Abstract
Mycoplasma pneumoniae is a human respiratory tract pathogen causing chronic bronchitis and atypical or “walking” pneumonia. The major surface protein P1 must form complexes with proteins P30 and P40/P90 in order to function in receptor binding and gliding motility, and variability in P1 and P40/P90 distinguishes the two major M. pneumoniae genotypes. Strains belonging to each genotype can be differentiated with high sensitivity and specificity by utilizing surface-enhanced Raman spectroscopy on silver nanorod arrays. Here we used the variable selection method of Variable Importance in Projection (VIP) to identify Raman bands important in M. pneumoniae strain classification. Furthermore, VIP analysis of mutants lacking P40/P90, or P1and P40/P90, correlated certain Raman bands important in distinguishing genotypes, with specific mycoplasma surface protein composition and presentation. Variable selection, and its correlation with specific mycoplasma surface components, is an important next step in developing this platform for M. pneumoniae detection and genotyping.
Keywords: Mycoplasma pneumoniae, surface-enhanced Raman spectroscopy, nanorod array, Variable Importance in Projection, genotyping
INTRODUCTION
Mycoplasmas are novel cell wall-less bacteria with characteristically minimal genomes and correspondingly limited biosynthetic capabilities. These limitations necessitate a symbiotic lifestyle, often as normal flora but in some cases as pathogens (Citti and Blanchard 2013). Mycoplasma pneumoniae is a respiratory tract pathogen in humans, causing tracheobronchitis and primary atypical or “walking” pneumonia (Atkinson and Waites 2014; Waites et al. 2008). M. pneumoniae accounts for 20–40% of all community-acquired pneumonia and is the leading cause of pneumonia in older children and young adults. Infections are typically chronic in nature and can precipitate or exacerbate asthma (Atkinson and Waites 2014; Waites et al. 2008).
M. pneumoniae infection requires the capacity to penetrate the gel layer mucous that lines the airways and adhere to the underlying epithelium (Prince et al. 2014; Szczepanek et al. 2012). Gliding motility and receptor binding are mediated by the terminal organelle, a polar, membrane-bound extension of the mycoplasma cell defined by its complex, electron-dense core (Balish 2014). The membrane protein P1 is a major surface component of the terminal organelle and is directly involved in receptor recognition and cell gliding (Baseman et al. 1982; Jacobs et al. 1989; Seto et al. 2005). P1 function in adherence and gliding requires several accessory proteins, and thus mutants lacking terminal organelle proteins P40 and P90 (P40/P90; mutant III-4) are non-adhering and non-motile despite the presence of P1 (Krause et al. 1982; Hasselbring et al. 2005). The introduction of the recombinant wild-type allele for P40/P90 into mutant III-4 by transposon delivery restores a wild-type phenotype (Waldo et al. 2005). Cross-linking and other biochemical analyses suggest that P1 and P40/P90 are components of an adhesin complex on the terminal organelle surface (Layh-Schmitt and Herrmann 1994; Layh-Schmitt and Harkenthal 1999; Layh-Schmitt et al. 2000; Nakane et al. 2011), and particles likely corresponding to such complexes are prominent on the terminal organelle by electron cryo-tomography (Henderson and Jensen 2006; Kawamoto et al. 2016).
Variability in the primary structures of P1 and P40/P90 accounts for much of the genetic diversity exhibited by M. pneumoniae (Spuesens et al. 2009; Fig. 1). Genome sequence analysis has established that MPN141 and MPN142, which encode P1 and P40/P90, respectively, are within the largest region of DNA sequence polymorphism between the two major genotypes to which clinical isolates can be generally categorized (Spuesens et al. 2009; Xiao et al. 2015). Switching in predominance between these genotypes has been documented (Lind et al. 1995), and recent studies reveal co-circulation of both genotypes (Diaz et al. 2015; Pereyre et al. 2012). Thus strain genotyping is important to better understand the biology of M. pneumoniae and the nature and basis of its epidemiology.
Figure 1.
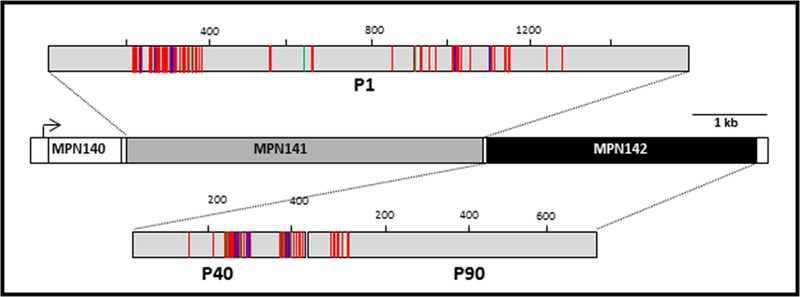
Schematic representation of amino acid sequence variability in M. pneumoniae proteins P1 and P40/90 between the genotype 1 reference strain M129 and the genotype 2 reference strain FH. P1 and P40/P90 are the products of MPN141 and MPN142, respectively. Red green, and blue lines indicate amino acid substitutions, insertions, and deletions, respectively, in FH relative to M129.
We recently demonstrated that vibrational spectroscopy can correctly classify globally diverse M. pneumoniae clinical isolates relative to genotype reference strains (Henderson et al. 2015). Several features of Raman spectroscopy make it particularly useful for analysis of biological samples, including narrow bandwidths, good spatial resolution, and the absence of interference by water. Traditional Raman spectroscopy is limited by characteristically weak signal strength. However, significant enhancement can be achieved from increased electromagnetic fields for molecules in close proximity to nanoscopically roughened metal surfaces, by surface-enhanced Raman spectroscopy (SERS; Tian et al. 2002). Given its inherently high specificity, Raman spectra can serve as a molecular fingerprint or barcode for pathogen identification (Patel et al. 2008). We employ a highly ordered silver nanorod array (NA) that provides consistent SERS enhancement factors of approximately 108 and have used this platform for highly reproducible detection of infectious agents, including mycoplasmas (Hennigan et al. 2010; Henderson et al. 2014).
SERS enhancement is, by definition, greatest for analyte components that are in closest proximity to the silver nanorod array, and thus we reasoned that mycoplasma surface components are probably responsible for the spectral features that distinguish M. pneumoniae genotypes by means of NA-SERS and partial least squares discriminant analysis (PLS-DA). We recently showed that the selection of a reduced wavenumber variable set by using simple analysis of variance aids in the chemometric differentiation of human mycoplasmas (Choi JY et al., in revision). Here we used the more sophisticated variable selection method of Variable Importance in Projection (VIP) to identify the Raman bands most important to classifying M. pneumoniae strains. VIP analysis was specifically developed for PLS-based variable selection and is particularly adapted to use in the PLS-DA methods that we employ for mycoplasma genotype discrimination (Eriksson et al. 2006; Wold et al. 1993). Furthermore, because genotype classification reflects differences in mycoplasma terminal organelle proteins P1 and P40/P90, we also predicted that VIP analysis of terminal organelle mutants lacking those proteins would correlate distinguishing Raman bands with specific mycoplasma surface protein composition and presentation. Variable selection is expected to be an important next step in developing SERS as a platform for M. pneumoniae detection and genotyping, and the ability to correlate distinguishing band numbers with specific surface proteins adds significant value to its molecular fingerprinting capabilities.
EXPERIMENTAL
Mycoplasma Strains and Culture
Wild type M. pneumoniae strains M129 (type 1) and FH (type 2), and the spontaneously arising terminal organelle mutant strains III-4, and IV-22, both in the M129 background, were used in this study (Henderson et al. 2015). Mutant III-4 has a frameshift in the gene encoding the surface proteins P40/P90 (MPN142), also referred to as proteins C and B, respectively (Krause et al. 1982; Waldo et al. 2005). The loss of P40/P90 appears to change the tertiary / quaternary structure of protein P1, based on its loss of function (Krause et al. 1982) and a dramatic shift in its accessibility to protease (Cloward and Krause 2011). Mutant IV-22 has a frameshift in the MP141 gene encoding P1 (Su et al. 1989), with loss of P1 also resulting in loss of P40/P90 (Krause et al. 1982). Twenty-four additional M. pneumoniae clinical isolates (13 genotype 1 strains and 11 genotype 2 strains) were provided by the Pneumonia Response and Surveillance Laboratory at the Centers for Disease Control and Prevention (Atlanta, GA USA). Their genotype groups were determined using DNA sequence analysis, quantitative real-time PCR in combination with high resolution melt curve analysis, and restriction fragment length polymorphism sequencing analysis (Henderson et al. 2015). Mycoplasmas grown in SP4 culture medium in tissue culture flasks at 37°C were prepared for NA-SERS as described by (Henderson et al. 2015).
Briefly, for harvest of M129, FH, and the 24 other clinical isolates, the spent growth medium was decanted and each flask was washed 4× with 0.1× volume of sterile chilled phosphate-buffered saline (PBS; pH 7.2) and then scraped into 1 ml sterile PBS and syringe-passaged 10× with a 25-gauge needle to disperse clumps. The mutant strains do not adhere to plastic and therefore were harvested from the spent SP4 medium by centrifugation and washed 3X with chilled PBS. Formaldehyde in sterile PBS (pH 7.0) was added to the mycoplasma suspensions to a final concentration of 4%, and samples were stored at 4°C until used for SERS. Growth medium negative control samples were prepared in parallel under the same conditions as the M. pneumoniae strains (Henderson et al. 2014; 2015).
SERS and Chemometric Analysis
NA-SERS substrates were cleaned for 5 min in a PDC-32G Ar+ plasma cleaner (Harrick Plasma, Ithaca, NY) to remove potential surface contamination, then patterned to create 3 mm diameter wells (Henderson et al. 2015). 1,2-bis(4-pyridyl)ethylene (10−4 M in methanol) was used to control for consistent spectrum enhancement between substrates. Raman spectra were acquired using a Renishaw inVia Reflex multi-wavelength confocal imaging microscope (Hoffman Estates, IL) and a Leica apochromatic 5× objective (Numerical Aperture 0.12), illuminating a 1265 μm2 area on the substrate to allow for spatial averaging (Henderson et al. 2015). Samples were irradiated with a 785-nm near-infrared diode laser (Renishaw) at 10% power capacity (28 mW), with spectra collected in 3 10-sec acquisitions. An internal silicon standard was included as a control for instrument performance.
Samples were applied to the NA substrates in 1-μl volumes per well in duplicate and dried overnight. Spectra were typically collected from five random locations per sample spot, for a total of 10 spectra per sample. SERS analysis of the clinical isolates was described previously (Henderson et al. 2015). For SERS analysis of the mutant strains, four separate biological replicates were examined totaling 60–75 spectra per strain. Raman spectra (400–1800 cm−1) were acquired using Renishaw WiRE 3.4 software, with instrument settings optimized to maximize signal and minimize saturation or sample degradation by the laser.
Spectra were averaged using GRAMS32/A1 software package (Galactic Industries, Nashua, NH) to assess signal-to-noise quality, and baseline-corrected with a concave rubberband algorithm (10 iterations on 64 points) for preliminary evaluation and peak assignment (OPUS, Bruker Optics, Inc., Billerica, MA). Chemometric analyses were carried out with MATLAB 7.10.0 (Mathworks, Inc., Natick, MA) and PLS-Toolbox 7.5.1 (Eigenvector Research Inc., Wenatchee, WA). Raw spectra were pre-processed using the first derivative of each and a 15-point, second order polynomial Savitsky-Golay algorithm, and each dataset was vector-normalized and mean-centered. Multivariate statistical analysis of the datasets was performed using the PLS-Toolbox software. Statistical models were cross-validated using a Venetian blinds algorithm with 10 data splits.
VIP Analysis
The complexity of the SERS spectra obtained from whole bacteria makes the extraction of relevant information and the interpretation of data challenging. The excellent reproducibility of SERS spectra allows the use of multivariate analysis methods for data analysis, classification, and identification. A variety of such methods have been used for evaluating biomedical vibrational spectra, and the statistical basis for application of these methods to vibrational spectroscopy is well established (Jarvis and Goodacre 2008). While unsupervised pattern recognition methods such as principal components analysis and hierarchical cluster analysis can be used for spectral interpretation, we have successfully employed the supervised method known as PLS-DA (Ballabio and Consonni 2013; Barker and Rayens 2003) for the analysis of SERS spectra of human and avian mycoplasma species (Henderson et al. 2015; Hennigan et al. 2010; 2012).
PLS-DA is a full-spectrum, multivariate, supervised method where previous knowledge of sample constituents is used to build a classification model for spectral differentiation. PLS-DA is a modification of partial least squares regression in which the variation within classes is minimized and the latent variables that best describe the spectral differences between classes are emphasized. The number of potential latent variables used in the PLS-DA model is directly related to the resolution of the Raman spectrum and the number of wavenumber data points collected therein, and can easily lead to thousands of potential latent variable descriptors of classification properties. However, in statistical modeling, it is common to assume that only a smaller number of variables are actually correlated to the discrimination ability and are relevant to the model. Therefore, an important component of multivariate statistical classification is feature selection, i.e. the objective identification of those variables that contribute useful information, while eliminating variables that contain mostly noise or unnecessary information (Xiaobo et al. 2010). Ideally, feature selection results in simpler models with higher accuracy in prediction results.
We used the VIP feature selection method (Eriksson et al. 2013) in this work. This filter-based variable selection method is based on canonical powered PLS regression methods (Indahl et al. 2009). VIP analysis assumes that in the overall data matrix x there is a subset of dimension A that contains all relevant information needed for predicting the desired properties y. The principle of VIP is to summarize the importance of each variable j reflected in the PLS weight vector w. The VIP score is for each variable j is calculated as follows:
| (1) |
where p is the total number of variables (wavenumbers), A the number of retained latent variables, waj, the PLS weight of the jth variable for the ath latent variable, and SSa the sum of squares explained by the ath component, i.e. the percentage of y explained by the ath latent variable. VIP scores above a defined threshold value are considered highly influential, while the individual cutoff criterion for selection of influential variables is a function of the data structure in a particular experiment.
RESULTS AND DISCUSSION
VIP analysis for M. pneumoniae Clinical Isolates Categorized by Genotype
We previously established the close similarity in Raman wavenumber location and intensity for baseline-corrected NA-SERS spectra of M. pneumoniae reference strain M129 and other genotype 1 clinical isolates, and reference strain FH and other genotype 2 clinical isolates, where PLS-DA modeling of the spectra differentiates these by major genotype with high specificity and sensitivity (Henderson et al. 2015). Here we examined VIP scores for those models in order to identify the key wavenumbers that discriminate the two genotypes. VIP scores for each variable above a predefined zero threshold were plotted for all clinical isolates relative to the genotype reference strains (Fig. 2). The higher the score, the greater the significance of those bands to the models. Given our focus here on only those wavenumbers having the greatest significance, we only considered scores ≥ 2 going forward. As expected, the VIP profiles for models based on a genotype 1 or genotype 2 reference were virtually identical and exhibited only a limited number of major discriminating Raman wavenumber regions (1659–1675, 1395–1400, 1388–1394, 960–971, and 947–957 cm−1; Fig. 2, yellow), plus a few additional minor discriminating wavenumbers (summarized in Table 1)
Figure 2.
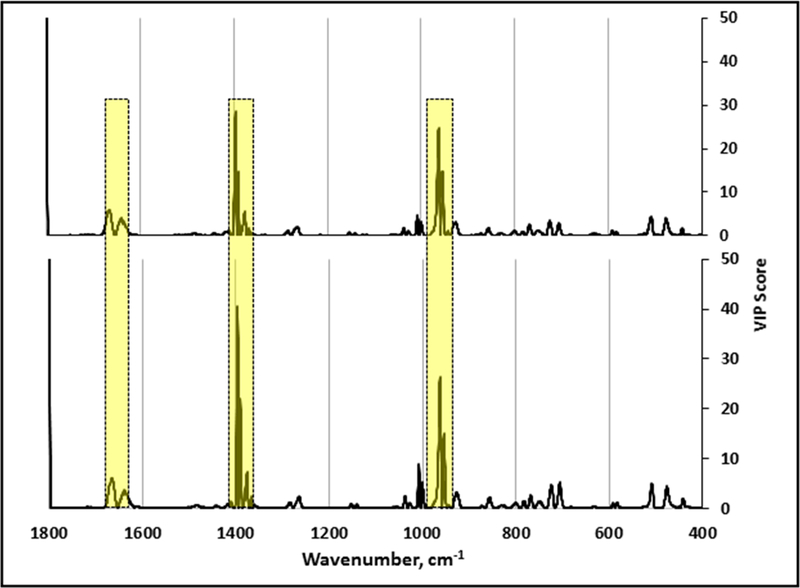
VIP analysis of PLS-DA models that discriminate diverse clinical isolates relative to the genotype 1 reference strain M129 (upper; n= 195 spectra) or the genotype 2 reference strain FH (lower; n=175 spectra). Regions highlighted by yellow boxes indicate major discriminating Raman band wavenumbers. The band at wavenumber 1800 cm-1 was variable in mycoplasma samples and growth medium controls and is likely a function of sample preparation. As there are no known biomolecules assigned to this wavenumber, this band was not considered further here.
Table 1.
Consensus wavenumbers having VIP scores > 0 from analysis of M. pneumoniae strains. High, > 5; Low, 2– 5; colors correspond to the highlighted peaks in figures 2 (yellow) and 5 (orange and green).
| Wavenumber | VIP scores: clinical strains vs type 1 reference M129 | VIP scores: clinical strains vs type 2 reference FH | VIP scores: M129 vs III-4 |
VIP scores: M129 vs IV-22 | VIP scores: III-4 vs IV-22 |
Tentative bond vibrational mode assignments |
|---|---|---|---|---|---|---|
| 1659–1675 | High | High | --- | --- | --- | 1675: Amide I a,b 1663, 1670, 1672: Amide I c |
| 1631–1647 | Low | Low | --- | --- | --- | 1646: Amide I a 1637: Amide I b, d |
| 1474–1485 | --- | --- | Low | Low | Low | --- |
| 1457–1467 | --- | --- | Low | Low | Low | 1440–1460: C-H2 def b, c |
| 1438–1451 | --- | --- | Low | High | Low | 1442–1443: CH2 scissoring a; 1440–1460: C-H2 def b, c ; 1347, 1350: CH2 bend (protein, lipid) d |
| 1395–1400 | High | High | --- | --- | --- | 1400: COO- symmetric stretching b |
| 1388–1394 | High | High | --- | --- | --- | 1392: COO- symmetric stretchinga |
| 1375–1381 | High | High | --- | --- | --- | C-H bend (protein) d |
| 1347–1352 | --- | --- | Low | --- | Low | 1350: Amide IIIb, Trpc |
| 1334–1341 | --- | --- | Low | --- | Low | 1335, 1338, 1339: Trp c
1337, 1341: C-H bend (protein) d |
| 1324–1330 | --- | --- | Low | --- | --- | 1330: Trpa; 1329: C-H bend (protein) d |
| 1262–1266 | Low | Low | --- | --- | --- | 1230–1295: Amide III b, c |
| 1051–1061 | --- | --- | High | High | High | 1061: Gln, C-N stretch b
1056: C-C stretch d |
| 1039–1050 | --- | --- | High | High | High | 1040: Phe c; 1043, C-C stretch d |
| 1006–1020 | Low | High | High | High | High | 1013,1019: Phea; 1009: Trpc |
| 997–1005 | Low | Low | High | High | High | 997: Phea;; 1004: Pheb; 1001–1003: Phe c |
| 960–971 | High | High | High | High | Low | 971: C-C stretch a ; 960: C-C stretch c; 961, 963, 966, 967: C-N stretch d |
| 947–957 | High | High | High | High | Low | --- |
| 933–943 | --- | --- | High | High | Low | 934: C-C stretch c, d |
| 923–931 | Low | Low | --- | High | Low | 924, 925, 931: C-COO- stretch d |
| 810–811 | --- | --- | --- | Low | Low | --- |
| 797–804 | --- | --- | High | High | --- | 801: -O-P-O d |
| 778–786 | --- | --- | Low | --- | Low | 785: Cytosine/Uracil stretchb |
| 766–770 | Low | Low | --- | --- | --- | --- |
| 738–744 | --- | --- | Low | Low | Low | 746: Trpa |
| 716–728 | Low | Low | High | High | Low | 720: C-H rocking; Adeb; 720–721: Trp c |
| 702–714 | Low | High | High | High | Low | --- |
| 581–583 | --- | Low | ||||
| 574–578 | --- | --- | Low | --- | Low | 575–577, 582:carbohydrated |
| 565–569 | --- | --- | Low | --- | --- | 568: Trp c |
| 505–513 | Low | Low | --- | --- | --- | 507–509, 513: S-S c |
| 498–500 | --- | --- | Low | --- | Low | |
| 472–480 | Low | Low | --- | --- | --- | --- |
NA-SERS Analysis of Wild-Type M. pneumoniae and Terminal Organelle Mutants
The M. pneumoniae genes encoding the major mycoplasma proteins P1 and P40/P90 are within the largest region of DNA sequence polymorphisms between strains of genotypes 1 and 2 (Xiao et al. 2015). These proteins are abundant and accessible on the mycoplasma cell surface (Layh-Schmitt and Herrmann 1994), where they likely contribute significantly to the M. pneumoniae signature from surface-enhanced Raman scattering, including wavenumber variables that are important in discriminating the two major genotypes. Analysis of mutants lacking these proteins should thus be informative in correlating spectral features with specific mycoplasma surface components. M. pneumoniae strains III-4 and IV-22 arose spontaneously from the wild-type M129 parent strain and have well defined mutations resulting in loss of proteins P40/P90 (mutant III-4) or P40/P90 and P1 (mutant IV-22) (Krause et al. 1982; Waldo et al. 2005; Su et al. 1989). Fig. 3 shows representative baseline-corrected NA-SERS spectra for M129 and mutants III-4 and IV-22. Despite their close similarity, PLS-DA modeling distinguished the mutants from the parent strain with >95% sensitivity and specificity, and from each other with >85% sensitivity and specificity (Fig. 4).
Figure 3.
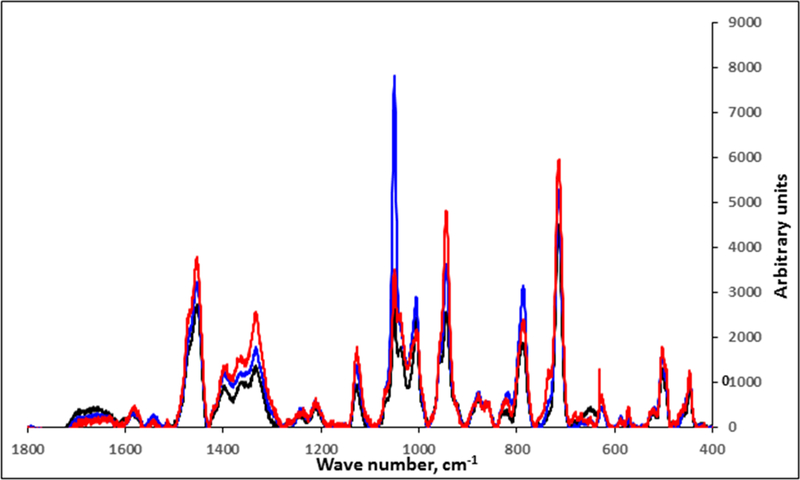
Baseline-corrected and averaged NA-SERS spectra for wild-type M129 (black; n=75) and mutants III-4 (red; n=68) and IV-22 (blue; n=67).
Figure 4.
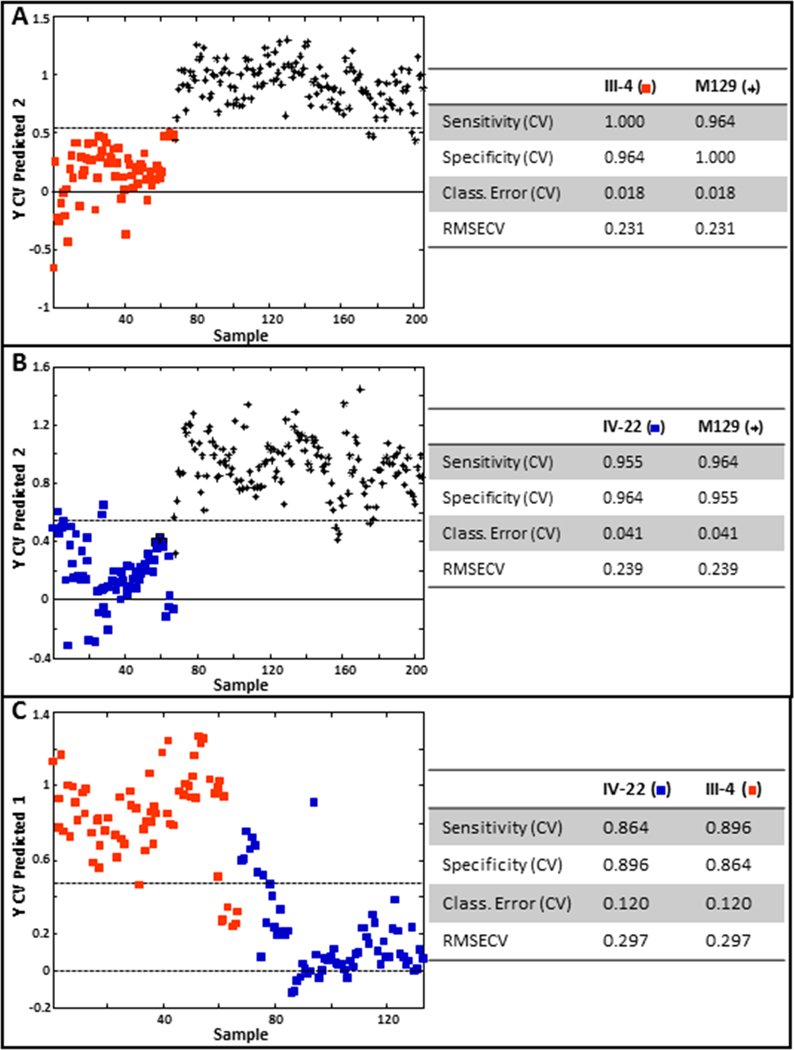
PLS-DA of mutant III-4 (A) or mutant IV-22 (B) relative to the wild-type parent strain M129, and of mutant III-4 relative to mutant IV-22. Stars, M129; red squares, mutant III-4; blue squares, mutant IV-22. Cross-validated statistics were obtained using Venetian blinds with 10 data splits to represent the prediction performance for each model. CV, cross-validated; RMSECV, root mean square error, cross-validated.
VIP Analysis of M. pneumoniae Mutants
The proximity of analyte to substrate is paramount to SERS amplification of scattering. Therefore, abundant mycoplasma cell-surface proteins such as P1 and P40/P90 are likely to contribute significantly to the SERS signature of intact M. pneumoniae cells. Furthermore, VIP scores from the PLS-DA models above that discriminated M129, III-4, and IV-22 will likely reflect variables specifically associated with P1 and P40/P90. Comparison of VIP scores from the pairwise modeling of mutant III-4 with M129, mutant IV-22 with M129, and mutant III-4 with mutant IV-22 revealed striking similarities (Fig. 5A; Table 1). In particular, VIP scores >5 for wavenumber regions 1006–1020, 997–1005, 960–971, 947–957, 933–943, 716–728, and 702–714 cm−1, were shared in common from PLS-DA analysis of each mutant with the wild-type parent strain. Moreover, VIP scores > 5 for wavenumber regions 1051–1061 and 1039–1050 cm−1 were shared in common from PLS-DA analysis of each mutant with the parent strain, as well as between mutants. Finally, because differences in P1 and P40/P90 differentiate the two major M. pneumoniae genotypes, we predicted that comparison of VIP scores from the genotype and mutant analyses would reveal commonalities, and this was indeed the case, most notably 947–974 cm−1 (Fig. 5B)
Figure 5.
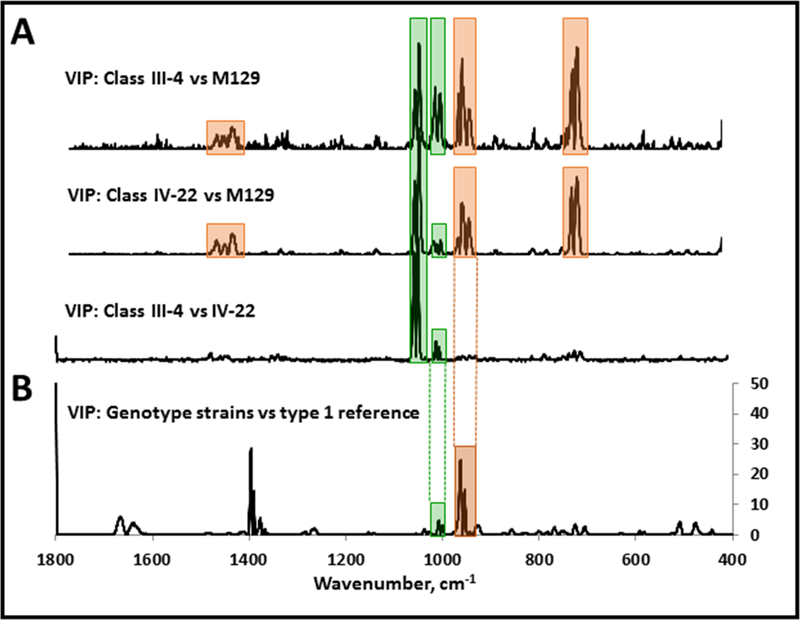
VIP analysis from pairwise PLS-DA modeling of (A) mutant III-4 with M129, mutant IV-22 with M129, and mutant III-4 with mutant IV-22, or (B) diverse clinical isolates relative to the genotype 1 reference strain M129. Orange, major band wavenumbers in common for VIP analysis of wild-type M129 and either mutant (A) and also from the genotype VIP analysis (B); green, major band wavenumbers in common for VIP analysis of wild-type M129 and either mutant, as well as VIP analysis of the two mutants (A) and also for the genotype VIP analysis (B).
Previous studies established proof-of-principle for the application of this bio-sensing platform for detection and typing of M. pneumoniae in clinical samples (Henderson et al. 2015; Hennigan et al. 2010). However, we anticipate that chemometric analysis of SERS data may ultimately require feature selection for this technology to be applied to the more diverse, biochemically complex backgrounds associated with clinical material such as human throat swab samples. With that in mind, one focus of the current study was to address what VIP analysis might reveal about the identity and relative importance of specific SERS variables in strain differentiation. For this purpose, we utilized two different M. pneumoniae populations sharing an important common denominator. The first population was diverse clinical isolates belonging to the two major genotypes and differing mainly in the primary structure of surface proteins P1, P40, and P90. The second population consisted of spontaneously arising mutants that differ specifically in the presence or absence of P1, P40, and P90. VIP analysis of the former population identified wavenumbers most important in genotyping, while VIP analysis of the latter population allowed us to test the hypothesis that commonalities would emerge and indirectly correlate certain wavenumbers with specific proteins differing between the major genotypes.
M. pneumoniae strains are generally categorized by genotype based on variation in the sequence of the MPN141 gene, encoding the major adhesin protein P1. Both MPN 141 and MPN142, encoding P1 and P40/P90, are located within the largest region of DNA sequence polymorphism between genotypes (Spoesens et al. 2009; Xiao et al. 2015). Strains of genotypes 1 and 2 differ in the architecture and robustness of the biofilms they form and in the distribution of an N-acetyl glucosamine-containing polysaccharide in their biofilm matrices (Simmons et al. 2013), but no differences have been established yet in their clinical outcomes (Nilsson et al. 2010). PLS-DA models of their NA-SERS signatures consistently distinguish genotypes 1 and 2 (Henderson et al. 2015). Differences in P1, P40, and / or P90 likely account in large part for the wavenumbers that contribute most significantly to their differentiation by PLS-DA, both as a direct function of primary structure and also likely reflecting distinct tertiary or quaternary structures. Our finding that only a few wavenumbers contribute significantly to genotype discrimination is consistent with the predominant role for only a limited number of proteins in differentiating the major genotypes.
Specific vibrational modes can be associated with individual Raman bands, and we previously noted the vibrational mode assignment reported by others that correspond to individual bands prominent in M. pneumoniae NA-SERS spectra (Henderson et al. 2014). Those mode assignments associated with Raman bands identified from VIP analysis in the current study are indicated in Table 1 and include bond vibrations present in amino acids and sugars, which is consistent with their strong correlation with proteins P1 and P40/P90. Given the high biochemical complexity of the mycoplasma surface, it is highly unlikely that vibrational mode assignment alone could be sufficient to associate large biomolecules such as P1, P40, or P90 specifically with wavenumbers identified by VIP analysis of the genotypes. For this reason, rather than vibrational mode assignment, we approached this correlation by utilizing mutants specifically lacking P1, P40, and P90 for VIP analysis. And as with the results from the genotype VIP analysis, based on what is known about these mutants it was not surprising that so few wavenumbers accounted for most of the discriminating ability of PLS-DA modeling.
A reasonable starting point in considering the VIP analyses further is the assumption that differences in P1, P40, and/or P90 account for the wavenumbers with the highest VIP scores, although SERS spectra could be impacted both directly (e.g. the presence or absence of these proteins), as well as indirectly (e.g. the failure of P1 to form a functional adhesin complex in the absence of P40 and P90) (Layh-Schmitt and Herrmann 1994; Layh-Schmitt and Harkenthal 1999; Layh-Schmitt et al. 2000; Nakane et al. 2011). Thus major spectral differences for wild-type M129 and mutant III-4 likely resulted from the absence of P40/ P90 and a change in P1 tertiary structure, as suggested by loss of function (Krause et al. 1982) and strongly supported by its strikingly greater accessibility to protease in mutant III-4 (Cloward and Krause 2011). Likewise, spectral differences for M129 and mutant IV-22 likely resulted from the complete absence of P40, P90, and P1 in mutant IV-22. In contrast, spectral differences for mutants III-4 and IV-22, which both lack P40 and P90, were likely limited to the presence (III-4) or absence (IV-22) of a non-functional P1. With that in mind, we speculate that the three wavenumber regions (1436–1485, 932–974, and 699–728 cm−1; Fig. 5, orange) having high VIP scores from PLS-DA modeling of each mutant with the wild-type parent strain but not the modeling of the two mutants, are likely specifically due to the presence or absence of P40/P90. Likewise, the wavenumber regions 1035–1062 cm−1 and to a lesser extent 996–1017 cm−1 (Fig. 5, green) were prominent for all three mutant PLS-DA models, and we speculate that these are likely associated with P1 in its non-functional configuration. Finally, if we extrapolated to the VIP analysis of the PLS-DA modeling of the genotype data, two regions stood out in particular. The first corresponds to 932–974 cm−1, which was prominent in the modeling of each mutant with wild-type M129, and which we speculate is likely due to P40/P90. The second corresponds to 996–1017 cm−1 and likely corresponds to P1-associated differences. Finally, it is noteworthy but not surprising that there are several wavenumbers having high VIP scores for the genotyping but not for any mutants, such as 1374–1401 cm−1, for example. This is to be expected given that the major genotypes differ in the nature of P1 and P40/P90 as part of a functional adhesion complex, while the mutants differ in the presence or absence of P40/P90 and the presence, absence, or nature of P1.
ACKNOWLEDGMENTS
This work was supported by the US National Institutes of Health (grants AI096364 to DCK and GM102546 to RAD). The funding source had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit this article for publication.
REFERENCES
- Atkinson TP, and Waites KB 2014. Mycoplasma pneumoniae infections in childhood. Pediatr. Inf. Dis. J. 33: 92–94. [DOI] [PubMed] [Google Scholar]
- Balish MF 2014. Mycoplasma pneumoniae, an underutilized model for bacterial cell biology. J. Bacteriol. 196: 3675–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio D and Consonni V 2013. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods 5: 3790–3798. [Google Scholar]
- Barker M and Rayens W 2003. Partial least squares for discrimination. J. Chemometr. 17: 166–173. [Google Scholar]
- Baseman JB, Cole RM, Krause DC, and Leith DK 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol 151: 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citti C and Blanchard A 2013. Mycoplasmas and their host: emerging and re-emerging minimal pathogens. Trends Microbiol 21: 196–203. [DOI] [PubMed] [Google Scholar]
- Cloward JM and Krause DC 2011. Loss of co-chaperone TopJ impacts adhesin P1 presentation and terminal organelle maturation in Mycoplasma pneumoniae. Molec. Microbiol 81: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culha M, Adigüzel A, Yazici MM, Kahraman M, Sahin F, and Güllüce M 2008. Characterization of thermophilic bacteria using surface-enhanced Raman scattering. Appl. Spectrosc 62: 1226–1232. [DOI] [PubMed] [Google Scholar]
- Diaz MH, Benitez AJ, and Winchell JW 2015. Investigations of Mycoplasma pneumoniae infections in the United States: trends in molecular typing and macrolide resistance from 2006–2013. J. Clin. Microbiol 53: 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, and Wold S, editors. 2006. Multi- and megavariate data analysis, Part 1, basic principles and applications. Umeå, Sweden: Umetrics AB. [Google Scholar]
- Erkisson L, Byrne T, Johansson E, Trygg J, and Vikstrom C, editors. 2013. Multi- and Megavariate Data Analysis: Basic Principles and Applications. 3rd edn. Malmo, Sweden: Umetrics Academy. [Google Scholar]
- Hasselbring BM, Jordan JL, and Krause DC. 2005. Mutant analysis reveals specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J. Bacteriol 187: 6281–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GP and Jensen GJ 2006. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol. Microbiol 60: 376–385. [DOI] [PubMed] [Google Scholar]
- Henderson KC, Sheppard ES, Rivera-Betancourt OE, Choi JY, Dluhy RA, Thurman KA, Winchell JM, and Krause DC 2014. The multivariate detection limit for Mycoplasma pneumoniae as determined by nanorod array-surface-enhanced Raman spectroscopy and comparison with limit of detection by qPCR. Analyst 139: 6426–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KC, Benitez AJ, Ratliff AE, Crabb DM, Sheppard ES, Winchell JW, Dluhy RA, Waites KB, Atkinson TP, and Krause DC 2015. Specificity and strain-typing capabilities of Nanorod Array-Surface Enhanced Raman Spectroscopy for Mycoplasma pneumoniae detection. PLoS ONE 10(6):e0131831. doi: 10.1371/journal.pone.0131831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan SL, Driskell JD, Dluhy RA, Zhao Y, Tripp RA, Waites KB, and Krause DC. 2010. Detection of Mycoplasma pneumoniae in simulated and true clinical throat swab specimens by Nanorod Array-Surface-Enhanced Raman Spectroscopy. PLoS One 5(10):e13633 http://dx.plos.org/10.1371/journal.pone.0013633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan SL, Driskell JD, Ferguson-Noel N, Dluhy RA, Zhao Y, Tripp RA, and Krause DC. 2012. Detection and Differentiation of Avian Mycoplasmas by Surface-Enhanced Raman Spectroscopy Based on a Silver Nanorod Array. Appl. Environ. Microbiol 78: 1930–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indahl UG, Liland KH, and Næs T. 2009. Canonical partial least squares—a unified PLS approach to classification and regression problems. J. Chemometr 23: 495–504. [Google Scholar]
- Jacobs E, Gerstenecker B, Mader B, Huang CH, Hu PC, Halter R, and Bredt W 1989. Binding sites of attachment-inhibiting monoclonal antibodies and antibodies from patients on peptide fragments of the Mycoplasma pneumoniae adhesin. Infect. Immun 57: 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis RM and Goodacre R 2008. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev 37: 931–936. [DOI] [PubMed] [Google Scholar]
- Kawamoto A, Matsuo L, Kato T, Yamamoto H, Namba K, and Miyata M. 2016. Periodicity in attachment organelle revealed by electron cryotomography suggests conformational changes in gliding mechanism of Mycoplasma pneumoniae. MBio 7: e00243–16. 10.1128/mBio.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Leith DK, Wilson RM, and Baseman JB 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun 35: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G and Herrmann R 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun 62: 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G and Harkenthal M 1999. The 40- and 90-kDa membrane proteins (ORF6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesion) association with the Triton shell. FEMS Microbiol. Lett 174: 143–149. [DOI] [PubMed] [Google Scholar]
- Layh-Schmitt G, Podtelejnikov A, and Mann M. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiol 146: 741–747. [DOI] [PubMed] [Google Scholar]
- Lind K, Benzon MW, Jensen S, and Clyde WA Jr. 1997. A seroepidemiological study of Mycoplasma pneumoniae infections in Denmark over the 50-year period 1946–1995. Eur. J. Epidemiol 13: 581–586. [DOI] [PubMed] [Google Scholar]
- Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, and Puppels GJ. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods. 51:255–271. [DOI] [PubMed] [Google Scholar]
- Nakane D, Adan-Kubo J, Kenri T, and Miyata M. 2011. Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J. Bacteriol 193: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AC, Björkman P, Welinder-Olsson C, Widell A, and Persson K. 2010. Clinical severity of Mycoplasma pneumoniae (MP) infection is associated with bacterial load in oropharyngeal secretions but not with MP genotype. BMC Infect. Dis 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel IS, Premasiri WR, Moir DT, and Ziegler LD 2008. Barcoding bacterial cells: A SERS based methodology for pathogen identification. J. Raman. Spectrosc 39: 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyre S, Charron A, Hidalgo-Grass C, Touati A, Moses AE, Nir-Paz R, and Bébéar C 2012. The spread of Mycoplasma pneumoniae is polyclonal in both an endemic setting in France and in an epidemic setting in Israel. PLoS One 7:e38585 10.1371/journal.pone.0038585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podstawka E, Ozaki Y, and Proniewicz LM 2004. Adsorption of S-S containing proteins on a colloidal silver surface studied by surface-enhanced Raman spectroscopy. Appl. Spectrosc 58: 1147–1156. [DOI] [PubMed] [Google Scholar]
- Prince OA, Krunkosky TM, and Krause DC 2014. In vitro spatial and temporal analysis of Mycoplasma pneumoniae colonization of human airway epithelium. Infect. Immun 82: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Kenri T, Tomiyama T, and Miyata M 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol 187: 1875–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WL, Daubenspeck JM, Osborne JD, Balish MF, Waites KB, and Dybvig K 2013. Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiol 159: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuesens EB, Oduber M, Hoogenboezem T, Sluijter M, Hartwig NG, van Rossum AMC, and Vink C 2009. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in M. pneumoniae. Microbiol 155: 2182–2196. [DOI] [PubMed] [Google Scholar]
- Su CJ, Chavoya A, and Baseman JB 1989. Spontaneous mutation results in loss of the cytadhesin (P1) of Mycoplasma pneumoniae. Infect. Immun 57: 3237–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek SM, Majumder S, Sheppard ES, Liao X, Rood D, Tulman ER, Wyand S, Krause DC, Silbart LK, and Geary SJ 2012. Vaccination of BALB/c mice with avirulent Mycoplasma pneumoniae P30 mutants results in disease exacerbation upon challenge with a virulent strain. Infect. Immun 80: 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z-Q, Ren B, and Wu D-Y 2002. Surface-enhanced Raman scattering: from Noble to transition metals and from rough surfaces to ordered nanostructures. J. Phys. Chem. B 106: 9463–9483. [Google Scholar]
- Waites KB, Balish MF, and Atkinson TP 2008. New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol 3: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo RH III, Jordan JL, and Krause DC 2005. Identification and complementation of a mutation associated with loss of Mycoplasma pneumoniae virulence-specific proteins B and C. J. Bacteriol. 187: 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Johansson A, and Cochi M, editors. 1993. PLS-partial least squares projections to latent structures. Leiden: ESCOM Science Publishers, p. 523–550. [Google Scholar]
- Xiao L, Ptacek T, Osborne JD, Crabb DM, Simmons WL, Lefkowitz EJ, Waites KB, Atkinson TP, and Dybvig K 2015. Comparative genome analysis of Mycoplasma pneumoniae. BMC Genomics 16: 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaobo Z, Jiewen Z, Povey MJ, Holmes M, and Hanpin M 2010. Variables selection methods in near-infrared spectroscopy. Anal. Chim. Acta 667: 14–32. [DOI] [PubMed] [Google Scholar]
- Yang X, Gu C, Qian F, Li Y, and Zhang JZ 2011. Highly sensitive detection of proteins and bacteria in aqueous solution using surface-enhanced Raman scattering and optical fibers. Anal. Chem 83: 5888–5894. [DOI] [PubMed] [Google Scholar]


