Abstract
Ewing Sarcoma was first described in 1921 in the Proceedings of the New York Pathological Society by an eminent American pathologist from Cornell named James R. Ewing as a “diffuse endothelioma of bone”. Since this initial description, more has been discovered regarding Ewing Sarcoma and in the 1980’s both Ewing Sarcoma and peripheral primitive neuroectodermal tumors due to their similar features and shared identical genetic abnormality were grouped into a class of cancers entitled Ewing Sarcoma Family of Tumors (ESFT). Ewing Sarcoma is the second most common pediatric osseous malignancy followed by osteosarcoma, with highest incidence among 10 to 20-year olds. Ewing Sarcoma is consistently associated with chromosomal translocation and functional fusion of the EWSR1 gene to any of several structurally related transcription factor genes of the E26 transformation-specific (ETS) family. These tumor-specific molecular rearrangements are useful for primary diagnosis, may provide prognostic information, and present potential therapeutic targets. Therefore, ways to rapidly and efficiently detect these defining genomic alterations is of clinical relevance. Within the past decade liquid biopsies, including extracellular vesicles (EVs), have emerged as a promising alternative and/or complimentary approach to standard tumor biopsies. It was recently reported that fusion mRNAs from tumor-specific chromosome translocations can be detected in Ewing Sarcoma cell-derived exosomes. Within this review, we overview the current advances in Ewing Sarcoma and the opportunities and challenges in exploiting circulating exosomes, primarily small bioactive EVs (30–180 nm), as developing sources of biomarkers for diagnosis and therapeutic response monitoring in children and young adult patients with ESFT.
Keywords: Circulating Exosomes, Liquid Biopsy, Ewing Sarcoma Family of Tumors
Introduction
In Dr. Ewing’s paper in 1921 (1), he reported a 14-year-old female who developed a tumor of the radius believed to be osteosarcoma, which, at that time, was already well known to clinicians and usually managed by amputation. For reasons unknown, this patient received therapies other than amputation. With no clinical improvement, she was then treated at Memorial Hospital with 12,760 miCu-hr of radium every two weeks for three doses, she obtained complete response by both examination and radiographic imaging. The responsiveness of this tumor to radiotherapy suggested to some clinicians that this was a distinct entity from osteosarcoma. Unfortunately, after tumor recurrence, a biopsy of the tumor was taken to settle differences in opinion regarding the tumor. Pathology confirmed that the tumor was unlike osteosarcoma, and rather, was described vaguely using the term “round cell sarcoma” by Dr. Ewing. He further noted that the tumor cells had the appearance of blood vessels of the bone and therefore characterized the tumor as an “endothelioma of bone”.
ESFTs are a clinicopathologic spectrum of the same neoplastic entity, consisting of osseous Ewing Sarcoma, extraosseous Ewing Sarcoma, peripheral primitive neuroectodermal tumor (PNET) and Askin tumor (Ewing Sarcoma arising from the chest wall). From a gross pathology standpoint, the ESFT have a gray-tannish appearance with infiltrative borders. Morphologically, ESFT are composed of sheets of small round blue cells with a high nuclear-to cytoplasmic ratio. While microscopically, these tumors are divided into three major histologic subtypes: classical Ewing Sarcoma, atypical Ewing Sarcoma (tumors arising from soft tissue) and peripheral primitive neuroectodermal tumor (PNET); with classical Ewing Sarcoma makes up the majority of cases. Strong cell-surface glycoprotein p30/32MIC2 (CD99) expression is characteristic of ESFT and diffuse membranous staining is present in 95%–100% of tumors (2). ESFTs are consistently associated with chromosomal translocation and functional fusion of the EWSR1 gene to any of several structurally related transcription factor genes of the E26 transformation-specific (ETS) family (3). EWS-Ets fusions have been seen in 80-95% of cases with Ewing Sarcoma, atypical and PNET (Figure 1) (4). It was first thought that the ESFTs were each distinct biological entity. However, based on their immunohistochemical, cytogenetic, molecular uniformity, range of neural differentiation (PNET being most differentiated) and their shared/similar response to chemotherapeutic regimens, it was determined that they are all related and originate from unique mesenchymal stem cells capable of multilineage differentiation (5).
Figure 1. Visual schematic of the EWS-Ets fusion gene.

(A) The EWS-Ets rearrangement is caused by the fusion of the N-terminal transactivation domain of the EWSR1 gene with the C-terminal DNA binding domain of a select member of the Ets gene family. (B) Approximately 90% or more of ETW-Ets fusions involve the FLI1 gene. ERG is the next most common fusion partner accounting for between 5 and 10 % of cases. Rarely, EWS has been observed to be paired with ETV1, ETV5, and FEV1, and these fusions account for less than 1% of total cases.
Diagnosis and Treatment of Ewing Sarcoma
ESFT is the second most common pediatric osseous malignancy (6) in children and young adults exceeded in prevalence only by osteosarcoma. The incidence of ESFT is approximately 200 new ESFT diagnoses annually in the United States and with the highest incidence among 10-20 year olds (7–9). Caucasians are more affected than Asians and especially African-Americans (9). It is an aggressive sarcoma within the pediatric population, and often presents with a high rate of metastases (10). Although the overall survival (OS) for patients with localized disease now approaches 65-75%, the acute and long-term toxicities of therapy are substantial. Lymph node involvement and staging for sarcomas as defined by the American Joint Committee on Cancer are not relevant in ESFT, since the rarity of nodal involvement. The strongest prognostic factor across different treatment strategies is presence of metastasis at time of diagnosis. Currently the five year overall survival remains <30% for patients with initially metastatic disease (10). However, those with isolated pulmonary metastases have better clinical outcomes than those with metastases at other sites (3 year Event Free Survival [EFS], 29%−52%) (11–14). Despite the current standard intensive therapy regimens and improved local control therapy, 30-40% of ESFT patients experience recurrence. The 5-year survival for children with progressive (refractory) or recurrent pediatric ESFT remains stagnant at 20% (15).
The clinical presentation of ESFT are usually nonspecific, with pain (82%–88% of patients) and a mass/swelling (60% of patients) being the most common signs or symptoms (16, 17). For this reason, diagnosis is often done by first ruling out more common diseases. When a child or young adult is diagnosed with a suspected form of bone cancer, the standard diagnostic assay is obtainment of a tissue biopsy for gross pathologic evaluation, immunohistochemistry staining and cytogenetic evaluation in order to confirm diagnosis of ESFT. Although it is the gold standard, tissue biopsies are, in general, invasive, costly, and painful (18). These factors as well as others, make it difficult to perform repeated biopsies on a pediatric patient, thus it remains impractical to monitor tumor changes to therapy over time (19).
A majority of patients with localized disease on initial presentation have subclinical micrometastatic disease, thus the treatment of ESFT relies on a multidisciplinary approach which combines risk-adapted chemotherapy and focal therapy (surgery, radiation or both) to minimize risk of long-term sequelae and maximize the chance of cure in all ESFT patients. The standard frontline therapy for these patients primarily involves the use of neoadjuvant chemotherapy with the goal of eliminating micrometastases and reducing the size/volume of the primary tumor. Further, local control consists of surgical resection, radiation therapy (RT), or both. A benefit of surgical resection after neoadjuvant chemotherapy is in the opportunity to gather information concerning amount of necrosis of the tumor. In the French Society of Paediatric Oncology (EW88) study, event free survival at 5 years for patients with >95% necrosis was 75%, 70-95% necrosis was 48%, and <70% necrosis was 20% within the resected tumors (20). Radiation for local control is recommended for non-resectable ESFT tumors as well as when there is poor likelihood of adequate surgical margins with resection alone. Diagnosis and monitoring of patients with ESFT tumors consist of imaging techniques such as radiography, magnetic resonance imaging (MRI) and computed tomographic (CT) scans. On radiography, ESFT of bone reveals aggressive features, reflecting the high-grade nature of this malignant lesion (21). Coupled with CT, Positron emission tomography with 2-deoxy-2-[fluorine-18]fluoro-D-glucose (FDG PET) has been shown to demonstrate sensitivity and specificity in the staging of ESFT (22). In regards to osseous metastases, FDG PET has been shown to be far superior to bone scintigraphy, while for primary bone lesions (18% not evident at bone scintigraphy when compared with FDG PET) (23). FDG PET is more useful in evaluation of tumor response to treatment, with its ability to depict molecular changes before morphologic abnormalities are evaluated on cross-sectional imaging. Macrometastasis, which is the presence of clinically apparent metastases, has prognostic significance, and staging of patients is based on imaging of the primary tumor and the sites of likely metastasis. Lab based studies include a complete blood count, baseline chemistries, erythrocyte sedimentation rate assay (increased in ~ 50% of patients) and serum lactate dehydrogenase measurement. Laboratory advancements and the improvements in tumor banking, have made way for an increase exploratory studies for biomarkers in ESFT. But, as to date there exists no readily available biomarkers for this disease.
Molecular Characterization of ESFT
The advent of cytogenetic evaluation of biopsied lesions demonstrate that ESFT tumors share a common karyotype abnormality. ESFT are characterized by specific rearrangements of one of the five alternative Ets family member genes, i.e, FLI1, ERG, ETV1, E1AF, and FEV with EWSR1 (Figure 1) (24). This recipient gene encodes for a multifunctional protein that is involved in various cellular processes, including gene expression, cell signaling, and RNA processing and transport. In approximately 90% of these lesions, the disease-associated genomic alteration is a translocation between the long arms of chromosomes 11 and 22 (t[11 ;22] [q24;q12]) (25–27). Upon cloning of the specific breakpoints, it was shown to involve the EWS gene (EWSR1) on chromosome 22 and the FLI1 gene on chromosome 11. This genetic mutation is thought to lead to the oncogenic conversion of EWS by exchanging its RNA binding domain with different DNA binding domains, thus generating tumor-specific fusions proteins. Subsequent studies found that many alternative forms of these two gene translocations exist, corresponding to variations in the locations of the EWS and FLI1 breakpoints. The most common form, “Type 1”, accounts for approximately 51% of cases and consists of the first seven exons of EWS joined to exons 6-9 of FLU; while “Type 2”, accounts for approximately 27% of cases and also includes FLI1 exon 5(28, 29). The second most common cytogenetic aberration after EWS-FL11, seen in only 5%–10% of ESFT, is the translocation t(21;22)(q22;q12), which is referred to as EWS-ERG (Figure 1) (30). The fusion protein derived from EWS-ERG is similar to that of the EWS-FLI1 gene product and thought to also be associated with the pathogenesis of the disease. Others have suggested, EWS-FLI1 alone cannot fully explain the pathogenesis of ESFT. Elzi and colleague suggested that FLI1-EWS, a fusion gene reciprocal to EWS-FLI1, is frequently expressed in Ewing Sarcoma and might also be required for Ewing sarcomagenesis (31). While the differences in the proliferation rates of ESFT had been previously thought to be assigned to distinct transcriptional activation potentials of EWS-FLI1 Type 1 and EWS-FLI1 Type 2 proteins (32), genomic expression studies have failed to demonstrate this relationship (33). A prospective evaluation of EWS-Ets transcript structure in more than 560 patients lacked statistical significance of prognostic benefit in patients with localized disease carrying Type 1 EWS-FLI1 expressing tumors. This evaluation did however, find that patients with non-type 1/non-type 2 EWS-FLI1 demonstrated a small but insignificant risk of progression or relapse than other fusion types (34).
Circulating Exosomes: Emerging Liquid Biopsy for Diagnosis of ESFT
Despite significant advancements in treating localized ESFT, the 5-year overall survival for patients with metastatic or relapsed/recurrent ESFT remains dismal. A possible contributor to these poor clinical outcomes is that, aside from clinical symptomatology and radiographic imaging, no suitable and readily available biomarker exists, that can specifically monitor disease progression (refractory) or detect recurrence in ESFT. Tissue biopsy is the gold standard, while itis highly invasive and not practical to collect tissue samples repeatedly on pediatric patients. Within the past decade liquid biopsies for the analysis of cell-free DNA, RNA, and soluble proteins have emerged as a much-heralded alternative and/or complimentary approach to the current standard tumor biopsies. They are often minimally invasive, quicker, and more easily repeatable because they collect needed information from a bio-fluid sample, such as blood, urine, saliva, or cerebrospinal fluid. Potentially revolutionary, liquid biopsy promises to dramatically improve disease diagnosis, surveillance, and survival prediction (35, 36). Despite the incredible promise, traditional liquid biopsies have not been able to entirely replace tissue biopsies (37). However, the discovery of small microvesicles termed exosomes, which are enriched in tumor-derived RNAs (including miRNAs) as well as an assortment of proteins, encapsulated within a lipid bilayer, are leading to way towards changing the liquid biopsy landscape.
We, and others, have shown that tumor cells constitutively release exosomes which are characteristic of the parental cell of origin (38–43). Virtually all tumor cells release exosomes, and the accumulation of these vesicles represent key features of tumor transformation to malignant status (44). The rate of exosomal release is significantly increased in most neoplastic cells and occurs continuously at all stages of tumor development (45). In addition, qualitative and quantitative changes in the cargo transported by these microvesicles are observed with tumor progression (46, 47). Extracellular shedding of exosomes occurs in other types of cells under specific physiological conditions and accumulation of exosomes from non-neoplastic cells is frequently observed in vivo (48). However, there are more tumor-derived exosomes accumulated in biologic fluids, including sera, malignant ascites, and pleural fluids (49–51). Cancer cell derived exosomes may facilitate the growth, invasion and metastasis of these malignant tumors due to their ability to provide autocrine, paracrine, and endocrine signaling, which promote tumorigenesis (52–56). Cancer cell derived exosomes may also contain oncogenic elements, which have the potential to initiate tumor pathway signaling and promote metastasis (39, 57). The constitutive release and enrichment of certain proteins and nucleic acids into exosomes prompts consideration of the use of these vesicles as diagnostic tools in cancer. While circulating tumor cells and cell-free DNA have taken prominent focus as predominant liquid biopsy-based biomarkers (58–61), exosomes offer a robust alternative since they are i) enriched with original cellular content (e.g., proteins, mRNAs and microRNAs), ii) actively released from viable cells instead of shed or apoptotic cells, iii) relatively abundant (e.g., 1×106 to 1×1011 particles per mL of blood), iv) very stable in bodily fluids (62–64), and v) representative of the heterogeneous tumor cell population.
To date, most efforts invested in exosome research have been focused on adult cancers and very limited progresses have been reported in childhood cancers. Recently, it has been reported that fusion mRNAs from tumor-specific chromosome translocations can be detected in Ewing Sarcoma cell-derived exosomes. The first group, Miller and colleagues spiked purified Ewing Sarcoma derived exosomes into healthy human plasma and were able to isolate and detect these exosomes bearing the genetic information of Ewing Sarcoma (65). A second group, Tsugita et. al., were able to detect the EWS-FLI1 mRNA in nanovesicles in four out of ten mice inoculated with the Ewing Sarcoma cell line A673 and three out of seven mice inoculated with the Ewing Sarcoma cell line TC135 (66). To advance our knowledge of tumor-derived exosomes and establish their utility as valuable tools for the diagnosis and therapeutic monitoring in ESFT patients, we recently used molecular approaches to profile the proteome and miRNA content of ESFT-derived exosomes. For example, global proteomic profiling via mass spectrometry identified approximately 1,200 exosomal associated proteins, including many with biomarker potential in ESFT. Two proteins identified are currently being used in conjunction with our microfluidic platforms (discussed below) to enrich for ESFT-associated exosomes from clinical samples using immuno-capture techniques. These approaches appear promising in that we are now able to routinely detect the EWS-Ets fusion transcripts in enriched exosomes from clinical samples (Samuel and Godwin, unpublished data). Such limited advancement in the studies of pediatric cancer-derived exosomes highlights the need of new enabling technologies to address current challenges in isolation and of exosomes in malignancies with reliable quality and substantial concentration and in molecular characterization of exosomal cargos in relation to their biological and pathological roles.
Reliable and efficient isolation of exosomes in malignancies is still a major challenge, especially within the pediatric population, due to: 1) size overlap with other EVs, including microparticles (200 nm - 1 μM) and apoptotic bodies, and lack of specific exosomal markers limiting purity of isolated exosomes, which interfere the accuracy of subsequent molecular diagnosis seriously (67); and 2) low isolation efficiency and poor sensitivity of conventional exosomal detection assays requiring large amount of bio-fluid samples (68), which within the pediatric population may be limited due to sample volumes that can be safely collected. Currently, a variety of exosome isolation techniques have been developed utilizing different physical and chemical properties of exosomes, such as size, density, morphology, membrane rigidity, and membrane composition (69). These techniques can be roughly classified into five categories, based on the mechanisms and the platforms: ultracentrifugation-based protocols, size exclusion and filtration, affinity capture, exosome precipitation, and microfluidics-based technologies. It is noted that microfluidic technologies may exploit the same principles as the bench-top methods, such as size exclusion and immunocapture. Several reviews have summarized prior technologies for exosome isolation and analysis (70–75). In the following sections we will focus on the state-of-the-art progress of exosome technologies, especially the microfluidics-based technologies, and discuss our outlook on the future development of exosome-based liquid biopsy for diagnosis and prognosis of pediatric cancer, including ESFTs. To this end, we will first briefly summarize existing conventional bench-top techniques to benchmark the performance of the newly emerged microfluidic platforms.
Conventional Exosome Isolation Techniques
There are a number of exosome isolation methods that have been extensively adapted in standard biology and clinical laboratory settings. Differential ultracentrifugation is the gold standard and one of the most extensively used methods for exosome isolation (76). Ultracentrifugation-based methods usually consist of a serial steps starting with low-speed centrifugation for a short time to remove large particles like cell debris, followed by higher speed (e.g., 10,000× g) for extended time period to remove large vesicles, and finalized by to pelleting and washing small exosomes by ultracentrifugation at >100,000× g. While this method suffers from low recovery efficiency (~5-20%) and poor purity, it shows that 81% of the worldwide researchers choose ultracentrifugation for their exosome isolation needs, according to a recent survey reported by Chris Gardiner in 2016 (77). This interesting observation could be attributed to several factors: 1) the approach is easy to use and robust, requiring very little technical expertise; 2) it requires minimal reagent and instrument cost when having the access to an ultracentrifuge; and 3) it affords flexibility to handle cell culture conditioned media and a range of bodily fluids, such as plasma, serum, urine, cerebrospinal fluid, and ascite fluids, with variable volumes from milliliters to liters. Variations on this method, such as adding in a sucrose gradient centrifugation step, lead to higher purity of extracted exosomes (78). In this case, one important factor that one should take into consideration is that the solution should be kept iso-osmotic throughout the gradient in order to preserve particle sizes.
Ultrafiltration and size-exclusion chromatography (SEC) isolate exosome and other EVs based on their sizes. Ultrafiltration uses nanoporous membranes with common cut-off pore sizes between 1-100 nm as the filter to extract exosomes of this size range (79). The recovery efficiency was found to depend on adherence interaction between exosomes surface proteins and the membranes. Therefore, the use of a certain type of hydrophilized membranes is recommended to enhance filtration efficiency. SEC resolves particles into bands of different sizes which are eluted out of the column sequentially. In contrast to ultrafiltration, SEC doesn’t enrich separated exosomes. These size-exclusion methods are faster than ultracentrifugation and do not require highly sophisticated equipment. However, forced filtration and shearing force may cause membrane fusion and loss of integrity, which will potentially interfere the results of downstream analysis.
In addition to the density- and size-based methods, exosome precipitate methods have gained increasing popularity. Exosomes can be precipitated out from biofluids by adding certain precipitants, such as water-excluding polymers (e.g., polyethylene glycol, dextrans, or polyvinyls), to change the solubility or dispersibility of exosomes, and then collected by low-speed centrifugation (80). Several exosome precipitation kits based on this mechanism have been commercialized, such as ExoQuick (System Bioscience) and Total Exosome Isolation Reagent (ThermoFisher). The major advantage of these kits is that they are very convenient to use and only require routine laboratory equipment (e.g., microcentrifuge). Nevertheless, a major drawback of this method arises from co-precipitation of other less soluble contaminants, such as proteins and polymeric materials, limiting the yield and purity of isolated exosomes from complex biological samples. Thus, additional sample preparation procedures are required to improve the purify of extracted materials, such as pre-isolation treatments to remove subcellular particles (e.g., lipoproteins) and/or post-isolation processing to remove the polymeric precipitants.
A promising route for harvesting highly purified exosomes is affinity-based capture that exploits specific antibody-antigen interactions or receptor-ligand binding. Immunomagnetic capture methods have been established, such as a commercial Exo-Flow Capture Kit (System Bioscience), to target either generic exosome markers (e.g., tetraspanin proteins CD9, CD63 and CD81) or specific proteins expressed on the membrane of exosomes for targeted and rapid isolation of exosomes. With this strategy, the selectivity of exosome isolation can be improved significantly, which confers a distinct advantage in some specific designs where enrichment of certain subpopulations of exosomes from biofluids is preferred. However, it is also important to note that this method needs extensive optimization for individual cases owing to the facts that exosomal expression of proteins is heterogeneous, targeted surface makers may be expressed in only a small fraction of exosomes, and the lack of clear understanding of exosome biology to define specific exosomal biomarkers (81). These limits could result in variable efficiency and specificity of exosome isolation, which interfere the accuracy of subsequent molecular profiling seriously. Meanwhile, immunoaffinity based methods are better suited for processing small-volume biofluid samples and can be expensive for large-scale preparation of exosomes (76). In addition to immunocapture, Wan et. al. recently reported a new lipid-affinity method for rapid enrichment of total EVs, which is a lipid nanoprobe composed of a lipid tail for EV membrane insertion and a biotin tag for collecting labelled EVs by NeutrAvidin-coated magnetic beads (82). This method permitted rapid isolation of EVs in 15 minutes, while yielding similar isolation efficiency and molecular composition as that of ultracentrifugation. Such isolation performance could facilitate the studies and application of exosomes, such as point-of-care cancer diagnostics.
The aforementioned techniques are the most commonly used exosome isolation methods and their performance was briefly summarized in Table 1. These method are coupled with conventional benchtop assays for downstream analysis, such as electron microscopy, Western blot, mass spectrometry, nanoparticle tracking analysis (NTA), and flow cytometry. The lack of standardized characterization methods for isolated exosomes and large differences in the types of bio-fluids make the cross-assay comparison difficult. Many studies on exosome isolation have been conducted in the past few years and it has been documented that isolation methods have significant impact on the observed molecular profiles of isolated exosomes as they result in vast difference in dependence on sample matrix, isolation yield, enrichment factor, exosome purity, and effects on subsequent extraction of proteins and RNAs (83–87). While there is still no consensus on the optimal exosome isolation method, the on-going efforts in systematic characterization of these methods and development of innovative techniques, along with rapid progress in exosome biology, would provide much better guidelines for users to select and optimize an isolation method that best fits their specific applications and needs (88–90).
Table 1.
A brief comparison of different conventional exosomes isolation methods.
| Method | Time | Advantages | Disadvantages |
|---|---|---|---|
| Ultracentrifugation | 5-10 hrs | + Robust and low reagent cost + Isolation of total EVs + Large-scale production + Vesicle structure maintained |
- Lengthy process - Low isolation efficiency - Co-isolation of contaminants |
| Sucrose density gradient | 16-90 hrs | + Vesicles divided into different populations | - Labor-intensive - Low yield - Long run time |
| Size exclusion/Ultrafiltration | 2-4 hrs | + Resolve vesicles of different sizes | - Impure factions: high pressure breaks larger vesicles into smaller ones |
| Polymer precipitation | 0.5-12 hrs | + No sophisticated equipment needed + Easy to use |
- Low purity - Alters functionality of vesicles - Require pre-/post-treatment cleanup |
| Immunoaffinity | 2-6 hrs | + High purity + Specific molecular selection of exosome subpopulations + Small sample volume + Compatibility with robotic liquid handling for high-throughput sample preparation |
- High cost - Antibody cross-reactivity - Lack of well-defined markers - Heterogeneity in surface marker expression limits isolation purity and reliability |
Microfluidic Exosome Isolation Techniques
Unique microscale fluid behaviors and properties, the ability to integrate functional microelements, and the amenability to circuit-level design make microfluidics a powerful platform for developing innovative exosome isolation strategies. Compared to conventional bench-top methods, microfluidic platforms offer significant reductions in sample volume, reagent consumption and isolation time, while tremendously improving both isolation yield and exosome quality (72, 74). Not only do microfluidic technologies implement the working mechanisms used in conventional methods, but also enable us to create innovative approaches leveraging on the unique flow behaviors at the micrometer scale and the inherent advantages in device integration and scaling. Herein we will discuss the recently reported microfluidic isolation methods that are roughly grouped into two categories: 1) have the same principal mechanism of isolation as in the conventional methods; and 2) utilize unique microscale flow properties and/or innovative microfabricated structures to create new or substantially improved exosome isolation strategies.
The microfluidic technologies in the first group primarily adopted the immumoaffinity capture and the size-based isolation mechanisms. Specific capture of exosomes by antibodies immobilized on solid surfaces represents the most commonly used approach because of its simplicity, compatibility with microfabrication, and ability to target specific subpopulations. In 2010, Chen et, al., reported the first microfluidic exosome isolation platform in which an anti-CD63 antibody-functionalized herringbone structure was used for immunocapture of exosomes from human sera for off-chip mRNA analysis (91). Similarly, an ExoChip device developed in the 96-well format also utilized the unstructured anti-CD63 antibody-functionalized surface of microchambers to capture exosomes, which were detected by non-specific fluorescent staining with a lipophilic membrane carbocyanine dye (DiO) (92). To further improve the capture efficiency and capacity, various nanomaterials or nanostructures have been investigated as nano-interfaces to immensely increase the binding surface area and capture probe density. We have developed a nano-IMEX chip (Figure 2a) in which a graphene oxide/polydopamine (GO/PDA) interface is incorporated by microfluidic layer-by-layer deposition of GO and polydopamine films which induces the formation of nanoporous structure of the PDA coating (93). Such nanostructured interface profoundly improves the immunocapture efficiency of exosomes to afford ultrahigh sensitivity for detection of exosomes in human plasma for cancer diagnosis. In these surface-based isolation chips, the capture efficiency is governed by the bulk-to-surface mass transfer and the capture capacity is largely determined by the pre-designed chip dimensions. In contrast, several microfluidic methods have been developed in which immunomagnetic microbeads are used for exosome capture (70, 94–97). The bead-based format offers several advantages, including near-solution binding kinetics, flexible capture capacity, and great scalability for large-volume samples. For instance, we have reported a simple and robust ExoSearch chip (Figure 2b) in which the sample and the suspension of immunomagnetic beads are co-flowed and passively mixed in a serpentine microchannel for rapid and efficient exosome capture (94). This ExoSearch chip features extremely simple and scalable design and the continuous-flow operation, thus providing highly expendable capture capacity for processing large quantities of biological fluids in a high throughput manner.
Figure 2. Microfluidic exosome isolation based on the immunocapture and size-exclusion mechanisms.

(a) Enhanced immunoaffinity capture of exosomes using a nanostructured GO/PDA interface in a nano-IMEX chip. Adapted with permission from reference (93). (b) Continuous flow mixing and immunomagnetic capture of exosomes using an ExoSearch chip. Adapted with permission from reference (94). (c) Trapping of exosome-like lipid vesicles on nanowire-on-micropillar arrays. Adapted with permission from reference (97). (d) Size-exclusion isolation of exosomes on an Exodisc platform. Adapted with permission from reference (98).
Size-based isolation of exosomes from other components in biological fluids is another useful approach because of its simplicity and potential for high-throughput sample processing. Wang and colleagues fabricated a porous nanowire-on-micropillar structure which is made of ciliated silicon micropillars (Figure 2c) (97). Exosomes with diameters between 40 and 100 nm were trapped onto these nanostructured micropillars and subsequently recovered via dissolution of the nanowires in PBS for 24 h. The double membrane based filtration approaches, driven by pressure or electrophoresis, have been studied extensively (99–101). In these cases, the first membrane was used to remove cells and other debris and the second one to collect exosomes. Woo et. Al. recently reported a “lab-on-a-disc” Exodisc system (Figure 2d), which integrates sample loading, double filtration, and exosomes recovery in a single centrifugal microfluidic disc chip (98). Driven by g-force of low-speed spinning, this Exodisc was found to remove >95% of protein contaminants, afford high recovery rate, and yield >100-fold higher concentration of exosomal mRNA than that produced with ultracentrifugation. Nevertheless, using the membranes of 20 and 600 nm pore sizes in the Exodics will likely lead to co-isolation of exosomes with larger EVs, including microvesicles and apoptotic bodies, which limits the exosome purity. Liu et. al. developed a modular microfluidic exosome total filtration unit, termed as ExoTIC (102). Several ExoTIC modules, each with a different membrane pore size (e.g200, 100, 80, 50, and 30 nm), can be connected in series to produce enriched exosomes at several specific, narrow size ranges. Not only does this tandem modular device increase the purity of isolated exosomes, but also enables well-defined fractionation of exosomes for exploring size-related molecular characteristics.
Beyond the conventional mechanisms, newly emerged microfluidic tools have expanded the spectrum of the properties of exosomes assessable for flow-mediated isolation, such as compressibility, surface charges, diffusivity, and dielectric properties of exosomes. Lee et. al. performed continuous and versatile sorting of exosomes (diameter <200 nm) from cell culture medium and other types of EVs of larger sizes from stored red blood cell products using an acoustic nanofilter system (103). To achieve this level of size resolution, interdigitated transducer (IDT) electrodes were used to produce a symmetric standing surface acoustic wave perpendicular to the flow direction, deflecting larger particles toward side outlets while concentrating small particles at the center outlet (Figure 3a). Wu et. al. further combined a cell-removal module with the exosome-isolation module to construct an integrated acoustofluidic chip which enables isolation of circulating exosomes from whole blood with a blood cell removal rate of >99.999% (104). Liu et. al. reported a viscoelasticity-based microfluidic system that separates exosomes from larger particles relying on the particle migration induced by size-dependent elastic lift forces in a viscoelastic medium (Figure 3b) (105). Satisfactory exosome purity (>90%) and recovery rate (>80%) were achieved, using a small amount of biocompatible poly(ethylene oxide) (PEO) as the additive in the media to control the viscoelastic forces exerted on exosomes. Different dielectric properties of particles and surrounding fluid also provides a useful route for particle separation and enrichment (106). Dielectric particles will experience a dielectrophoretic force (DEP) when subjected to a gradient electric field. Ibsen et. al. reported DEP-assisted enrichment and isolation of exosomes using an alternating current electrokinetic (ACE) microarray chip (Figure 3c) (107). The ACE microarray was used to generate dielectrophoretic force to trap small exosomes in the high-field regions around the circular microelectrodes within 15 min, while larger cells or smaller molecules remain relatively unaffected by the DEP field and can be washed away. Using this device, the authors demonstrated rapid isolation and on-chip fluorescence detection of glioblastoma (GBM) exosomes from undiluted human plasma samples in less than 30 min.
Figure 3. Hydrodynamic exosomes isolation in microchips based on various exosome properties.
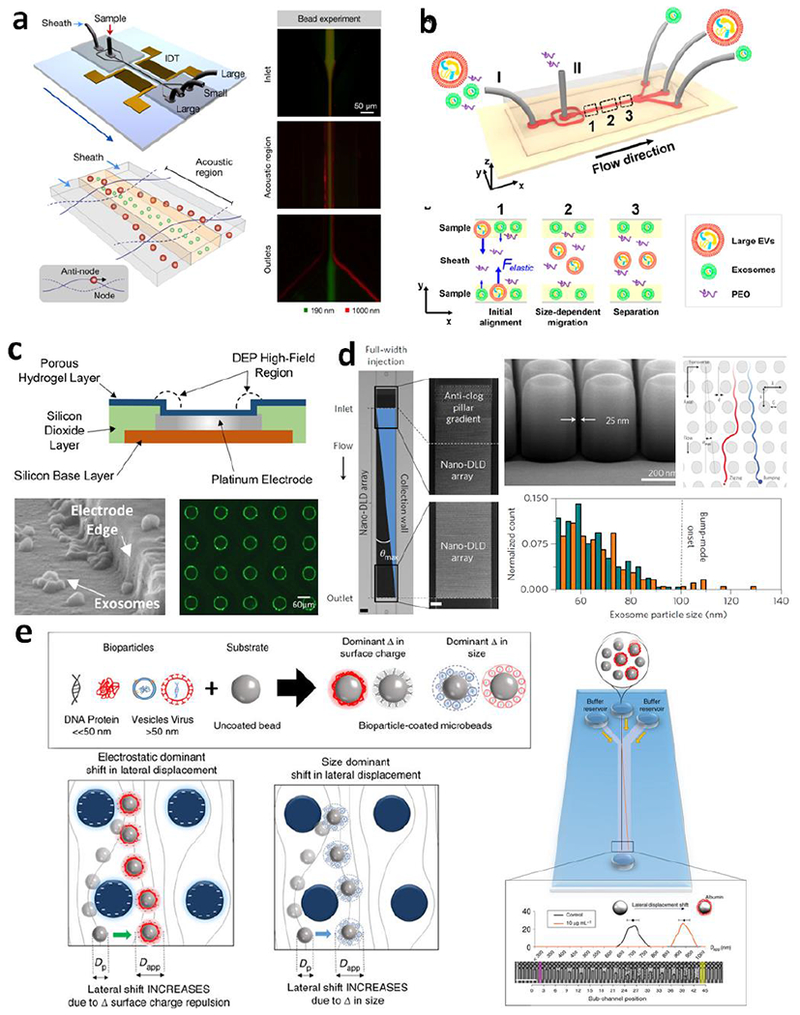
(a) Acoustic nanofilter system for exosomes isolation. Adapted with permission from Ref (103). (b) Isolation of exosomes by microfluidic viscoelastic flows. Adapted with permission from Ref (105). (c) Trapping of exosomes in ACE microarray chip. Adapted with permission from Ref (107). (d) Nano-DLD chips for size sorting of exosomes in laminar flow streams displaced by staggered nanopillars. Adapted with permission from Ref (108). (e) DLD fractionation of bioparticle-bead conjugates in an array of negatively charged micropillars based on the change of size and surface charge of micro-beads upon binding with bioparticles, including proteins and exosomes. Adapted with permission from Ref (109).
Deterministic lateral displacement (DLD) is a continuous-flow particle sorting mechanism enabled by using an array of laterally staggered micro-/nano-pillars to manipulate the laminar flow at the (sub)-micrometer scales. The geometry and arrangement of micropillars govern the paths of flow streamlines and determine a critical cutoff particle diameter DC. Particles with a diameter larger than DC will be displaced laterally following a bumping mode throughout the array, while smaller particles travel along with the streamlines without lateral displacement on average; thus particles of various sizes can be sorted into different streams in a continuous-flow fashion. Recently Wunsch et. al. first adapted this mechanism to develop a nano-DLD platform for separation of colloids and exosomes as small as 20 nm, by scaling the pillar array structure down to the nanometer size range (Figure 3d) (108). A potential limitation of this technology arises from the need of expensive and sophisticated nanolithography for device fabrication and extremely small volume capacity of the nanodevice, both of which could hamper practical applications of this technology. A clever solution to address these limitations was recently reported which makes it possible to use a regular microfluidic DLD device with micrometer-sized pillar arrays (Figure 3e)(109). In this approach, bioparticles are captured on the surface of 1 pm polymer beads and induces the change in size and/or electrostatic charges which cause the lateral displacement of the beads flowing through the DLD array. Interestingly, the degree of lateral shifts of the beads was found to be dependent on the amount of bioparticles bound to the beads, expanding the DLD chip from a separation device to a useful tool for label-free, quantitative measurement of the target bioparticles. Ko et. al. attempted to overcome the low throughput and susceptibility to clogging of nanofluidics for exosome isolation by developing an exosome track-etched magnetic nanopore (ExoTENPO) chip (110). This method used a commonly used nanopore membrane to form magnetic traps at the edges of pores for immunomagnetically labelled exosomes, creating a multiple-channel nanofluidic system for high-throughput, highly selective exosome isolation based on both size exclusion and surface protein expression. The authors demonstrated the potential applications of the ExoTENPO chip by isolating exosomes from murine and human plasma samples for downstream mRNA profiling by qPCR. Combined the mRNA profiling with a machine learning algorithm, this approach generated a set of optimized linear discriminators to identify samples from heterogeneous pancreatic cancer individuals, in both a murine model and in a clinical cohort. It is worth noting that most of these new exosome isolation strategies enabled by microfluidic technologies are contact free and label free, which ease the recovery of intact, active exosomes and increase the throughput of sample processing. While further optimization and improvement are needed, the emergence of these conceptually new exosome isolation approaches will open new opportunities to propel rapid progress in basic and translational exosome research towards ultimate clinical utilities.
Integrated Microfluidic Analysis of Exosomes towards Liquid Biopsy
Liquid biopsy analysis of tumors holds great promise to shift the paradigms in both cancer diagnosis and personalized therapy. As an emerging dimension of liquid biopsies of tumors, exosomes have attracted increasing importance during the progress towards precision medicine. Exosome secretion is a dynamic process, producing diverse populations with 5-fold differences in size and as much as a 104-fold differences in concentration between healthy and disease states (81, 111–113). Such heterogeneity hinders the sensitive and specific analysis of exosomes in bio-fluids. The aforementioned isolation technologies can be employed in a stand-alone manner, functioning as preparative front ends that interface with conventional downstream detection and analysis. Meanwhile, microfluidic platforms also hold potential to leverage or even transform quantitative detection and comprehensive molecular characterization of exosomes by virtue of offering an attractive combination of high throughput and sensitivity with low reagent consumption and the potential for portability. Over the past few years, researchers have developed a number of integrated microfluidic platforms for analysis of exosome levels, quantification of disease-specific subpopulations, and omics-level characterization of exosomal proteins and RNAs (97, 100, 114, 115). Herein we will focus on the microfluidic devices for the point-of-care (POC) and clinical applications and discuss the studies on microfluidic analysis of disease-associated exosomal biomarkers in bodily fluids (e.g., serum, plasma, and whole blood).
Vaidyanathan et. al., demonstrated a microfluidic device that produces a tunable alternating current electrohydrodynamic flow (nanoshearing) in microchannels to enhance the specificity and sensitivity of immunocapture and detection of exosomes (114). This multiplexed device permitted simultaneous colorimetric detection of multiple exosomal markers (HER2, PSA, and CD9) for breast cancer diagnosis. We have developed for the first time a cascading flow chip that enables analysis of intravesicular proteins in selected exosome subpopulations (116). This device integrates exosome immuno-isolation, enrichment, chemical lysis, protein immunoprecipitation, and sandwiched immunoassay assisted by chemifluorescence detection (Figure 4a). Using this technology, quantitative analysis of both type 1 insulin growth factor receptor (IGF-1R) and its phosphorylation status in plasma-derived circulating exosomes was demonstrated for detection of non-small-cell lung cancer patients from healthy individuals. To further improve the exosome detection sensitivity, we combined the GO/PDA nano-interfaced exosome capture chip with a sensitive and robust sandwich immunoassay targeting a combination of CD9, CD81 and EpCAM markers (93). This technology affords an extremely low limit of detection (LOD) of 50 exosomes per μL and the ability to detect circulating exosomes directly from minimal volume of human plasma for cancer detection. Shao et al. devised a microfluidic nuclear magnetic resonance system (pNMR) in which EVs bound with marker-specific magnetic nanoparticles resulted in faster decay of the NMR signal depending on the protein expression levels (117). The pNMR-based exosome sensing enabled diagnosis and monitoring treatment responses for glioblastoma patients by probing four GBM signature protein biomarkers (EGFR, EGFRvlll, PDPN, and IDH1 R132H), which offered >90% combined accuracy (AUC = 0.95). A nPLEX chip (Figure 4b), developed by the same group, consists of an array of nanohole lattice patterned on gold film-coated substrate of parallel microfluidic channels for label-free transmission Surface Plasmon Resonance (SPR) sensing of immunocaptured exosomes (118). In this study, ascites samples from ovarian cancer patients were studied by probing several exosomal protein markers selected by proteomic profiling of exosomes from 10 ovarian cancer cell lines. Combined expression levels of EpCAM and CD24 markers in ascites-derived exosomes were found to improve ovarian cancer diagnosis with an accuracy of 97%. Jeong et. al., reported a portable magneto-electrochemical device (iMEX) as shown in Figure 4c (119). Exosomes are immunomagnetically captured from patient samples and profiled through electrochemical reaction in eight parallel channels. This assay was validated to discriminate 11 ovarian cancer patients from 5 healthy controls, by probing the expression levels of EpCAM and CD24. The iMEX system offered an affordable and miniaturized device that can be readily translated for on-site exosome detection.
Figure 4. Examples of integrated microfluidic analysis of exosomes towards liquid biopsy.

(a) Integrated platform for intravesicular protein analysis of ovarian cancer exosomes. Adapted with permission from Ref (116). (b) Molecular analysis of EpCAM and CD24 specific exosomes from ovarian cancer on nPLEX chip. Adapted with permission from Ref (118). (c) Portable magneto-electrochemical device (iMEX) for molecular diagnosis of ovarian cancer. Adapted with permission from Ref (119). (d) iMER chip for integrated and multiplexed quantification of exosomal mRNA levels. Adapted with permission from Ref (95).
In addition to the protein contents, exosomes contain numerous types of nucleic acids, including mRNAs, microRNAs, as well as both single and double-stranded DNA, which add to the significant value of exosomes for disease diagnosis, prognosis, and therapeutic treatment. Shao et. al. developed an integrated and multiplexed iMER platform to detection mRNA levels in tumor-derived exosomes (Figure 4d) (95). On-chip exosome capture, mRNA extraction and RT-qPCR analyses were carried out on a single chip tandemly. The iMER analysis of exosomal mRNAs in sera of GBM patients and healthy controls revealed that combined exosomal mRNA levels of EPHA2, EGFR, and PDPN have a diagnostic accuracy of 90% for GBM (AUC = 0.945). Furthermore, longitudinal measurements of mRNA levels of MGMT and APNG, two important enzymes involved in repairing DNA damaged by the drug temozolomide (TMZ), demonstrated the feasibility of using the iMEX-based exosome analysis to monitor the development of drug resistance during treatment. Recently, Reategui and colleagues adapted a nano-engineered herringbone microchip initially developed for CTC capture to exosome analysis (120). This EVHB-Chip features an engineered nano-interface coated on the herringbone grooves to maximize exosome interactions with antibody-coated surfaces, thus improving the limit of detection (LOD) to a level of 100 vesicles per μL and largely expedite RNA isolation from tumor-specific EVs to facilitate comprehensive mRNA profiling off-chip.
There is a surge of microfluidic platforms in the last five years that have been developed for integrated exosome analysis, as summarized in Table 2. Most of these systems employed immunoaffinity isolation of exosomes, owing to its compatibility with microfabrication, high isolation efficiency and importantly, unmatched specificity to target tumor-related exosomes in complex biological samples. Meanwhile, miniaturization and integration of molecular detection methods with sample preparation steps are beneficial, because the analytical performance for measuring exosomal biomarkers can be substantially improved, while reducing the sample loss, cross-contamination, and analysis time, in comparison to conventional bench-top assays. From a perspective of translation studies, rapid exosome isolation and reliable molecular assessment from smaller ‘real life’ clinical samples poses a key bottleneck to moving exosome-based liquid biopsy towards clinical utilities. The advantage of microfluidics in small sample requirement makes it an inherent suitable tool to overcome this challenge.
Table 2.
Integrated microfluidic platforms for exosome analysis.
| Microfluidic platform | Isolation method | Detection method | Targeted disease | Measured biomarkers | Clinical sample | Limit of Detection | Reference |
|---|---|---|---|---|---|---|---|
| ExoChip | Immunoaffinity (CD63) | Fluorescent staining (Dio) | Pancreatic cancer | Overall exosome counts | Serum | 0.5 pM | Kanwar 2014 (92) |
| nano-IMEX | Immunoaffinity (CD81) | Enzymatic amplification of fluorescence | Ovarian cancer | EpCAM, CD9, CD81 | Plasma | 50 μL−1 | Zhang 2016 (93) |
| Exodisc | Double filtration | Colorimetric ELISA | Bladder cancer | CD9, CD81 | Urine | N/A | Woo 2017 (98) |
| RInSE | Immunoaffinity beads labeled (EpCAM) | Flow cytometry | Breast cancer | CD81 | Blood | N/A | Dudani 2015 (96) |
| μNMR | differential ultracentrifugation | Immunomagnetic tagging (CD63), μNMR detection | GBM | EGFRvIII, EGFR, PDPN, IDH1 R132H | Plasma | 104 μL−1 | Shao 2012 (117) |
| nPLEX | Immunoaffinity (CD63) | Nanohole array SPR | Ovarian cancer | EpCAM, CD24 | Ascites fluid | 3000 exosomes (670 aM) | Im 2014 (124) |
| iMEX | Magnetic immunoaffinity (CD63) | Electrochemical sensing | Ovarian cancer | EpCAM, CD24, CA125, HER2, MUC18, EGFR | Plasma | 104 μL−1 | Jeong 2016 (119) |
| ExoSearch | Immunoaffinity (CD9) | Multiplexed immunoassay | Ovarian cancer | CA125, CD24, EpCAM | Plasma | 750 μ−1 | Zhao 2016 (94) |
| Integrated flow-through Chip | Immunomagnetic capture (EpCAM) | On-chip exosome lysis and ELISA | NSCLC | IGF-1R, phospho-IGF-1 R | Plasma | N/A | He 2014 (116) |
| nPES | Immunoaffinity (CD81) | Nano-plasmon enhanced scattering | Pancreatic cancer | EphA2 | Plasma (1 μL) | N/A | Liang 2017 (125) |
| μMED | Magnetic immunoaffinity (CD81) | Fluorescence immunoassay | Concussion | GluR2 | Mouse serum | 104 μL−1 | Ko 2016 (73) |
| iMER | Magnetic immunoaffinity (EGFR and EGFRvIII) | On-chip exosome lysis and RT-qPCR | GBM | EPHA2, EGFR, PDPN, MGTM, APNG | Serum | N/A | Shao 2015 (95) |
| EVHB-Chip | Immunoaffinity (EGFR, EGFRvIII, ephA2, podoplanin, PDGFR, MCAM) | Off-chip exosome lysis and digital droplet PCR | GBM | mRNA profile (e.g., EGFR, EGFRvIII) | Serum, plasma | 100 μL−1 | Reategui 2018 (120) |
| ExoTENPO | immunomagnetic nanopore trapping (CD63, CD9, CD81, EpCAM) | Off-chip RT-qPCR | Pancreatic cancer | mRNA profiles (CK18, CD63, Erbb3, KRAS) | Mouse plasma | N/A | Ko 2017 (110) |
As discussed above, extensive studies have been conducted to investigate exosomal biomarkers for a variety of cancer types, ranging from pancreatic, ovarian, breast, lung, bladder and GBM, generating exciting evidences to support the promising potential of exosomes as a surrogate for tissue biopsy of tumors. However, these studies are all focused on adult cancers, and very limited efforts have been invested in childhood cancers. Compared to adult cancer, pediatric cancers present distinct challenges in the research, clinical trials and patient care. Pediatric cancers are often more aggressive and progressive (121) and the diagnosis mainly relies on either a surgical or needle biopsy to obtain sufficient tissue for the histological, molecular and cytogenetic workup. This highly invasive procedure is associated with potential risks of complications that can have a life-long impact on childhood patients, put economic burden on the family of a young patient, and has often no feasibility to repeat for accurate diagnosis and treatment monitoring to minimize the toxic side effects. Moreover, major ethical challenges arise, including clinical equipoise, when considering an invasive biopsy procedure for children (122, 123). Therefore, there is a pressing need in developing liquid biopsy-based biomarkers and tests to improve the disease management in pediatric cancer. Fortunately, the relatively slow progress in pediatric cancers has been recognized and childhood cancers have been listed as one of the ten research areas recommended for the Cancer Moonshot initiatives announced in 2016. These efforts will open tremendous opportunities to accelerating the assessment and translation of exosomes as liquid biopsy markers for pediatric cancers, including ESFTs. Ewing Sarcoma is an orphan disease that would greatly benefit from repetitive non-invasive tests, thus avoiding the risks and costs associated with repeated imaging and sedation and invasive biopsies in children. The lack of reliable blood biomarkers presents a serious obstacle to the treatment and management of Ewing Sarcoma, more importantly, recurrent or metastatic Ewing Sarcoma. Our laboratories are working closely to develop and validate our ESFT-exosome based assays, using pediatric clinical longitudinal samples, for early diagnosis, monitoring of disease burden and response to therapy, and detection of early recurrence of disease in children and adolescent with Ewing Sarcoma.
Conclusions:
Analysis of extracellular vesicles, primarily focused on exosomes, is a cornerstone of emerging liquid biopsy techniques, whose importance in diagnostics and precision medicine is only beginning to be fully understood. Efforts are underway to evaluate the clinical utility of circulating exosomes across diseases and clinical implications, especially in pediatric malignancies such as ESFT. But the development of technologies that can make routine analysis of liquid biopsy samples feasible is also critically important and presents a tremendous opportunity for microfluidic solutions. It is notable that most of the advancements highlighted here have occurred within the past 5 years, indicating that this field is still in its infancy. This body of literature lays a foundation for future progress by introducing new methods for exosome capture, detection, and analysis. These advancements also suggest directions for future work needed to realize the vision of exosome-based liquid biopsy applications.
ACKNOWLEDGMENTS
This work was supported by 1R21EB024101, 1R21CA186846, 1R33CA214333, and 1R21CA207816 from the NIH. Dr. Andrew K. Godwin is the Chancellors Distinguished Chair in Biomedical Sciences endowed Professor at University of Kansas Medical Center. All authors have read the journal’s policy on disclosure of potential conflicts of interest. The authors declare no competing financial interest. All authors have read the journal’s authorship agreement and the manuscript has been reviewed by and approved by all named authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ewing J The Classic: Diffuse endothelioma of bone. Proceedings of the New York Pathological Society 1921;12:17. [DOI] [PubMed] [Google Scholar]; Clin Orthop Relat Res. 2006;450:25–7. [DOI] [PubMed] [Google Scholar]
- 2.Ramani P, Rampling D, Link M. Immunocytochemical study of 12E7 in small round-cell tumours of childhood: an assessment of its sensitivity and specificity. Histopathology. 1993;23(6):557–61. [DOI] [PubMed] [Google Scholar]
- 3.Nunn MF, Seeburg PH, Moscovici C, Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306(5941):391–5. [DOI] [PubMed] [Google Scholar]
- 4.Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, et al. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331(5):294–9. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig JA. Ewing sarcoma: historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr Opin Oncol. 2008;20(4):412–8. [DOI] [PubMed] [Google Scholar]
- 6.Arndt CA, Rose PS, Folpe AL, Laack NN. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin Proc. 2012;87(5):475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein M, Kovar H, Paulussen M, Randall RL, Schuck A, Teot LA, et al. Ewing’s sarcoma family of tumors: current management. Oncologist. 2006;11(5):503–19. [DOI] [PubMed] [Google Scholar]
- 8.Esiashvili N, Goodman M, Marcus RB Jr. Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. 2008;30(6):425–30. [DOI] [PubMed] [Google Scholar]
- 9.Gurney JG SA, Bulterys M. Malignant Bone Tumors In: Ries LAG SM, Gurney JG, et al. , editor. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995. Bethesda, MD: National Cancer Institute SEER Program; 1999. p. 99–110. [Google Scholar]
- 10.Arvand A, Denny CT. Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene. 2001;20(40):5747–54. [DOI] [PubMed] [Google Scholar]
- 11.Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348(8):694–701. [DOI] [PubMed] [Google Scholar]
- 12.Paulussen M, Craft AW, Lewis I, Hackshaw A, Douglas C, Dunst J, et al. Results of the EICESS-92 Study: two randomized trials of Ewing’s sarcoma treatment--cyclophosphamide compared with ifosfamide in standard-risk patients and assessment of benefit of etoposide added to standard treatment in high-risk patients. J Clin Oncol. 2008;26(27):4385–93. [DOI] [PubMed] [Google Scholar]
- 13.Oberlin O, Rey A, Desfachelles AS, Philip T, Plantaz D, Schmitt C, et al. Impact of high-dose busulfan plus melphalan as consolidation in metastatic Ewing tumors: a study by the Societe Francaise des Cancers de l’Enfant. J Clin Oncol. 2006;24(24):3997–4002. [DOI] [PubMed] [Google Scholar]
- 14.Ladenstein R, Potschger U, Le Deley MC, Whelan J, Paulussen M, Oberlin O, et al. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. J Clin Oncol. 2010;28(20):3284–91. [DOI] [PubMed] [Google Scholar]
- 15.Barker LM, Pendergrass TW, Sanders JE, Hawkins DS. Survival after recurrence of Ewing’s sarcoma family of tumors. J Clin Oncol. 2005;23(19):4354–62. [DOI] [PubMed] [Google Scholar]
- 16.Potratz J, Jurgens H, Craft A, Dirksen U. Ewing sarcoma: biology-based therapeutic perspectives. Pediatr Hematol Oncol. 2012;29(1):12–27. [DOI] [PubMed] [Google Scholar]
- 17.Widhe B, Widhe T. Initial symptoms and clinical features in osteosarcoma and Ewing sarcoma. J Bone Joint Surg Am. 2000;82(5):667–74. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmidt S, Wirtz H. Lung Cancer: Current Diagnosis and Treatment. Deutsches Arzteblatt International. 2009;106(49):809–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantel K, Alix-Panabieres C. Real-time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Research. 2013;73(21):6384–8. [DOI] [PubMed] [Google Scholar]
- 20.Oberlin O, Deley MC, Bui BN, Gentet JC, Philip T, Terrier P, et al. Prognostic factors in localized Ewing’s tumours and peripheral neuroectodermal tumours: the third study of the French Society of Paediatric Oncology (EW88 study). Br J Cancer. 2001;85(11):1646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saifuddin A, Whelan J, Pringle JA, Cannon SR. Malignant round cell tumours of bone: atypical clinical and imaging features. Skeletal Radiol. 2000;29(11):646–51. [DOI] [PubMed] [Google Scholar]
- 22.Treglia G, Salsano M, Stefanelli A, Mattoli MV, Giordano A, Bonomo L. Diagnostic accuracy of (1)(8)F-FDG-PET and PET/CT in patients with Ewing sarcoma family tumours: a systematic review and a meta-analysis. Skeletal Radiol. 2012;41(3):249–56. [DOI] [PubMed] [Google Scholar]
- 23.Volker T, Denecke T, Steffen I, Misch D, Schonberger S, Plotkin M, et al. Positron emission tomography for staging of pediatric sarcoma patients: results of a prospective multicenter trial. J Clin Oncol. 2007;25(34):5435–41. [DOI] [PubMed] [Google Scholar]
- 24.Kovar H, Ban J, Pospisilova S. Potentials for RNAi in sarcoma research and therapy: Ewing’s sarcoma as a model. Semin Cancer Biol. 2003;13(4):275–81. [DOI] [PubMed] [Google Scholar]
- 25.Jedlicka P Ewing Sarcoma, an enigmatic malignancy of likely progenitor cell origin, driven by transcription factor oncogenic fusions. Int J Clin Exp Pathol. 2010;3(4):338–47. [PMC free article] [PubMed] [Google Scholar]
- 26.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29(32):4504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aurias A, Rimbaut C, Buffe D, Zucker JM, Mazabraud A. Translocation involving chromosome 22 in Ewing’s sarcoma. A cytogenetic study of four fresh tumors. Cancer Genet Cytogenet. 1984;12(1):21–5. [DOI] [PubMed] [Google Scholar]
- 28.Zucman-Rossi J, Legoix P, Victor JM, Lopez B, Thomas G. Chromosome translocation based on illegitimate recombination in human tumors. Proc Natl Acad Sci U S A. 1998;95(20):11786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoubek A, Dockhorn-Dworniczak B, Delattre O, Christiansen H, Niggli F, Gatterer-Menz I, et al. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J Clin Oncol. 1996;14(4):1245–51. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher BD, Hanna SL, Fairclough DL, Gronemeyer SA. Pediatric musculoskeletal tumors: use of dynamic, contrast-enhanced MR imaging to monitor response to chemotherapy. Radiology. 1992;184(1):243–8. [DOI] [PubMed] [Google Scholar]
- 31.Elzi DJ, Song M, Houghton PJ, Chen Y, Shiio Y. The role of FLI-1-EWS, a fusion gene reciprocal to EWS-FLI-1, in Ewing sarcoma. Genes Cancer. 2015;6(11-12):452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin PP, Brody RI, Hamelin AC, Bradner JE, Healey JH, Ladanyi M. Differential transactivation by alternative EWS-FLI1 fusion proteins correlates with clinical heterogeneity in Ewing’s sarcoma. Cancer Res. 1999;59(7):1428–32. [PubMed] [Google Scholar]
- 33.Aryee DN, Sommergruber W, Muehlbacher K, Dockhorn-Dworniczak B, Zoubek A, Kovar H. Variability in gene expression patterns of Ewing tumor cell lines differing in EWS-FLI1 fusion type. Lab Invest. 2000;80(12):1833–44. [DOI] [PubMed] [Google Scholar]
- 34.Le Deley MC, Delattre O, Schaefer KL, Burchill SA, Koehler G, Hogendoorn PC, et al. Impact of EWS-ETS fusion type on disease progression in Ewing’s sarcoma/peripheral primitive neuroectodermal tumor: prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. J Clin Oncol. 2010;28(12):1982–8. [DOI] [PubMed] [Google Scholar]
- 35.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature Reviews Clinical Oncology. 2013;10(8):472–84. [DOI] [PubMed] [Google Scholar]
- 36.Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Translational Cancer Research. 2015;4(3):280–90. [Google Scholar]
- 37.Ilie M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Translational Lung Cancer Research. 2016;5(4):420–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atay S, Wilkey DW, Milhem M, Merchant M, Godwin AK. Insights into the Proteome of Gastrointestinal Stromal Tumors-Derived Exosomes Reveals New Potential Diagnostic Biomarkers. Mol Cell Proteomics. 2018;17(3):495–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atay S, Banskota S, Crow J, Sethi G, Rink L, Godwin AK. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci U S A. 2014;111(2):711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget. 2017;8(7):11917–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14(19):3773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha S, Aranda E, Hayakawa Y, Bhanja P, Atay S, Brodin NP, et al. Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nature Communications. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atay S GA. Tumor-derived exosomes: A message delivery system for tumor progression. Commun Integr Biol. 2014;7(1):e28231 Epub 2014/04/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18(5):199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor DD, Lyons KS, Gercel-Taylor C. Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecologic oncology. 2002;84(3):443–8. [DOI] [PubMed] [Google Scholar]
- 46.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–46. [DOI] [PubMed] [Google Scholar]
- 47.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic oncology. 2008;110(1):13–21. [DOI] [PubMed] [Google Scholar]
- 48.Monleon I, Martinez-Lorenzo MJ, Monteagudo L, Lasierra P, Taules M, Iturralde M, et al. Differential secretion of Fas ligand- or APO2 ligand/TNF-related apoptosis-inducing ligand-carrying microvesicles during activation-induced death of human T cells. J Immunol. 2001;167(12):6736–44. [DOI] [PubMed] [Google Scholar]
- 49.Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128(7):1796–804. [DOI] [PubMed] [Google Scholar]
- 50.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31(1):114–21. [DOI] [PubMed] [Google Scholar]
- 51.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360(9329):295–305. [DOI] [PubMed] [Google Scholar]
- 52.Soung YH, Nguyen T, Cao H, Lee J, Chung J. Emerging roles of exosomes in cancer invasion and metastasis. BMB reports. 2016;49(1):18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorczynski RM, Erin N, Zhu F. Serum-derived exosomes from mice with highly metastatic breast cancer transfer increased metastatic capacity to a poorly metastatic tumor. Cancer Med. 2016;5(2):325–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez M, Silva J, Herrera A, Herrera M, Pena C, Martin P, et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget. 2015;6(38):40575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6(35):37151–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91(4):431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Loughlin AJ, Woffindale CA, Wood MJ. Exosomes and the emerging field of exosome-based gene therapy. Curr Gene Ther. 2012;12(4):262–74. [DOI] [PubMed] [Google Scholar]
- 59.Roberson CD, Atay S, Gercel-Taylor C, Taylor DD. Tumor-derived exosomes as mediators of disease and potential diagnostic biomarkers. Cancer Biomark. 2010;8(4-5):281–91. [DOI] [PubMed] [Google Scholar]
- 60.Peng P, You Y, Shen K. [Isolation, identification and clinical significance of ascites-derived exosomes from patients with ovarian epithelial cancer]. Zhonghua Fu Chan Ke Za Zhi. 2009;44(4):268–72. [PubMed] [Google Scholar]
- 61.Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep. 2011;25(3):749–62. [DOI] [PubMed] [Google Scholar]
- 62.Navabi H, Croston D, Hobot J, Clayton A, Zitvogel L, Jasani B, et al. Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells Mol Dis. 2005;35(2):149–52. [DOI] [PubMed] [Google Scholar]
- 63.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171–82. [DOI] [PubMed] [Google Scholar]
- 64.Li QL, Bu N, Yu YC, Hua W, Xin XY. Exvivo experiments of human ovarian cancer ascites-derived exosomes presented by dendritic cells derived from umbilical cord blood for immunotherapy treatment. Clin Med Oncol. 2008;2:461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller IV, Raposo G, Welsch U, Prazeres da Costa O, Thiel U, Lebar M, et al. First identification of Ewing’s sarcoma-derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol Cell. 2013;105(7):289–303. [DOI] [PubMed] [Google Scholar]
- 66.Tsugita M, Yamada N, Noguchi S, Yamada K, Moritake H, Shimizu K, et al. Ewing sarcoma cells secrete EWS/Fli-1 fusion mRNA via microvesicles. PLoS One. 2013;8(10):e77416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Journal of Neuro-Oncology. 2013;113(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furi I, Momen-Heravi F, Szabo G. Extracellular vesicle isolation: present and future. Annals of Translational Medicine. 2017;5(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khatun Z, Bhat A, Sharma S, Sharma A. Elucidating diversity of exosomes: biophysical and molecular characterization methods. Nanomedicine. 2016;11(17):2359–77. [DOI] [PubMed] [Google Scholar]
- 70.He M, Zeng Y. Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. Jala. 2016;21(4):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab on a Chip. 2017;17(21):3558–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gholizadeh S, Draz MS, Zarghooni M, Sanati-Nezhad A, Ghavami S, Shafiee H, et al. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: Current status and future directions. Biosensors & Bioelectronics. 2017;91:588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ko J, Carpenter E, Issadore D. Detection and isolation of circulating exosomes and microvesicles for cancer monitoring and diagnostics using micro-/nano-based devices. Analyst. 2016;141(2):450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liga A, Vliegenthart ADB, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab on a Chip. 2015;15(11):2388–94. [DOI] [PubMed] [Google Scholar]
- 75.Li P, Kaslan M, Lee SH, Yao J, Gao ZQ. Progress in Exosome Isolation Techniques. Theranostics. 2017;7(3):789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biological Chemistry. 2013;394(10):1253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. Journal of Extracellular Vesicles. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Scientific Reports. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, et al. Benchtop isolation and characterization of functional exosomes by sequential filtration. Journal of Chromatography A. 2014;1371:125–35. [DOI] [PubMed] [Google Scholar]
- 80.Rider MA, Hurwitz SN, Meckes DG Jr. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Scientific reports. 2016;6:23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yanez-Mo M, Siljander PRM, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan Y, Cheng G, Liu X, Hao SJ, Nisic M, Zhu CD, et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nature Biomedical Engineering. 2017;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. [DOI] [PubMed] [Google Scholar]
- 84.Andreu Z, Rivas E, Sanguino-Pascual A, Lamana A, Marazuela M, González-Alvaro I, et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. Journal of Extracellular Vesicles. 2016;5(1):31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. International journal of molecular medicine. 2017;40(3):834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Helwa I, Cai JW, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. Plos One. 2017;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of Extracellular Vesicles. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, et al. Methodological Guidelines to Study Extracellular Vesicles. Circulation research. 2017;120(10):1632–48. [DOI] [PubMed] [Google Scholar]
- 91.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab on a Chip. 2010;10(4):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab on a Chip. 2014;14(11):1891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang P, He M, Zeng Y Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab on a Chip. 2016;16(16):3033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab on a Chip. 2016;16(3):489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shao HL, Chung J, Lee K, Balaj L, Min C, Carter BS, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nature Communications. 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dudani JS, Gossett DR, Tse HTK, Lamm RJ, Kulkarni RP, Di Carlo D. Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics. 2015;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fang SM, Tian HZ, Li XC, Jin D, Li XJ, Kong J, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. Plos One. 2017;12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, et al. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. Acs Nano. 2017;11(2):1360–70. [DOI] [PubMed] [Google Scholar]
- 99.Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab on a Chip. 2012;12(24):5202–10. [DOI] [PubMed] [Google Scholar]
- 100.Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, Yu T, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Scientific Reports. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang J, Choi MK, Kim DH, Hyeon T Designed Assembly and Integration of Colloidal Nanocrystals for Device Applications. Advanced materials. 2016;28(6):1176–207. [DOI] [PubMed] [Google Scholar]
- 102.Liu F, Vermesh O, Mani V, Ge TJJ, Madsen SJ, Sabour A, et al. The Exosome Total Isolation Chip. Acs Nano. 2017;11(11):10712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee K, Shao HL, Weissleder R, Lee H. Acoustic Purification of Extracellular Microvesicles. Acs Nano. 2015;9(3):2321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu MX, Ouyang YS, Wang ZY, Zhang R, Huang PH, Chen CY, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(40):10584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu C, Guo JY, Tian F, Yang N, Yan FS, Ding YP, et al. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. Acs Nano. 2017;11(7):6968–76. [DOI] [PubMed] [Google Scholar]
- 106.Ramos A, Morgan H, Green NG, Castellanos A. Ac electrokinetics: a review of forces in microelectrode structures. Journal of Physics D-Applied Physics. 1998;31(18):2338–53. [Google Scholar]
- 107.Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY, Ong J, et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. Acs Nano. 2017;11(7):6641–51. [DOI] [PubMed] [Google Scholar]
- 108.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nature Nanotechnology. 2016;11(11):936–40. [DOI] [PubMed] [Google Scholar]
- 109.Zeming KK, Salafi T, Shikha S, Zhang Y Fluorescent label-free quantitative detection of nano-sized bioparticles using a pillar array. Nature Communications. 2018;9(1):1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T, et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. Acs Nano. 2017;11(11):11182–93. [DOI] [PubMed] [Google Scholar]
- 111.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology. 2013;200(4):373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hromada C, Muhleder S, Grillari J, Redl H, Holnthoner W. Endothelial Extracellular Vesicles-Promises and Challenges. Frontiers in Physiology. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kalra H, Drummen GPC, Mathivanan S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. International Journal of Molecular Sciences. 2016;17(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJA, et al. Detecting Exosomes Specifically: A Multiplexed Device Based on Alternating Current Electrohydrodynamic Induced Nanoshearing. Analytical Chemistry. 2014;86(22):11125–32. [DOI] [PubMed] [Google Scholar]
- 115.Ibn Sina AA, Vaidyanathan R, Dey S, Carrascosa LG, Shiddiky MJA, Trau M. Real time and label free profiling of clinically relevant exosomes. Scientific Reports. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab on a Chip. 2014;14(19):3773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shao HL, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nature Medicine. 2012;18(12):1835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Im H, Shao HL, Weissleder R, Castro CM, Lee H. Nano-plasmonic exosome diagnostics. Expert Review of Molecular Diagnostics. 2015;15(6):725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. Acs Nano. 2016;10(2):1802–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reategui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nature Communications. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boklan J Little patients, losing patience: pediatric cancer drug development. Molecular cancer therapeutics. 2006;5(8):1905–8. [DOI] [PubMed] [Google Scholar]
- 122.Schnepp RW, Bosse KR, Maris JM. Improving Patient Outcomes With Cancer Genomics: Unique Opportunities and Challenges in Pediatric Oncology. Jama. 2015;314(9):881–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berg SL. Ethical challenges in cancer research in children. The oncologist. 2007;12(11):1336–43. [DOI] [PubMed] [Google Scholar]
- 124.Im H, Shao HL, Park YI, Peterson VM, Castro CM, Weissleder R, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nature Biotechnology. 2014;32(5):490–U219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liang K, Liu F, Fan J, Sun DL, Liu C, Lyon CJ, et al. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nature Biomedical Engineering. 2017;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]


