Abstract
Autophagy is a conserved process that catabolizes intracellular components to maintain energy homeostasis and to protect cells against stress. Autophagy has crucial roles during development and disease, and evidence accumulated over the past decade indicates that autophagy also has a direct role in modulating ageing. In particular, elegant studies using yeasts, worms, flies and mice have demonstrated a broad requirement for autophagy-related genes in the lifespan extension observed in a number of conserved longevity paradigms. Moreover, several new and interesting concepts relevant to autophagy and its role in modulating longevity have emerged. First, select tissues may require or benefit from autophagy activation in longevity paradigms, as tissue-specific overexpression of single autophagy genes is sufficient to extend lifespan. Second, selective types of autophagy may be crucial for longevity by specifically targeting dysfunctional cellular components and preventing their accumulation. And third, autophagy can influence organismal health and ageing even non-cell autonomously, and thus, autophagy stimulation in select tissues can have beneficial, systemic effects on lifespan. Understanding these mechanisms will be important for the development of approaches to improve human healthspan that are based on the modulation of autophagy.
Autophagy is an evolutionarily conserved catabolic process that has an essential role in cellular homeostasis by facilitating lysosomal degradation and recycling of intracellular macromolecules and organelles (collectively referred to as autophagic cargo). Autophagy was first discovered as a survival mechanism in yeasts subjected to nutrient deprivation, a condition that potently stimulates the process over basal levels. Since then, studies in several different organisms have established critical roles for autophagy in a variety of biological processes ranging from development to ageing1. Interestingly, autophagy is often found perturbed in age-related disorders such as cancer, diabetes and neurodegenerative diseases2,3. Accordingly, autophagy is important for the maintenance of organismal health, which prominently declines with ageing.
Three types of autophagy have been distinguished on the basis of the mechanism of cargo sequestration: microautophagy (sequestration of cytoplasmic components directly into the lysosome, where acidic hydrolases mediate degradation), chaperone-mediated autophagy (selective degradation of unique, motif-containing cargo proteins recognized and delivered to the lyso-some by a chaperone complex) and macroautophagy (degradation of cytosolic material via sequestration into double-membrane vesicles called autophagosomes that subsequently fuse with lysosomes). This Review focuses on macroautophagy (hereafter termed autophagy), which has been extensively studied in the context of ageing, particularly in invertebrate models.
The autophagy process is mediated by a number of autophagy-related (ATG) proteins and can be divided into at least five sequential steps: first, initiation; second, double-membrane nucleation and formation of a preautophagosome or phagophore; third, phagophore elongation and sequestration of cytoplasmic cargo; fourth, fusion of the autophagosome (the fully enclosed phagophore) to a lysosome; and fifth, degradation of sequestered cargo in the autolysosome. Different ATG molecules are implicated in these steps4 (Fig. 1). Key upstream regulators of this multistep process include the highly conserved nutrient sensors mTOR and AMP-activated kinase (AMPK) — which notably are also critical longevity determinants (see BOX 1) — which directly phosphorylate Unc-51-like kinase 1 (ULK1; Atg1 in yeasts), a conserved kinase that serves as the key upstream initiator of autophagy5. Another set of key autophagy proteins is the family comprising microtubule-associated protein light chain 3 (LC3) proteins and γ-aminobutyric acid receptor-associated proteins (GABARAPs) in mammals (Atg8 in yeasts). Fluorescently tagged or endogenous LC3/GABARAP family proteins are commonly used as steady-state auto-phagy markers in many species to facilitate microscopic visualization of phagophores and autophagosomes in the cell6. LC3/GABARAP family proteins are proteolytically processed and attached to autophagosomal membranes, where they participate in cargo recognition and recruitment to the phagophore by interacting with various autophagy receptors or cargo receptors bound to proteins or organelles. Prominent examples of autophagy receptors are p62 (also known as SQSTM1), which recognizes ubiquitylated proteins or organelles targeted for degradation7, and BCL-2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), a receptor for mitochondria destined for degradation by mitophagy8. Such specific clearance of cargo, including organelles and macromolecules, is collectively referred to as selective autophagy. Notably, damaged macromolecules and organelles are known to accumulate over time, likely contributing to the functional decline experienced during ageing. Here, we discuss the current literature linking autophagy, including selective types of autophagy, to organismal, tissue and cellular ageing. These data have been accumulated from studies of model organisms, including yeasts, worms, flies and mice, showing conservation of autophagy as a molecular mechanism important for longevity, with potential implications for healthspan in humans.
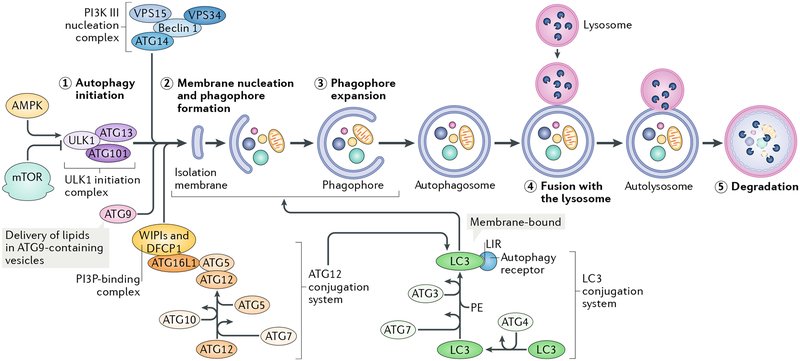
Fig. 1 |. The macroautophagy process.
A schematic depicting the process and main regulatory machinery of macroautophagy (referred to as autophagy) is shown. The conserved metabolic sensors and longevity determinants mTOR and AMP-activated kinase (AMPK) are the main regulators of autophagy, with mTOR acting as an inhibitor and AMPK as an activator. When autophagy is induced, cytoplasmic material (the autophagic cargo) is engulfed by double membranes, starting from the formation of a cup-shaped structure called the phagophore to the sequestration into double-membrane vesicles, called autophagosomes, which subsequently fuse with acidic lysosomes and form autolysosomes, where cargo is degraded. Autophagy is a multistep process that includes (1) initiation, (2) membrane nucleation and phagophore formation, (3) phagophore expansion, (4) fusion with the lysosome, and (5) degradation, which correspondingly are regulated by multiple proteins, referred to as autophagy-related proteins (ATGs). ATGs assemble into several complexes: the Unc-51-like kinase 1 (ULK1; Atg1 in yeasts) initiation complex, the class III PI3K nucleation complex and the phosphatidylinositol 3-phosphate (PI3P)-binding complex, which directs the distribution of the machinery that enables autophagosome formation, and includes the ATG12 and the microtubule-associated protein light chain 3/γ-aminobutyric acid receptor-associated proteins (LC3/GABARAPs; Atg8 in yeasts) conjugation systems (for simplicity, only LC3 is noted in the figure). In the ATG12 conjugation system, ATG12 is attached to ATG5, which is then attached to ATG16L1 (Atg16 in yeasts), followed by dimerization (not shown) and interaction with the PI3P-binding complex (formed by WD repeat domain phosphoinositide-interacting proteins (WIPIs; Atg18 in yeasts) and zinc-finger FYVE domain-containing protein 1 (DFCP1). The ATG12-ATG5-ATG16L1 complex then promotes conjugation of LC3 (or GABARAP), whereby LC3 is cleaved by the protease ATG4 to form LC3-I, which is then conjugated with phosphatidylethanolamine (PE) to form LC3-II. This conjugate is incorporated into pre-autophagosomal and autophagosomal membranes, where LC3 can interact with cargo receptors, which harbour LC3-interacting motifs (LIRs). Membranes for phagophore expansion are delivered, at least in part, by ATG9-containing vesicles. For simplicity, only the names of vertebrate ATGs are shown. VPS15, PI3K regulatory subunit 4 (also known as PIK3R4 in humans); VPS34, phosphatidylinositol 3-kinase catalytic subunit type 3 (also known as PIK3C3 in humans).
Box 1 |. Conserved longevity paradigms linked to autophagy.
Ageing is a complex physiological process characterized by the progressive failure of tissue and cellular functions, ultimately leading to death of the organism. Interestingly, extensive research efforts using model organisms ranging from yeasts to mice have identified a number of genetic pathways and environmental interventions that can delay ageing and thus extend organismal lifespan in a conserved fashion (see the table). These interventions are also referred to as longevity paradigms. the first example of such a longevity paradigm was described in the 1930s, when reduced food intake without malnutrition (called dietary restriction) was shown to extend the lifespan of rats, a treatment shown later to have beneficial effects in several other organisms130. similarly, reducing the activity levels of two major nutrient-sensing pathways, the mTOR131 and insulin/iGF1 (REF.132) signalling pathways, extends lifespan in a number of species, and overexpression of the nutrient sensor AMP-activated protein kinase (AMPK) extends lifespan in worms and flies133. Other interventions also extend lifespan in at least yeasts, worms and flies, including reduced levels of mitochondrial respiration134 and hormetic heat shock135. Lastly, a number of pharmacological interventions extend lifespan in a number of species, for example, the polyamine spermidine136 and the plant phenol resveratrol137. the study of these longevity paradigms has long focused on identifying the underlying molecular mechanisms mediating lifespan extension, including roles for different transcription factors126. A common theme is that all of the above-mentioned conserved longevity paradigms require autophagy-related and lysosomal genes for their lifespan extension in one or more organisms (indicated by * in the table; these links, and reports of lifespan extension by overexpression of autophagy genes, can be found in TABLE 1).
| Longevity paradigm | Organism | Refs |
|---|---|---|
| Genetic longevity paradigms | ||
| Dietary restriction |
|
130 |
| mTOR inhibition (for example, rapamycin) |
|
131 |
| Reduced insulin/IGF1 signalling |
|
132 |
| Increased AMPK activity |
|
133 |
| Reduced mitochondrial respiration |
|
134 |
| Hormetic heat shock |
|
135 |
| Germ-line removal |
|
64 |
| Reduced TGFβ/Activin signalling |
|
12,138 |
| Pharmacological longevity paradigms | ||
| Spermidine |
|
136 |
| Resveratrol |
|
137 |
| Urolithin A |
|
63 |
TGFβ, transforming growth factor-β.
Autophagy in organismal ageing
Different lines of evidence indicate that ageing modulates the autophagy process. Autophagy-reporter analyses and gene expression studies in different species indicate a decline in autophagy over time, whereas genetic experiments carried out in multiple short-lived model organisms to modulate autophagy gene activity indicate that autophagy activation can be used as a strategy to promote longevity, as summarized below.
Autophagy decline in ageing animal models.
Many organisms show signs of decreased autophagic capacity with age. For example, levels of lysosomal protease activity decline with age in the nematode Caenorhabditis elegans9; autophagy gene transcripts decrease with age in tissues of the fruitfly Drosophila melanogaster, including in the brain (Atg2, Atg8a (LC3/GABARAP in mammals), Atg18 (WIPI1 and WIPI2 in mammals) and bchs (ALFY in mammals))10 and in (Atg1 (ULK1 in mammals), Atg5, Atg6 (BECN1 in mammals), Atg7 and Atg8a11–12; protein levels of LC3 and ATG7 decline with age in mouse hypothalamus13 and in mouse and human muscle14; and lysosomal-associated membrane protein type 2a (LAMP2A) as well as chaperone-mediated autophagy decline in rat liver15. Consistent with such changes in the levels of key autophagy components, assays monitoring the autophagy process indicate a decline in autophagic capacity over time in several species. For example, a recent spatiotemporal analysis of autophagy in C. elegans using fluorescently tagged protein LGG-1 (LC3/GABARAP in mammals and Atg8 in yeasts) as a marker of autophagosomes and autolysosomes in combination with autophagy inhibitors (an approach known as a flux assay) shows an age-dependent increase in the number of autophagosomes and autolysosomes in four major tissues (intestine, body-wall muscle, pharyngeal muscle and neurons), with possible tissue-specific kinetic differences still to be determined. This accumulation of autophagic structures likely reflects impaired autophagic activity16. Another recent study similarly reported a reduction in autophagic activity in whole-body extracts of aged C. elegans17. Moreover, electron microscopy analysis of mouse and rat livers shows an accumulation of autophagic vacuoles with age, and chemical alteration of the process indicates that aged animals have a decreased ability to turn over autophagic vesicles18,19. Consistently, proteolysis of long-lived proteins is impaired in the livers of old rats18,20, whereas the lifespan-extending intervention of dietary restriction, that is, reduction in food intake without malnutrition (see BOX 1) prevents this decline 21,22, suggesting an age-dependent decline in autophagic function and lysosomal degradation. Thus, evidence from multiple model organisms shows that autophagy gene expression and protein levels decrease with age, at least in some contexts, causing an accumulation of autophagic structures and possibly limiting autophagic capacity to maintain cellular homeostasis. Further studies of tissue-specific and cell type-specific differences will be required to better understand the exact contribution of autophagy defects in each tissue to systemic ageing.
Genetic links of autophagy to ageing.
Autophagy, and more specifically, requirement of different ATG genes, has been directly linked to ageing via genetic experiments in multiple model organisms (TABLE 1; see also REF23 for additional genetic links between autophagy and specific long-lived C. elegans mutants), showing a broad and critical role for autophagy genes in several conserved longevity paradigms (BOX 1). Indeed, impairment of autophagy genes by RNAi in young adult animals abrogates lifespan extension in all long-lived mutants of any species tested so far (see BOX 1 for longevity paradigms and discussion below for the effects oflater-in-life impairments of autophagy on lifespan) but generally has small or no effects on the lifespan of normal invertebrate animals23–25. The latter observations likely reflect the fact that residual expression of autophagy genes in RNAi approaches is sufficient to support the basal auto-phagy required for fitness of wild-type animals. This is in contrast to the rapid lethality of mice following auto-phagy gene knockout in adulthood26 or the sickly and short-lived invertebrate animals resulting from auto-phagy impairments during development, irrespective of their genetic background. This reflects critical roles for autophagy in adult mammals, particularly as a buffer against neurodegeneration and various infections26, and important developmental roles for autophagy in invertebrates1,27. Notably, where analysed, long-lived invertebrate mutants also display increased steady-state markers of autophagy, indicating increased autophagic activity in these mutants and providing evidence that autophagy activation is generally associated with lifespan extension in various genetic backgrounds. This has been directly assessed by flux assays in long-lived C. elegans with reduced insulin/iGF1 signalling (daf-2 mutants, which carry mutations in the insulin/IGF1-like receptor) and in mutants lacking a germline (glp-1 mutants, which carry mutations in the Notch receptor); these mutants generally show increased autophagic capacity compared with wild-type C. elegans, yet with notable tissue-specific differences16 (see also below). Moreover, several long-lived worms and flies display increased expression of multiple ATG and lysosomal genes (reviewed in REF28). Collectively, these observations suggest a model in which increased autophagic activity has a causal role in promoting lifespan extension in long-lived animals. It should be noted, however, that for a proportion of the studies, especially the work in D. melanogaster, the conclusions are based on knockdown of single autophagy genes.
Table 1 |.
Summary of autophagy genes linked to organismal ageing in model organisms and to age-related disorders in humans
| Gene (mouse orthologue) | Function in autophagy | Association with lifespan determination or age-related diseases in humans | Refs |
|---|---|---|---|
| Saccharomyces cerevisiae | |||
| ATG1(Ulk1) | Autophagy initiation | Required for longevity induced by rapamycin | 142 |
| ATG11 | Autophagosome-vacuole fusion; selective autophagy | Required for longevity induced by rapamycin | 142 |
| ATG7 | El-like enzyme for the Atg5-Atg12 complex and the Atg8 conjugation systems | Required for longevity induced by dietary restriction, rapamycin and spermidine | 110,142,143 |
| ATG5 | Conjugated by Atg12 | Required for longevity induced by dietary restriction by methionine restriction | 143 |
| ATG8 | Phagophore elongation and cargo recruitment | Required for longevity induced by dietary restriction by methionine restriction | 143 |
| VAM3 | SNARE protein involved in vacuolar fusion | Required for longevity induced by dietary restriction | 144 |
| VAM7 | SNARE protein involved in vacuolar fusion | Required for longevity induced by dietary restriction | 144 |
| VMA2 | Vacuolar ATPase subunit | Required for longevity induced by dietary restriction | 123 |
| ATG15 | Putative lipase required for intravacuolar disintegration of autophagic bodies | Required for longevity induced by dietary restriction | 144 |
| Caenorhabditis elegans | |||
| unc-51 (Ulk1) | Autophagy initiation | Required for longevity induced by mTOR inhibition, dietary restriction, germline ablation and reduced mitochondrial respiration | 72,145 |
| bec-1(Becn1) | Allosteric regulator of VPS-34 | Required for longevity induced by mTOR inhibition, dietary restriction, germline ablation, reduced mitochondrial respiration, spermidine, resveratrol, urolithin A and other paradigmsa | 31,63,72,110,146–149 |
| vps-34 | Kinase that produces PI3P to enable recruitment of machinery that forms autophagosomes | Required for longevity induced by mTOR inhibition, dietary restriction, germline ablation, reduced mitochondrial respiration and urolithin A | 63,72,148,150 |
| atg-9 | Phagophore formation | Other paradigmsa | 151 |
| atg-18 (Wipl) | Phagophore formation | Required for longevity induced by inhibition of insulin/IGF1 signalling (M), dietary restriction (I, M and possibly N), germline ablation (I), reduced mitochondrial respiration, AMPK overexpressionc, inhibition of a downstream effector of the mTOR pathway, S6K and other paradigmsa (but overexpression from the endogenous promoter does not extend lifespan) | 16,72,94,97,152 |
| lgg-3 (Atg12) | Ubiquitin-like modifier of ATG-5 | Required for longevity induced by inhibition of insu1in/IGF1 signalling, dietary restriction and other paradigmsa | 147 |
| atg-7 | El-like enzyme for the ATG-5-ATG-12 complex and the ATG-8 conjugation systems | Required for longevity induced by inhibition of insu1in/IGF1 signalling | 147,150 |
| lgg-1 (Lc3/Gabarap family genes) | Phagophore elongation and cargo recruitment | Required for longevity induced by inhibition of insu1in/IGF1 signalling (M), germline ablation, mitochondrial respiration, AMPK overexpressionc and other paradigmsa (but overexpression from the endogenous promoter does not extend lifespan) | 72,148 |
| atg-4.1 | ATG-8 processing to make it conjugation- competent, and ATG-8 delipidation | Other paradigmsa | 151 |
| vha-16 | Subunit of vacuolar proton-translocating ATPase | Required for longevity by germline ablation | 31 |
| C08H9.1 | Lysosomal degradation | Required for longevity induced by inhibition of insu1in/IGF1 signalling | 153 |
| lipl-1 | Lysosomal lipolysis | Overexpression from the endogenous promoter extends lifespan | 74 |
| lipl-3 | Lysosomal lipolysis | Overexpression from the endogenous promoter extends lifespan | 74 |
| lipl-4 (Hla-l) | Lysosomal lipolysis |
|
71 |
| dct-1 (Bnip3L) | Mitochondrial receptor protein | Required for longevity induced by inhibition of insu1in/IGF1 signalling, dietary restriction, mitochondrial dysfunction, urolithin A and other paradigmsa | 45,60,63 |
| pink-1 | Kinase that enables mitophagy | Required for longevity induced by inhibition of insulin/IGF1 signalling, dietary restriction, mitochondrial dysfunction, urolithin A and other paradigmsa | 45,60,63 |
| sqst-1 (Sqstml) | Receptor protein | Required for longevity induced by mitochondrial dysfunction, urolithin A and other paradigmsa | 60,63 |
| hlh-30 (Tfeb) | Transcription factor regulating lysosomal biogenesis and autophagy |
|
31,152 |
| Drosophila melanogaster | |||
| Atg1 (Ulk1) | Autophagy initiation | Required for longevity induced by AMPK overexpression (N) (overexpression from a neuronal-specific promoter during adulthood extends lifespan); also required for longevity induced by overexpression of the mitochondrial protein dynamin-related protein 1 (Drp1) | 24,25 |
| Atg7 | E1-like enzyme for the Atg5-Atg12 complex and the Atg8 conjugation systems | Required for longevity induced by spermidine | 110 |
| Atg5 | Conjugated by Atg12 | Required for longevity induced by rapamycin | 154 |
| Atg8a (Lc3/Gabarap family genes) | Phagophore elongation and cargo recruitment | Overexpression from a neuronal-specific and a muscle-specific promoter extends lifespanb | 10,12 |
| Parkin | E3 ubiquitin ligase that facilitates mitophagy | Overexpression from ubiquitous and neuronal-specific promoters during adulthood extends lifespan | 61 |
| Drp1 | Dynamin-related protein that promotes mitochondrial fission and facilitates mitophagy | Overexpression from ubiquitous, intestine-specific and neuronal-specific promoters in midlife extends lifespan in an autophagy-dependent fashion | 25 |
| Mus musculusd | |||
| Atg7 | E1-like enzyme for the ATG5-ATG12 complex and the ATG8 conjugation systems | Depletion in the muscle impairs muscle function and shortens lifespan | 14 |
| Atg5 | Conjugated by ATG12 | Overexpression from a ubiquitous promoter extends lifespan | 29 |
| Human | |||
| BECN1 | Allosteric regulator of VPS34 | Variants have been associated with breast cancer prognosis | 155 |
| WDR45 | Phagophore formation | Mutations cause neurodegeneration with brain iron accumulation | 156,157 |
| ATG7 | E1-like enzyme for the ATG5-ATG12 complex and the ATG8 conjugation systems | Variants have been proposed to impact age at onset of Huntington disease | 158 |
| ATG5 | Conjugated by ATG12 | Mutations cause ataxia and developmental delay | 35 |
| ATG16L1 | LC3 lipidation | Mutation T300A increases risk of Crohn’s disease | 159,160 |
| TECPR2 | Interacts with LC3 | Mutations cause spastic paraparesis | 33 |
| ZFYVE26 | Autophagosome maturation | Mutations cause spastic paraplegia | 34 |
| EPG5 | Autophagosome-lysosome fusion | Mutations cause Vici syndrome | 161 |
| PRKN | E3 ubiquitin ligase that facilitates mitophagy | Mutations cause autosomal recessive Parkinson disease | 36 |
| PINK1 | Kinase that facilitates mitophagy | Mutations cause autosomal recessive Parkinson disease | 37 |
| SQSTM1 | Receptor protein | Mutations cause Paget disease of bone and motor neuron disease | 38,39 |
| TBK1 | Kinase that phosphorylates autophagy receptors | Mutations cause motor neuron disease | 40 |
The table summarizes autophagy-related (ATG) and lysosomal genes and their role in conserved longevity paradigms (in yeasts, worms, flies and mice) or in age-related diseases (in humans). AMPK, AMP-activated kinase; GABARAP, γ-aminobutyric acid receptor-associated protein; E1-like, ubiquitin-activating enzyme-like; I, intestinal-specific RNAi in adult animals; LC3, microtubule-associated protein light chain 3; M, muscle-specific RNAi in adult animals; N, neuronal-specific RNAi in adult animals; PI3P, phosphatidylinositol 3-phosphate; PRKN, Parkin; RSK, ribosomal protein S6 kinase; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; Sqstm1, sequestosome-1; VPS34, phosphatidylinositol 3-kinase catalytic subunit type 3.
Additional longevity paradigms, for example, calcineurin, frataxin and miR-34 depletion, require this autophagy gene in C. elegans (see REF.23 for additional links).
Moreover, reduced Activin signalling requires Atg8a is required for longevity associated with the loss of Activin signalling in muscle12.
Unpublished observations from M.H.’s laboratory.
See note added in proof and REF.162.
In further support of a direct role for autophagy genes in lifespan determination, overexpression of specific autophagy genes can extend lifespan in several species (TABLE 1). For example, overexpression of fly Atg8a in the nervous system10 or in the muscle12 is sufficient to extend fly lifespan. Similarly, neuron-specific overexpression of Atg1 in flies24, and ubiquitous overexpression of Atg5 in mice, is sufficient to stimulate autophagy, improve motor function and extend lifespan29. While all these lifespan extending manipulations are accompanied by increases in autophagy markers and improved healthspan parameters (see section on tissue-specific roles for auto-phagy below), it remains to be formally tested whether the observed longevity requires the autophagy process in all cases. In this regard, it is noteworthy that overexpression of the helix-loop-helix transcription factor hlh-30 (TFEB in mammals), a conserved regulator of many ATG and lysosomal genes28,30, extends lifespan in C. elegans in an autophagy-dependent fashion31. Collectively, these observations indicate, but do not prove, that upregulation of autophagy may be an effective approach to delay ageing and promote healthspan in diverse species including mammals.
Importantly, genetic and age-related loss of auto-phagic and lysosomal function has also been linked to the development of several age-related diseases, including neurodegenerative diseases and cancer (TABLE 1). For example, loss-of-function mutations of several ATG genes (for example, BECN1 (ATG6 in yeasts), ATG5 and ATG7) result in decreased autophagy along with accumulation of dysfunctional organelles and disordered and aggregated proteins in mammalian models of neurodegenerative disorders, including Huntington disease (Huntingtin (HTT) accumulation), Alzheimer disease (amyloid-β (Aβ) and Tau accumulation) and Parkinson disease (α-synuclein accumulation) (reviewed in REF 32). Importantly, Mendelian mutations in auto-phagy regulators can cause neurodegenerative diseases, including spastic paraplegia33,34 and ataxia35, and loss of activity of autophagy receptors as well as other regulators of selective autophagy (see also next section) can cause Parkinson disease36,37 or other forms of motor neuron disease38–40. Overall, accumulating evidence supports a beneficial role for autophagy in counteracting ageing and age-related diseases, although the underlying mechanisms are not fully understood.
Selective autophagy in ageing
While the above genetic links indicate involvement of the bulk autophagy in ageing processes, there is now evidence that the impairment of the turnover of specific cargoes via selective autophagy might have important roles in age-related pathologies and in ageing. Below, we discuss studies implicating the different types of selective autophagy in ageing and lifespan determination (FIG. 2).
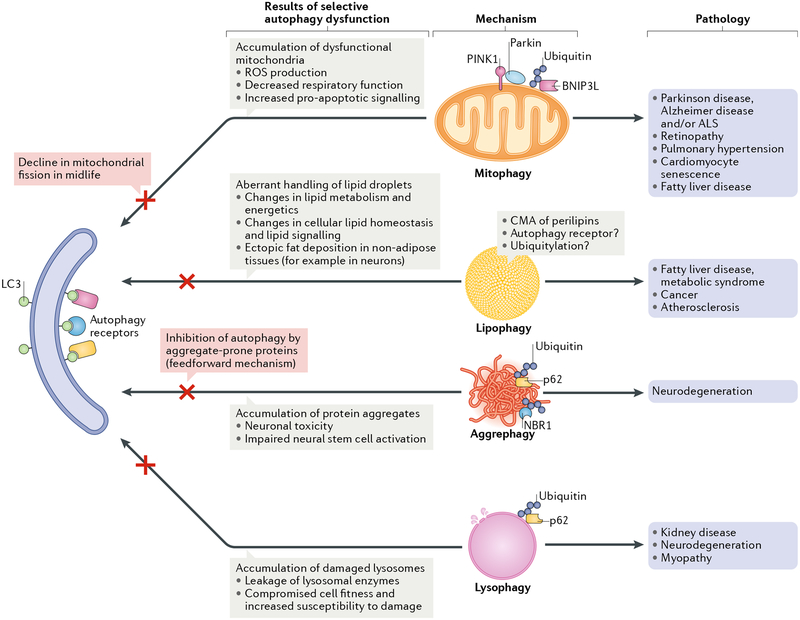
Fig. 2 |. Selective types of autophagy linked to organismal ageing.
A schematic summarizing selective types of autophagy linked to pathologies of ageing in model organisms. In these selective types of autophagy, autophagosomes recruit mitochondria (mitophagy), lipid droplets (lipophagy), aggregate-prone proteins (aggrephagy) and lysosomes (lysophagy). This is generally mediated by so-called autophagy receptors that bridge the cargo and the autophagy machinery (some examples of autophagy receptors that have been indicated to function in the context of ageing are depicted, but most likely other receptors are involved). Consequences of deficiencies in these types of selective autophagy and their links to age-related diseases are listed. Note that while this figure illustrates possible links between forms of selective autophagy and diseases, it is very challenging to demonstrate causality for the selective autophagy in disease in a direct sense, as opposed to links or associations. For example, PTEN-induced putative protein kinase 1 (PINK1), which is mutated in a rare form of recessive Parkinsonism, has been implicated in stress-induced mitophagy in tissue-culture models, leading to the assumption that loss of PINK1 causes disease via defects in mitophagy. However, recent work suggests that loss of PINK1 in mice does not affect mitophagy, thus challenging the model141. ALS, amyotrophic lateral sclerosis; BNIP3L, BCL-2/adenovirus E1B 19 kDa protein-interacting protein 3-Like; CMA, chaperone-mediated autophagy; LC3, microtubule-associated protein light chain 3; NBR1, next to BRCA1 gene 1 protein; ROS, reactive oxygen species.
Mitophagy.
The accumulation of dysfunctional mitochondria is a shared hallmark of ageing and numerous diseases of old age41–44. Although the underlying mechanisms that lead to age-related loss of mitochondrial function remain incompletely understood and may involve numerous processes, it has been suggested that a decline in mitophagy has a key role43–45. In mammals, the degradation of damaged mitochondria is mediated by a pathway comprising PTEN-induced putative protein kinase 1 (PINK1) and the E3 ubiquitin-protein ligase Parkin. In recent years, the molecular mechanisms of mitophagy have been elucidated in some detail from studies in mammalian cell culture and genetic studies in model organisms46–48. Disruptions in mitophagy have been implicated in the pathophysiology of age-related diseases such as heart disease49, retinopathy50, fatty liver disease51, pulmonary hypertension52, kidney disease53 and neurodegenerative disorders, including Parkinson disease, amyotrophic lateral sclerosis54 and Alzheimer disease46,55 (FIG. 2). However, as with all forms of selective autophagy described below, it is very challenging to demonstrate causality for the selective autophagy in human disease in a direct sense, as opposed to links or associations.
Studies in worms45, flies25, mice and humans56,57 have reported a decline in mitophagy markers in aged animals. This decline may be relevant to age-related pathologies, as loss of Pink1 or Parkin leads to early-onset behavioural decline and shortened lifespan in flies58,59. Two recent studies in C. elegans have investigated the importance of mitophagy in longevity assurance45,60. The gene dct-1 is a putative orthologue to the mammalian proteins BNIP3 and BNIP3-like (BNIP3L), which act as mitophagy receptors in mammals47. Inhibition of dct-1 leads to an increase in mitochondrial content, indicating that DAF-16/FOXO controlled germline tumour affecting-1 (DCT-1) is the nematode orthologue of BNIP3L and functions as a key regulator of mitophagy45. Moreover, inhibition of dct-1 or pink-1 shortens the lifespan of long-lived daf-2 mutants and eat-2 mutants (which recapitulate experimental dietary restriction paradigms45) and in several long-lived C. elegans models of moderate mitochondrial dysfunction (where mild mitochondrial stress signalling evokes beneficial effects on longevity)45,60 (TABLE 1). Collectively, these studies indicate that mitophagy indeed has a causal role in lifespan extension, at least in long-lived C. elegans models.
A number of studies have examined the impact of increased mitophagy on ageing and lifespan. Critically, ubiquitous or neuron-specific, adult-onset upregulation of Parkin extends the lifespan of D. melanogaster61 (TABLE 1). Moreover, it was recently reported that a midlife shift towards a more elongated mitochondrial morphology is linked to impaired mitophagy and the accumulation of dysfunctional mitochondria in the flight muscle of aged D. melanogaster25. Promoting dynamin-related protein 1 (Drp1)-mediated mitochondrial fission in midlife restores mitochondrial morphology to a youthful state, facilitates mitophagy and improves mitochondrial respiratory function. Importantly, transient, midlife induction of Drp1 improves markers of organismal health, delays age-onset gut pathology and prolongs fly lifespan in an Atg1-dependent fashion (TABLE 1). Furthermore, upregulating Drp1 specifically in neurons or the intestine (see also section below on tissue-specific effects of autophagy induction), from midlife onwards, is sufficient to prolong fly lifespan25. These findings indicate that a midlife decline in mitophagy, resulting at least in part from a shift in mitochondrial dynamics, contributes to ageing-related onset of mitochondrial dysfunction and limits lifespan, at least in D. melanogaster.
Given the findings above, it has been proposed that pharmacological interventions that stimulate mitophagy may prove effective in delaying the health decline associated with ageing62. Consistent with this model, dietary treatment of C. elegans with the human microflora-metabolite urolithin A induces mitophagy and prolongs worm lifespan63. More specifically, short-term urolithin A treatment in worms induces mitochondrial fragmentation and reduces mitochondrial content in an autophagy-dependent fashion. Urolithin A treatment improves a number of markers of C. elegans healthspan and maintains respiratory capacity of mitochondria during ageing. These lifespan-extending effects of urolithin A treatment require the mitophagy genes pink-1 and dct-1 (TABLE 1). Importantly, urolithin A treatment is also beneficial in rodents, where it was shown to improve exercise capacity in two different mouse models of age-related decline of muscle function, as well as in young rats63, overall suggesting conserved beneficial effects of inducing mitophagy for organismal fitness.
Lipophagy.
Studies in diverse organisms have suggested that specific alterations in lipid metabolism are associated with different pro-longevity interventions64. In recent years, the contribution of autophagy to intracellular lipid droplet degradation, referred to as lipophagy, has been identified65. The first clear demonstration that lipid droplets could be turned over via autophagy came from studies in cultured hepatocytes with reduced ATG5 levels66 (we note that chaperone-mediated autophagy has also been linked to lipid metabolism; reviewed in REF67). The fact that autophagy can regulate lipid metabolism expands the physiological relevance of autophagy to modulate the cellular energetic balance directly. Furthermore, alterations in lipophagy could impact cell physiology indirectly via alterations in the regulatory activities that lipids exert inside cells. As a result, it has been proposed that alterations in lipophagy may underlie the metabolic syndrome of ageing68 (FiG. 2), which comprises a constellation of features including obesity, dysregulated lipoprotein metabolism, abnormal glucose handling and high blood pressure. Lipophagy has also been linked to cancer69 and atherosclerosis70.
Recent studies in C. elegans have provided further evidence for the role of lipophagy in longevity. Specifically, lifespan extension in germline-less glp-1 mutants requires both the lysosomal lipase LIPase-like 4 (LIPL-4) (REF 71) and autophagy genes72, whereby autophagy-dependent lypolysis and LIPL-4-dependent lipolysis promote longevity independently72 (TABLE 1). Furthermore, increased lysosomal lipolysis has been directly linked to lifespan extension in worms71,73,74. Although the molecular mechanisms involved are not fully understood, overexpression of lipl-4 has been shown to induce nuclear translocation of a lysosomal lipid chaperone lipid-binding protein 8 (LBP-8), consequently promoting longevity by activating nuclear hormone receptor 49 (NHR-49) and NHR-80 (REF.75). Of further note, aged worms display a deposition of lipids in non-adipose tissues, including the nervous system76. Interestingly, dietary restriction, an intervention that promotes longevity, reduces this ectopic fat accumulation, whereas inhibition of ATG genes, namely, lgg-1 (LC3/GABARAP family in mammals, ATG8 in yeasts) and hlh-30 (homologue of mammalian TFEB), increases fat accumulation76, which is consistent with a role for aberrant lipophagy in ectopic fat deposition in C. elegans.
Aggrephagy.
Aggrephagy describes the selective recruitment of protein aggregates, or possibly oligomeric forms of proteins that are destined to form aggregates, to autolysosomes. Aggregation-prone proteins, including tau, α-synuclein and mutant HTT, accumulate as aggregates in ageing neurons, which is thought to contribute to toxicity in neurodegenerative diseases such as Alzheimer disease, Parkinson disease and Huntington disease (FiG. 2). These proteins are autophagy substrates77–79, and autophagy upregulation by chemical80, genetic81 or environmental means (by hormetic heat shock82) can ameliorate signs of these diseases in a wide range of animal models including C. elegans, D. melanogaster, zebrafish and mice (reviewed in REF.83). Furthermore, α-synuclein84 and many mutant polyglutamine-expanded proteins, like mutant HTT85, can inhibit autophagy. This could potentially introduce a feedforward loop supporting disease pathogenesis. Accordingly, inhibition of auto-phagy accelerates neurodegenerative disease by accumulating disease-causing aggregate-prone proteins, like mutant HTT and α-synuclein115. Thus, any age-dependent decrease in autophagy in the brain will have a major impact on protein aggregation in these mutant backgrounds. Indeed, age is a major risk factor in most neurodegenerative diseases.
Lysophagy.
The selective degradation of damaged lysosomes (lysophagy) appears to be an important mechanism, which has been suggested to shield the cytoplasmic contents from leakage of lysosomal hydrolases86, and lysophagy has been shown to be protective against acute kidney injury in mice86 (FiG. 2). It is interesting to speculate that any age-dependent loss of autophagy may reduce this mechanism of cellular protection against lysosomal enzyme leakage into the cytoplasm and would thus predispose to kidney damage and chronic renal failure, two age-related conditions. While compromised autophagy increases kidney damage with age in mice87, it is interesting that basal auto-phagy is increased in kidney proximal tubules in older versus younger mice87. However, starvation-induced autophagy in kidney proximal tubules is blunted in aged mice87. Interestingly, autophagy appears to be less active in podocytes (the most damage-prone cells in the glomerulus)88 than in cells in the proximal tubule, but no age-dependent change in podocyte autophagy was observed87. Thus, in this case, autophagic capacity may correlate inversely with cell type-specific vulnerability to damage in the kidneys and may suggest that autophagy is a protective process in this organ. While these differences in susceptibility to damage may be mediated in part by differential capacity for lysophagy, it is likely that altered clearance of other autophagy substrates may contribute, including removal of damaged mitochondria. The links of lysophagy to physiology and disease are still largely limited to studies in the kidney, likely owing to recent characterization of lysophagy, although the most recent studies have also proposed links between decreased lysophagy and muscle disease as well as neurodegeneration89. However, it is possible that efficient clearance of dysfunctional lysosomes may have much broader importance for determining cellular fitness that is progressively compromised upon accumulation of defects in autophagic clearance during ageing and that aberrations in lysophagy may be relevant to many other organ systems.
Tissue-specific autophagy and ageing
While ageing is linked to a decline in physiological functions at both the tissue and the organismal level, it remains unclear how ageing of individual tissues may limit the lifespan of the organism. Thus, it is of interest to understand tissue-specific roles for autophagy in ageing, including selective types of autophagy in individual tissues and cell types of model organisms (TABLE 2), as reviewed below.
Table 2 |.
Tissue-specific functions of autophagy in ageing
| Intestine | Muscle | Nervous system |
|---|---|---|
| Links between autophagy genes and ageing have been demonstrateda | ||
| Yes (worms and flies) | Yes (worms, flies and mice) | Yes (worms and flies) |
| Functions of autophagy in the tissue related to ageing | ||
| Maintenance of intestinal barrier function (worms and flies)24,94 | Supporting learning and memory processes (flies)111,112 | |
| Longevity paradigms in which tissue-specific autophagy has a role | ||
| Tissue-specific mechanisms of autophagy linked to ageing | ||
| Maintenance of tight junctions and regulation of tight junction permeability (human intestinal cell line)103 | ||
| Inter-tissue communication and systemic effects of autophagy on ageing | ||
| Communication to the intestine: improved intestinal barrier function (worms and potentially also flies)24,94 | ||
| Potential mechanisms supporting non-tissue autonomous effects of autophagy on ageing | ||
| Not known | Decrease in insulin-like peptide secretion and reduced systemic insulin-like peptide signalling (flies)11,12 | Sustained neurotransmitter release from chemosensory neurons (worms)97 and reduced levels of neuropeptides and/or insulin-like peptides (worms97 and flies24) |
Atg1, autophagy-related gene 1 (ULK1 in mammals); Atg8a, autophagy-related 8a; Drp1, dynamin-related protein 1; FOXO, forkhead box protein.
See TABLE 1.
Intestine.
Intestinal barrier dysfunction, whereby the ability to prevent the passage of harmful intraluminal entities is compromised, is a common feature of ageing organisms and has been linked to a number of human diseases90. In D. melanogaster, ageing-related intestinal barrier dysfunction is linked to microbial dysbiosis, increased immune gene expression, loss of motor activity and systemic metabolic defects and is a harbinger of mortality91,92. Together with data showing that the intestine represents a critical target organ for genetic interventions that prolong lifespan93, these findings support the idea that maintaining intestinal integrity during ageing is critical for organismal health and viability. Dietary restriction delays the onset of intestinal barrier dysfunction in both C. elegans94 and D. melanogaster92,95; likewise, short-term protein restriction has recently been linked to improved intestinal barrier function in adult pigs96. Although direct tests are needed, improved intestinal barrier function may be a phenotype shared by multiple conserved longevity paradigms. In support of this, long-lived C. elegans daf-2 mutants also have improved intestinal barrier function94.
Intestine-specific expression of autophagy genes has been shown to be critical for lifespan extension observed in several longevity paradigms (TABLES 1,2), including dietary restriction in C. elegans94. Indeed, various reporters of autophagic activity and flux analyses indicate that autophagy is induced in the intestine of long-lived eat-2 mutants. Furthermore, intestine-specific RNAi of two LC3/GABARAP family homologues, lgg-1 and lgg-2 or of the WIPI homologue atg-18 significantly decreases the extended lifespan of these mutant worms. Consistent with a role for intestinal autophagy in longevity mediated by dietary restriction, atg-18 mutants do not display lifespan extension upon food dilution (a direct dietary restriction protocol), and intestine-specific expression of atg-18 in these mutants restores dietary restriction-mediated lifespan extension97. Moreover, either whole-body or intestine-specific RNAi of autophagy genes abrogates the improvements in age-related intestinal barrier function in eat-2 mutants94. Collectively, these findings suggest that autophagy induction in the intestine of animals subjected to dietary restriction can act to maintain intestinal barrier function during ageing and that this is important for lifespan extension. Even though dietary restriction has been reported to increase autophagy markers in multiple tissues and organs of mice98–100, it remains unknown whether modulation of autophagy, systemically or in specific organ systems, such as the intestine, has a causal role in dietary restriction-mediated lifespan extension in mammals. How may intestinal autophagy, induced by, for example, dietary restriction, improve intestinal barrier function? In D. melanogaster, intestines of aged animals are characterized by altered expression and localization of septate junction proteins, which may contribute to the impairment of epithelial integrity and increased tissue permeability and consequently may be linked to ageing-related intestinal barrier dysfunction101. Although a direct role for autophagy impairment has not yet been shown to promote intestinal barrier dysfunction in organisms other than C. elegans, defects in ATG proteins have been linked to the pathogenesis of Crohn’s disease, which is characterized by intestinal barrier dysfunction in humans102. Moreover, it has been shown that autophagy selectively reduces epithelial tight junction permeability by lysosomal degradation of the tight junction protein claudin 2 (REF.103). Therefore, it is possible that investigating the interplay between autophagy, epithelial junction proteins and ageing may provide novel therapeutic approaches to maintain intestinal health during ageing.
Additional links exist between the intestine, auto-phagy and longevity. For example, flux analysis in germline-lessglp-1 C. elegans mutants indicates induced autophagy in the intestine of these animals, and RNAi-mediated knockdown of atg-18 in the intestine of adult animals abrogates the lifespan extension observed in glp-1 mutants16 (TABLE 1). Curiously, the same RNAi treatment did not significantly shorten the lifespan of C. elegans daf-2 mutants, indicating that intestinal autophagy may not be contributing to the lifespan extension resulting from reduced insulin/IGF1 signalling16. However, in another recent study it was found that intestinal reintroduction of ATG-18 into daf-2 and atg-18 double mutants extended lifespan of these animals to match that of daf-2 mutants97. Notably, this latter study does not discriminate between the role of AT G-18 during development versus during ageing, and more experiments are needed to fully address the role of intestinal autophagy in daf-2 mutants. Consistent with these observations linking intestinal autophagy to longevity in C. elegans, intestinal overexpression of AMPKalpha (AMPK catalytic subunit, hereafter referred to as AMPK) in D. melanogaster induces markers of autophagy and autophagy gene expression in the intestine and extends fly lifespan24, collectively indicating an important role for intestinal autophagy in lifespan determination of flies.
Interestingly, intestinal overexpression of AMPK promotes autophagy not only in the intestine itself but also in the fly brain24. Similarly, neuronal overexpression of Atg1, or of AMPK (see also next subsection), is associated with increased autophagy markers and autophagy gene expression in the fly intestine24. Importantly, neuronal Atg1 overexpression also improves intestinal barrier function, indicating that autophagy can act non-cell autonomously and even across tissues to promote longevity. However, mechanisms of inter-tissue pro-longevity effects remain to be elucidated. One observation is that neuronal Atg1 overexpression is associated with reduced insulin-like peptide levels in the brain24. It is also worth considering that ablation of the insulin-like peptide-producing median neurosecretory cells in the brain lowers systemic insulin signalling and can prolong fly lifespan104. Therefore, it is possible that the pro-longevity effects of autophagy stimulation in neurons may involve decreased insulin-like peptide signalling.
It is interesting to speculate as to whether induction of intestinal or neuronal autophagy can impact systemic autophagy levels to prolong lifespan, for example, upon dietary restriction. Inhibition of autophagy genes in the intestine of worms considerably impairs organismal motility, which is presumed to be a marker of neuro-muscular function, in C. elegans eat-2 mutants94. However, the question of whether intestine-specific inhibition of autophagy genes, under dietary restriction, impacts autophagy in neurons or muscle remains unanswered. Collectively, these studies indicate an important role for autophagy in the intestines of multiple organisms; it will therefore be interesting to investigate the requirement for autophagy in the intestines of ageing mammals, including the role of autophagy in intestinal integrity.
Nervous system.
Several studies have implicated the nervous system in modulating lifespan, yet the cellular mechanisms involved are not well understood105,106. There are a number of suggestions that autophagic activity is compromised with age in the brains of different species. As noted above, autophagy is decreased in mouse hypothalamic neurons13,107, and the mRNA levels of a number of ATG genes are decreased in aged human brains108. However, further work is required to more rigorously test the hypothesis that autophagic activity may decline in an age-dependent fashion in the nervous system.
Neuronal autophagy has been linked to lifespan in several studies in flies, in which overexpression of single autophagy genes has been shown to promote longevity (TABLES 1,2). Specifically, pan-neuronal over-expression of Atg8a throughout life extends the lifespan of D. melanogaster and improves neuronal proteostasis and organismal response to oxidative stress10. Likewise, adult-onset, pan-neuronal overexpression of Atg1 extends fly lifespan24. As noted above, such adult-onset induction of Atg1 or AMPK in the fly nervous system prevents intestinal barrier dysfunction during ageing, and both the cell-autonomous and the non-cell autonomous effects of Atg1 and AMPK overexpression on the intestine are linked to an increase in autophagy markers and ATG gene expression in the intestinal epithelium24.
In C. elegans, restoring expression of atg-18 in neurons of atg-18;daf-2 double mutants fully rescues the short lifespan of these animals97. Moreover, expression of atg-18 exclusively in chemosensory neurons is sufficient to unleash the beneficial effects of reduced insulin/IGF1 signalling in daf-2 mutants. Interestingly, in the same study, it was reported that atg-18 expression in chemosensory neurons alone does not rescue the shortened lifespan of atg-18 only mutants97. This suggests that atg-18 expression in chemosensory neurons is important for lifespan extension in response to metabolic alterations but is not sufficient to support normal animal lifespan. Although it was shown that extended lifespan resulting from the rescue of atg-18 expression in chemosensory neurons in atg-18;daf-2 mutants depends genetically on the release of neurotransmitters from these neurons, the underlying physiological mechanisms for how rescuing autophagy in only a subpopulation of neurons can impact the lifespan of the entire organism remain to be investigated.
How may autophagy contribute to brain function during ageing? An age-related decline in memory formation has been reported in both model organisms and humans109, yet the underlying mechanisms are not well understood. Interestingly, recent studies have linked autophagy to cognitive functions in D. melanogaster treated with polyamines such as spermidine and putrescine. These compounds promote lifespan in diverse species by augmenting autophagy110 (TABLE 1). Notably, dietary spermidine suppresses age-induced memory impairment in an autophagy-dependent manner in flies111. Most recently, it has been reported that spermidine counteracts age-related alterations in the size and function of a specific synaptic compartment, the presynaptic active zone, to maintain memory in aged flies112 (TABLE 2). Together, these findings support a model in which an age-dependent decline in autophagy contributes to cognitive ageing. Autophagy may impact many processes in the central nervous system and other tissues that contribute to ageing, including degradation of aggregate-prone cytoplasmic proteins (aggrephagy) and dysfunctional mitochondria (mitophagy), as discussed in detail above.
Muscle.
Recent work in mammals and flies indicates that maintaining muscle integrity and function is critical for counteracting systemic ageing and for lifespan determination113. Although the mechanisms involved in muscle-mediated modulation of systemic ageing are not fully understood, emerging evidence suggests that muscle-derived growth factors and cytokines, known as myokines, can modulate systemic physiology113. Interestingly, of the four major tissues examined in C. elegans, the greatest change in autophagy markers occurs in the body-wall muscle16, potentially reflecting muscle as a tissue with especially high autophagic activity.
Muscle-specific autophagy has been linked to longevity in studies in C. elegans and in D. melanogaster. In worms, inhibition of lgg-1 and atg-18 in the body-wall muscle of adult animals is sufficient to shorten the lifespan of both eat-2 mutants94 and daf-2 mutants16. In turn, overexpression of Atg8a in fly muscle increases lifespan12 (TABLE 1). Furthermore, in D. melanogaster, overexpression of the transcription factor forkhead box, subgroup O (FOXO) maintains proteostasis during muscle ageing, at least in part by promoting autophagy, which is linked to extended lifespan11,12.
In mice, muscle-specific ATG7 deficiency causes impaired muscle function and reduced lifespan14. While interpreting lifespan-shortening interventions can prove challenging114, the latter study suggests that loss of muscle function during ageing, owing to impaired autophagy, limits lifespan in mice.
How could autophagy be important for muscle function? The muscle of animals is critical for mobility, which declines with age owing to sarcopenia, or age-related muscle loss115. A recent study in mice lacking Atg7 in the muscle showed that autophagy has a critical role in maintaining the neuromuscular junction and muscle strength, at least in part by increasing the number and improving the function of mitochondria, which are highly abundant at neuromuscular junctions14 (TABLE 2). Indeed, overexpression of Atg7 in muscle prevents age-associated myofibre degeneration14.
Autophagy may also have important roles in specialized cells of the muscle. Mammalian muscle contains muscle stem cells, also referred to as satellite cells. Satellite cells are usually present in a quiescent state but require autophagy to become activated, resulting in their proliferation and differentiation into muscle fibres. Autophagy likely provides nutrients for this metabolically demanding event116,117. Of note, the ability to activate satellite cells declines with ageing, and impaired autophagy was recently shown to have a causal role in this phenotype. Specifically, autophagy is used to maintain stemness of satellite cells by preventing cellular senescence, likely via mechanisms that at least in part relate to mitochondrial maintenance118 (TABLE 2). Notably, induction of autophagy by the mTOR inhibitor rapamycin can reverse senescence and restore regenerative functions of both aged murine and human satellite cells118. In conclusion, autophagy has important protective roles in the ageing muscle, and it will be interesting to investigate whether boosting autophagy can alleviate sarcopenia and improve mobility in aged animals.
The blood and the immune system.
One aspect of blood ageing is related to the decline in immunity. Immune cell senescence that progressively occurs during ageing is associated with decreased immune surveillance and is a risk factor for numerous ageing-related diseases, including cancer. Recent studies have revealed that autophagy-deficient immune cells show numerous ageing phenotypes and that autophagy-inducing agents can improve the immune responses in elderly people119. Hence, autophagy has emerged as a novel target to treat late-onset diseases associated with immune cell senescence. To this point, it is interesting to note that autophagic capabilities appear to be better maintained in immune cells of exceptionally long-lived humans than in the cells of those with an average lifespan120. At present, however, it is not known whether it is possible to induce autophagy specifically in immune cells and improve immune function.
Another blood compartment that displays changes in autophagy over time is that of the haematopoietic stem cells (HSCs). These stem cells differentiate into multiple types of blood cells in vertebrates throughout their lives but progressively lose this regenerative capacity over time. Recent studies have implicated autophagy as a key pathway in homeostasis of the blood system121. Further, it has been reported that autophagy is essential for maintaining the quiescence of HSCs throughout life, as it restricts their proliferation and prevents premature exhaustion by limiting the number of active mitochondria122. It remains to be tested whether the autophagy processes in the immune system and HSCs are directly linked to organismal lifespan.
Conclusions and perspectives
Evidence has been mounting over the past decade that autophagy and ageing are closely linked. In particular, work in model organisms ranging from yeasts to mice has shown that multiple ATG genes are required for the long lifespan induced by conserved longevity paradigms. Combined with gene expression data and autophagy-marker analyses generally indicating that long-lived animals also have increased levels of autophagy, these observations indicate that increased autophagy contributes to extended lifespan. In turn, autophagy appears to become limited with normal ageing, possibly occurring in a tissue-specific fashion and involving selective types of autophagy.
Why would autophagy decline with age in most tissues, and what could be the mechanism for this decline? One general mechanism could involve alterations in the activity of key regulators of autophagy, such as the nutrient sensor mTOR. The activity of mTOR, which negatively regulates autophagy, has been reported to increase over time in at least some tissues of mice, but with some notable exceptions122,123. In tissues where mTOR activity may be increased with age, it remains to be investigated which mTOR-controlled steps of autophagy, prominently including phosphorylation of ULK1 and regulation of TFEB, are altered with ageing and whether these alterations are directly linked to mTOR upregulation. Lastly, it will be important to address what the consequences of these changes in autophagy are and what exact step(s) of the autophagy process may ultimately become limiting for lifespan. One possibility could be impairment of lysosomal acidification, as observed in yeasts124. Consistent with this idea, the activity of several lysosomal proteases decrease with age in C. elegans9. Another possibility might involve age-dependent impairment of autophagic vesicle transport, as observed in neurons125. Regardless of the mechanism, stalled autophagy could become detrimental to cells, as certain types of cellular components might accumulate to potentially toxic levels and possibly in a tissue-specific fashion. As an example, accumulation of autophagosomes or the autophagic machinery has been observed in aged C. elegans16,17 and in livers from aged mice18. It is an important objective for future research to understand the regulation of age-associated changes in autophagic activity. Because autophagy is tightly linked to another major proteostatic process, the ubiquitin-proteasomal system, it will likely also be important to understand how the coordination of these two systems changes over time.
As noted above, mounting evidence suggests that different longevity paradigms require functional autophagy. Because at least two such conserved paradigms, namely, reduced insulin/IGF1 signalling and germline removal, affect autophagy regulation differently at the tissue-specific level in C. elegans16, there may be multiple ways to increase lifespan by autophagy modulation. Given that several of the longevity paradigms are additive for lifespan extension126, it will be interesting to investigate whether combining the paradigms can produce synergistic results with respect to induction of the autophagy process. Moreover, it will be valuable to further examine the cell-autonomous and non-cell autonomous roles of autophagy in long-lived animals. To this end, it will be important to address tissue requirements for autophagy in longevity models in mammalian systems.
Both the ageing and the autophagy research fields have been driven forward tremendously by the use of genetically tractable model organisms, with many new concepts emerging from research in these systems. Although a lot of research is still needed to consolidate these new and exciting findings and to investigate their conservation across species, it is interesting to consider future prospects. Surely, it is possible that more types of specific autophagic cargo in addition to those discussed here (mitochondria, protein aggregates, lipid droplets and lysosomes) will prove to be relevant to ageing, particularly because selective autophagy has been shown to be a widespread process that and also includes eRphagy, ribophagy, xenophagy and nucleophagy. In addition, it will be important to fully address the functional role of auto-phagy not only in different tissues but also over time. To this end, a very recent study surprisingly reported that late-life inhibition of ATG genes with functions in the phagophore nucleation complex causes a potent lifespan extension in wild-type C. elegans17. This is in stark contrast to the effects of inhibiting the same genes early in adult life, which has no or small lifespan-shortening effects in wild-type C. elegans23. Thus, the multistep autophagy process may affect ageing in a much more complex manner than previously anticipated, and future experiments are needed to address this important point. Another interesting avenue to explore further may be the interplay between autophagy, commensal bacteria and their homeostasis and organismal health and ageing, given recent attention regarding links between microbiota dynamics and host ageing127,128. Likewise, as non-conventional roles for ATG genes, for example, in protein secretion, are becoming increasingly recognized129, it also remains to be addressed whether any of these non-conventional functions may be linked to ageing. Finally, it will be very attractive to explore the growing number of pharmacological interventions that can induce autophagy with possible effects on longevity (BOX 2), opening up the possibilities for lifespan extension in humans.
Box 2 |. Autophagy inducers relevant to lifespan extension.
Pharmacological agents that increase autophagy by inducing autophagosome biogenesis can be considered in small-molecule and non-small-molecule categories. In the former, one can divide such agents into those acting via inhibition of the nutrient sensor and major autophagy regulator mTOR and those acting via mTOR-independent pathways. Rapamycins, which target mTOR, have shown lifespan benefits in model organisms ranging from yeasts to mice139, and it is possible that some of these are via effects on autophagy. while there are side effects caused by rapamycins, such as immunosuppression and poor glucose tolerance, and they are large molecules that do not penetrate the blood-brain barrier well, intermittent mTOR inhibition with agents that may be able to selectively target mTOR and get into the brain may be a feasible approach for lifespan extension. Numerous mTOR-independent molecules with autophagy-inducing effects have been described elsewhere and include metformin and trehalose83. While the general impact of such drugs and their target pathways have not been widely studied in model organisms and in relation to conserved longevity paradigms, such experiments may be useful and informative, particularly because autophagy induction with ATG5 overexpression, which is a more specific genetic approach, lengthens lifespan in mice29.
Autophagy can also be induced with non-small-molecule approaches, including an autophagy-inducing peptide based on beclin 1 (REF140); it will be interesting to test whether this peptide modulates organismal lifespan.
In addition to targeting pathways impacting autophagosome biogenesis, it may be beneficial to identify drugs that act at the level of the lysosome. For example, transcription factor EB (TFEB), which is a master regulator of lysosomal function, also regulates autophagy30. The Caenorhabditis elegans orthologue of TFEB positively regulates lifespan, at least in part via autophagy31. Thus, it will be interesting to identify drugs targeting this transcription factor and to investigate their effects on autophagy and longevity across species.
Note added in proof: A recent study showed that decreased interaction between Beclin1 and its negative regulator Bcl-1increases autophagic flux and extends health- and lifespan in mice162.
Acknowledgements
The authors apologize to the colleagues whose work they were unable to discuss owing to space limitations. The authors are grateful to the assistance of C. Karabiyc with the figures. Work in M.H.’s laboratory is funded by the National Institute on Aging (R01AG038664) and National Institute of General Medical Sciences (R01GM117466). Work in D.W.W.’s laboratory is funded by the National Institute on Aging (R01AG037514, R01AG049157 and R01AG040288). M.H. and D.W.W. are both Julie Martin Mid-Career Awardees in Aging Research supported by The Ellison Medical Foundation and the American Federation for Aging Research. Work in D.C.R.’s laboratory is funded by the UK Dementia Research Institute (funded by the Medical Research Council, Alzheimer Disease Research UK and the Alzheimer Disease Society), the Wellcome Trust (Principal Research Fellowship to D.C.R. (095317/Z/11/Z)), the Rosetrees Trust, Strategic Grant to Cambridge Institute for Medical Research (100140/Z/12/Z), Alzheimer Disease Research UK, the Tau Consortium and the Biomedical Research Centre at Addenbrooke’s Hospital.
Footnotes
Competing interests
D.C.R. is a consultant for E3Bio and has consulted for GlaxoSmithKline and AstraZeneca. D.C.R. has grant support from AstraZeneca and AbbVie.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Autophagosomes
Cytosolic double-membrane-bound vesicles capable of sequestering cytoplasmic inclusions and organelles destined for degradation in the autolysosome.
Autolysosome
A cytosolic vesicle resulting from fusion between an autophagosome and acidic lysosomes in which degradation of the inner membrane and sequestered autophagosomal material takes place.
mTOR
An evolutionarily conserved protein kinase that negatively regulates autophagy.
Autophagy receptors
Proteins that facilitate recruitment of cargo to the phagophore for subsequent lysosomal degradation.
Hypothalamus
A region of forebrain that coordinates the autonomic nervous system and the activity of the pituitary, which controls various homeostatic systems, including body temperature.
Lysosomal-associated membrane glycoprotein 2a
(LAMP2A). A lysosomal protein with a key role in chaperone-mediated autophagy.
Insulin/IGF1 signalling
A multi-component signalling pathway that regulates metabolism and longevity in a conserved fashion.
Spastic paraplegia
An inherited disorder characterized by spasticity of the legs.
Ataxia
Loss of control over bodily movements.
Mitochondrial fission
The separation of a single mitochondrion into two or more daughter organelles.
Urolithin A
A metabolite produced by gut microorganisms from ellagic acid. Urolithin A induces mitophagy.
Lipid droplet
A cellular organelle that regulates the storage and hydrolysis of neutral lipids.
Lipid chaperone
A fatty acid-binding protein important in the transport of lipids inside and between cells.
Hormetic heat shock
An aspect of hormesis, meaning beneficial effects of a treatment for which higher intensity is harmful. During hormetic heat shock, non-lethal exposure to elevated temperature induces a response that results in increased stress resistance and longevity.
Proximal tubules
The segment of the kidney that is responsible for reabsorption of nearly two-thirds of all filtered water, sodium and chloride.
Podocytes
Highly specialized cells of the kidney glomerulus that wrap around capillaries.
Glomerulus
A key structure of a nephron, the functional unit of the kidney.
Microbial dysbiosis
The condition of having imbalances in the microbial communities either in or on the body.
Septate junction
An intercellular occluding junction found in invertebrate epithelia.
Crohn’s disease
A chronic inflammatory bowel disease.
Insulin-like peptide
A type of peptide with homology to insulin, a hormone produced in the pancreas that regulates glucose levels in the blood.
Chemosensory neurons
Sensory neurons responsive to chemical stimuli.
Presynaptic active zone
The part of the nerve terminal from which neurotransmitters are released by synaptic vesicle exocytosis.
Sarcopenia
The degenerative loss of muscle mass, quality and strength associated with ageing.
Senescence
Loss of the ability of a cell to divide, differentiate and grow.
Beclin 1
Mammalian orthologue of yeast autophagy-related 6 (Atg6), which forms part of the class iii phosphatidylinositol 3-kinase complex involved in activating autophagy.
ERphagy
The selective degradation of the endoplasmic reticulum by autophagy.
Ribophagy
The selective degradation of ribosomes by autophagy.
Xenophagy
The selective degradation of intracellular pathogens by autophagy; xenophagy is part of the cell-autonomous innate immunity defence.
Nucleophagy
The selective removal of nuclear material from a cell by autophagy.
References
- 1.Levine B & Kroemer G Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J & Klionsky DJ Autophagy and human disease. Cell Cycle 6,1837–1849 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Dikic I & Elazar Z Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, He D, Yao Z & Klionsky D J The machineryof macroautophagy. Cell Res 24, 24–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan D, Kim J, Shaw RJ & Guan KL The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 7, 643–644 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12,1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansen T & Lamark T Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaminets A, Behl C & Dikic I Ubiquitin- dependent and independent signals in selective autophagy. Trends Cell Biol 26, 6–16 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Sarkis GJ, Ashcom JD, Hawdon JM & Jacobson LA Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech. Ageing Dev 45,191–201 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Simonsen A et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4,176–184 (2008).This study is the first to show that overexpression of an autophagy gene is sufficient to extend lifespan (neuronal overexpression of Atg8 in D. melanogaster).
- 11.Demontis F & Perrimon N FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 (2010).This study is the first to link loss of proteostasis during muscle ageing to autophagy and organismal ageing.
- 12.Bai H, Kang P, Hernandez AM & Tatar M Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLOS Genet 9, e1003941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik S et al. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep 13, 258–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnio S et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep 8,1509–1521 (2014).This study shows that autophagy inhibition in the muscle of mice impairs muscle function, possibly via impairing mitochondrial function, and shortens lifespan.
- 15.Cuervo AM & Dice JF Age-related decline in chaperone-mediated autophagy. J. Biol. Chem 275, 31505–31513 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Chang JT, Kumsta C, Heilman AB, Adams LM & Hansen M Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 6, e18459 (2017).This study is the first comprehensive spatiotemporal analysis of autophagy activity in a living organism, C. elegans. It indicates an age- dependent decrease in autophagy as well as tissue- specific regulation and functions of autophagy in two different longevity models.
- 17.Wilhelm T et al. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev 31, 1561–1572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Roso A et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp. Gerontol 38, 519–527 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Terman A The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 41(Suppl. 2), 319–326 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Donati A et al. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J. Gerontol. A Biol. Sci. Med. Sci 56, B288–B293 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Cavallini G, Donati A, Gori Z, Pollera M & Bergamini E The protection of rat liver autophagic proteolysis from the age-related decline co-varies with the duration of anti-ageing food restriction.Exp. Gerontol 36, 497–506 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Donati A et al. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J. Gerontol. A Biol. Sci. Med. Sci 56, B375–B383 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Hansen M in Ageing: Lessons from C. elegans Ch.15 (eds Olsen A & Gill MS (Springer, Cham, Switzerland, 2016). [Google Scholar]
- 24.Ulgherait M, Rana A, Rera M, Graniel J & Walker DW AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep 8, 1767–1780 (2014).This study is the first to show that neuronal Atg1 overexpression improves intestinal barrier function and extends D. melanogaster lifespan.
- 25.Rana A et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun 8, 448 (2017).This study shows that increased expression of Drp1 in midlife facilitates mitophagy and extends D. melanogaster lifespan.
- 26.Karsli-Uzunbas G et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. CancerDiscov 4, 914–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H et al. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy 11,9–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapierre LR, Kumsta C, Sandri M, Ballabio A & Hansen M Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 11,867–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyo JO et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun 4,2300 (2013).This is the first study to show that overexpression of an autophagy gene (ATG5) can extend mammalian lifespan.
- 30.Settembre C et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapierre LR et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun 4, 2267 (2013).This study shows that overexpression of hlh-30 (TFEB in mammals) is sufficient to extend the lifespan of C. elegans in an autophagy-dependent manner.
- 32.Hara T et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Oz-Levi D et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am. J. Hum. Genet 91,1065–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanein S et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am. J. Hum. Genet 82, 992–1002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5, e12245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitada T et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Attardo G et al. [The follow-up of malformation uropathies diagnosed “in utero”]. Pediatr. Med. Chir 14, 119–126 (1992). [PubMed] [Google Scholar]
- 38.Laurin N, Brown JP, Morissette J & Raymond V Recurrent mutation of the gene encoding sequestosome 1(SQSTM1/p62) in Paget disease of bone. Am. J. Hum. Genet 70,1582–1588 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubino E et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology 79,1556–1562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freischmidt A et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci 18, 631–636 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Wallace DC A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet 39, 359–407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Otin C, Blasco MA, Partridge L, Serrano M & Kroemer G The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun N, Youle RJ & Finkel T The mitochondrial basis of aging. Mol. Cell 61,654–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green DR, Galluzzi L & Kroemer G Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palikaras K, Lionaki E & Tavernarakis N Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015).This study is the first to indicate a requirement for mitophagy in several conserved longevity paradigms in C. elegans.
- 46.Pickrell AM & Youle RJ The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashrafi G & Schwarz TL The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 20, 31–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youle R.j. & Narendra DP Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol 12, 9–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutta D, Calvani R, Bernabei R, Leeuwenburgh C & Marzetti E Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circ. Res 110,1125–1138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh LP, Devi TS & Yumnamcha T The role of Txnip in mitophagy dysregulation and inflammasome activation in diabetic retinopathy: a new perspective. JOJ Ophthalmol 4, 555–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang L et al. Differential effects of reticulophagy and mitophagy on nonalcoholic fatty liver disease. Cell Death Dis 9, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall JD, Bazan L Zhang Y, Fares WH & Lee PJ Mitochondrial dysfunction and pulmonary hypertension: cause, effect or both. Am. J. Physiol. Lung Cell. Mol. Physiol 10.1152/ajplung.00331.2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen K et al. Optineurin-mediated mitophagy protects renal tubular epithelial cells against accelerated senescence in diabetic nephropathy.Cell Death Dis 9,105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khalil B & Lievens JC Mitochondrial quality control in amyotrophic lateral sclerosis: towards a common pathway? Neural Regen Res 12, 1052–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Vicente M Neuronal mitophagy in neurodegenerative diseases. Front. Mol. Neurosci 10, 64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond MJ et al. Downregulation of E3 ubiquitin ligases and mitophagy-related genes in skeletal muscle of physically inactive, frail older women: a cross-sectional comparison. J. Gerontol. A Biol. Sci. Med. Sci 69,1040–1048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun N et al. Measuring in vivo mitophagy. Mol. Cell 60, 685–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark IE et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Greene JC et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA 100, 4078–4083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiavi A et al. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr. Biol 25,1810–1822 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Rana A, Rera M & Walker DW Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc. Natl Acad. Sci. USA 110, 8638–8643 (2013).This study is the first to show a direct link between Parkin, a key regulator of mitophagy, and longevity by showing that parkin overexpression is sufficient to extend D. melanogaster lifespan.
- 62.Palikaras K, Daskalaki I, Markaki M & Tavernarakis N Mitophagy and age-related pathologies: development of new therapeutics by targeting mitochondrial turnover. Pharmacol. Ther 178, 157–174 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Ryu D et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med 22, 879–888 (2016).This study describes a natural compound that induces mitophagy and promotes health in diverse species.
- 64.Hansen M, Flatt T & Aguilaniu H Reproduction, fat metabolism, and life span: what is the connection? Cell Metab 17,10–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schulze RJ, Sathyanarayan A & Mashek DG Breaking fat: the regulation and mechanisms of lipophagy. Biochim. Biophys. Acta 1862, 1178–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh R et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaushik S & Cuervo AM The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol 19, 365–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh R & Cuervo AM Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol 2012, 282041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maan M, Peters JM, Dutta M & Patterson AD Lipid metabolism and lipophagy in cancer. Biochem. Biophys. Res. Commun 10.1016/j.bbrc.2018.02.097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen K, Yuan R, Zhang Y, Geng S & Li L Tollip deficiency alters atherosclerosis and steatosis by disrupting lipophagy. J. Am. Heart Assoc 6, e004078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang MC, O’Rourke EJ & Ruvkun G Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lapierre LR, Gelino S, Melendez A & Hansen M Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol 21,1507–1514 (2011).This study is the first to indicate a role for selective autophagy, namely, lipophagy, in organismal lifespan (C. elegans).
- 73.O’Rourke EJ, Kuballa P, Xavier R & Ruvkun G Omega-6 polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev 27, 429–440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Rourke EJ & Ruvkun G MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol 15, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Folick A et al. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347, 83–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palikaras K et al. Ectopic fat deposition contributes to age-associated pathology in Caenorhabditis elegans. J. Lipid Res 58, 72–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ravikumar B, Duden R & Rubinsztein DC Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet 11,1107–1117 (2002).This study is the first to show that aggregate-prone proteins causing neurodegeneration are autophagy substrates.
- 78.Webb JL, Ravikumar B, Atkins J, Skepper JN & Rubinsztein DC Alpha-synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem 278, 25009–25013 (2003). [DOI] [PubMed] [Google Scholar]
- 79.Berger Z et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet 15, 433–442 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Ravikumar B et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet 36, 585–595 (2004).This is the first report suggesting that autophagy upregulation can ameliorate neurodegenerative disease caused by mutant HTT in model organisms like D. melanogaster and mice.
- 81.Lopez A et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebrafish model by autophagy induction. Brain 140, 1128–1146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumsta C, Chang JT, Schmalz J & Hansen M Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun 8,14337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Menzies FM et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Winslow AR et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J. Cell Biol 190, 1023–1037 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashkenazi A et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maejima I et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32, 2336–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto T et al. Time-dependent dysregulation of autophagy: implications in aging and mitochondrial homeostasis in the kidney proximal tubule. Autophagy 12, 801–813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lenoir O, Tharaux PL& Huber TB Autophagy in kidney disease and aging: lessons from rodent models. Kidney Int 90, 950–964 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Papadopoulos C et al. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J 36, 135–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu DJ & Jasper H Epithelia: understanding the cell biology of intestinal barrier dysfunction. Curr. Biol 27, R185–R187 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Clark RI et al. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep 12, 1656–1667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rera M, Clark RI & Walker DW Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl Acad. Sci. USA 109, 21528–21533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rera M, Azizi MJ & Walker DW Organ-specific mediation of lifespan extension: more than a gut feeling? Ageing Res. Rev 12, 436–444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gelino S et al. Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction. PLOS Genet 12, e1006135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Regan JC et al. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. Elife 5, e10956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fan P, Liu P, Song P, Chen X & Ma X Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep 7, 43412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minnerly J, Zhang J, Parker T, Kaul T & Jia K The cell non-autonomous function of ATG-18 is essential for neuroendocrine regulation of Caenorhabditis elegans lifespan. PLOS Genet 13, e1006764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grumati P et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med 16,1313–1320 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Wohlgemuth SE, Seo AY, Marzetti E, Lees HA & Leeuwenburgh C Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp. Gerontol 45,138–148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Donati A, Recchia G, Cavallini G & Bergamini E Effect of aging and anti-aging caloric restriction on the endocrine regulation of rat liver autophagy. J. Gerontol. A Biol. Sci. Med. Sci 63, 550–555 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Resnik-Docampo M et al. Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nat. Cell Biol 19, 52–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spalinger MR, Rogler G & Scharl M Crohn’s disease: loss of tolerance or a disorder of autophagy? Dig. Dis 32, 370–377 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Nighot PK, Hu CA & Ma TY Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J. Biol. Chem 290, 7234–7246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Broughton SJ et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl Acad. Sci. USA 102, 3105–3110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Broughton S & Partridge L Insulin/IGF-like signalling, the central nervous system and aging. Biochem. J 418, 1–12 (2009). [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 548, 52–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saha S et al. Mutations in LRRK2 potentiate age-related impairment of autophagic flux. Mol. Neurodegener 10, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lipinski MM et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 107, 14164–14169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bishop NA, Lu T & Yankner BA Neural mechanisms of ageing and cognitive decline. Nature 464, 529–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eisenberg T et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol 11, 1305–1314 (2009).This study shows beneficial effects of the autophagy inducer spermidine on longevity in multiple organisms.
- 111.Gupta VK et al. Restoring polyamines protects from age-induced memory impairment in an autophagy- dependent manner. Nat. Neurosci 16,1453–1460 (2013) [DOI] [PubMed] [Google Scholar]
- 112.Gupta VK et al. Spermidine suppresses age-associated memory impairment by preventing adverse increase of presynaptic active zone size and release.PLOS Biol 14, e1002563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Demontis F, Piccirillo R, Goldberg AL (& Perrimon, N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell 12, 943–949 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miller RA ‘Accelerated aging’: a primrose path to insight? Aging Cell 3, 47–51 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Nair KS Aging muscle. Am. J. Clin. Nutr 81, 953–963 (2005). [DOI] [PubMed] [Google Scholar]
- 116.Tang AH & Rando TA Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J 33, 2782–2797 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fiacco E et al. Autophagy regulates satellite cell ability to regenerate normal and dystrophic muscles. Cell Death Differ 23,1839–1849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Garcia-Prat L et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). [DOI] [PubMed] [Google Scholar]
- 119.Zhang H, Puleston DL& Simon AK Autophagy and immune senescence. Trends Mol. Med 22, 671–686 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Raz Y et al. Activation-induced autophagy is preserved in CD4+ T-cells in familial longevity. J. Gerontol. A Biol. Sci. Med. Sci 72,1201–1206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Doulatov S & Daley GQ Autophagy: it’s in your blood. Dev. Cell 40, 518–520 (2017). [DOI] [PubMed] [Google Scholar]
- 122.Ho TT et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baar EL, Carbajal KA, Ong IM & Lamming DW Sex- and tissue-specific changes in mTOR signaling with age in C57BU6J mice. Aging Cell 15,155–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hughes AL & Gottschling DE An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maday S & Holzbaur EL Autophagosome assembly and cargo capture in the distal axon. Autophagy 8, 858–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kenyon CJ The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 127.Heintz C & Mair W You are what you host: microbiome modulation of the aging process. Cell 156, 408–411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clark RI & Walker DW Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell. Mol. Life Sci 75, 93–101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martinez J et al. Molecular characterization of LC3- associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Lee C & Longo V Dietary restriction with and without caloric restriction for healthy aging. F1000Res 5, 117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kennedy BK & Lamming DW The mechanistic target of rapamycin: the grand conducTOR of metabolism and aging. Cell. Metabolism 23, 990–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Piper MD, Selman C, McElwee JJ & Partridge L Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J. Intern. Med 263, 179–191 (2008). [DOI] [PubMed] [Google Scholar]
- 133.Burkewitz K, Weir HJ & Mair WB AMPK as a pro-longevity target. EXS 107, 227–256 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Munkacsy E & Rea SL The paradox of mitochondrial dysfunction and extended longevity. Exp. Gerontol 56, 221–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gems D & Partridge L Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab 7, 200–203 (2008). [DOI] [PubMed] [Google Scholar]
- 136.Madeo F, Eisenberg T, Pietrocola F & Kroemer G Spermidine in health and disease. Science 359, eaan2788 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Pallauf K, Rimbach G, Rupp PM, Chin D & Wolf IM Resveratrol and lifespan in model organisms. Curr. Med. Chem 23, 4639–4680 (2016). [DOI] [PubMed] [Google Scholar]
- 138.Shaw WM, Luo S, Landis J, Ashraf J & Murphy CT The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr. Biol 17, 1635–1645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saxton RA & Sabatini DM mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shoji-Kawata S et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.McWilliams TG et al. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab 27, 439–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alvers AL et al. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 5, 847–849 (2009).This is the first study to show a requirement for autophagy genes in a conserved longevity paradigm in yeasts.
- 143.Ruckenstuhl C et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLOS Genet 10, e1004347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tang F et al. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy 4, 874–886 (2008). [DOI] [PubMed] [Google Scholar]
- 145.Toth ML et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338 (2008). [DOI] [PubMed] [Google Scholar]
- 146.Melendez A et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003).This study is the first to show a direct role for an autophagy gene in a conserved longevity paradigm in C. elegans (bec-1 (Becn1 in mammals and ATG6 in yeasts) in long-lived daf-2 mutants).
- 147.Hars ES et al. Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95 (2007). [DOI] [PubMed] [Google Scholar]
- 148.Hansen M et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLOS Genet 4, e24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morselli E et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis 1, e10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jia K & Levine B Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 3, 597–599 (2007). [DOI] [PubMed] [Google Scholar]
- 151.Yang J et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 35,11–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McQuary PR et al. C. elegans S6K mutants require a creatine-kinase-like effector for lifespan extension. Cell Rep 14, 2059–2067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McColl G et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab 12, 260–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bjedov I et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster.Cell Metab 11, 35–46 (2010).This study is the first to show a requirement for autophagy in a conserved longevity paradigm in D. melanogaster through mTOR inhibition.
- 155.Tang H et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine 2, 255–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Haack TB et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet 91,1144–1149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saitsu H et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet 45, 445–449 (2013). [DOI] [PubMed] [Google Scholar]
- 158.Metzger S et al. The V471A polymorphism in autophagy-related gene ATG7 modifies age at onset specifically in Italian Huntington disease patients. PLOS ONE 8, e68951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hampe J et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1.Nat. Genet 39, 207–211 (2007). [DOI] [PubMed] [Google Scholar]
- 160.Rioux JD et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet 39, 596–604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cullup T et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet 45, 83–87 (2013).This study shows how a mutation in an autophagy regulator causes a Mendelian neurodegenerative disease.
- 162.Fernandez AF et al. Disruption of the beclin 1-BCL2 autophagy-regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018).This study shows that decreased interaction between Beclin1 and its negative regulator Bcl-1 increases autophagic flux and extends healthspan and lifespan in mice.