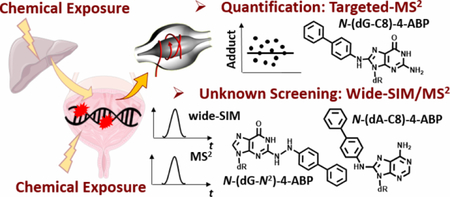
Abstract
Epidemiological studies have linked aromatic amines (AAs) from tobacco smoke and some occupational exposures with bladder cancer risk. Several epidemiological studies have also reported a plausible role for structurally related heterocyclic aromatic amines present in tobacco smoke or formed in cooked meats with bladder cancer risk. DNA adduct formation is an initial biochemical event in bladder carcinogenesis. We examined paired fresh-frozen (FR) and formalin-fixed paraffin-embedded (FFPE) non-tumor bladder tissues from 41 bladder cancer patients for DNA adducts of 4-aminobiphenyl (4-ABP), a bladder carcinogen present in tobacco smoke, and 2-amino-9H-pyrido[2,3-b]indole, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline, possible human carcinogens, which occur in tobacco smoke and cooked meats. These chemicals are present in urine of tobacco smokers or omnivores. Targeted DNA adduct measurements were done by ultra-performance liquid chromatography-electrospray ionization multi-stage hybrid Orbitrap MS. N-(2′-Deoxyguanosin-8-yl)-4-ABP (N-(dG-C8)-4-ABP) was the sole adduct detected in FR and FFPE bladder tissues. Twelve subjects (29%) had N-(dG-C8)-4-ABP levels above the limit of quantification, ranging from 1.4 to 33.8 adducts per 109 nucleotides (nt). DNA adducts of other human AA bladder carcinogens, including 2-naphthylamine (2-NA), 2-methylaniline (2-MA), 2,6-dimethylaniline (2,6-DMA), and lipid peroxidation (LPO) adducts were screened for in bladder tissue, by our untargeted data-independent adductomics method, termed wide-selected ion monitoring (wide-SIM)/MS2. Wide-SIM/MS2 successfully detected N-(dG-C8)-4-ABP, N-(2′-deoxyadenosine-8-yl)-4-ABP and the presumed hydrazo linked adduct, N-(2′-deoxyguanosin-N2-yl)-4-ABP, and several LPO adducts in bladder DNA. Wide-SIM/MS2 detected multiple DNA adducts of 2-NA, 2-MA and, 2,6-DMA, when calf thymus DNA was modified with reactive intermediates of these carcinogens. However, these AA-adducts were below the limit of detection in unspiked human bladder DNA (< 1 adduct per 108 nt). Wide-SIM/MS2 can screen for many types of DNA adducts formed with exogenous and endogenous electrophiles and will be employed to identify DNA adducts of other chemicals that may contribute to the etiology of bladder cancer.
TOC
Introduction
Bladder cancer (BC) is one of the ten most common forms of cancer worldwide with a very high rate of recurrence.1,2 The risk factors for developing BC include genetic and molecular abnormalities, chronic infection of the bladder, and chemical exposures.2 The elevated risk of BC in factory workers of the dye, textile, and rubber industries has been linked to the occupational exposures of AAs, such as 4-ABP and 2-naphthylamine (2-NA).3 Both chemicals were used as antioxidants in the rubber industry and as dye intermediates prior to their banning by regulatory legislation.4 Benzidine and 4,4′-methylenebis-2-chloroaniline (MOCA), two other aromatic amines used as industrial chemicals, have also been linked to BC.4 Many epidemiology studies have consistently shown that cigarette smoking is an important risk factor for BC.5,6 4-ABP and 2-NA are present in mainstream tobacco smoke at levels ranging up to several ng per cigarette.7 The major sources of exposure to 4-ABP are thought to be cigarette smoking,8 followed by combustion of fossil fuels,9 textile and printing related industries,10 and through the use of some hair dye and cosmetic products.11,12 2-MA and 2,6-DMA are other AAs formed in tobacco smoke and may contribute to BC.13
4-ABP has been widely used as a prototypical AA for mechanistic studies on chemical carcinogenesis of the bladder.14,15 The carcinogenic metabolites of 4-ABP, N-hydroxy-4-ABP (HONH-4-ABP) and its N-glucuronide conjugate formed in the liver, circulate in the blood stream and reach the bladder, where they can undergo solvolysis to form the reactive nitrenium ion species that covalently binds to the DNA of the uroepithelium (Figure 1).16 Alternatively, cytochrome P450s expressed in the uroepithelium can bioactivate 4-ABP and possibly other AAs to their genotoxic HONH-AA metabolites, which are capable of reacting with DNA to form DNA adducts and induce mutations.17 The major DNA adduct of 4-ABP is N-(dG-C8)-4-ABP, with lesser amounts of N-(2′-deoxyadenosine-8-yl)-4-ABP (N-(dA-C8)-4-ABP) and N-(2′-deoxyguanosin-N2-yl)-4-ABP (N-(dG-N2)-4-ABP) also formed.14
Figure 1.
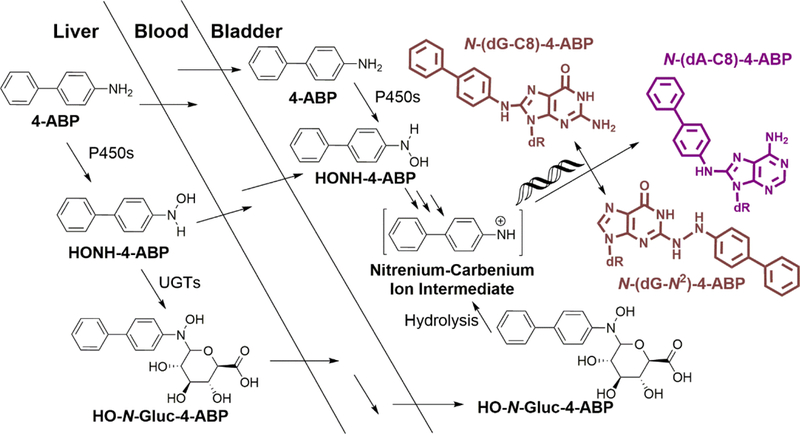
Scheme of the metabolic activation of 4-ABP leading to DNA adduct formation in bladder. 4-ABP is first oxidized to the HONH-4-ABP by cytochrome P450 (P450) in liver. HONH-4-ABP may undergo circulation and reach the bladder to form DNA adducts. As a conjugation pathway, UDP-glucuronosyltransferases (UGTs) catalyze the glucuronidation of HONH-4-ABP to form N-Gluc-HONH-4-ABP, which undergoes circulation through the blood and collected in the bladder. HO-N-Gluc-4-ABP can undergo hydrolysis in urine at acidic pH and forms a nitrenium-carbenium ion intermediate that can damage DNA in the bladder epithelial cells. A small portion of unmetabolized 4-ABP can also reach bladder through circulation in the blood and undergo oxidation by P450 expressed in the bladder epithelium to form HONH-4-ABP which reacts with DNA.
While tobacco smoking is a well-known risk factor for BC,18–20 it is uncertain which of the chemicals present in tobacco smoke are principally responsible for DNA damage of the bladder and the development of BC. AAs, including 4-ABP, are assumed to contribute to the pathogenesis of BC in smokers; however, the levels of 4-ABP in mainstream tobacco smoke occur only at 1 – 3 ng per cigarette.7 In a perspective written more than 20 years ago, Poirier and Beland concluded that “exposure to 4-ABP through tobacco smoke and environment pollution are too low to account for BC risk, and factors other than 4-ABP-DNA adducts, such as adducts of other carcinogens, the influence of promoters, and synergistic effects of these factors contribute substantially to smoking-related BC in humans.”21 There are at least 26 AAs and structurally related HAAs present in tobacco smoke;7,8 yet, our knowledge about these and other chemicals in tobacco smoke that damage DNA of the bladder are largely limited to 4-ABP and more recently to studies conducted on the electrophile acrolein.22,23
Non-tobacco associated BC risks have been linked to several classes of carcinogens in the diet. Well-done meat consumption has been identified as a risk factor for BC, based on the European Prospective Investigation into Cancer and Nutrition and American Association of Retired Persons Diet and Health Studies,24–26 and case-control studies conducted in Texas and Uruguay.27,28 Some studies have reported that red and processed meats containing HAAs but also N-nitroso compounds may be causative agents of BC.24–26,29–32 However, other case-control studies in Spain,33 Sweden,34 and the USA35 did not find a link between meat consumption and BC risk.
Cooked meats contain more than 20 genotoxic HAAs,36 which are structurally related to AAs and undergo similar pathways of metabolism and bioactivation as AAs.37 All HAAs assayed thus far in rodent bioassays have been shown to be carcinogenic, inducing tumors at multiple sites.36 Smokers excrete elevated levels of mutagenic compounds in urine compared to nonsmokers, and a significant portion of the mutagenicity is thought to be attributed to AAs and/or HAAs, based on the high induction of mutations in S. typhimurium strains TA98 and YG1024 that are sensitive to these classes of chemicals.38–40 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) was reported as a major DNA-damaging agent in urine from smokers of black tobacco.41 Other HAAs, including 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) and 2-amino-9H-pyrido[2,3-b]indole (AαC) have been detected in urine of smokers or omnivores.37,42,43 4-ABP, 2-NA, and methylated anilines, also have been detected in urine of smokers and nonsmokers and occupational factory workers exposed to diesel exhaust.44–47 These chemicals could undergo bioactivation by P450s expressed in the bladder epithelium.17
There are a limited number of reports in the literature on the detection of DNA adducts of 4-ABP in human bladder tissues (Table 1). Talaska was the first to detect 4-ABP-DNA adducts in human bladder biopsy samples, by employing 32P-postlabeling.48 The levels of the presumed N-(dG-C8)-4-ABP was 16 ± 3 adducts per 108 nt in current smokers (N = 13), and it was detected at significantly higher adduct levels (~4-fold) than that estimated in current non-smokers (N = 20). Thereafter, studies employing 32P-postlabeling,23,49 immunohistochemistry (IHC) of paraffin blocks of transurethral resection from bladder cancer patients50 or exfoliated urothelial cells,51 gas chromatography with negative ion chemical ionization mass spectrometry (GC-NICI-MS),52,53 and high-performance liquid chromatography (HPLC)-triple quadrupole (QqQ)-MS have reported the measurement of 4-ABP-DNA adducts in bladder.22 There is little information regarding the capacity of monocyclic AAs54 and HAAs to form DNA adducts in human bladder, although AαC forms appreciable levels of DNA adducts in mouse bladder.55
Table 1.
4-ABP-DNA adducts detected in human bladder tissues*
| Method of Detection |
Tissue Sources | Smoking Status | Sample Preparation |
LOD or LOQ |
Adduct ranges | Correlation of Adduct Levels to the Smoke Status |
Ref |
|---|---|---|---|---|---|---|---|
| IHC | FFPE of transurethral resection of bladder cancer patients (N = 46) |
24 current smokers, 22 non-smokers, |
Monoclonal antibody against 4-ABP-DNA adducts |
N.R. (relative staining) |
100% positivity, 1 to 7 x 10−6 nt |
Significantly higher relative staining intensity in current smokers |
50 |
| Exfoliated urinary cells (N = 20) |
20 current smokers | Same as above | N.R. | N.A. (relative staining) | No correlation between no. of cigarette smoked to adduct levels |
51 | |
|
32P-post- labeling |
Human urinary bladder biopsy samples (N = 42) |
13 current smokers, 9 non-smokers, 20 former smokers |
Nuclease P1 treatment or 1- butanol extraction |
5 × 10−10 nt | 100% positivity, 0.1 to 1.6 × 10−8 nt (total-4- ABP-DNA adducts) |
Significantly higher (4-fold) adducts in current smokers |
48 |
| Normal human urothelial mucosa (N = 19) |
3 current smokers, 11 non-smokers, 4 former smokers, 1 N.A. |
1-Butanol extraction |
N.R. | 100% positivity, 1.7 to 7.5 × 10−8 nt (normal) and 2.3 to 10.4 × 10−8 nt (tumor) (total 4-ABP- DNA adducts) |
No difference in adduct levels between normal and tumor bladder tissues |
23 | |
| Bladder tumor tissues (N = 10) |
3 current smokers, 6 former smokers, 1 N.A. |
||||||
| GC-NICI- MS |
Human lung (N = 11) Human bladder urinary mucosa (N = 8) |
6 current smokers, 1 non smoker, 1 former smoker, 3 N.A. |
Alkaline and Thermal hydrolysis |
0.32 × 10−8 nt |
91% positivity, < 0.3 to 49.5 × 10−8 nt (lung). 62.5% positivity, <0.3 to 3.9 × 10−8 nt (bladder) |
No correlation between adduct levels and number of cigarette smoked |
52 |
| Bladder cancer biopsies (N = 75) |
46 current smokers, 8 non-smokers, 17 former smokers, 4 N.A. |
Alkaline and Thermal hydrolysis (same as above) |
3.3 × 10−8 nt (100 µg DNA) |
49% positivity, ND to 1.1 x 10−5 nt) (total 4-ABP- adducts) |
Strong correlation between adduct levels and overall tumor grades, and between adduct levels and current smoking status in higher tumor grades |
53 | |
| Normal epithelial and submucosal balder tissues (N = 46) and bladder tumors in urothelial carcinoma patients (N = 12) |
(Bladder tumor patients) 5 current smokers,** 3 non-smokers, 4 former smokers* |
Acid and thermal hydrolysis |
N.R. | 30% positivity, N.D. to 1.4 × 10−7 nt (tumor) |
No correlation between adduct levels and smoking status |
54 | |
| HPLC-QqQ- MS |
Bladder cancer patients (N = 27) |
1 current smoker, 25 non- smokers, 1 not available |
Enzymatic hydrolysis and immunoaffinity chromatography |
2 × 10−9 nt (200 µg DNA) |
44% positivity, N.D. to 8 x 10−8 nt (N-(dG-C8)-4- ABP) |
No correlation between adduct levels and smoking status |
22 |
| nanoUPLC- ESI- Orbitrap- MS2 |
Tumor adjacent normal tissues from bladder cancer patients (N = 41) |
6 current smokers, 11 non-smokers, 24 former smokers |
Enzymatic hydrolysis and online trapping |
1.4 × 10−9 nt (20 µg DNA) |
29% positivity, 1.4 to 33.8 × 10−9 nt (N-(dG- C8)-4-ABP) |
No correlation between adduct levels and smoking status |
This study |
: The reported adduct levels from the cited papers were converted and expressed as adducts per nucleotides (nt).
: Current smoker: continued smoking up to at least 4 weeks before surgery; former smokers stopped smoking 4 weeks or more prior to surgery
N.R. Not reported.
N.A. Not available.
N.D. Not detected.
We have developed online DNA adduct enrichment and ultra-performance liquid chromatography-electrospray ionization multi-stage ion trap MS (UPLC-ESI-IT-MSn) to quantify and characterize DNA adducts of 4-ABP and HAAs in rodent and human tissues, and human saliva.56,57 We have also successfully employed FFPE tissues to screen for DNA adducts of aristolochic acid-I58 and PhIP59 in paired FR and FFPE human renal and prostate tissues, respectively, by high-resolution accurate mass (HRAM) nanoUPLC-ESI-Orbitrap-MS2. The method has a limit of quantification (LOQ) of 1 – 2 adducts per 109 nt, when employing 10 – 20 µg DNA.59 We also recently developed an untargeted, unbiased data-independent screening method, termed wide-SIM/MS2, which can screen for a wide-range of DNA adducts in human tissues.60 In this study, we have characterized and quantified DNA adducts of 4-ABP in DNA from FR and FFPE bladder tissue of BC patients, and employed wide-SIM/MS2 60 to screen for other DNA adducts that may form with 4-ABP, and other AAs, including 2-NA, 2-MA, 2,6-DMA, and several prominent HAAs, including PhIP, MeIQx and AαC, which may damage bladder DNA.
Experimental Procedures
Materials.
Calf thymus DNA (CT DNA), DNase I (Type IV, bovine pancreas), Benzonase nuclease ultrapure, alkaline phosphatase (Escherichia coli), nuclease P1 (from Penicillium citrinum), RNase A (bovine pancreas), RNase T1 (Aspergillus oryzae), proteinase K (Tritiachium album), C2H5OH for molecular biology (200 proof), Tris-HCl, BisTris, EDTA, Na2HPO4, citric acid, β-mercaptoethanol (βME), 4-ABP, 2-nitrosotoluene, 1,3-dimethyl-2-nitrobenzene, pyruvonitrile, triethylamine (NEt3), Pd/C (10%), hydrazine hydrate, Zn dust, NH4Cl, tetrahydrofuran (THF) stabilized with butylated hydroxytoluene (BHT) (250 ppm), hexane, ethyl acetate (EtOAc), p-xylene, HPLC grade NH4CH3CO2, isopropanol and silica thin layer chromatography (TLC) plates, 250 µm, were purchased from Sigma-Aldrich (St. Louis, MO). Phosphodiesterase I (Crotalus adamanteus venom) was purchased from Worthington Biochemicals Corp. (Newark, NJ). CH2Cl2, 10% neutral buffered formalin, Optima™ LC-MS grade HCO2H, CH3CN, CH3OH, and H2O, were purchased from Thermo Fisher Scientific (Waltham, MA). HAAs were purchased from Toronto Research Chemicals (North York, Canada). [13C10]-dG was purchased from Cambridge Isotope Laboratory (Tewksbury, MA). N-(dG-C8)-4-ABP and N-([13C10]-dG-C8)-4-ABP, N-(2′-deoxyguanosin-8-yl)-AαC (N-(dG-C8)-AαC) and N-([13C10]-dG-C8)-AαC, N-(2′-deoxyguanosin-8-yl)-MeIQx (N-(dG-C8)-MeIQx) and N-([2H3C]-dG-C8)-MeIQx, N-(2′-deoxyguanosin-8-yl)-PhIP (N-(dG-C8)-PhIP) and N-([13C10]-dG-C8)-PhIP, [2H11]-6-(1-hydroxyhexanyl)-8-hydroxy-1,N2-propano-deoxyguanosine ([2H11]-dG-HNE-I) were synthesized as described.49,61–64 Microliter CapLC vials with silylated inserts were purchased from Wheaton (Millville, NJ). N-hydroxy-2-naphthylamine (HONH-2-NA) had been provided by the late Dr. Fred F. Kadlubar (National Center for Toxicological Research, Jefferson, AR). N-(2′-Deoxyguanosin-8-yl)-2-NA (N-(dG-C8)-2-NA),65 1-(2′-deoxyguanosin-N2-yl)-2-NA (1-(dG-N2)-2-NA), and 1-(2′-deoxyadenosin-N6-yl)-2-NA (1-(dA-N6)-2-NA) were synthesized as previously reported.66,67 1H-NMR chemical shifts of 2-NA adducts are provided in supporting information Table S1 and their high resolution accurate mass spectra are discussed in the Results. PhIP- and 4-ABP-modifed CT DNA49,63,68 were kindly provided by Dr. Frederick A. Beland (National Center for Toxicological Research, Jefferson, AR). The N-(2′-deoxyguanosin-8-yl)-2-MA (N-(dG-C8)-2-MA) and N-(2′-deoxyguanosin-8-yl)-2,6-DMA (N-(dG-C8)-2,6-DMA) synthetic standards were provided by Dr. Gabriele Sabbioni (Institute of Environmental and Occupational Toxicology, Airolo, Switzerland),69 and 4-oxo-(2E)-nonenal (ONE) adducts dG-ONE, dA-ONE, Dr. Ian Blair (University of Pennsylvania, Philadelphia, PA).
Human Bladder Sample Collection and DNA Extraction.
The research protocol was reviewed and approved by the Institutional Review Board at University of Minnesota. De-identified human normal tumor-adjacent bladder mucosa tissues were obtained during bladder cancer surgery and snapped-frozen in liquid nitrogen and stored at −80 °C. Portions of normal, non-proliferating human bladder mucosa, located at least 2 cm away from the tumor were dissected by certified pathologists’ assistants from cystectomy specimens obtained during bladder cancer surgery. The normal tumor-adjacent bladder mucosa tissues were de-identified, snap-frozen in liquid nitrogen and stored at −80° C. The mirror section of the frozen tissue was fixed in 10% neutral buffered formalin for 24 hours at room temperature. Tissues underwent serial dehydration with C2H5OH followed by p-xylene wash, and embedded in paraffin by a Sakura Tissue Tech VIP 2000 (Torrance, CA).
The procedures of DNA isolation have been published.70 Briefly, the freshly frozen bladder tissues were thawed on ice and homogenized in TE buffer (50 mM Tris-HCl buffer, pH 8.0, and 10 mM EDTA) containing 10 mM βME by a PRO 200, PRO Scientific homogenizer equipped with a 5 mm saw tooth type blade (Oxford, CT). The homogenized tissues (equivalent of 25 mg of wet tissue weight) were centrifuged at 3000 g for 10 min at 4 °C. DNA was isolated from the nuclear pellet by the phenol/chloroform extraction method, followed by digestion with RNase A and RNase T1 at 37 °C for 1.5 h and proteinase K digestion at 37 ˚C for 2 h.58 To extract DNA from FFPE blocks, the whole tissues were carefully removed from the paraffin block with a scalpel, deparaffinized with p-xylene, and rehydrated in a gradient of C2H5OH. After homogenization in TE buffer with 10 mM βME, an equivalent of 25 mg dry weight of tissue was processed with the ZR FFPE DNA MiniPrep™ Kit (Zymo Research, Irvine, CA), following the manufacturer’s instructions with modifications.58 An overnight digestion of tissue with proteinase K (0.2 mg) was performed at 50 °C to ensure complete reversal of the cross-linked DNA. The DNA concentration and purity were measured by UV spectrophotometry.
Preparation of 2-NA, 2-MA, and 2,6-DMA-modified CT DNA
2-NA-modified CT DNA.
CT DNA solution (0.5 ml of 1 mg/mL in 10 mM sodium phosphate-citrate buffer, pH 5.0) was purged under Ar, and 5 µL of freshly prepared N-hydroxy-2-naphthylamine (HONH-2-NA, 1 mg/mL in DMSO:C2H5OH (3:1, v:v)) was added.71 The mixture was incubated at 37 °C for 1 h, followed by three extractions with equal vol. of EtOAc to remove the unreacted HONH-2-NA. The modified CT DNA was precipitated by the addition of 0.1 vol. of 5 M NaCl, followed by two vol. of cold isopropanol. The DNA pellet washed with 70% C2H5OH and reconstituted in LC-MS grade H2O.
N-Hydroxy-2-methylaniline (HONH-2-MA).
THF stabilized with BHT (250 ppm) was purged under Ar, and then 1 mL was added to 1 mg Pd/C (10%) powder. The mixture was mixed gently and stored at – 20 °C for 20 min. Hydrazine of 20 µL (0.35 mmol) was added to the solution, which was mixed and kept at – 20 °C for 5 min.72 2-Nitrosotoluene (2.6 mg, 21.5 µmol) was added to the mixture and the reaction was kept at – 20 °C for 2 h with occasional mixing. The progress of the reaction was monitored by HPLC-UV using an Aquasil C18 HPLC column (4.5 × 250 mm, Thermo Fisher Scientific, Waltham, MA) with a 20 min linear gradient starting from 1% and ending at 99% B (solvent A, 10 mM NH4CH3CO2 in H2O, pH 6.8, and B, CH3CN) at 1 mL/min flow rate, monitoring at wavelengths 260, 290 and 340 nm. After the completion of the reaction, the Pd/C pellet was removed by centrifugation, and the supernatant was used immediately for CT DNA modification (vide infra).
N-Hydroxy-2,6-dimethylaniline (HONH-2,6-DMA).
NH4Cl (40 mg, 0.75 mmol) was dissolved in 1.6 mL of 50% CH3OH in a 50 °C water bath. The solution was stirred vigorously before the addition of 41.6 µL 1,3-dimethyl-2-nitrobenzene (0.31 mmol) and 40 mg Zn dust (0.63 mmol).73 The reaction was monitored by TLC with hexane:EtOAc (1:1, (v;v)). After 10 min, the insoluble Zn particles were removed by centrifugation, and the supernatant was concentrated under a stream of N2 to remove CH3OH. The HONH-2,6-DMA was extracted with an equal vol of CH2Cl2 three times. The combined CH2Cl2 extract was dried with MgSO4, and the solvent was concentrated to dryness under N2. THF stabilized with BHT (250 ppm) was purged under Ar and 0.5 mL was added to dissolve the HONH-2,6-DMA.
2-MA- and 2,6-DMA-modified CT DNA.
A 1.1 mol. ratio of NEt3 was added to HONH-2-MA or HONH-2,6-DMA (20 µmol) in THF (0.5 mL). The solution was cooled to – 30 °C in a dry ice bath containing CH3CN. A 1.1 mol. ratio of pyruvonitrile was added to convert HONH-AA to N-acetoxy-AA.69,73 The solution was mixed and kept at – 30 °C for 2 h. CT DNA solution (1 mg/mL in 10 mM BisTris with 0.1 mM EDTA, pH 7.1) of 0.5 mL was purged under Ar, and the THF solution of N-acetoxy-AA was added and the mixture incubated at 37 °C overnight. Unreacted N-acetoxy-AAs and its decomposition products were extracted with equal vol. of EtOAc. DNA was isolated as described (vide supra).
Enzymatic Digestion of DNA.
For quantitative measurements, human bladder DNA (20 μg) was spiked with stable, isotopically labeled DNA adduct internal standards at levels of 1.25 adducts per 108 nt. The N-([13C10]-dG-C8)- adducts of 4-ABP, AαC, PhIP, and N-([2H3]-dG-C8)-MeIQx were added just prior to the enzymatic digestion. A mixture of PhIP- and 4-ABP-modifed CT DNA of known adduct levels were used as positive controls.49,63,68 DNA samples were incubated at 37 °C with DNase I (2 Kunitz units) (or Benzonase, 300 U for samples analyzed by wide-SIM/MS2, see below) and nuclease P1 (0.1 U) for 3.5 h, followed by phosphodiesterase 1 (3.2 mU) and alkaline phosphatase (40 mU) at 37 °C overnight. The digestion solutions were dried by vacuum centrifugation and reconstituted in 50% DMSO (30 µL). The reconstituted solutions were sonicated, centrifuged at 21000 g for 5 min, and the supernatants were transferred to a silylated borosilicate glass inserts for LC-MS analysis.
For the characterization of DNA adducts by wide-SIM/MS2, bladder DNA was spiked at a level of 4 adducts per 108 nt (N-([13C10]-dG-C8)- adducts of 4-ABP, AαC, PhIP, N-([2H3C]-dG-C8)-MeIQx) and 8 adducts per 108 nt ([2H11]-HNE-dG) prior to the enzymatic digestion of DNA.
DNA Adducts Quantification (nanoUPLC-ESI-Orbitrap-MS2), Characterization (nanoUPLC-ESI-Orbitrap-MS3), and Screening (wide-SIM/MS2).
An UltiMate 3000 RSLC nano UHPLC System interfaced with an Orbitrap Fusion™ Tribrid™ MS (Thermo Fisher Scientific, Waltham, MA) was used for the analysis. Chromatography was performed using a New Objectives (Woburn, MA) PicoFrit emitter (75 µm x 200 mm, 10 ± 1 μm orifice) custom packed with a Luna C18 5µm stationary phase (Phenomenex Corp. Torrance, CA) and mounted in a Nanospray Flex™ ion source (Thermo Fisher Scientific, Waltham, MA). An Acclaim PrepMap trap cartridge RP C18 (0.3 × 5 mm, 5 μm, 100 Å, Thermo Fisher Scientific, Waltham, MA) was employed for online DNA adduct enrichment. The LC solvents were (A) 0.05% HCO2H in H2O and (B) 0.05% HCO2H in 95% CH3CN. The DNA digests (8 μL) were injected onto the trap column and washed with solvent A for 4 min at a flow rate of 12 µL/min by the loading pump. This procedure removed the salt, polar components in the digest, and non-modified nucleosides. After trapping, adducts were back-flushed onto the analytical column with a linear gradient: 1 to 99% B in 15 min (0.3 µL/min) (targeted-MS2 and targeted-MS3),59 or 1 to 30% B over 4 min (0.6 µL/min), followed by 30 to 99% B over 30 min (0.2 µL/min), then 0.6 µL/min for the sequential column washing and equilibrating (wide-SIM/MS2).60
The quantification of the DNA adducts was conducted at the MS2 scan stage by fragmentation of the parent ions and monitoring of the respective aglycone ions ([M+ H - 116.0473]+), and their corresponding internal standards ([M + H - 116.0473]+) for N-([2H3C]-dG-C8)-MeIQx, or [M + H - 121.0641]+ for [13C10] labeled internal standards with a 5 ppm mass tolerance). Adduct structures were confirmed at the MS3 scan stage.70 The wide-SIM/MS2 method contained a total of 20 Orbitrap scan events (with a resolution of 60,000 at m/z 200, all resolution values reported here are full width at half maximum (FWHM)). Each wide-SIM event covers a mass of 30 m/z ± 2 m/z edge overlapping (details reported below) with quadrupole isolation, and its corresponding MS2 fragmentation event isolates the same mass range and a product ion detection range of m/z 100 to 550. With the 10 wide-SIM events (and the 10 corresponding MS2 events), a mass range of m/z 330–630 is analyzed. A second wide-SIM/MS2 method was used with a resolution of 500,000 and a single 30 m/z wide-SIM and MS2 pair with a center mass of m/z 420. The DNA adducts screened for were tentatively identified as “present” when the manually extracted ion chromatogram (EIC) peaks of the precursor ions ([M + H]+) co-eluted with the corresponding EIC peaks of their aglycone ions ([M + H - 116.0473]+ or [M + H - 121.0641]+). The identities of the adducts were confirmed by targeted MS3 analyses (vide infra).70 The MS was operated in the positive ionization mode with a 2200 V spray voltage and 300 ºC ion transfer tube temperature. Other MS parameters are as follows: quadrupole isolation, 2 m/z (targeted-MSn) or 34 m/z (wide-SIM/MS2, 30 m/z with ±2 m/z edge overlapping); RF-lens, 90%; Orbitrap resolution, 30,000 (targeted-MSn), 60,000 (wide-SIM/MS2), or 500,000 (wide-SIM/MS2); HCD collision energy, 25% (MS2) and 45% (MS3); maximum injection time, 100 ms or 1014 ms (wide-SIM/MS2 at 500,000 resolution); data type, profile; AGC target, 5E4 (targeted-MSn, SIM in wide-SIM/MS2 at 60,000 resolution), 1E5 (MS2 in wide-SIM/MS2), or 2E5 (SIM in wide-SIM/MS2 at 500,000 resolution). Xcalibur™ version 3.0.63 (Thermo Scientific) was used for data acquisition and analysis. Theoretical m/z of precursors, aglycones and fragments were generated based on their structures in ChemBioDraw Ultra version 13.0.2. The EICs were manually generated with a 5 ppm mass tolerance using Xcalibur’s Qualbrowser module. The MS was externally calibrated by Pierce™ LTQ ESI Positive Ion Calibration Solution (Thermo Fisher Scientific, Waltham, MA) and the internal calibrant of fluoranthene (m/z 202.0777) was employed as a lock mass and applied during the data acquisition.
Results
We sought to determine if the AA and HAA carcinogens present in tobacco smoke and/or cooked meat had formed DNA adducts in bladder tissues of bladder cancer patients. The major dG-C8 adducts formed with 4-ABP, AαC, MeIQx, and PhIP were measured in non-tumor adjacent bladder epithelium by an HR-targeted-MS2 method. The method was previously validated and has an LOQ value of 1 – 2 adducts per 109 nt, employing 20 µg DNA.59 We also employed our newly developed wide-SIM/MS2 scanning technology,60 to screen for multiple DNA adducts formed with several other potential AA bladder carcinogens.
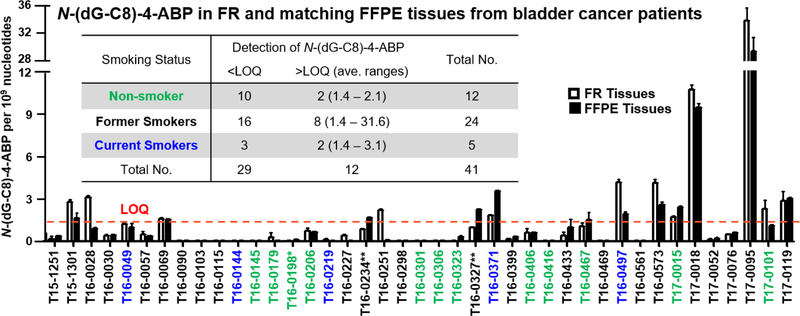
The adduct levels in paired FR and FFPE tissues of each subject, together with their demographics are listed in Table 2. Among the 41 patients, 24 were self-reported former smokers, including one current e-cigarette smoker (T16–0234) and one current smokeless tobacco user (T16–0327), and 5 subjects were current smokers. Twelve subjects had never smoked tobacco, but one subject used smokeless tobacco (T16–0198). N-(dG-C8)-4-ABP was the only adduct detected in the bladder DNA, with comparable levels found in the bladder DNA of FR and matching FFPE tissues (Table 2 and Figure 2). We previously reported that levels of DNA adducts of aristolochic acid I in the cortex of kidney from patients with upper urinary tract cancer,58 and DNA adducts of PhIP in the prostate genome of prostate cancer patients of paired FR and FFPE were largely within 20% of each other.59 Our current study shows that the N-(dG-C8)-4-ABP adduct is also relatively stable towards the formalin fixation and the crosslink reversal procedures. Collectively, these findings demonstrate the great potential for FFPE tissues to serve as biospecimens for the biomonitoring of environmental chemical exposures through the measurement of the relevant DNA adducts.
Table 2.
Demographic of the bladder cancer patients and N-(dG-C8)-4-ABP levels (per 108 nt). LOQ is 1.4 adducts per 109 nt.
| Subject Code |
Age | Sex | Race | Smoking Status | Cigarette Consumption, ppd |
Smoking Duration, y |
Quit Time |
N-(dG-C8)-4-ABP level ± SD, FR tissues |
N-(dG-C8)-4-ABP level ± SD, FFPE tissues |
Average N-(dG- C8)-4-ABP level |
|---|---|---|---|---|---|---|---|---|---|---|
| T15–1251 | 77 | M | C | Former Smoker | 0.5 | NA | 2006 | 0.2 ± 0.2 | 0.4 ± 0.00 | <LOQ |
| T15–1301 | 60 | F | C | Former Smoker | NA | NA | 2007 | 2.8 ± 0.2 | 1.6 ± 0.4 | 2.2 |
| T16–0028 | 56 | F | C | Former Smoker | 0.5 | 30 | 2015 | 3.1 ± 0.1 | 0.9 ± 0.1 | 2.0 |
| T16–0030 | 79 | M | C | Former Smoker | 6 times/d | 60 | 2015 | 0.4 ± 0.0 | 0.5 ± 0.0 | <LOQ |
| T16–0049 | 60 | M | C | Current Smoker | 0.5 | 35 | -- | 1.2 ± 0.0 | 0.9 ± 0.3 | <LOQ |
| T16–0057 | 70 | M | C | Former Smoker | 1 | 25 | 1995 | 0.5 ± 0.2 | 0.4 ± 0.0 | <LOQ |
| T16–0069 | 60 | M | C | Former Smoker | 1.5 | 45 | 2015 | 1.6 ± 0.1 | 1.5 ± 0.0 | 1.6 |
| T16–0090 | 74 | F | C | Former Smoker | 1.5 | 40 | 2012 | N. D. | N. D. | <LOQ |
| T16–0103 | 47 | M | C | Former Smoker | NA | NA | 2016 | N. D. | N. D. | <LOQ |
| T16–0115 | 82 | M | C | Former Smoker | 2 | 2 | NA | N. D. | N. D. | <LOQ |
| T16–0144 | 47 | M | C | Current Smoker | 1 | 30 | -- | N. D. | N. D. | <LOQ |
| T16–0145 | 77 | M | C | Non-smoker | -- | -- | -- | N. D. | N. D. | <LOQ |
| T16–0179 | 65 | M | C | Non-smoker | -- | -- | -- | 0.3 ± 0.3 | N. D. | <LOQ |
| T16–0198a | 59 | M | C | Non-smoker | Smokeless tobacco | -- | -- | N. D. | 0.1 ± 0.0 | <LOQ |
| T16–0206 | 51 | M | C | Non-smoker | -- | -- | -- | 0.7 ± 0.2 | 0.7 ± 0.0 | <LOQ |
| T16–0219 | 81 | M | C | Current Smoker | 1–2 | 30 | -- | 0.1 ± 0.1 | N. D. | <LOQ |
| T16–0227 | 87 | M | C | Former Smoker | NA | NA | 2006 | 0.4 ± 0.1 | N. D. | <LOQ |
| T16–0234b | 60 | M | C | Former Smoker | 1.5 | 30 | 2014 | 0.8 ± 0.0 | 1.7 ± 0.1 | <LOQ |
| T16–0251 | 66 | M | C | Former Smoker | 1 | 29 | 1995 | 2.2 ± 0.1 | 0.1 ± 0.0 | <LOQ |
| T16–0298 | 65 | M | C | Former Smoker | NA | 20 | NA | N. D. | N. D. | <LOQ |
| T16–0301 | 37 | M | C | Non-smoker | -- | -- | -- | N. D. | N. D. | <LOQ |
| T16–0306 | 81 | M | C | Non-smoker | -- | -- | -- | N. D. | N. D. | <LOQ |
| T16–0323 | 70 | M | C | Non-smoker | -- | -- | -- | N. D. | 0.3 ± 0.1 | <LOQ |
| T16–0327c | 78 | M | C | Former Smoker | 1 | 24 | 1977 | 1.0 ± 0.1 | 2.2 ± 0.0 | 1.6 |
| T16–0371 | 69 | M | C | Current Smoker | 0.25 | NA | -- | 1.9 ± 0.0 | 3.6 ± 0.0 | 2.8 |
| T16–0399 | 59 | M | C | Former Smoker | 1.5 | 32 | 1973 | 0.1 ± 0.1 | 0.3 ± 0.1 | <LOQ |
| T16–0406 | 67 | M | C | Non-smoker | -- | -- | -- | 0.6 ± 0.3 | 0.6 ± 0.0 | <LOQ |
| T16–0416 | 59 | M | C | Non-smoker | -- | -- | -- | N. D. | N. D. | <LOQ |
| T16–0433 | 40 | M | AA | Former Smoker | 1 | 30 | 2004 | 0.4 ± 0.3 | 1.0 ± 0.5 | <LOQ |
| T16–0467 | 70 | M | C | Non-smoker | -- | -- | -- | 1.1 ± 0.2 | 1.5 ± 0.5 | <LOQ |
| T16–0469 | 78 | M | C | Former Smoker | 1 | 20 | 1980 | N. D. | N. D. | <LOQ |
| T16–0497 | 62 | M | C | Current Smoker | 1 | 35 | -- | 4.2 ± 0.2 | 1.9 ± 0.2 | 3.1 |
| T16–0561 | 69 | M | C | Former Smoker | 1 | 20 | 1980 | N. D. | N. D. | <LOQ |
| T16–0573 | 72 | M | C | Former Smoker | 1 | 10 | 1996 | 4.1 ± 0.2 | 2.6 ± 0.2 | 3.4 |
| T17–0015 | 45 | M | C | Non-smoker | -- | -- | -- | 1.7 ± 0.1 | 2.4 ± 0.1 | 2.1 |
| T17–0018 | 62 | M | C | Former Smoker | NA | NA | NA | 10.7 ± 0.4 | 9.5 ± 0.3 | 10.1 |
| T17–0052 | 75 | M | C | Former Smoker | NA | NA | NA | 0.1 ± 0.1 | 0.2 ± 0.1 | <LOQ |
| T17–0076 | 72 | M | C | Former Smoker | 1 | 40 | 2009 | 0.5 ± 0.0 | 0.6 ± 0.0 | <LOQ |
| T17–0095 | 76 | M | C | Former Smoker | 0.25 | 60 | NA | 33.8 ± 1.9 | 29.3 ± 2.1 | 31.6 |
| T17–0101 | 73 | M | C | Non-smoker | -- | -- | -- | 2.3 ± 0.6 | 1.1 ± 0.1 | 1.7 |
| T17–0119 | 56 | M | C | Former Smoker | 0.55 | 30 | 2014 | 2.8 ± 0.7 | 3.0 ± 0.1 | 2.9 |
: Subject T16–0198 reported to use only smokeless tobacco products, and not cigarettes.
: Subject T16–0234 reported to be a former smoker but currently uses e-cigarette.
: Subject T16–0327 reported to be a former smoker but currently uses smokeless tobacco.
Figure 2.
The detection of N-(dG-C8)-4-ABP adducts in tumor-adjacent “normal” bladder tissues from bladder cancer patients using nanoUPLC-ESI-Orbitrap-MS2. Subjects that are self-reported “current smokers” are colored in blue, and non-smokers are labeled in green, including a smokeless tobacco user (T16–0198*). The remaining patients are former smokers (black), including one who is a current e-cigarette user (T16–0234**), and one smokeless tobacco user (T16–327**). The insert table summarizes the number of subjects, their smoking status, and N-(dG-C8)-4-ABP levels in FR and matching FFPE tissues. LOQ of N-(dG-C8)-4-ABP is 1.4 adducts per 109 nt.
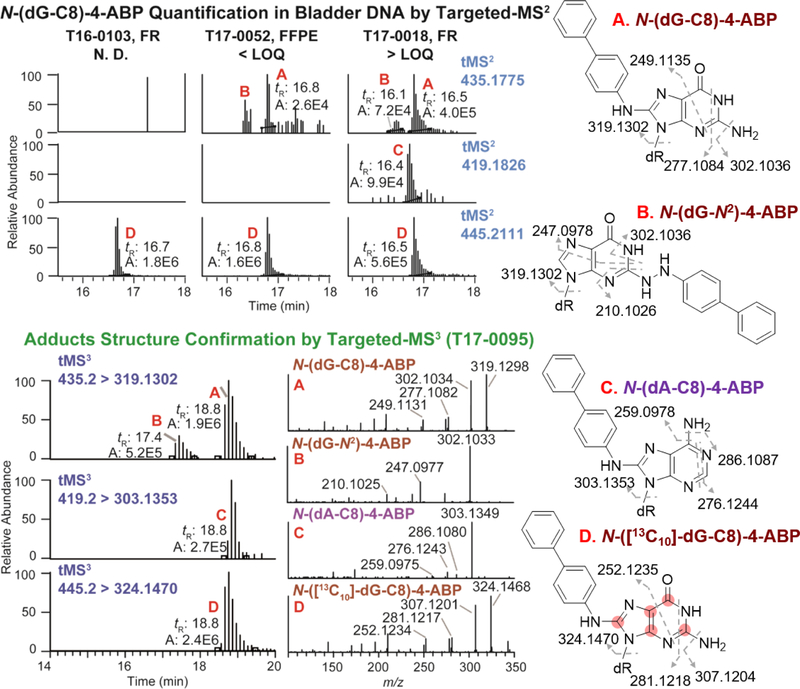
Twelve subjects were positive for N-(dG-C8)-4-ABP with levels ranging from the LOQ (1.4 adducts per 109 nt) up to 31.6 per 109 nt (averaged between FR and FFPE tissues). The two subjects (T17–0095 and T17–0018) who had the highest levels of N-(dG-C8)-4-ABP, at 31.6 and 10.1 adducts per 109 nucleosides, were former smokers. The putative hydrazo adduct N-(dG-N2)-4-ABP,74 an isomer of N-(dG-C8)-4-ABP, and N-(dA-C8)-4-ABP were detected by the targeted-MS2 analysis in these two subjects (Figure 3). These adducts were previously detected in human hepatocytes and in bladder tissue of canines treated with 4-ABP.14,61,75 The product ion spectra acquired at the MS3 scan stage are consistent with the proposed structures (Figure 3). Isotopically labeled internal standards of N-(dG-N2)-4-ABP and N-(dA-C8)-4-ABP were not available for quantification, but their levels were estimated to be ~10 – 20% of N-(dG-C8)-4-ABP, based on the MS responses and assuming similar ionization efficiencies (Figure 3).
Figure 3.
The EIC of 4-ABP DNA adducts measured by nanoUPLC-ESI-Orbitrap-MS2, with structure confirmation by targeted-MS3. Adducts shown are (A), N-(dG-C8)-4-ABP; (B), N-(dG-N2)-4-ABP; (C) N-(dA-C8)-4-ABP; (D), N-([13C10]-dG-C8)-4-ABP. The fragmentation schemes are shown in Figure 6. The ions used to construct EIC of targeted-MS2 were m/z 319.1302 (N-(dG-C8)-4-ABP and N-(dG-N2)-4-ABP), 303.1353 (N-(dA-C8)-4-ABP), 324.1470 (N-([13C10]-dG-C8)-4-ABP); and those of targeted-MS3 were reported in the MS3 spectra. All ions were extracted with theoretical mass at 5 ppm mass tolerance. The internal standard N-([13C10]-dG-C8)-4-ABP contains a total of 10 13C atoms on the molecule (five on guanine and five on dR)
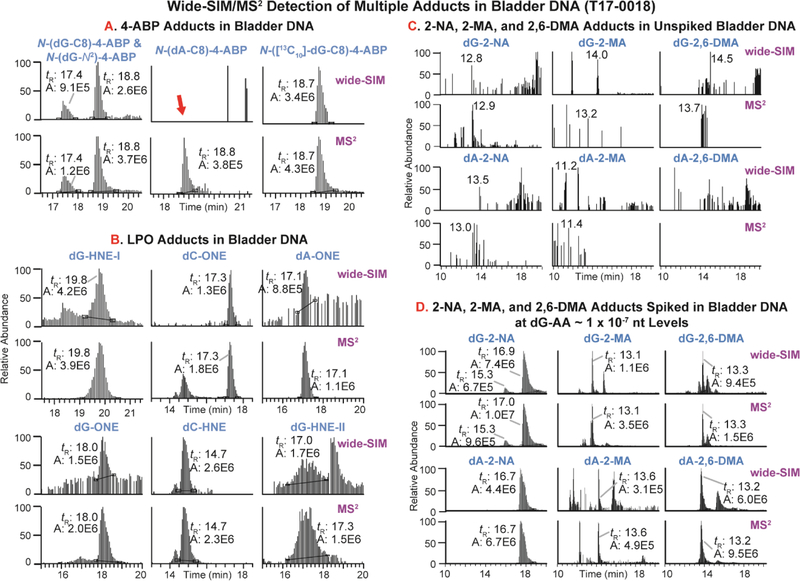
Our wide-SIM/MS2 scanning methodology60 was employed to screen for DNA adducts formed from reactive intermediates of several other AAs, which are possible bladder carcinogens. The scanning method allows for monitoring the frequently observed neutral loss of the 2′-deoxyribose moiety (dR, 116.0473 Da) upon collision-induced dissociation (CID) of the 2′-deoxynucleosides.76 Due to the limiting quantity of tissue, the only DNA tested was that of subjects T17–0018 and T17–0095, who had the highest levels of N-(dG-C8)-4-ABP. The adducts investigated through the generation of the parent and neutral loss EICs included: N-(dG-C8)-4-ABP and its ring-opened oxidized products,70 N-(dA-C8)-4-ABP, dG- and/or dA- adducts of 2-NA,77 2-MA,73 2,6-DMA,78 benzidine,79 and MOCA80 (Table S2). Wide-SIM/MS2 detected N-(dG-C8)-4-ABP and N-(dG-N2)-4-ABP in bladder DNA (Figure 4A); however, DNA adducts of the other listed AAs were not detected (Figure 4C).
Figure 4.
DNA adducts detected in bladder DNA (T17–0018) by wide-SIM/MS2. The co-elution of precursor ions in wide-SIM and aglycone ions in MS2 suggests the presence of (A) 4-ABP and (B) several LPO adducts. The false negative detection of the precursor N-(dA-C8)-4-ABP (indicated by the red arrow) in wide-SIM is explained in the main text. (C) 2-NA, 2-MA, and 2,6-DMA adducts are not detected in human bladder DNA, except when mixed with modified-CT DNA, at (D) dG-AA ~ one per 107 nt level or lower.
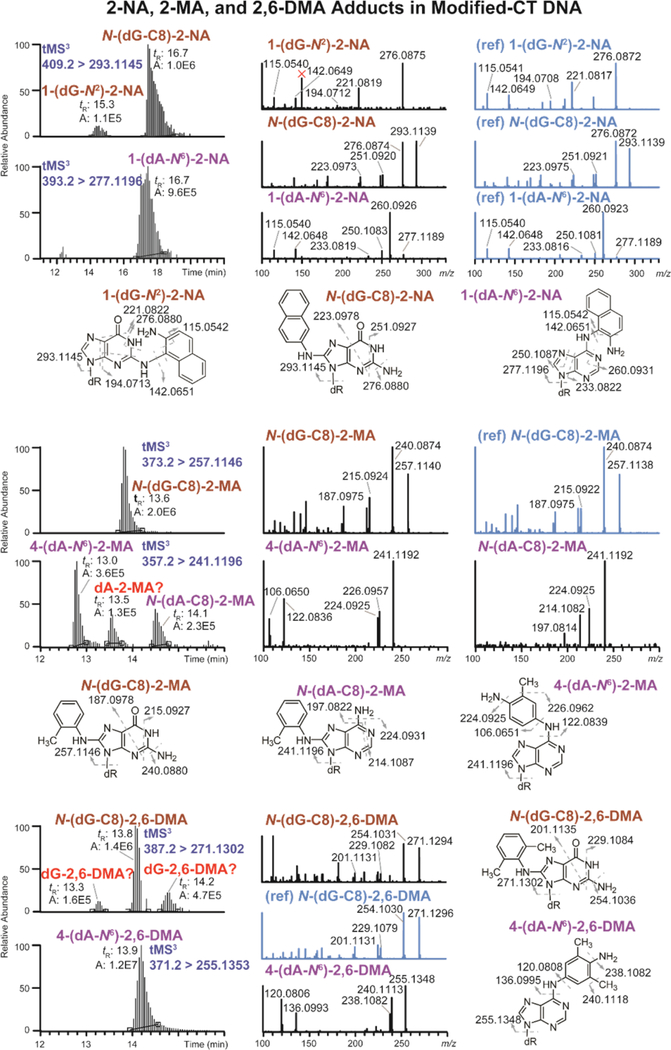
Previous studies using NMR have characterized the adduction products formed between the N-acetoxy intermediates of 2-MA and 2,6-DMA with dG and dA, and some of these adducts have also been observed in CT DNA treated with these N-acetoxy intermediates.73,78,81 However, relatively few AA-DNA adducts have been characterized by mass spectrometric analysis.82,83 In this study, we have characterized the adducts of 2-NA, 2-MA and 2,6-DMA formed with CT DNA. 2-NA, 2-MA, and 2,6-DMA adducts were biomimetically synthesized to evaluate the capacity of wide-SIM/MS2 to screen for these arylamine adducts. The adducts were produced by the reaction of CT DNA under mildly acidic pH conditions with HONH-2-NA to generate its nitrenium ion,71 or by chemical derivatization of other HONH-AAs with pyruvonitrile to generate reactive N-acetoxy intermediates that undergo heterolytic cleavage to produce the nitrenium ions (vide supra).69
The EIC, MS3 spectra, proposed adduct structures, and CID fragmentation are shown in Figure 5. Two isomeric adducts were detected for dG-2-NA, and three isomeric adducts were detected with dA-2-MA, and dG-2,6-DMA. 2-HONH-NA reacts with DNA to form 1-(2′-deoxyguanosin-N2-yl)-2-naphthylamine (1-(dG-N2)-2-NA), 1-(2′-deoxyadenosin-N6-yl)-2-naphthylamine (1-(dA-N6)-2-NA) and a ring-opened form of a purine ring-opened derivative of N-(2′-deoxyguanosin-8-yl)-2-naphthylamine (N-(dG-C8)-2-NA), tentatively identified as l-[5-(2,6-diamino-4-oxopyrimidinyl-N6-deoxyriboside)]-3-(2-naphthyl)urea.71 The product ion spectra that we acquired at the MS3 scan stage are consistent with the structures of the previously reported 2-NA adducts, except that we detected the intact N-(dG-C8)-2-NA and not a ring-opened form of the adduct. The product ion spectra of 2-NA, 2-MA, and 2,6-DMA are discussed below.
Figure 5.
EIC and MS3 spectra of the putative DNA adducts from the 2-NA, 2-MA, and 2,6-DMA-modified CT DNA. dG-AA adducts were estimated at ~one per 107 nt level relative to the response of N-([13C10]-dG-C8)-4-ABP. The synthetic adduct standards (ref), 1-(dG-N2)-2-NA, N-(dG-C8)-2-NA, 1-(dA-N6)-2-NA, N-(dG-C8)-2-MA, and N-(dG-C8)-2,6-DMA are highlighted in blue. The adducts at tR13.5 in dA-2-MA transition, and adducts at tR 13.1 and 14.2 in dG-2,6-DMA transition were not assigned (the MS3 spectra are shown in Figure S2).
The accurate mass of the fragment ions and observed neutral losses measured at the MS3 scan stage by Orbitrap, and the spectral pattern among the structurally similar adducts can be utilized to facilitate the assignment of the proposed structures of the AA DNA adducts. We have tabulated the characteristic fragment ions of the proposed adducts (fragmentation schemes are shown in Figure 6), and reported the measured mass to those of theoretical ones, and reported the relative mass differences in ppm (Table S3). The aglycones of all N-(dG-C8)-AA adducts undergo CID to form the following fragment ions: [A + H – NH3]+ ([A + H – 17.0266]+), [A + H – NH2CN]+ ([A + H – 42.0218]+), and [A + H – NH2CN-CO]+ ([A + H – 70.0167]+),61,83 with a mass accuracy below 5 ppm. Similarly, N-(dA-C8)-AA adducts with common fragment ions of [A + H – NH3]+ ([A + H – 17.0266]+), [A + H – HCN]+ ([A + H – 27.0109]+), and [A + H – NH3 – HCN]+ ([A – + H – 44.0375]+) 61 were observed for 4-ABP, and 2-MA adducts, but not for 2-NA and 2,6-DMA. 4-ABP is the only chemical that appears to form a hydrazo adduct (N-(dG-N2)-4-ABP).14 The assignment of 4-(dA-N6)-2-MA and 4-(dA-N6)-2,6-DMA are discussed in S1 in the supporting information.
Figure 6.
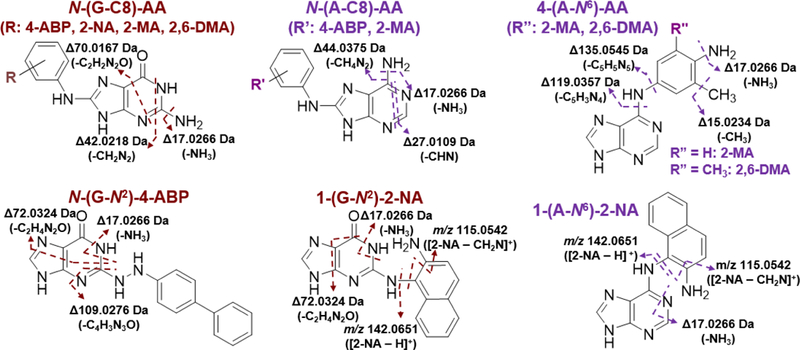
The proposed fragmentation scheme of the aglycone ions of the AA-DNA adducts. The MS3 spectra of the individual adducts are shown in Figure 3 and Figure 5.
On the basis of MS3 spectra assignments, the following adducts are observed: N-(dG-C8)-2-NA, 1-(dG-N2)-2-NA, and 1-(dA-N6)-2-NA; adducts of 2-MA, N-(dG/dA-C8)-2-MA and 4-(dA-N6)-2-MA; adducts of 2,6-DMA, N-(dG-C8)-2,6-DMA and 4-(dA-N6)-2,6-DMA. The MS3 spectra of the synthetic standards of N-(dG-C8)-2-NA, 1-(dG-N2)-2-NA, 1-(dA-N6)-2-NA, N-(dG-C8)-2-MA, and N-(dG-C8)-2,6-DMA (Figure 5) closely match those spectra obtained from chemically modified DNA (highlighted in blue in Figure 5) The relative amount of the major dG vs. dA adducts formed from the incubation of CT DNA at pH 5 with N-hydroxy-2-NA, or CT DNA at pH 7.1 with N-acetoxy-2-MA/2,6-DMA, were dG-2-NA ≈ dA-2-NA, dG-2-MA >> dA-2-MA (about two-fold), whereas dA-2,6-DMA >> dG-2,6-DMA (about 5 times), judging by the MS response and assuming a similar ionization efficiency among all AA-adducts.
Thereafter, the AA-treated CT DNA at levels of ~one arylamine adduct per 107 (shown in Figure 4D), or at levels of ~one arylamine adduct per108 nt (Figure S1B) was mixed with human bladder DNA. The levels of 2-NA-, 2-MA-, and 2,6-DMA DNA adducts were estimated based on their EICs of the dG-C8-AAs at the MS2 scan stage, relative to that of N-([13C10]-dG-C8)-4-ABP. The wide-SIM/MS2 chromatograms reveal the presence of multiple isomeric AA adducts formed with dG and dA, which were further characterized by targeted-MS3. Thus, wide-SIM/MS2 is capable of screening for a wide array of AA-DNA adducts in human bladder.
Wide-SIM/MS2 scanning is a robust screening method for identification of DNA adducts,60 however; false negative detections of adducts can occur, particularly in the wide-SIM scan of the precursor ions. The Orbitrap MS system utilizes automatic gain control (AGC) to tightly regulate the number of ions or charges entering the Orbitrap to avoid space-charging effects that would cause a deterioration in mass resolution and a drop-off in sensitivity.84 When background ions are abundant, the AGC limit is rapidly reached and the signals of the DNA adducts can fail to reach a sufficient abundance for detection. Alternatively, when the resolution of the Orbitrap is insufficient to resolve the adduct ion from an adjacent background ion, the resultant merged signal will be beyond the specified mass tolerance and lead to a false negative detection.60 An example is shown in Figure 4A, where the precursor ion of N-(dA-C8)-4-ABP in the bladder specimen was not detected in the wide-SIM scan at 60,000 resolution, even though its aglycone was a prominent feature in the MS2 scan. This interference also occurred in CT DNA modified with HONH-4-ABP (Figure 7A1 and 7A2).
Figure 7.
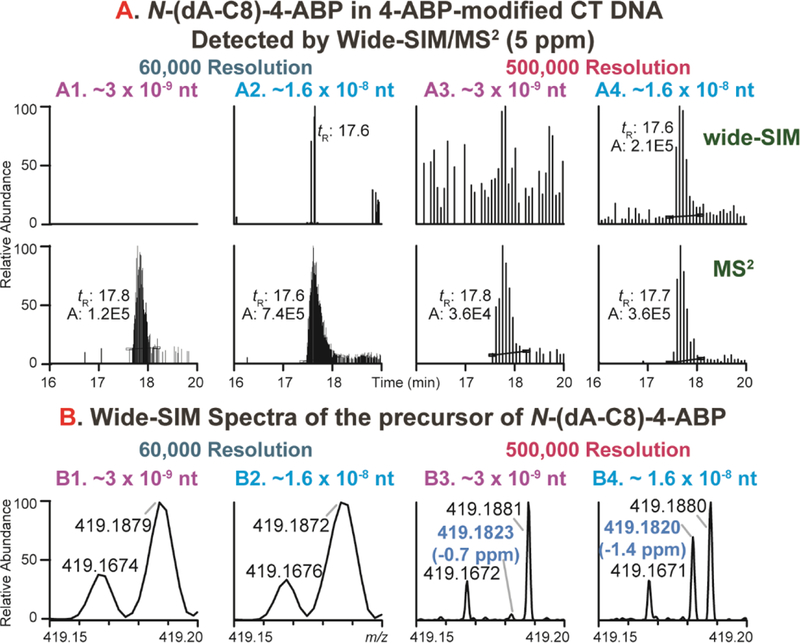
Wide-SIM/MS2 detection of N-(dA-C8)-4-ABP from 4-ABP-modified CT DNA at N-(dA-C8)-4-ABP levels ~3 adduct per 109 and ~1.6 adduct per 108 nt. Data were acquired at 60,000 or 500,000 resolution. (A1 - A4) the EICs of dA-C8–4-ABP precursor (m/z 419.1826) in wide-SIM and aglycones in MS2 at 5 ppm mass tolerance, while (B1 – B4) shows the MS spectra of wide-SIM scan averaged from tR 17.55 to 17.85 min between m/z 419.15 to 419.20. The peak of N-(dA-C8)-4-ABP is highlighted in blue.
We compared the signals of the precursor ion [M + H]+ (theoretical m/z = 419.1826) of N-(dA-C8)-4-ABP in wide-SIM with a mass tolerance of 5 ppm at 60,000 or 500,000 resolution, using 4-ABP-modified CT DNA with N-(dA-C8)-4-ABP at ~3 adducts per 109 and ~1.6 per 108 nt levels (Figure 7). A reproducible signal with a Gaussian chromatographic peak was obtained only at the level of 1.6 adduct per 108 nt with 500,000 resolution (Figure 7A4). The averaged MS spectrum of the precursor ion from tR 17.55 to 17.85 min revealed the presence of an interfering peak at m/z 419.1880 (500,000 resolution, Figure 7B3 and 7B4 13 ppm apart from dA-C8–4-ABP), which requires a resolving power of ~100,000 (FWHM at m/z 200) to achieve a 10% valley separation. We constructed EIC of the ions for dA-C8–4-ABP and the interferent at 5, 10, and 15 ppm mass tolerances, which further underscores the adverse effect of the isobaric interference on detection of N-(dA-C8)-4-ABP (Figure S3).
The overlaid MS spectra of N-(dA-C8)-4-ABP in wide-SIM scan at 60,000 and 500,000 resolution are shown in Figure 8 (1.6 per 108 nt level). At the apex of the chromatographic peak (tR 17.64 – 17.66 min, Figure 8A), the intensity of N-(dA-C8)-4-ABP (m/z = 419.1826) was similar to that of the interfering ion (red trace, 500,000 resolution). Thus, at 60,000 resolution (teal trace), the detected m/z was at the middle point of the two ions (reported as m/z 419.1853). In contrast, at the tail of the chromatographic peak (tR 17.84 – 17.86 min, Figure 8B), the intensity of the N-(dA-C8)-4-ABP was 20% of the interfering ion (red trace, 500,000 resolution), and the reported m/z at 60,000 resolution shifted towards the interfering ion (m/z 419.1880). Therefore, the inability to detect the precursor ion of N-(dA-C8)-4-ABP in SIM was mainly due to insufficient resolution to separate it from the interfering ion.
Figure 8.
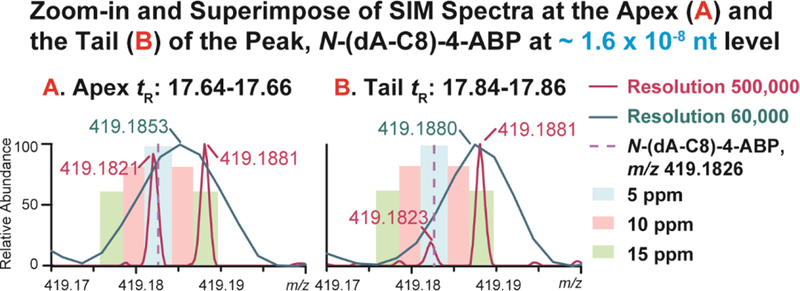
The overlaid MS spectra of the chromatographic peak of N-(dA-C8)-4-ABP precursor in wide-SIM scan at the apex (A, tR 17.64–17.66 min) and at the tail (B, tR 17.84–17.86 min) under 60,000 resolution (teal trace) or 500,000 resolution (red trace). The theoretical m/z of N-(dA-C8)-4-ABP, m/z 419.1826, is illustrated by the red dash line. The light blue box indicated the 5 ppm mass tolerance, the pink box, 10 ppm. The interfering ion, m/z 419.1880, is 13 ppm away from N-(dA-C8)-4-ABP and falls in the green box, which corresponds to 15 ppm mass tolerance.
An increase in the AGC can improve the intrascan dynamic range (the maximum abundance ratio between the most abundant and the least abundant signal observable within a given spectrum) and the LOD/LOQ,85 but the slower transient length will result in a reduced number of scans across the peak. When the AGC value was increased from 5E4 to 2E5 in the SIM scan of the wide-SIM/MS2 event and the maximum injection time was increased from 100 ms to 1014 ms to match the transient length required for 500,000 resolution, the measured injection times for the wide-SIM data acquisition, encompassing the N-(dA-C8)-4-ABP mass, ranged between 200 – 300 ms across the peak, enabled the detection of N-(dA-C8)-4-ABP at a level of 1.6 per 108 nt (Figure 7A2 and 7A4). However, the DNA adduct was not detected at a level of modification of 3 adducts per 109 nt at 60,000 resolution (Figure 7A1), and the peak was just above the background signal at a resolution of 500,000 (Figure 7A1 and 7A3), indicating that the limit of detection had been reached in wide-SIM.
Wide-SIM/MS2 also detected several DNA adducts of endogenous LPO products in bladder DNA. The dG-HNE-I, (m/z 424.2191), dG-HNE-II (m/z 422.2034), dC-HNE (m/z 382.1973), dG-ONE (m/z 404.1929), dA-ONE (m/z 388.1079), and dC-ONE (m/z 364.1867) adducts were detected (Figure 4B). These findings are consistent with our DNA adduct analysis of LPO adducts in human prostate tissues.59 The structures of the adducts are supported by the product ion spectra acquired at the MS3 stage (Figure S4). We are in the process of determining the potential for artifactual formation of LPO-DNA adducts generated during isolation and enzymatic digestion of DNA. Hence, any interpretation about the biological significance of the levels and the types of LPO adducts formed in bladder should be made with caution.
Discussion
We used a stable isotope dilution HR-targeted-MS2 method to quantify N-(dG-C8)-4-ABP adduct in non-tumor-adjacent bladder tissues of bladder cancer patients. We have observed that N-(dG-C8)-4-ABP, a major adduct of 4-ABP,14 is present at about 5 to 10 times higher levels than those of the tentatively assigned N-(dG-N2)-4-ABP and N-(dA-C8)-4-ABP adducts (assuming similar ionization efficiencies and responses of these adducts). The range of N-(dG-C8)-4-ABP levels observed is similar to those values reported by Talaska48 and later by Lee using a non-specific 32P-postlabeling,23 and by Lin and Bohm, employing GC-NICI-MS.52,54 However, the levels of adducts reported are much lower than those values reported by Curigliano using IHC,50 or by Airoldi with GC-NICI-MS,53 and Zayas by HPLC-QqQ-MS22 (Table 1). In our pilot study, we found highly variable levels of N-(dG-C8)-4-ABP adduct among current smokers, former smokers, and non-smokers.
The dG/dA adducts of multiple AAs were successfully screened by wide-SIM/MS2 and targeted-MS3 analysis with CT DNA modified with the reactive AA intermediates. The AA-modified DNA formed in vitro was spiked into human bladder DNA to confirm wide-SIM/MS2 was able to detect these adducts in a human DNA matrix. The adducts could be measured at levels approaching 1 adduct per 108 nt. Wide-SIM/MS2 also detected LPO adducts in human bladder DNA. The infrequent detection and the low levels of 4-ABP adducts and the absence of DNA adducts of alkylanilines may be attributed in part to inaccurate self-reporting of tobacco usage at the time of bladder surgery. The smoking status, including number of cigarettes smoked per day, the duration of smoking, and the time of quitting for former smokers were self-reported (Table 2). We do not know the time interval that the current smokers had refrained from smoking cigarettes before surgery, and DNA repair may account for the low levels of 4-ABP adducts and the absence of alkylaniline DNA adducts. The bladder tissues in this study came from a biobank, and a more accurate assessment of tobacco usage at the time of surgery, such as by measurement of nicotine or cotinine in plasma or urine, should be included in future studies to better correlate DNA adduct levels and smoking status among the subjects. Moreover, a dietary questionnaire was not available, and thus, exposures to HAAs through the diet are uncertain. The measurements of urinary AA and HAA biomarkers, or possibly hair biomarkers for HAAs would provide a better understanding of meat consumption.86
The formation of DNA adducts of 14C-labeled alkylanilines (2,6-DMA, 3,5-dimethylaniline, 3-ethylaniline) has been reported in liver and bladder of mice dosed with these chemicals, as measured by accelerator mass spectrometry;87 however, the identities of the DNA adducts are unknown due to the nature of the detection method. Jones treated mice with 4-ABP, 4-nitrobiphenyl and a variety of methylated anilines; however, only 4-ABP and 4-nitrobiphenyl were reported to form DNA adducts.69 In contrast, all dosed compounds formed sulfinamide adducts with hemoglobin (Hb), demonstrating that AAs underwent bioactivation through N-oxidation in rodents. Similarly, Hb sulfinamide adducts of methylated anilines have been detected in humans.88 To our knowledge, there are no reports in the literature on the measurements of DNA adducts of alkylanilines in human bladder by LC/MS methods. There is one report on the detection of a putative 2-MA adduct in human bladder. The method of analysis employed acid hydrolysis of DNA to liberate 2-MA, followed by GC-MS, yielding remarkably frequent detection and high adduct levels (92% positivity, 1.3 to 4.6 adducts per 106 nt).89 However, this method of detection has limitations in specificity because the mass spectrometer used was a single sector instrument with nominal mass resolution.
Recent studies propose that DNA adduct damage induced by alkylanilines may not occur through direct DNA adduct formation with the HONH-AA metabolites, but rather through their para-aminophenol metabolites, which undergo redox cycling via their quinone imine intermediates, producing reactive oxygen species (ROS) as by-products that can damage DNA and induce mutations.90–92 It is noteworthy that we detected several lipid peroxidation DNA adducts in bladder. Some of these adducts may be produced by ROS in tobacco smoke, or possibly by reactive alkylaniline quinone imine intermediates formed in the bladder, and will be the subject of further study.
Our untargeted wide-SIM/MS2 scanning method in conjunction with targeted-MSn are powerful approaches to screen for DNA adducts and will be further employed to advance our understanding of hazardous agents that form DNA adducts in the bladder and contribute to bladder carcinogenesis.
Supplementary Material
Acknowledgement
We thank Dr. Frederick A. Beland from the National Center for Toxicology Research/US FDA for providing 4-ABP treated CT-DNA, Dr. Gabriele Sabbioni from the Institute of Environmental and Occupational Toxicology in Switzerland for the synthetic N-(dG-C8)-2-MA, N-(dG-C8)-2,6-DMA adduct standards, Dr. Ian Blair from the University of Pennsylvania for the dG-ONE and dA-ONE synthetic standards, and Dr. Cole Drifka and the staff from BioNet Tissue Procurement for collecting human bladder samples.
Funding Support
This work is funded by R33CA18679 and R01CA220367 from the National Cancer Institute, R01ES019564 from the National Institute of Environmental Health Sciences, and by the National Center for Advancing Translational Sciences of the of the National Institutes of Health award number UL1TR000114. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, University of Minnesota, funded in part by Cancer Center Support Grant CA-077598. Salary support for P.W.V was provided by the National Cancer Institute of the National Institutes of Health under Award Number R50CA211256.
Abbreviations
- AA
aromatic amine
- HAAs
heterocyclic aromatic amines
- FFPE
formalin-fixed paraffin-embedded
- 4-ABP
4-aminobiphenyl
- AαC
2-amino-9H-pyrido[2,3-b]indole
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- N-(dG-C8)-4-ABP
N-(2′-deoxyguanosin-8-yl)-4-ABP
- N-(dA-C8)-4-ABP
N-(2′-deoxyadenosin-8-yl)-4-ABP
- N-(dG-N2)-4-ABP
N-(2′-deoxyguanosin-N2-yl)-4-ABP
- N-(dG-C8)-AαC
N-(2′-deoxyguanosin-8-yl)-AαC
- N-(dG-C8)-MeIQx
N-(2′-deoxyguanosin-8-yl)-MeIQx
- N-(dG-C8)-PhIP
N-(2′-deoxyguanosin-8-yl)-PhIP
- 2-NA
2-nephthylamine
- 2-MA
2-methylaniline
- 2,6-DMA
2,6-dimethylaniline
- LPO
lipid peroxidation
- SIM
selected ion monitoring
- BC
bladder cancer
- IHC
immunohistochemistry
- GC-NICI-MS
gas chromatography with negative ion chemical ionization mass spectrometry
- HPLC
high-performance liquid chromatography
- QqQ
triple quadrupole
- nanoUPLC-ESI-Orbitrap-MSn
nano flow ultra-performance liquid chromatography-electrospray ionization-Orbitrap-multi-stage MS
- HRAM
high-resolution accurate mass
- CT DNA
calf thymus DNA
- βME
β-mercaptoethanol
- EtOAc
ethyl acetate
- ONE
4-oxo-(2E)-nonenal
- FWHM
full width at half maximum
- HCD
high energy collision-induced dissociation
- 1-(dG-N2)-2-NA
1-(2′-deoxyguanosin-N2-yl)-2-naphthylamine
- 1-(dA-N6)-2-NA
1-(2′-deoxyadenosin-N6-yl)-2-naphthylamine
- N-(dG-C8)-2-NA
N-(2′-deoxyguanosin-8-yl)-2-naphthylamine
- N-(dG-C8)-2-MA
N-(2′-deoxyguanosin-8-yl)-2-methylaniline
- N-(dA-C8)-2-MA
N-(2′-deoxyadenosin-8-yl)-2-methylaniline
- 4-(dA-N6)-2-MA
4-(2′-deoxyadenosin-N6-yl)-2-methylaniline
- N-(dG-C8)-2,6-DMA
N-(2′-deoxyguanosin-8-yl)-2,6-dimethylaniline
- 4-(dA-N6)-2,6-DMA
4-(2′-deoxyadenosin-N6-yl)-2,6-dimethylaniline
- ROS
reactive oxygen species
References
- (1).Ferlay J, Shin HR, Bray F, Forman D, Mathers C, and Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917. [DOI] [PubMed] [Google Scholar]
- (2).Kaufman DS, Shipley WU, and Feldman AS (2009) Bladder cancer. Lancet 374, 239–249. [DOI] [PubMed] [Google Scholar]
- (3).Jemal A, Bray F, Center MM, Ferlay J, Ward E, and Forman D (2011) Global cancer statistics. CA Cancer J. Clin 61, 69–90. [DOI] [PubMed] [Google Scholar]
- (4).(2010) International Agency for Research on Cancer. IARC Monographs: Some aromatic amines, organic dyes, and related exposures. Vol. 99, International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- (5).Zeegers MPA, Tan FES, Dorant E, and van den Brandt PA (2000) The impact of characteristics of cigarette smoking on urinary tract cancer risk - A meta-analysis of epidemiologic studies. Cancer 89, 630–639. [DOI] [PubMed] [Google Scholar]
- (6).Murta-Nascimento C, Schmitz-Drager BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, and Malats N (2007) Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J. Urol 25, 285–295. [DOI] [PubMed] [Google Scholar]
- (7).Hoffmann D, Hoffmann I, and El-Bayoumy K (2001) The less harmful cigarette: a controversial issue. a tribute to Ernst L. Wynder. Chem. Res. Toxicol 14, 767–790. [DOI] [PubMed] [Google Scholar]
- (8).(1986) International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking International Agency for Research on Cancer, Lyon, France. [PMC free article] [PubMed] [Google Scholar]
- (9).Tokiwa H, Nakagawa R, and Horikawa K (1985) Mutagenic/carcinogenic agents in indoor pollutants; the dinitropyrenes generated by kerosene heaters and fuel gas and liquefied petroleum gas burners. Mutat. Res 157, 39–47. [DOI] [PubMed] [Google Scholar]
- (10).Cioni F, Bartolucci G, Pieraccini G, Meloni S, and Moneti G (1999) Development of a solid phase microextraction method for detection of the use of banned azo dyes in coloured textiles and leather. Rapid Commun. Mass Spectrom 13, 1833–1837. [DOI] [PubMed] [Google Scholar]
- (11).Turesky RJ, Freeman JP, Holland RD, Nestorick DM, Miller DW, Ratnasinghe DL, and Kadlubar FF (2003) Identification of aminobiphenyl derivatives in commercial hair dyes. Chem. Res. Toxicol 16, 1162–1173. [DOI] [PubMed] [Google Scholar]
- (12).Garrigos MC, Reche F, Pernias K, and Jimenez A (2000) Optimization of parameters for the analysis of aromatic amines in finger-paints. J. Chromatogr. A 896, 291–298. [DOI] [PubMed] [Google Scholar]
- (13).(2012) International Agency for Research on Cancer. IARC Monographs: Chemical Agents and Related Occupations. Vol. 100F, International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- (14).Beland FA, Beranek DT, Dooley KL, Heflich RH, and Kadlubar FF (1983) Arylamine-DNA adducts in vitro and in vivo: their role in bacterial mutagenesis and urinary bladder carcinogenesis. Environ. Health Perspect 49, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Feng Z, Hu W, Rom WN, Beland FA, and Tang MS (2002) N-hydroxy-4-aminobiphenyl-DNA binding in human p53 gene: sequence preference and the effect of C5 cytosine methylation. Biochemistry 41, 6414–6421. [DOI] [PubMed] [Google Scholar]
- (16).Kadlubar FF (1991) Carcinogenic aromatic amine metabolism and DNA adduct detection in humans. Xenobiotics and Cancer, 329–338. [PubMed]
- (17).Nakajima M, Itoh M, Sakai H, Fukami T, Katoh M, Yamazaki H, Kadlubar FF, Imaoka S, Funae Y, and Yokoi T (2006) CYP2A13 expressed in human bladder metabolically activates 4-aminobiphenyl. Int. J. Cancer 119, 2520–2526. [DOI] [PubMed] [Google Scholar]
- (18).Yu MC, Skipper PL, Taghizadeh K, Tannenbaum SR, Chan KK, Henderson BE, and Ross RK (1994) Acetylator phenotype, aminobiphenyl-hemoglobin adduct levels, and bladder cancer risk in white, black, and Asian men in Los Angeles, California. J. Natl. Cancer Inst 86, 712–716. [DOI] [PubMed] [Google Scholar]
- (19).Castelao JE, Yuan JM, Skipper PL, Tannenbaum SR, Gago-Dominguez M, Crowder JS, Ross RK, and Yu MC (2001) Gender- and smoking-related bladder cancer risk. J. Natl. Cancer Inst 93, 538–545. [DOI] [PubMed] [Google Scholar]
- (20).Yu MC, Skipper PL, Tannenbaum SR, Chan KK, and Ross RK (2002) Arylamine exposures and bladder cancer risk. Mutat. Res 506–507, 21–28. [DOI] [PubMed] [Google Scholar]
- (21).Poirier MC, and Beland FA (1997) Aromatic amine DNA adduct formation in chronically-exposed mice: considerations for human comparison. Mutat. Res 376, 177–184. [DOI] [PubMed] [Google Scholar]
- (22).Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper P, Yu MC, Tannenbaum SR, and Wogan GN (2007) Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis 28, 342–349. [DOI] [PubMed] [Google Scholar]
- (23).Lee HW, Wang HT, Weng MW, Hu Y, Chen WS, Chou D, Liu Y, Donin N, Huang WC, Lepor H, Wu XR, Wang H, Beland FA, and Tang MS (2014) Acrolein- and 4-aminobiphenyl-DNA adducts in human bladder mucosa and tumor tissue and their mutagenicity in human urothelial cells. Oncotarget 5, 3526–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lumbreras B, Garte S, Overvad K, Tjonneland A, Clavel-Chapelon F, Linseisen JP, Boeing H, Trichopoulou A, Palli D, Peluso M, Krogh V, Tumino R, Panico S, Bueno-De-Mesquita HB, Peeters PH, Lund E, Martinez C, Dorronsoro M, Barricarte A, Chirlaque MD, Quiros JR, Berglund G, Hallmans G, Day NE, Key TJ, Saracci R, Kaaks R, Malaveille C, Ferrari P, Boffetta P, Norat T, Riboli E, Gonzalez CA, and Vineis P (2008) Meat intake and bladder cancer in a prospective study: a role for heterocyclic aromatic amines? Cancer Causes Control 19, 649–656. [DOI] [PubMed] [Google Scholar]
- (25).Ferrucci LM, Sinha R, Ward MH, Graubard BI, Hollenbeck AR, Kilfoy BA, Schatzkin A, Michaud DS, and Cross AJ (2010) Meat and components of meat and the risk of bladder cancer in the NIH-AARP Diet and Health Study. Cancer 116, 4345–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Michaud DS, Holick CN, Giovannucci E, and Stampfer MJ (2006) Meat intake and bladder cancer risk in 2 prospective cohort studies. Am. J. Clin. Nutr 84, 1177–1183. [DOI] [PubMed] [Google Scholar]
- (27).Lin J, Forman MR, Wang J, Grossman HB, Chen M, Dinney CP, Hawk ET, and Wu X (2012) Intake of red meat and heterocyclic amines, metabolic pathway genes and bladder cancer risk. Int. J. Cancer 131, 1892–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Balbi JC, Larrinaga MT, De SE, Mendilaharsu M, Ronco AL, Boffetta P, and Brennan P (2001) Foods and risk of bladder cancer: a case-control study in Uruguay. Eur. J. Cancer Prev 10, 453–458. [DOI] [PubMed] [Google Scholar]
- (29).Grieb SM, Theis RP, Burr D, Benardot D, Siddiqui T, and Asal NR (2009) Food groups and renal cell carcinoma: results from a case-control study. J. Am. Diet. Assoc 109, 656–667. [DOI] [PubMed] [Google Scholar]
- (30).Catsburg CE, Gago-Dominguez M, Yuan JM, Castelao JE, Cortessis VK, Pike MC, and Stern MC (2014) Dietary sources of N-nitroso compounds and bladder cancer risk: findings from the Los Angeles bladder cancer study. Int. J. Cancer 134, 125–135. [DOI] [PubMed] [Google Scholar]
- (31).Faramawi MF, Johnson E, Fry MW, Sall M, and Zhou Y (2007) Consumption of different types of meat and the risk of renal cancer: meta-analysis of case-control studies. Cancer Causes Control 18, 125–133. [DOI] [PubMed] [Google Scholar]
- (32).Melkonian SC, Daniel CR, Ye Y, Tannir NM, Karam JA, Matin SF, Wood CG, and Wu X (2016) Gene-environment interaction of genome-wide association study-identified susceptibility loci and meat-cooking mutagens in the etiology of renal cell carcinoma. Cancer 122, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Garcia-Closas R, Garcia-Closas M, Kogevinas M, Malats N, Silverman D, Serra C, Tardon A, Carrato A, Castano-Vinyals G, Dosemeci M, Moore L, Rothman N, and Sinha R (2007) Food, nutrient and heterocyclic amine intake and the risk of bladder cancer. Eur. J. Cancer 43, 1731–1740. [DOI] [PubMed] [Google Scholar]
- (34).Augustsson K, Skog K, Jagerstad M, Dickman PW, and Steineck G (1999) Dietary heterocyclic amines and cancer of the colon, rectum, bladder, and kidney: a population-based study. Lancet 353, 703–707. [DOI] [PubMed] [Google Scholar]
- (35).Wu JW, Cross AJ, Baris D, Ward MH, Karagas MR, Johnson A, Schwenn M, Cherala S, Colt JS, Cantor KP, Rothman N, Silverman DT, and Sinha R (2012) Dietary intake of meat, fruits, vegetables, and selective micronutrients and risk of bladder cancer in the New England region of the United States. Br. J. Cancer 106, 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sugimura T, Wakabayashi K, Nakagama H, and Nagao M (2004) Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci 95, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Turesky RJ, and Le Marchand L (2011) Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem. Res. Toxicol 24, 1169–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Peters U, DeMarini DM, Sinha R, Brooks LR, Warren SH, Chatterjee N, and Rothman N (2003) Urinary mutagenicity and colorectal adenoma risk. Cancer Epidemiol. Biomarkers Prev 12, 1253–1256. [PubMed] [Google Scholar]
- (39).Smith CJ, McKarns SC, Davis RA, Livingston SD, Bombick BR, Avalos JT, Morgan WT, and Doolittle DJ (1996) Human urine mutagenicity study comparing cigarettes which burn or primarily heat tobacco. Mutat. Res 361, 1–9. [DOI] [PubMed] [Google Scholar]
- (40).DeMarini DM (2004) Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat. Res 567, 447–474. [DOI] [PubMed] [Google Scholar]
- (41).Peluso M, Castegnaro M, Malaveille C, Friesen M, Garren L, Hautefeuille A, Vineis P, Kadlubar F, and Bartsch H (1991) 32P Postlabelling analysis of urinary mutagens from smokers of black tobacco implicates 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) as a major DNA-damaging agent. Carcinogenesis 12, 713–717. [DOI] [PubMed] [Google Scholar]
- (42).Turesky RJ, Yuan J-M, Wang R, Peterson S, and Yu MC (2007) Tobacco smoking and urinary levels of 2-amino-9H-pyrido[2,3-b]indole in men of Shanghai, China. Cancer Epidemiol. Biomarkers Prev 16, 1554–1560. [DOI] [PubMed] [Google Scholar]
- (43).Konorev D, Koopmeiners JS, Tang Y, Franck Thompson EA, Jensen JA, Hatsukami DK, and Turesky RJ (2015) Measurement of the heterocyclic amines 2-amino-9H-pyrido[2,3-b]indole and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in urine: effects of cigarette smoking. Chem. Res. Toxicol 28, 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).el-Bayoumy K, Donahue JM, Hecht SS, and Hoffmann D (1986) Identification and quantitative determination of aniline and toluidines in human urine. Cancer Res 46, 6064–6067. [PubMed] [Google Scholar]
- (45).Grimmer G, Dettbarn G, Seidel A, and Jacob J (2000) Detection of carcinogenic aromatic amines in the urine of non-smokers. Sci. Total Environ 247, 81–90. [DOI] [PubMed] [Google Scholar]
- (46).Seidel A, Dahmann D, Krekeler H, and Jacob J (2002) Biomonitoring of polycyclic aromatic compounds in the urine of mining workers occupationally exposed to diesel exhaust. Int. J. Hyg. Environ. Health 204, 333–338. [DOI] [PubMed] [Google Scholar]
- (47).Riedel K, Scherer G, Engl J, Hagedorn HW, and Tricker AR (2006) Determination of three carcinogenic aromatic amines in urine of smokers and nonsmokers. J. Anal. Toxicol 30, 187–195. [DOI] [PubMed] [Google Scholar]
- (48).Talaska G, al-Juburi AZ, and Kadlubar FF (1991) Smoking related carcinogen-DNA adducts in biopsy samples of human urinary bladder: identification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl as a major adduct. Proc. Natl. Acad. Sci. U. S. A 88, 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO Jr., and Kadlubar FF (1992) Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem. Res. Toxicol 5, 691–697. [DOI] [PubMed] [Google Scholar]
- (50).Curigliano G, Zhang YJ, Wang LY, Flamini G, Alcini A, Ratto C, Giustacchini M, Alcini E, Cittadini A, and Santella RM (1996) Immunohistochemical quantitation of 4-aminobiphenyl-DNA adducts and p53 nuclear overexpression in T1 bladder cancer of smokers and nonsmokers. Carcinogenesis 17, 911–916. [DOI] [PubMed] [Google Scholar]
- (51).Hsu TM, Zhang YJ, and Santella RM (1997) Immunoperoxidase quantitation of 4-aminobiphenyl- and polycyclic aromatic hydrocarbon-DNA adducts in exfoliated oral and urothelial cells of smokers and nonsmokers. Cancer Epidemiol. Biomarkers Prev 6, 193–199. [PubMed] [Google Scholar]
- (52).Lin D, Lay JO Jr., Bryant MS, Malaveille C, Friesen M, Bartsch H, Lang NP, and Kadlubar FF (1994) Analysis of 4-aminobiphenyl-DNA adducts in human urinary bladder and lung by alkaline hydrolysis and negative ion gas chromatography-mass spectrometry. Environ. Health Perspect 102 Suppl 6, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Airoldi L, Orsi F, Magagnotti C, Coda R, Randone D, Casetta G, Peluso M, Hautefeuille A, Malaveille C, and Vineis P (2002) Determinants of 4-aminobiphenyl-DNA adducts in bladder cancer biopsies. Carcinogenesis 23, 861–866. [DOI] [PubMed] [Google Scholar]
- (54).Bohm F, Schmid D, Denzinger S, Wieland WF, and Richter E (2011) DNA adducts of ortho-toluidine in human bladder. Biomarkers 16, 120–128. [DOI] [PubMed] [Google Scholar]
- (55).Tang Y, Kassie F, Qian X, Ansha B, and Turesky RJ (2013) DNA adduct formation of 2-amino-9H-pyrido[2,3-b]indole and 2-amino-3,4-dimethylimidazo[4,5-f]quinoline in mouse liver and extrahepatic tissues during a subchronic feeding study. Toxicol. Sci 133, 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, and Turesky RJ (2010) Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem. Res. Toxicol 23, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Guo J, and Turesky RJ (2016) Human biomonitoring of DNA adducts by ion trap multistage mass spectrometry. Curr. Protoc. Nucleic Acid Chem 66, 7 24 21–27 24 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yun BH, Rosenquist TA, Nikolic J, Dragicevic D, Tomic K, Jelakovic B, Dickman KG, Grollman AP, and Turesky RJ (2013) Human formalin-fixed paraffin-embedded tissues: An untapped specimen for biomonitoring of carcinogen DNA adducts by mass spectrometry. Anal. Chem 85, 4251–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Xiao S, Guo J, Yun BH, Villalta PW, Krishna S, Tejpaul R, Murugan P, Weight CJ, and Turesky RJ (2016) Biomonitoring DNA adducts of cooked meat carcinogens in human prostate by nano liquid chromatography-high resolution tandem mass spectrometry: Identification of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA adduct. Anal. Chem 88, 12508–12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Guo J, Villalta PW, and Turesky RJ (2017) Data-independent mass spectrometry approach for screening and identification of DNA adducts. Anal. Chem 89, 11728–11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, and Turesky RJ (2009) Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal. Chem 81, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Turesky RJ, Rossi SC, Welti DH, Lay JO Jr., and Kadlubar FF (1992) Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol 5, 479–490. [DOI] [PubMed] [Google Scholar]
- (63).Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, and Doerge DR (2005) High-performance liquid chromatography electrospray ionization tandem mass spectrometry for the detection and quantitation of benzo[a]pyrene-DNA adducts. Chem. Res. Toxicol 18, 1306–1315. [DOI] [PubMed] [Google Scholar]
- (64).Kozekov ID, Turesky RJ, Alas GR, Harris CM, Harris TM, and Rizzo CJ (2010) Formation of deoxyguanosine cross-links from calf thymus DNA treated with acrolein and 4-hydroxy-2-nonenal. Chem. Res. Toxicol 23, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Elmquist CE, Stover JS, Wang Z, and Rizzo CJ (2004) Site-specific synthesis and properties of oligonucleotides containing C8-deoxyguanosine adducts of the dietary mutagen IQ. J. Am. Chem. Soc 126, 11189–11201. [DOI] [PubMed] [Google Scholar]
- (66).De Riccardis F, Bonala RR, and Johnson F (1999) A general method for the synthesis of the N2- and N6- carcinogenic amine adducts of 2 ‘-deoxyguanosine and 2 ‘-deoxyadenosine. J. Am. Chem. Soc 121, 10453–10460. [Google Scholar]
- (67).Takamura-Enya T, Enomoto S, and Wakabayashi K (2006) Palladium-catalyzed direct N-arylation of nucleosides, nucleotides, and oligonucleotides for efficient preparation of dG-N2 adducts with carcinogenic amino-/nitroarenes. J. Org. Chem 71, 5599–5606. [DOI] [PubMed] [Google Scholar]
- (68).Beland FA, Doerge DR, Churchwell MI, Poirier MC, Schoket B, and Marques MM (1999) Synthesis, characterization, and quantitation of a 4-aminobiphenyl-DNA adduct standard. Chem. Res. Toxicol 12, 68–77. [DOI] [PubMed] [Google Scholar]
- (69).Jones CR, and Sabbioni G (2003) Identification of DNA adducts using HPLC/MS/MS following in vitro and in vivo experiments with arylamines and nitroarenes. Chem. Res. Toxicol 16, 1251–1263. [DOI] [PubMed] [Google Scholar]
- (70).Guo J, Yun BH, Upadhyaya P, Yao L, Krishnamachari S, Rosenquist TA, Grollman AP, and Turesky RJ (2016) Multiclass carcinogenic DNA adduct quantification in formalin-fixed paraffin-embedded tissues by ultraperformance liquid chromatography-tandem mass spectrometry. Anal. Chem 88, 4780–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Kadlubar FF, Unruh LE, Beland FA, Straub KM, and Evans FE (1981) Formation of DNA adducts by the carcinogen N-hydroxy-2-naphthylamine. Natl. Cancer Inst. Monogr, 143–152. [PubMed]
- (72).Westra JG (1981) A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis 2, 355–357. [DOI] [PubMed] [Google Scholar]
- (73).Marques MM, Mourato LL, Santos MA, and Beland FA (1996) Synthesis, characterization, and conformational analysis of DNA adducts from methylated anilines present in tobacco smoke. Chem. Res. Toxicol 9, 99–108. [DOI] [PubMed] [Google Scholar]
- (74).Swaminathan S, and Hatcher JF (2002) Identification of new DNA adducts in human bladder epithelia exposed to the proximate metabolite of 4-aminobiphenyl using 32P-postlabeling method. Chem. Biol. Interact 139, 199–213. [DOI] [PubMed] [Google Scholar]
- (75).Kadlubar FF, Beland FA, Beranek DT, Dooley KL, Heflich RH, and Evans FE (1982) Arylamine-DNA adduct formation in relation to urinary bladder carcinogenesis and Salmonella typhimurium mutagenesis, In Environmental Mutagens and Carcinogens (Sugimura T, Kondo S, and Takebe J, Eds.) pp 385–396, Alan R. Liss, New York. [Google Scholar]
- (76).Wolf SM, and Vouros P (1994) Application of capillary liquid chromatography coupled with tandem mass spectrometric methods to the rapid screening of adducts formed by the reaction of N-acetoxy-N-acetyl-2-aminofluorene with calf thymus DNA. Chem. Res. Toxicol 7, 82–88. [DOI] [PubMed] [Google Scholar]
- (77).Kadlubar FF, Unruh LE, Beland FA, Straub KM, and Evans FE (1980) Invitro reaction of the carcinogen, N-hydroxy-2-naphthylamine, with DNA at the C-8 and N2 atoms of guanine and at the N6 atom of adenine. Carcinogenesis 1, 139–150. [DOI] [PubMed] [Google Scholar]
- (78).Marques MM, Mourato LL, Amorim MT, Santos MA, Melchior WB Jr., and Beland FA (1997) Effect of substitution site upon the oxidation potentials of alkylanilines, the mutagenicities of N-hydroxyalkylanilines, and the conformations of alkylaniline-DNA adducts. Chem. Res. Toxicol 10, 1266–1274. [DOI] [PubMed] [Google Scholar]
- (79).Yamazoe Y, Zenser TV, Miller DW, and Kadlubar FF (1988) Mechanism of formation and structural characterization of DNA adducts derived from peroxidative activation of benzidine. Carcinogenesis 9, 1635–1641. [DOI] [PubMed] [Google Scholar]
- (80).Kaderlik KR, Talaska G, DeBord DG, Osorio AM, and Kadlubar FF (1993) 4,4’-Methylene-bis(2-chloroaniline)-DNA adduct analysis in human exfoliated urothelial cells by 32P-postlabeling. Cancer Epidemiol. Biomarkers Prev 2, 63–69. [PubMed] [Google Scholar]
- (81).Beyerbach A, Farmer PB, and Sabbioni G (1996) Synthesis and analysis of DNA adducts of arylamines. Biomarkers 1, 9–20. [DOI] [PubMed] [Google Scholar]
- (82).Chiarelli MP, Wu HP, Antunes AM, and Branco PS (1999) Product ion studies of some novel arylamine adducts of deoxyguanosine by matrix-assisted laser desorption/ionization and post-source decay. Rapid Commun. Mass Spectrom 13, 2004–2010. [DOI] [PubMed] [Google Scholar]
- (83).Li L, Chiarelli MP, Branco PS, Antunes AM, Marques MM, Goncalves LL, and Beland FA (2003) Differentiation of isomeric C8-substituted alkylaniline adducts of guanine by electrospray ionization and tandem quadrupole ion trap mass spectrometry. J. Am. Soc. Mass Spectrom 14, 1488–1492. [DOI] [PubMed] [Google Scholar]
- (84).Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, and Horning S (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Kaufmann A, and Walker S (2017) Comparison of linear intrascan and interscan dynamic ranges of Orbitrap and ion-mobility time-of-flight mass spectrometers. Rapid Commun. Mass Spectrom 31, 1915–1926. [DOI] [PubMed] [Google Scholar]
- (86).Turesky RJ, Liu L, Gu D, Yonemori KM, White KK, Wilkens LR, and Le Marchand L (2013) Biomonitoring the cooked meat carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in hair: impact of exposure, hair pigmentation, and cytochrome P450 1A2 phenotype. Cancer Epidemiol. Biomarkers Prev 22, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Skipper PL, Trudel LJ, Kensler TW, Groopman JD, Egner PA, Liberman RG, Wogan GN, and Tannenbaum SR (2006) DNA adduct formation by 2,6-dimethyl-, 3,5-dimethyl-, and 3-ethylaniline in vivo in mice. Chem. Res. Toxicol 19, 1086–1090. [DOI] [PubMed] [Google Scholar]
- (88).Bryant MS, Vineis P, Skipper PL, and Tannenbaum SR (1988) Hemoglobin adducts of aromatic amines: associations with smoking status and type of tobacco. Proc. Natl. Acad. Sci. U.S.A 85, 9788–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Bohm F, Schmid D, Denzinger S, Wieland WF, and Richter E (2010) DNA adducts of ortho-toluidine in human bladder. Cancer Res 70. [DOI] [PubMed] [Google Scholar]
- (90).Skipper PL, Kim MY, Sun HL, Wogan GN, and Tannenbaum SR (2010) Monocyclic aromatic amines as potential human carcinogens: old is new again. Carcinogenesis 31, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Chao MW, Erkekoglu P, Tseng CY, Ye W, Trudel LJ, Skipper PL, Tannenbaum SR, and Wogan GN (2014) Intracellular generation of ROS by 3,5-dimethylaminophenol: persistence, cellular response, and impact of molecular toxicity. Toxicol. Sci 141, 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Chao MW, Erkekoglu P, Tseng CY, Ye W, Trudel LJ, Skipper PL, Tannenbaum SR, and Wogan GN (2015) Protective effects of ascorbic acid against the genetic and epigenetic alterations induced by 3,5-dimethylaminophenol in AA8 cells. J. Appl. Toxicol 35, 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.