatment clearance of the mAb, subsequent PET images were acquired with 18F-AlF-NOTA-ZEGFR:03115 (12 μg, 1.5–2 MBq/mouse). In addition, to demonstrate a lack of competition between the Affibody and the cetuximab remaining in the blood circulation, an extra group of mice (n = 3) was coinjected with cetuximab (3.75 μg, 26 pmol)—an amount that was estimated to still be present in the blood 13 d after completion of treatment, considering a mAb biologic half-life of 40 h. Densitometric analysis of the tumor tissue lysates was performed using ImageJ (NIH). Data are presented as mean ± SEM (blots, n = 2).
Statistical Analysis
Statistical analyses were performed Prism software.
Significance was determined using the unpaired 2-tailed Student t test with Welch correction. To determine statistical significance between uptake in the different xenograft models, a 2-way ANOVA with Tukey correction was used. Correlation analysis was performed using Spearman rank correlation, with linear regression and 95% confidence intervals. Statistically significant differences between groups were assumed if the P value was 0.05 or less. No data were excluded from the analysis.
RESULTS
ZEGFR:03115 Targeting Properties
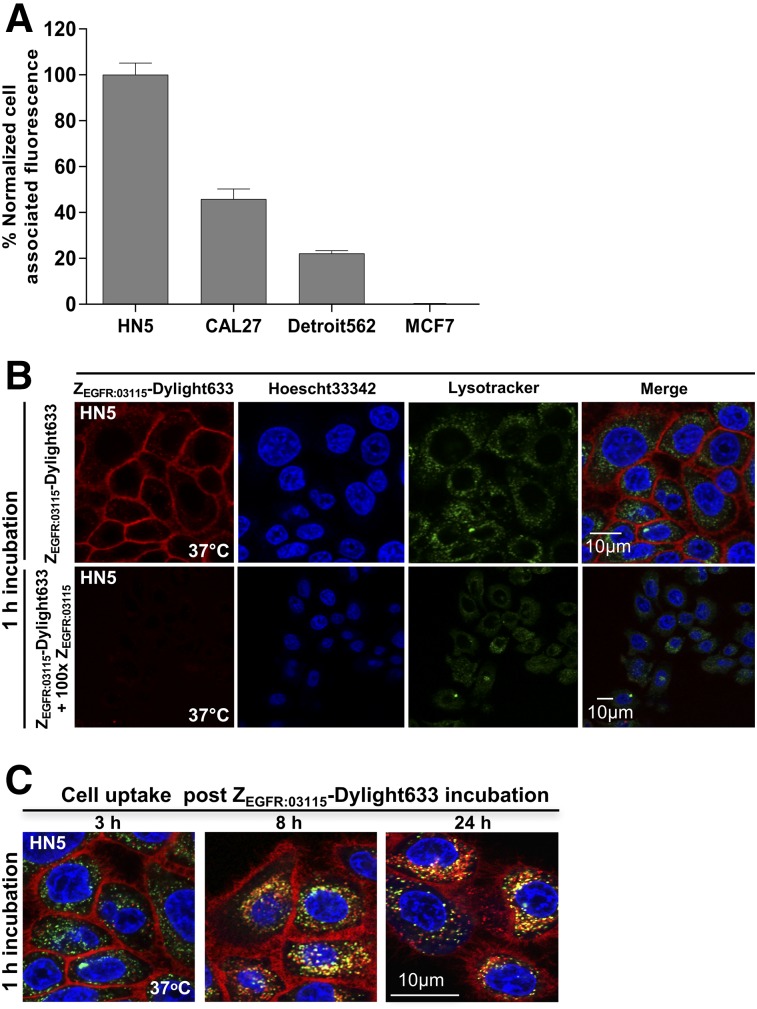
Initially, EGFR expression was determined in the selected cancer cell lines by Western blotting and flow cytometry. The Western blotting results revealed varying levels of EGFR expression, from high, HN5 (EGFR++++); to medium, CAL27 (EGFR+++); to low, Detroit562 (EGFR++); to negligible, MCF7 (EGFR+) (Supplemental Fig. 1), as was concordant with the flow cytometry data using ZEGFR:03115-Dylight633 (Fig. 1A).
FIGURE 1.
(A) EGFR expression as determined by flow cytometry in selected cancer cell lines. (B) In vitro binding specificity of ZEGFR:03115-Dylight633 in HN5 cells as shown by confocal microscopy. (C) Internalization studies of ZEGFR:03115-Dylight633 3, 8, and 24 h after 1 h of incubation in HN5 cells.
To demonstrate that ZEGFR:03115 specifically targets EGFR, HN5 cells were incubated with ZEGFR:03115-Dylight633 and the cell-associated fluorescence was visualized by confocal microscopy. After a 1-h incubation at 37°C, an intense fluorescent signal was found on the cell membrane (Fig. 1B). Images acquired 3, 8, and 24 h later showed an intracellular accumulation of the conjugate, although most still remained bound to the membrane, even 24 h after incubation (Fig. 1C). Additionally, incubating the cells with a 100-fold excess of unlabeled ZEGFR:03115 markedly decreased the fluorescent signal (Fig. 1B, bottom), demonstrating that binding is EGFR-specific and receptor-mediated.
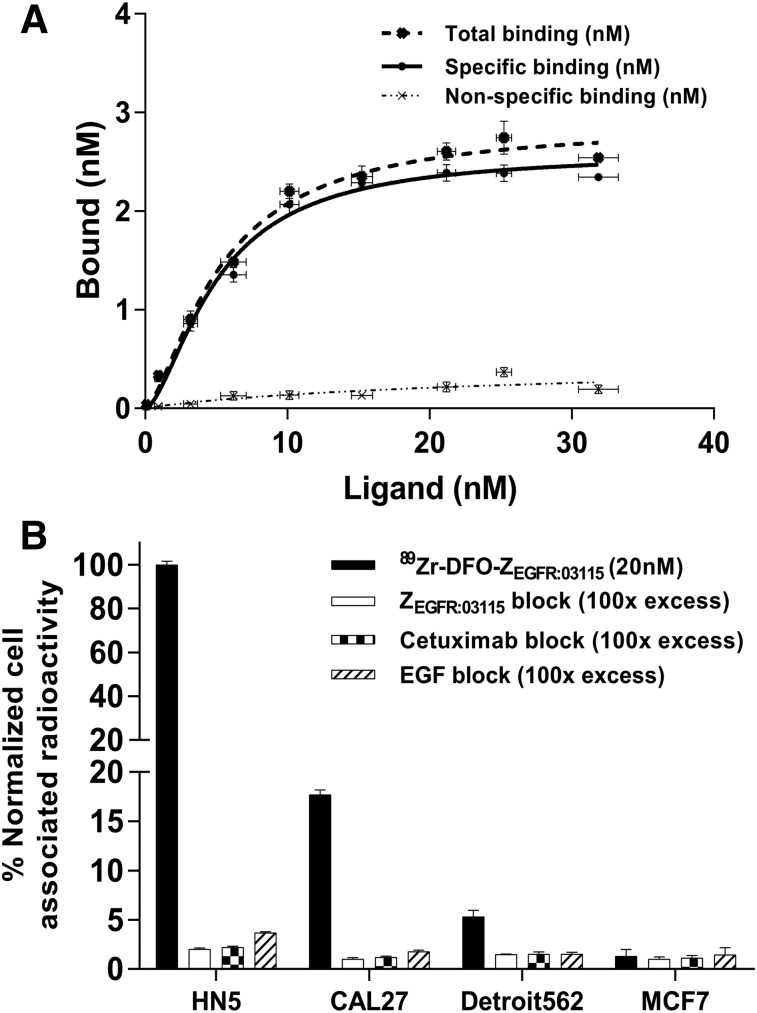
In Vitro Binding of 89Zr-DFO-ZEGFR:03115
The Kd and maximum number of binding sites of 89Zr-DFO-ZEGFR:03115 were determined by a cell-based saturation assay using CAL27 cells. The calculated Kd and maximum number of binding sites were 5.00 ± 0.3 nM and (3.71 ± 0.05) × 106 sites per cell, respectively (Fig. 2A). The target-binding specificity of 89Zr-DFO-ZEGFR:03115 was evaluated in a panel of cancer cell lines with different degrees of EGFR expression, demonstrating that the cell-associated radioactivity was consistent with the measured total EGFR protein expression level in each cell line (Figs. 1A and 2B). Importantly, blocking the receptor with a 100-fold excess of nonlabeled ZEGFR:03115, cetuximab, or the natural ligand EGF, which were demonstrated to compete with the Affibody molecule for the same binding site, significantly reduced radioactivity signal, further confirming the specificity of 89Zr-DFO-ZEGFR:03115 for the target (Fig. 2B).
FIGURE 2.
(A) Saturation curve obtained for CAL27 cells incubated with increasing concentrations of 89Zr-DFO-ZEGFR:03115 bound vs. 89Zr-DFO-ZEGFR:03115 incubation concentration. (B) In vitro binding specificity of 89Zr-DFO-ZEGFR:03115 in selected cell lines with and without blocking using unlabeled Affibody, cetuximab, or EGF. Data are normalized to maximum cell-associated radioactivity per experiment.
Protein Dose-Escalation Studies
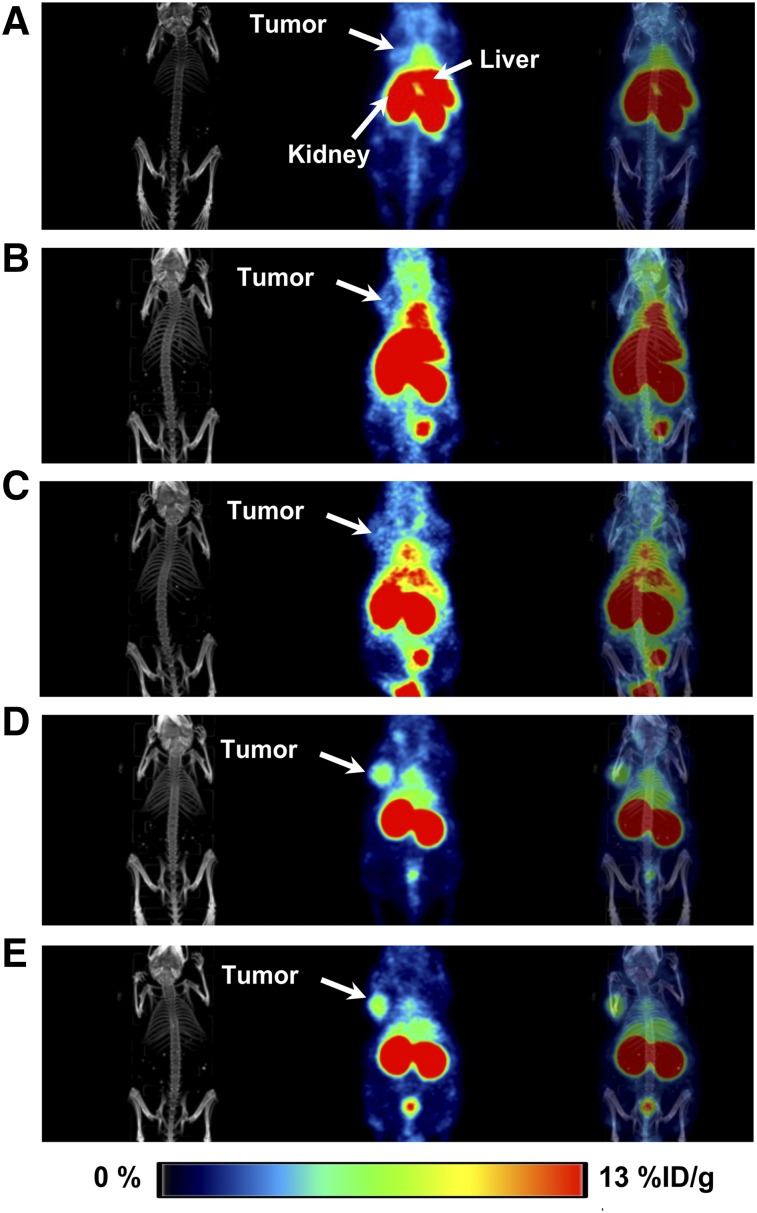
Protein dose-escalation studies were initially performed to test the bioavailability of the radioconjugate for tumor targeting. By adding increasing amounts of nonlabeled Affibody molecule (1, 5, 10, 15, and 20 μg) to the 89Zr-DFO-ZEGFR:03115 (2 μg, 2.5–3.4 MBq/mouse), the radioactivity signal in the liver significantly decreased from 20.74 ± 8.31 to 3.19 ± 0.14 %ID/g (Table 1). Concurrently, tumor uptake increased from 1.75 ± 0.21 to 3.69 ± 1.19 %ID/g (Table 1; Fig. 3). Importantly, an insignificant uptake was achieved when the nonspecific Affibody-based radioconjugate (89Zr-DFO-ZTaq) was injected in CAL27 tumors (0.26 ± 0.05 %ID/g) (Table 1; Supplemental Fig. B2). Also, no significant differences in tumor uptake (3.69 ± 1.19 %ID/g vs. 3.88 ± 0.46 %ID/g) were found when nonlabeled ZEGFR:03115 was either injected 30 min before the radioconjugate or coinjected with the 89Zr-DFO-ZEGFR:03115 (Supplemental Fig. 2). Therefore, the protein dose of 10 μg was administered alongside the radioconjugate for further studies, as it provided the best contrast between targeted and nontargeted tissues (tumor-to-background ratio of 10.25 at 3 h after injection).
TABLE 1.
Ex Vivo Biodistribution 3 Hours After Intravenous Administration of Increasing Amounts of Nonlabeled ZEGFR:03115 30 Minutes Before 2 μg of 89Zr-DFO-ZEGFR:03115 or 2 μg of 89Zr-DFO-ZTaq in CAL27 Xenografts
| 2 μg 89Zr-DFO-ZEGFR:03115 |
||||||
| Organ | 0 μg ZEGFR:03115 | 1 μg ZEGFR:03115 | 5 μg ZEGFR:03115 | 10 μg ZEGFR:03115 | 20 μg ZEGFR:03115 | 2 μg 89Zr-DFO-ZTaq (10 μg ZEGFR:03115) |
| Blood | 3.52 ± 1.42 | 6.01 ± 0.18 | 3.87 ± 0.30 | 4.46 ± 1.65 | 3.06 ± 0.20 | 0.64 ± 0.04 |
| Heart | 0.89 ± 0.22 | 1.53 ± 0.05 | 0.95 ± 0.09 | 1.03 ± 0.29 | 0.77 ± 0.06 | 0.25 ± 0.03 |
| Lungs | 1.45 ± 0.45 | 2.55 ± 0.23 | 1.84 ± 0.50 | 2.05 ± 0.1.06 | 1.45 ± 0.08 | 0.47 ± 0.05 |
| Kidney | 37.14 ± 1.17 | 54.52 ± 17.13 | 73.26 ± 12.91 | 140.84 ± 47.70 | 109.14 ± 22.30 | 172.04 ± 20.07 |
| Spleen | 0.91 ± 0.10 | 1.58 ± 0.23 | 0.94 ± 0.20 | 1.28 ± 0.13 | 0.94 ± 0.13 | 0.37 ± 0.08 |
| Liver | 20.74 ± 8.31 | 10.36 ± 1.49 | 3.82 ± 0.39 | 4.25 ± 1.55 | 3.19 ± 0.14 | 0.82 ± 0.22 |
| Pancreas | 0.73 ± 0.19 | 1.62 ± 0.24 | 0.71 ± 0.12 | 0.69 ± 0.13 | 0.46 ± 0.04 | 0.14 ± 0.03 |
| Tumor | 1.75 ± 0.21 | 1.70 ± 0.68 | 1.87 ± 0.58 | 3.69 ± 1.19 | 2.59 ± 0.48 | 0.26 ± 0.05 |
| Bone | 1.35 ± 0.24 | 1.24 ± 0.21 | 0.81 ± 0.15 | 0.87 ± 0.14 | 0.66 ± 0.09 | 0.21 ± 0.05 |
| Intestine | 0.59 ± 0.28 | 1.62 ± 0.57 | 0.90 ± 0.11 | 0.89 ± 0.11 | 0.81 ± 0.05 | 0.19 ± 0.03 |
| Muscle | 0.24 ± 0.01 | 0.35 ± 0.07 | 0.37 ± 0.09 | 0.36 ± 0.05 | 0.33 ± 0.04 | 0.08 ± 0.03 |
| Ratio | ||||||
| Tumor-to-blood | 0.50 | 0.28 | 0.48 | 0.83 | 0.85 | 0.41 |
| Tumor-to-muscle | 7.29 | 4.86 | 5.05 | 10.25 | 7.85 | 3.15 |
| Tumor-to-liver | 0.08 | 0.16 | 0.49 | 0.87 | 0.81 | 0.32 |
Data are mean ± SD (n ≥ 3) %ID/g.
FIGURE 3.
Whole-body coronal PET/CT images acquired 3 h after 89Zr-DFO-ZEGFR:03115 administration spiked with different amounts of nonlabeled ZEGFR:03115 in mice bearing CAL27 tumors: 2 μg of 89Zr-DFO-ZEGFR:03115 injected (A); 1 μg of ZEGFR:03115 injected 30 min before injection of 2 μg of 89Zr-DFO-ZEGFR:03115 (B); 5 μg of ZEGFR:03115 injected 30 min before injection of 2 μg of 89Zr-DFO-ZEGFR:03115 (C); 10 μg of ZEGFR:03115 injected 30 min before injection of 2 μg of 89Zr-DFO-ZEGFR:03115 (D); 10 μg of ZEGFR:03115 coinjected with 2 μg of 89Zr-DFO-ZEGFR:03115 (E).
89Zr-DFO-ZEGFR:03115 Pharmacokinetics
The pharmacokinetics of the 89Zr-DFO-ZEGFR:03115 were evaluated by performing biodistribution studies at 3, 24, and 48 h after injection of 2 μg (2.5–3.4 MBq) of radioconjugate with 10 μg of ZEGFR:03115. The tumor-to-muscle and tumor-to-blood ratios at 3 h were 10.31 and 1.04, respectively. Because of clearance from nonspecific organs, both values increased at 24 and 48 h after injection, with tumor-to-muscle ratios of 17.35 and 13.56, respectively, and tumor-to-blood ratios of 2.79 and 9.34, respectively (Table 2). At the same time, tumor uptake decreased from 3.88 ± 0.46 %ID/g at 3 h to 2.43 ± 0.27 %ID/g and 2.13 ± 0.12 %ID/g at 24 and 48 h, respectively. The higher tumor uptake would allow for better differentiation between the xenograft tumor types, so we therefore decided to perform the subsequent studies at 3 h after injection. As expected, the kidney uptake remained relatively high over time (104.85 ± 11.10 %ID/g even after 48 h) as a result of glomerular filtration of the Affibody molecule followed by reabsorption, degradation, and retention in proximal tubular cells. Additionally, as expected when using 89Zr-labeled agents, radioactivity accumulation in the bone at 24 h (2.28 ± 0.27 %ID/g) and 48 h (3.44 ± 0.59 %ID/g) was detected.
TABLE 2.
Ex Vivo Biodistribution 3, 24, and 48 Hours After Intravenous Injection of 2 μg of 89Zr-DFO-ZEGFR:03115 Coinjected with 10 μg of Nonlabeled ZEGFR:03115 in Mice Bearing CAL27 Xenografts
| Organ | 3 h | 24 h | 48 h |
| Blood | 3.70 ± 0.55 | 0.87 ± 0.32 | 0.23 ± 0.06 |
| Heart | 0.99 ± 0.19 | 0.37 ± 0.12 | 0.31 ± 0.04 |
| Lungs | 2.74 ± 0.93 | 0.88 ± 0.13 | 0.70 ± 0.08 |
| Kidney | 130.42 ± 22.25 | 108.80 ± 19.60 | 104.85 ± 11.10 |
| Spleen | 1.14 ± 0.18 | 0.72 ± 0.20 | 0.79 ± 0.37 |
| Liver | 4.99 ± 0.85 | 4.31 ± 1.00 | 4.11 ± 1.41 |
| Pancreas | 0.67 ± 0.14 | 0.36 ± 0.10 | 0.35 ± 0.25 |
| Tumor | 3.88 ± 0.46 | 2.43 ± 0.27 | 2.13 ± 0.12 |
| Bone | 0.99 ± 0.28 | 2.28 ± 0.27 | 3.44 ± 0.59 |
| Intestine | 1.09 ± 0.16 | 0.42 ± 0.03 | 0.40 ± 0.02 |
| Muscle | 0.38 ± 0.09 | 0.14 ± 0.04 | 0.16 ± 0.07 |
| Ratio | |||
| Tumor-to-blood | 1.04 | 2.79 | 9.34 |
| Tumor-to-muscle | 10.31 | 17.35 | 13.56 |
| Tumor-to-liver | 0.78 | 0.56 | 0.52 |
Data are mean ± SD (n ≥ 3) %ID/g.
Correlation of 89Zr-DFO-ZEGFR:03115 Tumor Uptake and EGFR Expression
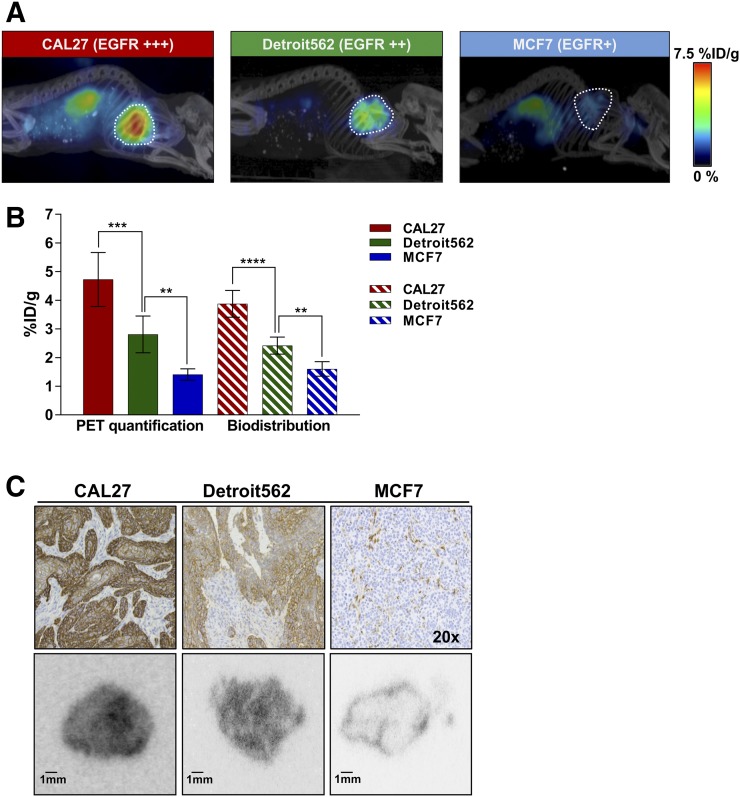
To evaluate whether 89Zr-DFO-ZEGFR:03115 could distinguish between tumors with varying levels of EGFR expression, mice bearing CAL27, Detroit562, and MCF7 xenografts received the radiotracer (2 μg coinjected with 10 μg of ZEGFR:03115; 2.4–3.4 MBq/mouse) and were imaged 3 h after injection using PET/CT. The quantified PET imaging data indicated that the highest levels of radioconjugate accumulation were in CAL27 tumors (4.73 ± 0.90 %ID/g) (Figs. 4A and 4B). Importantly, in vivo specificity of ZEGFR:03115 was confirmed by the lower 89Zr-DFO-ZEGFR:03115 uptake in low–EGFR-expressing MCF7 xenografts (1.41 ± 0.20 %ID/g). These data were then corroborated by the corresponding biodistribution results (Fig. 4B). Furthermore, tumor targeting by the radioconjugate correlated with EGFR expression measured ex vivo by Western blot analysis and immunohistochemical staining (Fig. 4C, top; Supplemental Fig. 3). In addition, the autoradiography of tissue slices from CAL27 and Detroit562 tumors confirmed marked differences in 89Zr-DFO-ZEGFR:03115 uptake as compared with MCF7 tumors (Fig. 4C, bottom).
FIGURE 4.
Radioconjugate uptake in xenografts with varying EGFR expression. (A) Representative whole-body sagittal PET/CT images acquired 3 h after injection. (B) PET quantification of radiotracer uptake in tumors (outlined on image) 3 h after injection in comparison with data obtained from biodistribution studies. Data are mean ± SD. (C) Histopathologic analysis of EGFR expression in xenograft models (top), and representative autoradiography tumor sections 3 h after radioconjugate administration (bottom). **P < 0.01. ***P < 0.001. ****P < 0.0001.
Monitoring Cetuximab-Induced In Vivo EGFR Downregulation
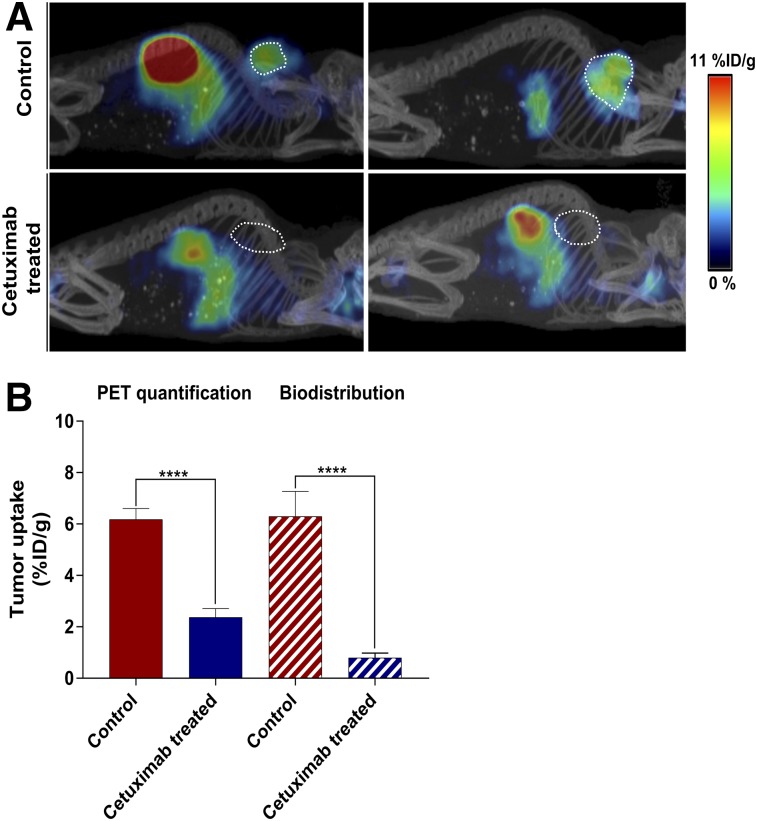
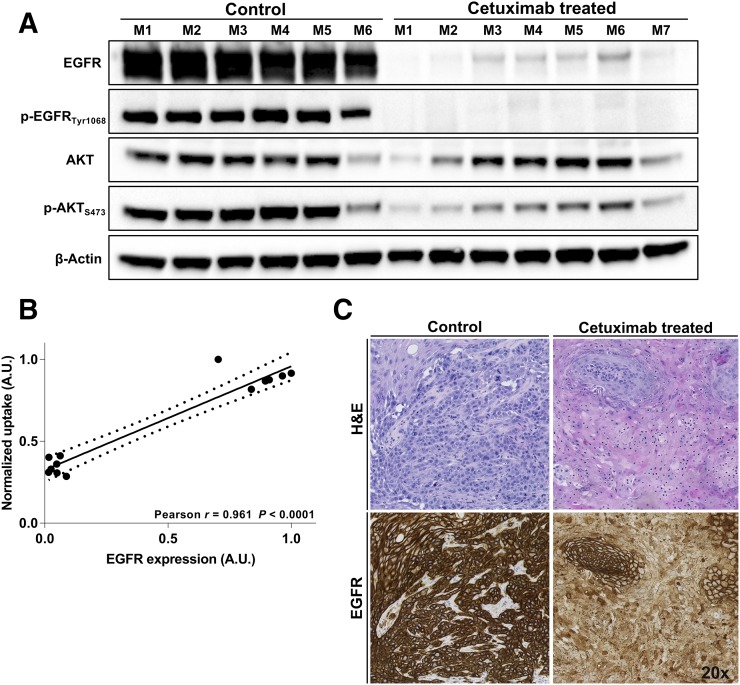
To monitor the response to cetuximab treatment, the long-lived isotope, 89Zr (half-life, 78.4 h), was replaced with 18F (half-life, 108 min). In vitro characterization of 18F-AlF-NOTA-ZEGFR:03115 confirmed the high affinity (Kd, 5.4 ± 1.1 nM) and specificity of the radioconjugate for EGFR (Supplemental Fig. 4). After 18F-AlF-NOTA-ZEGFR:03115 intravenous administration to mice bearing HN5 tumors, a significantly lower tumor uptake was observed in cetuximab-treated mice than in control HN5 tumors, as shown both by the PET image quantification (6.29 ± 0.89 %ID/g vs. 2.37 ± 0.31 %ID/g) and by the biodistribution data (6.18 ± 0.38 %ID/g vs. 0.80 ± 0.17 %ID/g) (Fig. 5; Supplemental Fig. 5A). PET quantification of the cetuximab-treated mice yielded a slightly higher %ID/g than the biodistribution studies, probably because of the relatively high radioactivity in the blood at 1 h after injection (Supplemental Figs. 5B and 5C). However, when background subtraction was applied, only a small amount of the activity was present in the treated tumors (0.28 ± 0.26 %ID/g) (Supplemental Fig. 5D). This significant decrease in the radioconjugate accumulation correlated with a downregulation of EGFR expression after cetuximab treatment as assessed by Western blotting (r = 0.961, P < 0.0001) (Figs. 6A and 6B; Supplemental Figs. 5E and 5F). Furthermore, discernible differences in phosphorylated EGFR and AKT were observed between control and treated mice (Fig. 6A). Additionally, a cetuximab-induced decrease in EGFR expression was confirmed by the immunohistochemistry (Fig. 6C). No significant change in tumor uptake was measured when a group of mice was injected with 18F-AlF-NOTA-ZEGFR:03115 and a quantity of cetuximab that was estimated to be still circulating in the blood 13 d after treatment (Supplemental Fig. 5F). This finding confirmed that the decrease in tumor uptake was not due to competition of the mAb for the same epitope on the receptor. Additionally, no significant decrease in tumor uptake was measured (Supplemental Fig. 5G).
FIGURE 5.
18F-AlF-NOTA-ZEGFR:03115 uptake assessed 1 h after injection. (A) Representative sagittal whole-body PET/CT images of mice bearing HN5 tumors (outlined on image) with or without treatment with cetuximab. (B) PET quantification in control and cetuximab-treated HN5 tumors and corresponding biodistribution %ID/g values. Data are mean ± SD (n ≥ 6). ****P < 0.0001.
FIGURE 6.
(A) Western blot of tumor tissue lysates from control and cetuximab-treated mice demonstrating EGFR protein expression, activation, and downstream signaling. (B) Spearman rank correlation analysis for EGFR expression as determined by Western blot against 18F-AlF-NOTA-ZEGFR:03115 tumor uptake as quantified by PET image analysis. Dashed lines represent 95% confidence levels. (C) Histopathologic analysis of EGFR expression and hematoxylin and eosin (H&E) staining in HN5 xenografts in both control and cetuximab-treated mouse. A.U. = arbitrary units.
DISCUSSION
Despite evidence demonstrating an important prognostic role for EGFR in HNSCC, receptor expression has not been predictive of response to EGFR-targeted therapy. This finding was potentially due to evaluation of EGFR expression in the major clinical trials by an immunohistochemistry-based scoring system, which may not reflect the heterogeneous receptor expression of the whole tumor mass and associated regional lymph node metastases. The integration of molecular imaging biomarkers into standard clinical protocols after injection could address these limitations, providing a global representation of tumor target expression and accessibility, enabling image-guided selection of patients for EGFR-targeted therapies.
Recently, therapeutic mAbs targeting EGFR have been radiolabeled with multiple radioisotopes to provide information about tumor targeting, pharmacokinetics, and accumulation in critical normal organs (23–25). Even et al. reported that 89Zr-cetuximab PET shows large interpatient variability in locally advanced HNSCCs and provides additional information about the accessibility of the drug into the tumor when compared with 18F-FDG PET and EGFR expression evaluated by ex vivo methods (26). However, Niu et al. have demonstrated in different HNSCC xenografts that poor 64Cu-DOTA-panitumumab delivery may result in lack of correlation between PET quantification and the EGFR protein expression (27).
We therefore used a low-molecular-weight targeting vector (i.e., Affibody molecule), because the rapid extravasation from the blood vessels and enhanced tumor mass penetration would provide favorable pharmacokinetics for imaging applications and facilitate visualization of tumor regions inaccessible to mAbs, particularly at early time points. In fact, van Dijk et al. have shown high and specific uptake of 111In-cetuximab-F(ab′)2 as early as 4 h after injection, with high imaging contrast at 24 h in FaDu xenografts. The authors additionally reported that the radioconjugate can monitor the effects of EGFR inhibition combined with irradiation in head and neck carcinoma models (23,28). However, EGFR-targeting mAb fragments do not bind to murine EGFR, limiting proper evaluation of the real tumor-to-background signal of these agents in preclinical models. To address this limitation, several recent studies have investigated whether EGFR-specific Affibody molecules, which cross-react with EGFR of mouse origin, can delineate EGFR-positive tumors and provide high-contrast tumor imaging in the presence of endogenous background levels of EGFR (15–17). In our study, we investigated the binding of ZEGFR:03115 in a panel of HNSCC models. The in vitro data clearly demonstrated that the fluorescent (ZEGFR:03115-Dylight633) and radiolabeled (89Zr-DFO-ZEGFR:03115, 18F-AlF-NOTA-ZEGFR:03115) Affibody-based conjugates maintained a high binding affinity for EGFR and enabled detection of differences in EGFR expression. The specificity for EGFR binding was confirmed by blocking assays with cetuximab and the natural ligand, EGF. To study the pharmacokinetics of ZEGFR:03115 at later time points, we radiolabeled the Affibody molecule with a long-lived PET radioisotope (17). 89Zr was seen as a better-suited radionuclide than 124I for such studies because radiometals are retained more readily than radiohalogens in cells.
Numerous studies have highlighted the importance of optimizing the protein dose when targeting EGFR in order to partially saturate endogenous EGFR expression in the liver and, subsequently, increase the tumor uptake (29,30). Therefore, we performed a protein dose-escalation study and found that we can clearly visualize the tumor 3 h after coinjecting 10 μg of ZEGFR:03115 alongside 89Zr-DFO-ZEGFR:03115 (tumor-to-muscle ratio of 10.31). Furthermore, radioconjugate tumor accumulation was confirmed to be EGFR-specific, as PET imaging of MCF-7 (EGFR+) showed much lower accumulation of the radioconjugate than tumors with higher receptor expression. Moreover, the 89Zr-DFO-ZTaq–related radioactivity in the CAL27 tumor (EGFR++++) was negligible. Importantly, the accumulation of 89Zr-DFO-ZEGFR:03115 in the tumor correlated with EGFR protein expression level, assessed ex vivo by tumor tissue lysates and EGFR staining of tumor sections derived from xenograft models with various levels of receptor expression. Our results successfully demonstrated the advantage of performing imaging at later time points with the Affibody molecule, as exemplified by the increase in tumor-to-background ratios. However, the decrease in tumor accumulation after 3 h after injection would limit sensitivity for discerning subtle changes in EGFR expression. Therefore, the 3-h time point was selected to compare the receptor levels between the xenograft models. Furthermore, the clinical use of an Affibody conjugate radiolabeled with 18F would be more suitable than that of an Affibody conjugate radiolabeled with 89Zr because of a lower radiation exposure burden to the patient, a more desirable positron branching ratio, and a shorter radioactive half-life (108 min vs. 78.4 h). We postulated that 18F-AlF-NOTA-ZEGFR:03115 could measure dynamic changes in receptor expression in response to EGFR inhibition.
This ability may provide useful information for adaptive treatment schedules with anti-EGFR mAbs, since current dosing of cetuximab is based on the patient’s weight and the only confirmed clinical variable that predicts response to this agent is the grade of skin toxicity that occurs after treatment initiation (31,32). Notably, after cetuximab treatment, we observed a significantly lower radioconjugate uptake in the group of mice treated with this mAb. Moreover, this change was in line with decreased EGFR protein levels evaluated by Western blot and immunohistochemistry. These results, together with the fact that no significant change in tumor volume was observed during the treatment, highlight the potential for using EGFR imaging as a tool for assessing cetuximab efficacy based on the receptor level rather than relying purely on anatomic imaging. Of note, EGFR downregulation, because of the internalization and subsequent degradation of EGFR in lysosomes, has recently been reported to be an important determinant of the efficacy of cetuximab treatment for colorectal cancer (33). Our data suggest that it may be possible to evaluate this effect noninvasively using PET-based imaging with radiolabeled Affibody molecules.
Furthermore, immunostaining of tumor sections revealed the presence of residual EGFR-positive cells that directly reflected their viability after cetuximab treatment. These results were corroborated by incomplete inhibition of p-AKT signaling. The surviving cell populations were likely inaccessible to the bulky antibody but potentially may have evaded cetuximab treatment by activating compensatory signaling pathways. However, we recognize that further investigations are needed to verify these findings, which may point the way to further treatment options for such tumors. In the future, 18F-AlF-NOTA-ZEGFR:03115 could also be used for the identification of residual EGFR-positive cells that may be responsible for subsequent cancer relapse.
CONCLUSION
Our results demonstrate that noninvasive molecular imaging of EGFR may open the door to guiding the selection and monitoring of anti-EGFR targeted therapy before it is possible to detect tumor response by changes in volume on conventional cross-sectional imaging modalities. Such an approach may spare patients unnecessary toxicities and improve EGFR-targeted therapy by tailoring a more personalized treatment.
DISCLOSURE
This research was supported in part by the Cancer Research U.K. Cancer Imaging Centre (C1060/A16464) and EPSRC (EP/H046526/1). This report is independent research funded by the National Institute for Health Research. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
Acknowledgments
We thank Affibody AB for supplying the Affibody molecules, and we thank the Pathology Core Facility for providing immunohistochemistry assistance.
REFERENCES
- 1.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Dijk LK, Boerman OC, Kaanders JH, Bussink J. PET imaging in head and neck cancer patients to monitor treatment response: a future role for EGFR-targeted imaging. Clin Cancer Res. 2015;21:3602–3609. [DOI] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. [DOI] [PubMed] [Google Scholar]
- 4.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24:2666–2672. [DOI] [PubMed] [Google Scholar]
- 5.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12:11–26. [DOI] [PubMed] [Google Scholar]
- 6.Rao SD, Fury MG, Pfister DG. Molecular-targeted therapies in head and neck cancer. Semin Radiat Oncol. 2012;22:207–213. [DOI] [PubMed] [Google Scholar]
- 7.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. [DOI] [PubMed] [Google Scholar]
- 8.Ang KK, Zhang Q, Rosenthal DI, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32:2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen MH, Chen H, Shord S, et al. Approval summary: cetuximab in combination with cisplatin or carboplatin and 5-fluorouracil for the first-line treatment of patients with recurrent locoregional or metastatic squamous cell head and neck cancer. Oncologist. 2013;18:460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asgeirsson KS, Agrawal A, Allen C, et al. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007;9:R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fracasso PM, Burris H, Arquette MA, et al. A phase 1 escalating single-dose and weekly fixed-dose study of cetuximab: pharmacokinetic and pharmacodynamic rationale for dosing. Clin Cancer Res. 2007;13:986–993. [DOI] [PubMed] [Google Scholar]
- 12.van Dongen GAMS, Visser GWM, Hooge MNLD, De Vries EG, Perk LR. Immuno-PET: a navigator in monoclonal antibody development and applications. Oncologist. 2007;12:1379–1389. [DOI] [PubMed] [Google Scholar]
- 13.Löfblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY. Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010;584:2670–2680. [DOI] [PubMed] [Google Scholar]
- 14.Miao Z, Ren G, Liu H, Qi S, Wu S, Cheng Z. PET of EGFR expression with an 18F-labeled affibody molecule. J Nucl Med. 2012;53:1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su X, Cheng K, Jeon J, et al. Comparison of two site-specifically 18F-labeled affibodies for PET imaging of EGFR positive tumors. Mol Pharm. 2014;11:3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summer D, Garousi J, Oroujeni M, et al. Cyclic versus noncyclic chelating scaffold for 89Zr-labeled ZEGFR:2377 affibody bioconjugates targeting epidermal growth factor receptor overexpression. Mol Pharm. 2018;15:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burley T, Martins CD, Da Pieve C, et al. 89Zr-affibody for specific detection of EGFR expression in HNSCC [abstract]. Eur J Nucl Med Mol Imaging. 2015;42(suppl):S247. [Google Scholar]
- 18.Cheng Q, Wallberg H, Grafstrom J, et al. Preclinical PET imaging of EGFR levels: pairing a targeting with a non-targeting Sel-tagged Affibody-based tracer to estimate the specific uptake. EJNMMI Res. 2016;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garousi J, Andersson KG, Mitran B, et al. PET imaging of epidermal growth factor receptor expression in tumours using 89Zr-labelled ZEGFR:2377 affibody molecules. Int J Oncol. 2016;48:1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer-Marek G, Kiesewetter DO, Capala J. Changes in HER2 expression in breast cancer xenografts after therapy can be quantified using PET and 18F-labeled affibody molecules. J Nucl Med. 2009;50:1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Workman P, Aboagye EO, Balkwill F, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeckx C, Op de Beeck K, Wouters A, et al. Overcoming cetuximab resistance in HNSCC: the role of AURKB and DUSP proteins. Cancer Lett. 2014;354:365–377. [DOI] [PubMed] [Google Scholar]
- 23.van Dijk LK, Hoeben BA, Kaanders JH, Franssen GM, Boerman OC, Bussink J. Imaging of epidermal growth factor receptor expression in head and neck cancer with SPECT/CT and 111In-labeled cetuximab-F(ab′)2. J Nucl Med. 2013;54:2118–2124. [DOI] [PubMed] [Google Scholar]
- 24.Aerts HJWL, Dubois L, Perk L, et al. Disparity between in vivo EGFR expression and 89Zr-labeled cetuximab uptake assessed with PET. J Nucl Med. 2009;50:123–131. [DOI] [PubMed] [Google Scholar]
- 25.Song IH, Noh Y, Kwon J, et al. Immuno-PET imaging based radioimmunotherapy in head and neck squamous cell carcinoma model. Oncotarget. 2017;8:92090–92105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Even AJ, Hamming-Vrieze O, van Elmpt W, et al. Quantitative assessment of zirconium-89 labeled cetuximab using PET/CT imaging in patients with advanced head and neck cancer: a theragnostic approach. Oncotarget. 2017;8:3870–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu G, Li Z, Xie J, Le QT, Chen X. PET of EGFR antibody distribution in head and neck squamous cell carcinoma models. J Nucl Med. 2009;50:1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk LK, Boerman OC, Franssen GM, Kaanders JH, Bussink J. 111In-cetuximab-F(ab′)2 SPECT and 18F-FDG PET for prediction and response monitoring of combined-modality treatment of human head and neck carcinomas in a mouse model. J Nucl Med. 2015;56:287–292. [DOI] [PubMed] [Google Scholar]
- 29.Tolmachev V, Rosik D, Wallberg H, et al. Imaging of EGFR expression in murine xenografts using site-specifically labelled anti-EGFR 111In-DOTA-Z EGFR:2377 Affibody molecule: aspect of the injected tracer amount. Eur J Nucl Med Mol Imaging. 2010;37:613–622. [DOI] [PubMed] [Google Scholar]
- 30.de Souza ALR, Marra K, Gunn J, et al. Fluorescent Affibody molecule administered in vivo at a microdose level labels EGFR expressing glioma tumor regions. Mol Imaging Biol. 2017;19:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orditura M, De Vita F, Galizia G, et al. Correlation between efficacy and skin rash occurrence following treatment with the epidermal growth factor receptor inhibitor cetuximab: a single institution retrospective analysis. Oncol Rep. 2009;21:1023–1028. [DOI] [PubMed] [Google Scholar]
- 32.Bar-Ad V, Zhang Q, Harari PM, et al. Correlation between the severity of cetuximab-induced skin rash and clinical outcome for head and neck cancer patients: the RTOG experience. Int J Radiat Oncol Biol Phys. 2016;95:1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada Y, Kimura T, Nakagawa T, et al. EGFR downregulation after anti-EGFR therapy predicts the antitumor effect in colorectal cancer. Mol Cancer Res. 2017;15:1445–1454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.