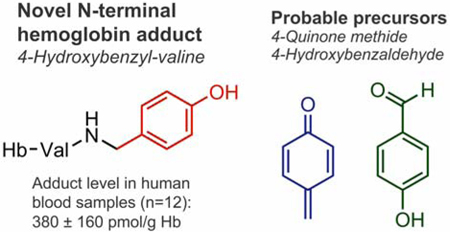
Abstract
Humans are exposed to a wide range of electrophilic compounds present in our diet and environment or formed endogenously as part of normal physiological processes. These electrophiles can modify nucleophilic sites of proteins and DNA to form covalent adducts. Recently, powerful untargeted adductomic approaches have been developed for systematic screening of these adducts in human blood. Our earlier untargeted adductomics study detected 19 unknown adducts to N-terminal valine in hemoglobin (Hb) in human blood. We now describe a full characterization of one of these adducts, which corresponds to the addition of a 4-hydroxybenzyl (4-OHBn) group to N-terminal valine in Hb to form N-(4-hydroxybenzyl)valine (4-OHBn-Val). The adduct structure was determined by comparison of its accurate mass, HPLC retention time, and MS/MS fragmentation to that of authentic standards prepared by chemical synthesis. Average 4-OHBn-Val adduct concentrations in 12 human blood samples were estimated to 380 ± 160 pmol/g Hb. Two possible routes of 4-OHBnVal adduct formation are proposed using two different precursor electrophiles: 4-quinone methide (4-QM) and 4-hydroxybenzaldehyde (4-OHBA). We found that 4-QM reacts rapidly with valine to form the 4-OHBn-Val adduct; however, the quinone methide is unstable under physiological conditions due to hydrolysis. It was shown that 4-OHBA forms reversible Schiff base adducts with valine, which can be stabilized via reduction in blood generating the 4-OHBn-Val adduct. In addition, trace amounts of isomeric 2-hydroxybenzyl-valine (2-OHBn-Val) adducts were detected in 12 human blood samples (estimated mean adduct level, 5.0 ± 1.4 pmol/g Hb). Further studies are needed to quantify the contributions from identified possible precursor electrophiles to the observed hydroxybenzyl adducts in humans.
Keywords: adducts, hemoglobin adducts, quinone methide, benzaldehyde, adductomics
Graphical Abstract
INTRODUCTION
DNA and protein adducts are formed when endogenous and exogenous electrophilic compounds react with nucleophilic sites of cellular biomolecules.1 Levels of formed adducts increase with the dose of electrophiles in vivo, which is associated with an increased risk of health effects, as has been demonstrated for genotoxic compounds/metabolites in animal cancer tests.2,3 The formation of covalent adducts, especially with DNA, is associated with an increased risk of cancer4−6 and other chronic diseases. As such, there is increasing evidence that endogenous and exogenous exposures, which also includes electrophilic compounds/intermediates, are equally important to disease development as genetic factors.7−9 Since many electrophilic compounds that form adducts with nucleophilic sites in DNA can also form adducts with hemoglobin (Hb), which is highly abundant and more available, electrophile-induced Hb adducts have been used as biomarkers for electrophile-induced DNA adduct formation.1,10,11
In previous studies, sensitive mass spectrometry (MS)-based targeted screening methods have been developed to detect structurally defined albumin and Hb adducts in human blood, making it possible to characterize and measure human exposures to known electrophiles.1,10,11 In recent years, such methodology has been expanded to allow for screening of unknown adducts to human serum albumin and Hb via liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based untargeted adductomics, making it possible to characterize the part of the human exposome containing electrophilic species.12,13 Unlike targeted analyses, untargeted screening does not require a priori knowledge of adduct structures, allowing for the detection and identification of unknown exposures.
The screening methodology developed by us is based on the FIRE procedure, which employs modified Edman degradation with fluorescein isothiocyanate (FITC) to selectively detach and derivatize the adducted N-terminal amino acid of Hb.14 The resulting fluorescein isothiohydantoin (FTH)-Val derivatives are analyzed by LC-MS/MS. In our first untargeted adductomic screening of Hb adducts in human blood, in addition to known adducts, 19 unknown adducts were detected, which initiated the challenging work of structurally identifying these adducts and their precursors. We have previously reported the identification of six of these adducts with human Hb and their probable precursors: ethylation, ethyl-vinyl ketone, acrylic acid, glyoxal, methylglyoxal, and 1-octen-3-one.13,15,16 The most abundant N-terminal Hb adduct in human blood measured was the previously identified methyl modification, followed by carboxymethylation.17 High levels of protein methylation in human blood can be explained by the endogenous methylating agent S-adenosylmethionine.18 The carboxymethyl adducts are formed as a result of degradation of glycated Hb (HbA1c),19,20 or from reaction with glyoxal, which comes mostly from dietary sources.21 However, many of the unknown adducts have yet to be identified.
The third most abundant N-terminal Hb adduct in human blood samples was an unknown adduct with m/z of 595.153 of the FTH derivative, which corresponds to an added mass of 106.042 Da from the adduct to Val,13,16 that is, an elemental composition of C7H6O. To accommodate this molecular formula, the only option is an aromatic adduct (see ref 13 ) for the strategy used to generate hypotheses on adduct identities using adductome LC-MS/MS data). Considering all possible molecular structures available, we hypothesized that the most likely precursors of this adduct were 2- or 4-quinone methide (2-QM and 4-QM, respectively, Scheme 1A). QMs are conjugated organic compounds that contain a cyclohexadiene moiety with a carbonyl and a conjugated exocyclic methylene group. QMs have been implicated as intermediates in a variety of biological processes22 and are responsible for the toxicity of many drugs. They are highly electrophilic Michael acceptors containing α,β-unsaturated carbonyls, which readily react with nucleophiles such as amino acid side chains of proteins15,23 and thus are anticipated to readily form adducts to N-terminal amino groups of Hb. Alternative precursors of the unknown adduct with added mass of m/z 106.042 include hydroxybenzaldehydes, which may form reversible Schiff base conjugates with the N-terminal amino group, followed by reduction to produce structurally identical hydroxybenzyl adducts (Scheme 1B).
Scheme 1. Proposed Formation of N-Terminal Valine-Quinone Methide Adducts in Hb Followed by Derivatization via FIRE Procedure13 and Cleavage during Acid Workup.
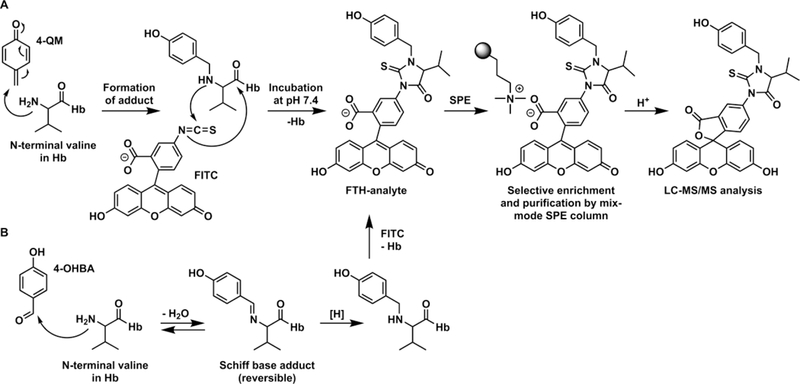
(A) 4-Quinone methide forms an adduct at the N-terminal valine in Hb. Derivatization with FITC cleaves the adducted valine from Hb, forming the FTH-analyte. FTH-analytes are enriched by MAX SPE and cyclized by acid work-up. (B) 4-OHBA forms an adduct at the N-terminal valine in Hb, resulting in a Schiff base. Reduction of this adduct forms the stable 4-OHBn-Val-Hb adduct. This then undergoes FITC derivatization, SPE, and acid work-up as shown in (A).
In the present work, we describe complete structural identification of the unknown Hb adduct with an added mass of 106.042 Da as N-(4-hydroxybenzyl)valine (4-OHBn). This structural assignment was made using liquid chromatography (LC) - high-resolution mass spectrometry (HRMS) on an Orbitrap mass spectrometer and comparisons with authentic reference adducts synthesized in our laboratory. The adduct was quantified in blood of smokers and nonsmokers, with no significant differences between the two groups. Possible precursors of the novel adduct in humans are discussed.
MATERIALS AND METHODS
Caution:
Fluorescein isothiocyanate (FITC) is toxic and should be handled with care.
Chemicals.
Fluorescein-5-isothiocyanate (isomer I, Reagent grade, FITC) was obtained from Karl Industries (Aurora, OH, USA). l-Valine p-nitroanilide hydrochloride (H-ValpNA) was obtained from Bachem (Bubendorf, Switzerland). d,l-Valine, sodium cyanoborohydride, benzaldehyde (BA), 2-hydroxybenzaldehyde (2-OHBA), 3-hydroxybenzaldehyde (3-OHBA), and 4-hydroxybenzaldehyde (4-OHBA) were purchased from Sigma-Aldrich Sweden AB. ortho-Quinone methide precursor (2-QMP, compound 5, see Scheme 2) was generously provided by Professor Steven Rokita (John Hopkins University).24−26 Acetonitrile (ACN), dimethylformamide (DMF), methanol, water and formic acid were obtained from VWR Chemicals. All solvents were of HPLC grade. All other chemicals and solvents were of analytical grade or higher.
Scheme 2. (A) Synthesis of 4-QM Precursor and Its Activation in the Presence of KF and (B) Activation of 2-QM Precursor in the Presence of KF.
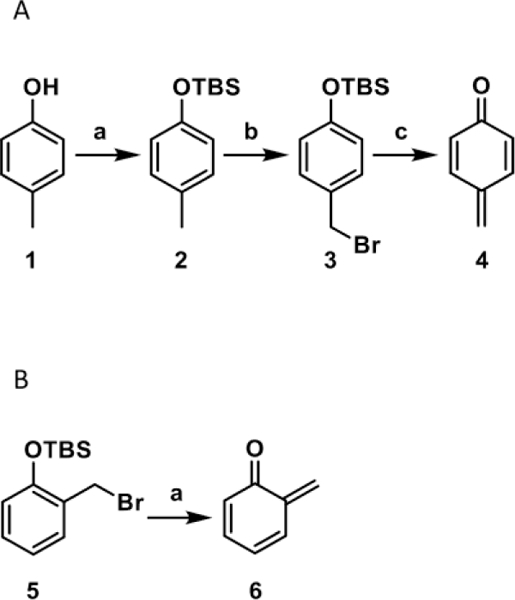
(A) (a) TBS-Cl, imidazole, in DMF, under nitrogen, at room temperature overnight; (b) NBS, AIBN, in CCl4, 85°C, 35 min; (c) KF, in phosphate buffer (pH 7.4) and ACN, room temperature, 30 min. (B) (a) KF, in phosphate buffer (pH 8.4) and water, 37°C, 30 min.
Instruments and Equipment
NMR Instrumentation and Mode of Analysis.
1H and HSQC spectroscopy characterization and quantification studies were performed on either a Bruker 500-MHz or a Bruker 700-MHz spectrometer using CDCl3, D2O, CD3OD, or DMSO-d6 as solvent.
Equipment for Incubation, Derivatization and Cleanup of Blood Samples.
A thermomixer comfort and a 5804 R centrifuge with rotor F-45–30-11 (Eppendorf Nordic, Denmark) were used for incubations and derivatization of blood samples. Oasis Max solid-phase extraction (SPE) cartridges (3 cc, 60 mg, 60 μm; mixed mode anion exchange) were obtained from Waters (Milford, MA, USA). The Hb analyzer (Hb 201+) was obtained from HemoCue (Ängelholm, Sweden).
Analytical HPLC System A.
Our analytical HPLC system A consisted of an Agilent Technologies HPLC System (1100 model) equipped with a UV detector and an autosampler. Chromatographic separation was performed using a Luna C18 (2) column (150 × 4.6 mm, 5 μm) and a gradient of 0.1% formic acid in water (A) and ACN (B) at a flow rate of 1 mL/min. Solvent composition was initially held at 10% B for 5 min, followed by a linear increase to 56% B in 13 min, followed by an increase to 95% B in 3 min, which was held for another 2 min before re-equilibration of the column for 6 min. UV absorbance was monitored at 254 nm.
Analytical HPLC System B.
The analytical HPLC system B consisted of a Shimadzu LC-system with two pumps (LC 10AD), an auto injector (SIL HTC), and a UV detector (SPD 10A). The column was a C18 reversed-phase column (Ace 5 C18, 250 × 2.1 mm, 5 μm particles) from Advanced Chromatography Technologies, and the detection wavelength was set at 274 nm. The HPLC mobile phases consisted of ACN and water containing 0.1% formic acid, and the flow rate was 0.3 mL/min. The program was isocratic for 0.5 min with 5% ACN, followed by a linear increase to 90% ACN over 18 min, after which it was lowered back to 5% ACN in 2 min and kept at 5% ACN until the program was stopped after 25 min.
Semipreparative HPLC.
The analytical HPLC system B, with exception of the column, was also used for the semipreparative purifications. The column used was a semipreparative Hichrom column (KR100–5C18–25098). The compounds were eluted isocratically with 35% ACN in water with 0.2% formic acid at a flow rate of 4 mL/min.
HPLC-MS.
The HPLC-ESI-HRMS system consisted of a Dionex UltiMate 3000 LC system interfaced to an Orbitrap Q Exactive HF mass spectrometer (Thermo Fisher Scientific, MA, USA). Mobile phase A consisted of 0.1% formic acid in H2O/ACN (95:5) and mobile phase B consisted of 0.1% formic acid in H2O/ACN (5:95). The MS instrument settings were the same as described previously,27 except that the m/z range of 550–600 was used in the data-independent acquisition DIA mode instead of the full 500–700 m/z range used previously.27 The target compounds (m/z 595) were further studied using the parallel reaction monitoring (PRM) mode.
Method 1.
Chromatographic separation was performed using a Discovery HS C18 column (3.0 μm, 2.1 × 150 mm) with a Discovery HS C18 guard column (3.0 μm, 2.1 × 20 mm) from Supelco Analytical. The gradient used started at 20% B and increased to 100% B in 25 min. It was thereafter held at 100% B for another 5 min, followed by re-equilibration at 20% B for 5 min. The flow rate used was 120 μL/min, and the injection volume was 20 μL.
Method 2.
To improve the chromatographic separation and at the same time decrease the analysis time, a new method was developed using an Aquity UPLC HSS C18 column (2.1 × 100 mm, 1.8 μm) from Waters. The gradient started at 20% B for 0.5 min, proceeded with an increase to 50% B in 3.5 min, then by an increase to 70% B in 3.5 min, and finally increased to 100% B in 1.5 min. Solvent composition was held at 100% B for 1 min before re-equilibration for 2.5 min. The flow rate used was 300 μL/min, and the injection volume was 20 μL.
Syntheses of QM Precursors, Authentic OHBn-valine Adducts, and FTH Derivatives
tert-Butyldimethyl(p-tolyloxy)silane (Compound 2 in Scheme 2A).
4-Cresol (compound 1, 2.0 g, 18.5 mmol) was dissolved in DMF (10 mL) under nitrogen. tert-Butyldimethylsilyl chloride (6.9 g, 46.2 mmol) and imidazole (6.2 g, 92.4 mmol) were added, and the reaction mixture was stirred at room temperature overnight. After completion of the reaction (as monitored by TLC), the reaction mixture was quenched by addition of H2O (50 mL). The mixture was extracted with ethyl acetate (3 × 40 mL), and the combined organic phases were washed with brine, dried over Na2SO4, and concentrated under reduced pressure to yield a crude product as a colorless oil. The desired compound (2) was purified by silica gel flash column chromatography (hexane/ethyl acetate step gradient of 99:1, 49:1, and finally 19:1) and isolated as a colorless oil (3.97 g, 96%). 1H NMR (CDCl3, 500 MHz): δ 0.00 (s, 6H), 0.80 (s, 9H), 2.09 (s, 3H), 6.54–6.55 (d, J = 5.0 Hz, 2H), 6.82–6.84 (d, J = 10.0 Hz, 2H).
(4-(Bromomethyl)phenoxy)(tert-butyl)dimethylsilane (Compound 3 in Scheme 2A).
N N-bromosuccinimide (0.80 g, 4.5 mmol) was added to a silyl ether-4-methyl-O-tert-butyldimethylsilylphenol (compound 2, 1.0 g, 4.5 mmol) solution in CCl4 (10 mL). The solution was heated to reflux for 10 min, and 2,2′-azobis(2-methylpropionitrile) (AIBN) (0.026 g, 0.04 mmol) was added. The reaction mixture was further refluxed for 35 min, cooled to room temperature, and filtered. The filtrate was washed with water, dried with Na2SO4, and concentrated under reduced pressure to yield compound 3 as a colorless oil (0.72 g, 53%). The purity of this compound was confirmed by MS infusion and 1H NMR (Supplemental Figure 1). 1H NMR (CDCl3, 500 MHz): δ 0.01 (s, 6H), 0.75 (s, 9H), 4.49 (s, 2H), 6.62–6.64 (d, J = 10 Hz, 2H), 7.14–7.15 (d, J = 5 Hz, 2H).
2-OHBn-ValpNA (Compound 7 in Scheme 3).
Scheme 3. Synthesis of ortho-Quinone Methide-ValpNA Adduct (7) and para-Quinone Methide-ValpNA Adduct (9) and Their Corresponding FTH Derivatives (compounds 8 and 10) from Their Respective Precursor Quinone Methides, 2-QM (6) and 4-QM (4).
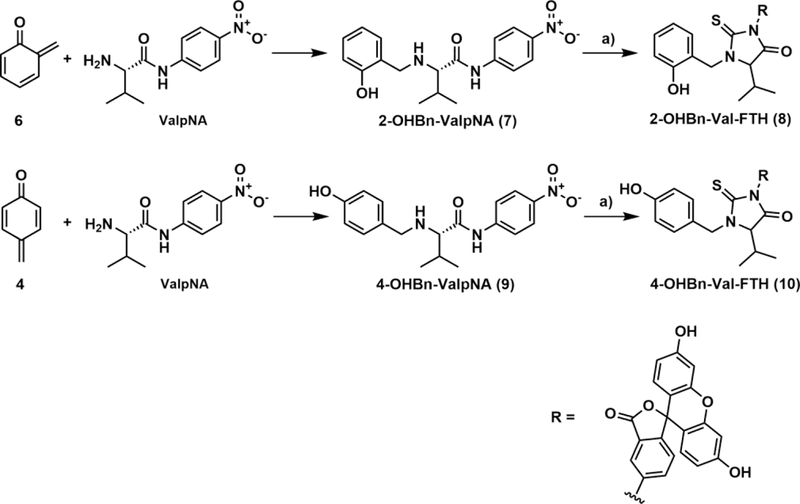
(a) FIRE derivatization: 15 μL of 1M KHCO3 and 5 mg of FITC added to 250 μL human whole blood samples spiked with compound 7 or 9, incubated overnight in a Thermomixer (37°C, 750 rpm).
Stock solutions of ValpNA (Scheme 3, 183 μM), 2-QMP (compound 5 in Scheme 2B, 83 mM), and KF (33 mM) were made in water. A solution of 50 μL of 183 μM ValpNA, 50 μL of 33 mM KF, 50 μL of 100 mM phosphate buffer (pH 8.4), and 300 μL water was prepared. To start the reaction, 50 μL of 83 mM 2-QMP was added, and the solution was incubated at 37 °C for 30 min with continuous mixing. The presence of the 2-OHBn-ValpNA adduct was confirmed by LC-MS (full scan in positive mode).
4-OHBn-ValpNA (Compound 9 in Scheme 3).
To synthesize the 4-OHBn-ValpNA, two different synthetic procedures were used, one small scale and one larger scale. For the small scale reaction, ValpNA (Scheme 3, 33.2 nmol) and KF (33.2 nmol) were dissolved in 78 μL of phosphate buffer (pH 7.4) and added to 622 μL of ACN. To start the reaction, 0.1 mg of 4-QMP (compound 3, 332 nmol) dissolved in 800 μL of ACN was added to this solution. The presence of the 4-OHBn-ValpNA adduct was confirmed by LC-MS (full scan in positive mode).
For the larger scale reaction, the amount of H2O in the reaction mixture was greatly reduced in order to minimize hydrolysis of the activated quinone methide. 4-QMP (compound 3 in Scheme 2A, 46.5 μmol, 10 μL) and ValpNA (4.6 μmol) were dissolved in anhydrous DMSO (77 μL). To start the reaction, 18 μL of KF (4.6 μmol) in H2O was added to this solution. The reaction was allowed to proceed for 1 h, after which the presence of the 4-OHBn-ValpNA adduct was confirmed by LC-MS (full scan in positive mode). The reaction mixture was diluted with H2O, filtered using a Costar Spin-X filter, and separated using HPLC system A. The 4-OHBn-ValpNA adduct eluted as a small peak at ∼12.0 min, which was collected and solvent evaporated to dryness. The purity of this peak was confirmed by MS infusion and 1H and HSQC NMR (Supplemental Figure 2). 4-OHBn-ValpNA: 1H NMR (DMSO-d6, 700 MHz): δ 0.96–0.97 (m, 3H), 0.99–1.0 (m, 3H), 2.26 (br s, 1H), 3.73 (s, 1H), 4.00 (br s, 1H), 4.10 (br s, 1H), 6.75 (d, 2H), 7.26 (d, 2H), 7.81 (d, 2H), 8.26 (d, 2H), 9.13–9.27 (m, 2H), 9.72 (br s, 1H), 11.03 (s, 1H).
N-Benzylvaline, N-(2-hydroxybenzyl)valine, N-(3-hydroxybenzyl)valine and N-(4-Hydroxybenzyl)valine (Compounds 11–14 in Scheme 4).
Scheme 4.
Structures of Valine Adducts Formed from Benzaldehyde and Hydroxybenzaldehydes.
Approximately 2.0 mmol of the respective benzaldehydes (BA, 2-, 3- and 4-OHBA, respectively), 2.2 mmol valine, and 2.5 mmol of sodium cyanoborohydride together with 100 mL methanol were placed in a 250 mL round-bottom flask. The reaction solution was kept at a temperature of 40 °C with stirring, and the progress of the respective reaction was followed by HPLC system B. When all benzaldehyde had reacted (after 4–48 h), the methanol in the reaction solution was removed with a rotary evaporator. Recrystallization from an ethanol–water mixture produced the N-benzylvalines as white crystals in yields ranging from 50 to 60%. The purity and structures of these compounds were confirmed by 1H and HSQC NMR (Supplemental Figures 3–6).
N-Benzylvaline (Bn-Val):
1H NMR (D2O, 500 MHz): δ 7.40–7.43 (m, 5H), 4.22 (d, J = 13 Hz, 1H), 4.08 (d, J = 13 Hz, 2H), 3.34 (d, J = 4.5 Hz, 1H), 2.06–2.13 (m, 1H), 0.93 (d, J = 7 Hz, 3H), 0.87 (d, J = 7 Hz, 3H).
N-(2-Hydroxybenzyl)valine (2-OHBn-Val):
1H NMR (CD3OD, 500 MHz): δ 0.25–0.31 (m, 6H), 1.50–1.53 (m, 1H), 2.52–2.57 (m, 1H), 3.41 (d, J = 13 Hz, 1H), 3.48 (d, J = 13 Hz, 1H), 6.15–6.19 (m, 2H), 6.54–6.59 (m, 2H).
N-(3-Hydroxybenzyl)valine (3-OHBn-Val):
1H NMR (DMSO-d6, 500 MHz): δ 0.88–0.92 (m, 6H), 1.90–1.94 (m, 1H), 2.86 (d, J = 5 Hz, 1H), 3.57 (d, J = 13.5 Hz, 1H), 3.80 (d, J = 13.5 Hz, 1H), 6.66 (d, J = 8 Hz, 1H), 6.75–6.79 (m, 2H), 7.10–7.13 (m, 1H).
N-(4-Hydroxybenzyl)valine (4-OHBn-Val):
1H NMR (D2O, 500 MHz): δ 0.74 (d, J = 7 Hz, 3H), 0.79 (d, J = 7 Hz, 3H), 1.76–1.80 (m, 1H), 2.91 (d, J = 5 Hz, 1H), 3.47 (d, J = 13 Hz, 1H), 3.67 (d, J = 12.5 Hz, 1H), 6.56 (d, J = 8 Hz, 2H), 7.02 (d, J = 8 Hz, 2H).
FTH derivatives of N-benzylvalines.
Approximately 0.12 mmol of the respective N-benzylvalines and 0.10 mmol of FITC were dissolved in 20 mL of phosphate buffer (0.1 M, pH 7.4). The solutions were kept at 60 °C, and the progress of the reactions was followed by HPLC system B. When all FITC had reacted, the reaction mixtures were evaporated to dryness in a rotary evaporator. Thereafter, the crude products were dissolved in methanol and purified by semipreparative HPLC, which afforded the FTH of the N-benzylvalines as orange-brown powders in yields of 30–40%.
N-Benzylvaline FTH (Bn-Val-FTH):
HRMS (Orbitrap ESI) calculated for C33H27N2O6S+ 579.1584, found 579.1585.
N-(2-Hydroxybenzyl)valine FTH (2-OHBn-Val-FTH):
HRMS (Orbitrap ESI) calculated for C33H27N2O7S+ 595.1534, found 595.1538.
N-(3-Hydroxybenzyl)valine FTH (3-OHBn-Val-FTH):
HRMS (Orbitrap ESI) calculated for C33H27N2O7S+ 595.1534, found 595.1536.
N-(4-Hydroxybenzyl)valine FTH (4-OHBn-Val-FTH):
HRMS (Orbitrap ESI) calculated for C33H27N2O7S+ 595.1534, found 595.1535.
Qualitative and Quantitative Analysis of Hb Adducts in Human Blood
Blood Samples and Study Population:
Commercial human blood used for incubation experiments was obtained from Komponentlab at Karolinska University Hospital Huddinge (Stockholm, Sweden). Bovine blood (with citrate) was obtained from Håtunalab AB (Bro, Sweden). Blood samples from six smokers and six nonsmokers, collected with approval from the Regional Ethical Review Board in Stockholm, Sweden (no 96–312), were also analyzed. Upon arrival blood samples were separated into red blood cells (RBCs) and plasma (except the samples used as whole blood for incubation). The RBCs were washed with 0.9% (w/v) sodium chloride and lysed with distilled water. The hemolyzed RBC samples were stored at −20 °C until the day of analysis.
FIRE Method for Derivatization and Cleanup of Blood Samples:
According to the FIRE procedure,14,28 blood samples (250 μL each) were derivatized by addition of KHCO3 (1M, 15 μL) and FITC (5 mg) to each sample, followed by incubation overnight in a Thermomixer (37 °C, 750 rpm). Thereafter, ACN (1.4 mL) was added to precipitate out the proteins, and the samples were centrifuged (10 min at 11,000 rpm). The SPE columns were conditioned with ACN (one column volume) and 0.01 M ammonium hydroxide (1/3 column volume). Next, 25 μL of 1 M ammonium hydroxide was added to the supernatant before transferring to the conditioned SPE columns. After loading, columns were washed with ACN and H2O (one column volume each) followed by 0.25% (w/v) cyanoacetic acid in H2O (1/2 column volume). Analytes were then eluted with 1.2 mL of 0.25% (w/v) cyanoacetic acid in ACN. Samples were dried under nitrogen and then reconstituted in 100 μL of H2O/ACN (3:2, v/v) in preparation for HPLC-ESI-HRMS analysis.
Spiking of Blood Samples with 4-OHBn-ValpNA and 2-OHBn-ValpNA:
Aliquots of the 4-OHBn-ValpNA (compound 9) reaction mixture were added (4.4, 22, and 88 μL, respectively) to three human whole blood samples. Aliquots of the 2-OHBn-ValpNA reaction mixture (compound 7, 100 and 200 μL) were added to another two human whole blood samples. Three additional human whole blood samples and one bovine RBC sample were used as controls. All seven samples were treated according to the FIRE procedure and analyzed with LC-HRMS according to Method 1.
Incubation of Fresh Whole Blood with BA, 2-OHBA, 3-OHBA, and 4-OHBA:
Aliquots of stock solutions of BA, 2-OHBA, 3-OHBA, and 4-OHBA (50 mM in ethanol (95%), 10 μL) were added to fresh human whole blood (990 μL), resulting in final concentrations of 0.50 mM. Two control whole blood human samples (990 μL), to which ethanol only was added (95%, 10 μL), were prepared. The samples were incubated overnight in a Thermomixer (37 °C, 500 rpm). The following day, the tubes were centrifuged (2000 × g, 4 °C, 15 min) and the top-layers, containing plasma, were removed. The remaining RBC fractions were washed with 0.9% (w/v) sodium chloride by gentle mixing followed by centrifugation (2000 × g, 4 °C, 10 min). The wash procedure was repeated twice. Thereafter, all six samples (incubated samples and controls) were derivatized according to the FIRE procedure described above with the exception that all volumes and amounts were scaled up to accommodate the larger blood volume used. The samples were analyzed with LC-HRMS according to Method 2.
Analysis of 2-OHBn-Val and 4-OHBn-Val in Blood Samples from Smokers and Nonsmokers:
The levels of 2-OHBn and 4-OHBn adducts to N-terminal valine in Hb were measured in blood samples from six smokers and six nonsmokers using the FIRE procedure. Prior to derivatization with FITC, Hb contents were measured in the samples. After incubations with FITC but before cleanup, 3-OHBn-Val-FTH (3.5 pmol) was added as an internal standard in order to allow for quantification of 2- and 4-OHBn-Val-FTH. For quantification, calibration curves were established from synthesized standards of 2- and 4-OHBn-Val-FTH in eluent H2O/ACN (3:2, v/v). The samples were analyzed with LC-HRMS according to Method 2.
RESULTS
Structural Identification of the Unknown Hb Adduct
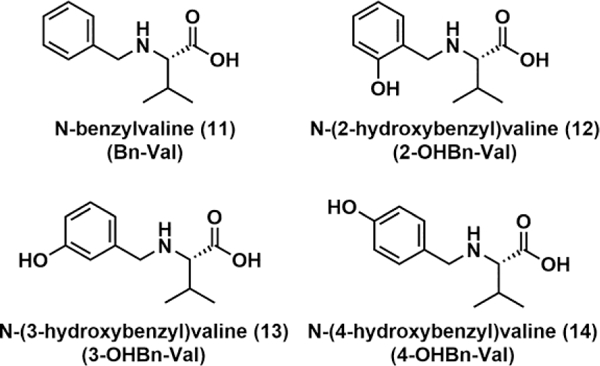
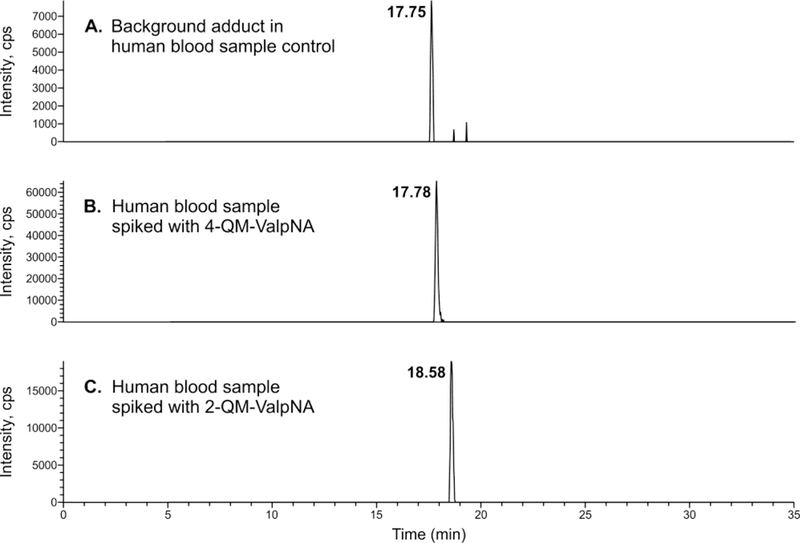
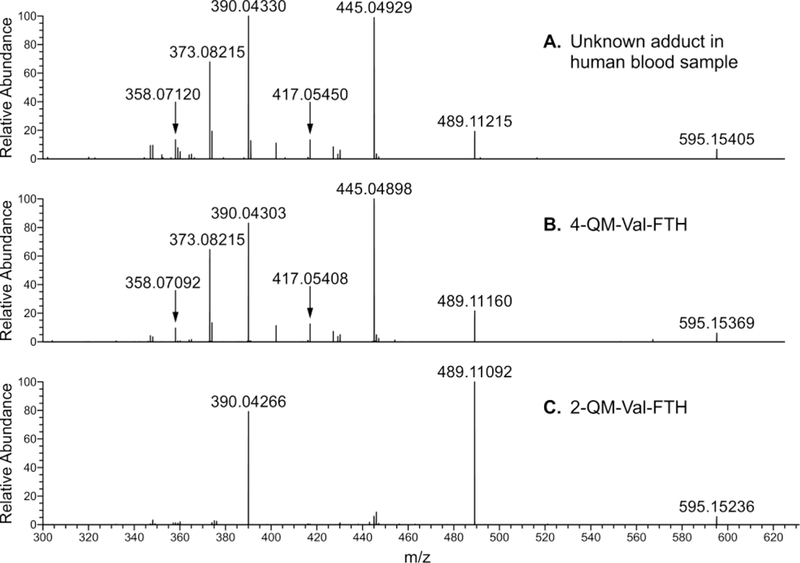
The unknown N-terminal Val Hb adduct derivative with molecular ion [M + H]+ at m/z 595.153 was detected in untargeted adductomics screening of N-terminal valine adducts in Hb using the FIRE procedure.13,27 In our HPLC-ESI-MS/MS experiments, this unknown eluted at 17.75 min (Figure 1A) and exhibited major MS/MS fragments at m/z 489.1121, 445.0492, 390.0433, 373.0821 (Figure 2A). Based on the accurate mass of 595.154 (corresponding to a mass addition of 106.042 Da to N-terminal valine) and HPLC retention time, the adduct was proposed to be N-(hydroxybenzyl)valine (OHBn-Val) derived from 2- or 4-QM (Scheme 1).
Figure 1.
HPLC chromatograms showing the exact mass (m/z = 595.15181–595.15539) of the FTH derivative of the identified adduct in (A) an unspiked sample of human blood, (B) a human blood sample spiked with 4-OHBn-ValpNA (compound 9), and (C) a human blood sample spiked with 2-OHBn-ValpNA (compound 7).
Figure 2.
MS2 spectra of (A) unknown adduct in human blood sample, (B) authentic standard 4-OHBn-Val-FTH (compound 10), and (C) authentic s tandard 2-OHBn-Val-FTH (compound 8).
2- and 4-QM are extremely reactive molecules that are readily hydrolyzed at ambient conditions.25,29 Our initial efforts to generate 2-QM in situ through oxidation of o-cresol using a previously published method30 failed, with no adduct formation seen by LC-MS infusion. Therefore, we employed an elegant strategy developed by the Rokita lab to generate chemically stable QM precursors (QMPs).25 QMP activation to quinone methides is achieved by the addition of KF.25
Authentic 2- or 4-QM Val adducts, that is, 2- and 4-OHBn-ValpNA, were generated using the p-nitrophenyl derivative of Val (ValpNA, Scheme 3), as a model of N-terminal valine in Hb, as described in our previous publications.15 ValpNA is cleaved to reveal free Val during the FIRE procedure (Scheme 3). The standards of 2-OHBn-ValpNA and 4-OHBn-ValpNA (compounds 7 and 9 in Scheme 3, respectively) were prepared using 2-QM (compound 6) and 4-QM (compound 4) and ValpNA (Scheme 3). QMs were generated in situ from QMP precursors in the presence of ValpNA, where they immediately reacted to form the adducts. 2-OHBn-ValpNA (compound 7) and 4-OHBn-ValpNA (compound 9) were isolated by HPLC, and their structures were verified by LC-MS/MS and 1H and HSQC NMR (Supplemental Figure 2).
To examine potential formation of QM-Val adducts in human blood, authentic 2-OHBn-ValpNA (compound 7, derived from 2-QM) and 4-OHBn-ValpNA (compound 9, derived from 4-QM) (Scheme 3) were spiked into human blood. These, along with unspiked human blood samples, underwent derivatization with FITC, followed by LC-HRMS analysis. The accurate mass, retention times, and fragmentation patterns of 2-OHBn-Val-FTH and 4-OHBn-Val-FTH from the spiked blood samples were compared to the unknown adduct seen in the unspiked blood samples. While the accurate mass of the 2-OHBn-Val-FTH adduct (compound 8, m/z 595.1524) matched the unknown (m/z 595.1540), the retention times differed (17.75 vs 18.58 min, see Figure 1A,C). Furthermore, the fragmentation patterns of the unknown adduct at m/z 595.1540 and authentic 2-OHBn-Val-FTH differ significantly, with fragments at m/z 445.0492 and 373.0821 not observed for 2-OHBn-Val-FTH (Figures 2A,C). Authentic 4-OHBn-Val-FTH (compound 10) eluted at the same retention time as the unknown adduct (17.78 min, Figure 1A,B) and displayed the same accurate mass (m/z 595.1537) and MS/MS fragmentation pattern (Figure 2A,B). Proposed structures of the highest abundance fragments can be found in the Supplemental Figure 7. Based on this evidence, the unknown adduct was identified as N-(4-hydroxybenzyl)valine (4-OHBn-Val) (Figure 1).
Alternative Formation of N-(4-hydroxybenzyl)valine Adducts from 4-OH-benzaldehyde
In addition to the 4-QM-mediated mechanism shown in Scheme 1A, the N-(4-hydroxybenzyl)valine adduct may also originate from reaction with 4-OH-benzaldehyde, followed by reduction (Scheme 1B). To explore this possibility, valine was reacted with benzaldehyde (BA), 2-OHBA, 3-OHBA, and 4-OHBA, followed by reduction with sodium cyanoborohydride and derivatization with FITC. The structures of these standard compounds were confirmed by 1H and HSQC NMR (Supplemental Figures 3–6). As expected, NMR spectrum, accurate mass, and MS/MS fragmentation of the FTH derivative of 4-OHBA-Val adduct matched those of 4-OHBn-Val-FTH derived from 4-QM, suggesting that 4-OHBA could be another potential precursor to the identified 4-OHBn-Val-FTH adduct (Scheme 1).
To determine whether the Schiff base formed between 4-OHBA and the N-terminal valine of Hb can be reduced in blood, fresh human blood was incubated with BA, 2-OHBA, 3-OHBA, and 4-OHBA, followed by incubation with FITC and LC-HRMS analysis. The valine adducts from all four benzaldehydes (Scheme 4) were detected (as their FTH derivatives), and the adducts were the same as those produced synthetically using reduction with sodium cyanoborohydride. Therefore, the reductive capacity of blood appears to be sufficient for 4-OHBn-Val to be formed from the Schiff base of 4-OHBA to N-terminal valine (Scheme 1B). By comparison of the spiked blood with the unspiked blood, relatively large amounts of the 4-OHBn-Val-FTH and minor amounts of 2-OHBn-Val-FTH could be detected in the unspiked blood, whereas 3-OHBn-Val-FTH and Bn-Val-FTH could not be detected in the unspiked blood.
Quantification of N-(4-hydroxybenzyl)valine Adduct Levels in Human Blood
The 4-OHBn-Val adduct was quantified in Hb samples from six nonsmoker and six smoker blood samples. As no background levels of 3-OHBn-Val-FTH (Scheme 4) were detected, this compound was used as an internal standard. For quantification, calibration curves were established from synthesized standards of 3-OHBn-Val-FTH (R2 = 0.984, five concentrations (0.001–0.005 pmol/μL) in duplicate) and 4-OHBn-Val-FTH (R2 = 0.987, five concentrations (0.01–0.05 pmol/μL) in duplicate). The 4-OHBn-Val adduct levels in human samples ranged from 140 to 650 pmol/g Hb, with an average adduct level of 380 ± 160 pmol/g Hb (Table 2). There were no significant differences between the smoker and nonsmoker samples (p value for two-sided t test was 0.48).
Table 2.
Quantified Adduct Levels of 4-OHBn-Val and 2-OHBn-Val Adducts in Nonsmokers and Smokers.
| Nonsmokers (pmol/g Hb) (n = 6) |
Smokers (pmol/g Hb) (n = 6) |
|||
|---|---|---|---|---|
| Adduct | Range | Mean ± SD | Range | Mean ± SD |
| 4-OHBA | 143–618 | 374 ± 179 | 247–651 | 379 ± 149 |
| 2-OHBA | 2.0–6.7 | 4.7 ± 1.6 | 3.6–6.1 | 5.3 ± 1.3 |
Quantification of 2-OHBn-Val isomer was also attempted in the same blood samples with the use of a calibration curve established by synthesized 2-OHBn-Val-FTH (R2 = 0.983, five concentrations (0.001–0.005 pmol/μL) in duplicate). The estimated level of 2-OHBn-Val adducts was much lower than 4-OHBn-Val, ranging from 2.0 to 6.7 pmol/g Hb, with an average adduct level of 5.0 ± 1.4 pmol/g Hb (Table 2). The levels are below the lowest point in the calibration curve used so they are rough estimations only.
DISCUSSION
Identification and Quantification of the 4-OHBn Adduct in Human Blood.
The FTH-derivative of the unknown adduct with [M + H]+ m/z 595.154 found in human blood samples was identified as 4-OHBn-Val-FTH using authentic standards prepared independently. Both the retention times and the accurate mass for synthetic 4-OHBn-Val-FTH and the FTH-derivative of the unknown adduct present in human samples matched (Figure 1, Figure 2, Table 1). Adduct identity was further confirmed by comparing MS/MS fragmentation patterns. In contrast, HPLC retention time and fragmentation of the unknown adduct differed from that of the synthetic 2-OHBn-Val-FTH isomer, despite having the same exact mass.
Table 1.
Comparison of Fragmentation Patterns of Adduct 595 for (A) the Unknown Analyte, (B) the Synthesized 4-OHBn-Val in an Incubated Blood Sample, and (C) the Synthesized 2-OHBn-Val in an Incubated Blood Sample.
| Relative Intensity of Fragments m/z (%) |
|||||||
|---|---|---|---|---|---|---|---|
| Sample | RT (min) | 358 | 373 | 390 | 417 | 445 | 489 |
| A | 17.75 | 16 | 65 | 100 | 13 | 98 | 21 |
| B | 17.78 | 17 | 48 | 100 | 9 | 73 | 16 |
| C | 18.58 | 2 | 0 | 59 | 0 | 6 | 100 |
Using a calibration curve generated from the synthesized standard matching the FTH derivative of the unknown adduct, 4-OHBn-Val-FTH, and synthesized 3-OHBn-Val-FTH as an internal standard, we quantified the levels of the background adduct at m/z 595.154 in human blood as approximately 380 pmol/g Hb. This adduct level corresponds to approximately seven OHBn-Val adducts per 106 Hb molecules, indicating the importance of the FIRE procedure to enrich these adducts. The mean adduct level and the large range in adduct level (143–651 pmol/g Hb) is similar to that of our previous studies with quantification without synthesized standard and a less suitable internal (surrogate) standard of this unknown adduct.13,27
Potential Sources for the Formation of the 4-OHBn Adduct.
Once the identity of this unknown adduct was confirmed, we sought to identify its possible sources. In addition to the human blood sample controls that were investigated, a bovine blood sample was also analyzed. While the level of the 4-OHBn adduct detected in the bovine sample was far lower than the level found in human samples, its presence shows that this adduct is not unique to humans (as indicated earlier).13 The adduct levels quantified in smokers and nonsmokers were not significantly different, thus the source of the adduct is highly unlikely to come from smoking. Additionally, this adduct was present in all blood samples analyzed in this study and in our previous studies using the FIRE procedure, indicating that the source may be endogenous or present in common foods. Independently, Rappaport’s research group has observed an adduct of the same added mass to Cys34 in human serum albumin in adductomics screening of different sample materials. It has been suggested to correspond to an adduct from benzaldehyde,31 and just recently quinone methide was also suggested as a precursor.32 These suggestions were not verified through comparison with reference compounds, but indicate that the same adduct has been observed to another nucleophilic site in serum albumin. Considering the relatively high abundance of this adduct observed to Hb and the nucleophilic strength of the respective nucleophilic sites, modification of serum albumin by the same precursor seems likely.
QMs can be formed in vivo through two electron oxidation reactions of 4-alkyl-substituted phenols catalyzed by cytochrome P450 enzymes or peroxidases.33 Substitutions, especially at the 2- and 6-positions of the phenol ring, strongly influence the reactivity of the QM. In general, the larger the substitutions, the lower the reactivity of the QM, due to steric effects.30,34 This would imply that a completely unsubstituted QM would have the highest reactivity.35 However, due to this high reactivity, unsubstituted QMs have been more difficult to study than more substituted ones. The Rokita lab has previously shown that unsubstituted ortho-quinone methides can form adducts at nucleophilic positions in DNA, including the N1 and N6 position of dA, N3 position of dC, and N1, N2, and N7 positions of dG.26 They also found that these adducts are reversible,36,37 though the stability of these adducts was highly dependent on the substituents on the quinone methide.38 Additionally, Bolton et al. found that substituted para-quinone methides of differing reactivity all formed adducts with the α-amino groups of lysine, histidine, tyrosine, and serine, and with the thiol of cysteine.39 The most electrophilic of these substituted quinone methides was also able to alkylate the side chain nitrogens of lysine and histidine.39 Due to the high electrophilicity of the unsubstituted 4-QM, it is likely capable of not only alkylating proteins at positions other than N-terminal valine but also nucleophilic positions in DNA.
This high reactivity of QMs was supported by our experiments examining ValpNA reactivity toward 4-QM, as the reaction was complete within an hour. However, para-quinone methides are also rapidly hydrolyzed, making it unlikely that the observed Hb adduct is formed by free 4-QM in human blood. In order to form a large enough amount of 4-OHBn-ValpNA adduct to be quantified by LC-MS, the reactions had to be carried out in DMSO in order to limit the hydrolysis of 4-QM. Reactions performed under physiological conditions only produced barely detectable amounts of Val adducts (results not shown). Thus, we hypothesize that there must be an alternative origin of the adduct.
One hypothesis is that this adduct is formed as a byproduct of tyrosine degradation by the [FeFe] hydrogenase maturase, HydG.40 HydG degrades tyrosine in order to form CO and CN– ligands for the enzyme’s [FeFe] cluster, resulting in the generation of 4-oxidobenzyl radical as a byproduct. While this radical is usually further degraded to form 4-cresol as the end product, it is possible that it could instead react to form the 4-OHBn adduct we observed.
Another possible source of this adduct is 4-OHBA. NMR of the synthesized reaction product between 4-OHBA and valine showed that the same 4-OHBn-Val adduct was formed as for 4-QM and valine. However, in order to form the 4-OHBn-Val adduct from 4-OHBA, the (reversible) Schiff base adduct, which is formed first, must be reduced (and stabilized). To explore if this may happen in vivo, 4-OHBA was incubated with fresh whole blood overnight, and indeed the reduced Schiff base adduct (4-OHBn-Val adduct) was detected (after derivatization to FTH with the FIRE procedure). Thus, it appears as if 4-OHBA could be a possible precursor for the detected 4-OHBn-Val adduct. 4-OHBA has previously been shown to be formed naturally. For instance, it is an active component of Gastrodia elata, a Chinese herbal medicine used to treat headaches and migraines, and of Dendrocalamus asper bamboo shoots.41,42 In addition, 4-OHBA could be formed in natural processes from tyrosine,43 including in the Maillard reaction that occurs during cooking of food.44 It is also a precursor to vanillin that is produced in vanilla beans45 and is an important volatile component of vanilla aroma and flavor.46,47 To be able to make a quantitative conclusion about the precursor of this adduct in humans, further studies must be done.
2-Hydroxybenzaldehyde (salicylaldehyde, 2-OHBA), which can form an adduct with the same structure as the adduct from 2-QM (2-OHBn-Val), is a naturally occurring component in buckwheat,48 cinnamon, and tea and is found in many foods. It is also structurally related to salicylic acid, one of the most commonly ingested analgesics and anti-inflammatory agents. Low levels of 2-OHBn-Val adduct were detected in both smokers and nonsmokers in this study (estimated levels: 5.0 ± 1.4 pmol/g Hb).
Conclusions
The unknown human adduct with m/z 595.154 was identified as an adduct corresponding to the addition of a 4-hydroxybenzyl group to N-terminal valine in Hb. By reaction with FITC, according to the FIRE procedure, N-(4-hydroxybenzyl)valine FTH (4-OHBn-Val-FTH) is formed. Two probable precursors have been identified: one being para-quinone methide (4-QM), which can form the 4-OHBn-Val adduct via a Michael addition with the N-terminal valine, and the second possible precursor is 4-hydroxybenzaldehyde (4-OHBA), which can form the same adduct via a Schiff base formation with the N-terminal valine followed by reduction. It is essential to establish which of the two probable precursors is the main contributor to the observed adduct to assess its toxicological relevance.
Supplementary Material
Acknowledgements
We thank Professor Steven Rokita (John Hopkins University) for kindly providing us with synthetic 2-QMP. We thank Robert Carlson (University of Minnesota Masonic Cancer Center) for preparing figures and schemes for this manuscript.
Funding
We acknowledge Sw. Research Council and Stockholm University for financial support. A.D. was partially supported by the American Chemistry Council and a grant from the National Cancer Institute (P01 CA138338).
Abbreviations
- 2-OHBA
2-hydroxybenzaldehyde
- 2-OHBn
2-hydroxybenzyl
- 2-QM
ortho-quinone methide
- 3-OHBA
3-hydroxybenzaldehyde
- 4-OHBA
4-hydroxybenzaldehyde
- 4-OHBn
4-hydroxybenzyl
- 4-QM
para-quinone methide
- ACN
acetonitrile
- AIBN
azobisisobutyronitrile
- BA
benzaldehyde
- DIA
data-independent acquisition
- DMF
dimethylformamide
- DMSO
dimethyl sulfoxide
- FITC
fluorescein isothiocyanate
- FTH
fluorescein isothiohydantoin
- Hb
hemoglobin
- HPLC-ESI+-MS/MS
high-pressure liquid chromatography tandem mass spectrometry
- HRMS
high-resolution mass spectrometry
- PRM
parallel reaction monitoring
- QM
quinone methide
- QMP
quinone methide precursor
- RBC
red blood cell
- SPE
solid-phase extraction
- TBS-Cl
tert-butyldimethylsilyl chloride
- TLC
thin-layer chromatography
- Val
valine
- ValpNA
l-valine p-nitroanilide
Footnotes
Supporting Information
1H and 135DEPT NMR spectra of the synthesized 4-QM precursor, 1H and HSQC NMR spectra of synthesized 4-OHBn-ValpNA, Bn-Val, 2-OHBn-Val, 3-OHBn-Val, 4-OHBn-Val, and proposed structures of MS fragments of 4-OHBn-Val-FTH (PDF).
The authors declare no competing financial interest.
References
- 1.Törnqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, and Rydberg P (2002) Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 778, 279–308, DOI: 10.1016/S1570-0232(02)00172-1 [DOI] [PubMed] [Google Scholar]
- 2.Granath FN, Vaca CE, Ehrenberg LG, and Tornqvist MA (1999) Cancer risk estimation of genotoxic chemicals based on target dose and a multiplicative model. Risk Anal 19, 309–320, DOI: 10.1023/A:1006933913194 [DOI] [PubMed] [Google Scholar]
- 3.Fred C, Tornqvist M, and Granath F (2008) Evaluation of cancer tests of 1,3-butadiene using internal dose, genotoxic potency, and a multiplicative risk model. Cancer Res. 68, 8014–8021, DOI: 10.1158/0008-5472.CAN-08-0334 [DOI] [PubMed] [Google Scholar]
- 4.Tretyakova N, Villalta PW, and Kotapati S (2013) Mass Spectrometry of Structurally Modified DNA. Chem. Rev. 113, 2395–2436, DOI: 10.1021/cr300391r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boysen G, Pachkowski BF, Nakamura J, and Swenberg JA (2009) The formation and biological significance of N7-guanine adducts. Mutat. Res, Genet. Toxicol. Environ. Mutagen. 678, 76–94, DOI: 10.1016/j.mrgentox.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwa Yun B, Guo J, Bellamri M, and Turesky RJ (2018) DNA adducts: Formation, biological effects, and new biospecimens for mass spectrometric measurements in humans. Mass Spectrom. Rev. 1–28, DOI: 10.1002/mas.21570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rappaport SM (2016) Genetic Factors Are Not the Major Causes of Chronic Diseases. PLoS One 11, e0154387, DOI: 10.1371/journal.pone.0154387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, and Hemminki K (2000) Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85, DOI: 10.1056/NEJM200007133430201 [DOI] [PubMed] [Google Scholar]
- 9.Wiseman M (2008) The second World Cancer Research Fund/American Institute for Cancer Research expert report. Food, nutrition, physical activity, and the prevention of cancer: a global perspective. Proc. Nutr. Soc. 67, 253–256, DOI: 10.1017/S002966510800712X [DOI] [PubMed] [Google Scholar]
- 10.Rubino FM, Pitton M, Di Fabio D, and Colombi A (2009) Toward an ″omic″ physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom. Rev. 28, 725–784, DOI: 10.1002/mas.20207 [DOI] [PubMed] [Google Scholar]
- 11.Sabbioni G and Turesky RJ (2017) Biomonitoring Human Albumin Adducts: The Past, the Present, and the Future. Chem. Res. Toxicol. 30, 332–366, DOI: 10.1021/acs.chemrestox.6b00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigoryan H, Edmands W, Lu SS, Yano Y, Regazzoni L, Iavarone AT, Williams ER, and Rappaport SM (2016) Adductomics Pipeline for Untargeted Analysis of Modifications to Cys34 of Human Serum Albumin. Anal. Chem. 88, 10504–10512, DOI: 10.1021/acs.analchem.6b02553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlsson H, von Stedingk H, Nilsson U, and Törnqvist M (2014) LC-MS/MS screening strategy for unknown adducts to N-terminal valine in hemoglobin applied to smokers and nonsmokers. Chem. Res. Toxicol. 27, 2062–2070, DOI: 10.1021/tx5002749 [DOI] [PubMed] [Google Scholar]
- 14.von Stedingk H, Rydberg P, and Törnqvist M (2010) A new modified Edman procedure for analysis of N-terminal valine adducts in hemoglobin by LC-MS/MS. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 878, 2483–2490, DOI: 10.1016/j.jchromb.2010.03.034 [DOI] [PubMed] [Google Scholar]
- 15.Carlsson H, Motwani HV, Osterman Golkar S, and Tornqvist M (2015) Characterization of a Hemoglobin Adduct from Ethyl Vinyl Ketone Detected in Human Blood Samples. Chem. Res. Toxicol. 28, 2120–2129, DOI: 10.1021/acs.chemrestox.5b00287 [DOI] [PubMed] [Google Scholar]
- 16.Carlsson H and Tornqvist M (2016) Strategy for identifying unknown hemoglobin adducts using adductome LC-MS/MS data: Identification of adducts corresponding to acrylic acid, glyoxal, methylglyoxal, and 1-octen-3-one. Food Chem. Toxicol. 92, 94–103, DOI: 10.1016/j.fct.2016.03.028 [DOI] [PubMed] [Google Scholar]
- 17.Carlsson H and Törnqvist M (2017) An Adductomic Approach to Identify Electrophiles In Vivo. Basic Clin. Pharmacol. Toxicol. 121, 44–54, DOI: 10.1111/bcpt.12715 [DOI] [PubMed] [Google Scholar]
- 18.Törnqvist M, Svartengren M, and Ericsson CH (1992) Methylations in hemoglobin from monozygotic twins discordant for cigarette smoking: hereditary and tobacco-related factors. Chem.-Biol. Interact. 82, 91–98, DOI: 10.1016/0009-2797(92)90016-E [DOI] [PubMed] [Google Scholar]
- 19.Shimada S, Tanaka Y, Ohmura C, Tamura Y, Shimizu T, Uchino H, Watada H, Hirose T, Nakaniwa T, Miwa S, and Kawamori R (2005) N-(carboxymethyl)valine residues in hemoglobin (CMV-Hb) reflect accumulation of oxidative stress in diabetic patients. Diabetes Res. Clin. Pract. 69, 272–278, DOI: 10.1016/j.diabres.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 20.Uchimura T, Nakano K, Hashiguchi T, Iwamoto H, Miura K, Yoshimura Y, Hanyu N, Hirata K, Imakuma M, Motomiya Y, and Maruyama I (2001) Elevation of N-(carboxymethyl)valine residue in hemoglobin of diabetic patients. Its role in the development of diabetic nephropathy. Diabetes Care 24, 891–896, DOI: 10.2337/diacare.24.5.891 [DOI] [PubMed] [Google Scholar]
- 21.Poulsen MW, Hedegaard RV, Andersen JM, de Courten B, Bugel S, Nielsen J, Skibsted LH, and Dragsted LO (2013) Advanced glycation endproducts in food and their effects on health. Food Chem. Toxicol. 60, 10–37, DOI: 10.1016/j.fct.2013.06.052 [DOI] [PubMed] [Google Scholar]
- 22.Peter MG (1989) Chemical Modifications of Biopolymers by Quinones and Quinone Methides. Angew. Chem., Int. Ed. Engl. 28, 555–570, DOI: 10.1002/anie.198905551 [DOI] [Google Scholar]
- 23.Enoch SJ, Ellison CM, Schultz TW, and Cronin MT (2011) A review of the electrophilic reaction chemistry involved in covalent protein binding relevant to toxicity. Crit. Rev. Toxicol. 41, 783–802, DOI: 10.3109/10408444.2011.598141 [DOI] [PubMed] [Google Scholar]
- 24.Huang C, Liu Y, and Rokita SE (2016) Targeting duplex DNA with the reversible reactivity of quinone methides. Signal transduction and targeted therapy 1, 16009, DOI: 10.1038/sigtrans.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rokita SE, Yang J, Pande P, and Greenberg WA (1997) Quinone Methide Alkylation of Deoxycytidine. J. Org. Chem. 62, 3010–3012, DOI: 10.1021/jo9700336 [DOI] [PubMed] [Google Scholar]
- 26.Weinert EE, Frankenfield KN, and Rokita SE (2005) Time-dependent evolution of adducts formed between deoxynucleosides and a model quinone methide. Chem. Res. Toxicol. 18, 1364–1370, DOI: 10.1021/tx0501583 [DOI] [PubMed] [Google Scholar]
- 27.Carlsson H, Aasa J, Kotova N, Vare D, Sousa PFM, Rydberg P, Abramsson-Zetterberg L, and Törnqvist M (2017) Adductomic Screening of Hemoglobin Adducts and Monitoring of Micronuclei in School-Age Children. Chem. Res. Toxicol. 30, 1157–1167, DOI: 10.1021/acs.chemrestox.6b00463 [DOI] [PubMed] [Google Scholar]
- 28.Rydberg P (2005) Patent: Method for Analyzing N-terminal Protein Adducts Using Isothiocyanate Reagents. International Publication Number: WO2005101020AI.
- 29.( (2009)Quinone Methides< (Rokita SE) John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 30.Bolton JL, Comeau E, and Vukomanovic V (1995) The influence of 4-alkyl substituents on the formation and reactivity of 2-methoxy-quinone methides: evidence that extended pi-conjugation dramatically stabilizes the quinone methide formed from eugenol. Chem.-Biol. Interact. 95, 279–290, DOI: 10.1016/0009-2797(94)03566-Q [DOI] [PubMed] [Google Scholar]
- 31.Lu SS, Grigoryan H, Edmands WM, Hu W, Iavarone AT, Hubbard A, Rothman N, Vermeulen R, Lan Q, and Rappaport SM (2017) Profiling the Serum Albumin Cys34 Adductome of Solid Fuel Users in Xuanwei and Fuyuan, China. Environ. Sci. Technol. 51, 46–57, DOI: 10.1021/acs.est.6b03955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Grigoryan H, Edmands WMB, Dagnino S, Sinharay R, Cullinan P, Collins P, Chung KF, Barratt B, Kelly FJ, Vineis P, and Rappaport SM (2018) Cys34 Adductomes Differ between Patients with Chronic Lung or Heart Disease and Healthy Controls in Central London. Environ. Sci. Technol. 52, 2307–2313, DOI: 10.1021/acs.est.7b05554 [DOI] [PubMed] [Google Scholar]
- 33.Thompson DC, Thompson JA, Sugumaran M, and Moldeus P (1993) Biological and toxicological consequences of quinone methide formation. Chem.-Biol. Interact. 86, 129–162, DOI: 10.1016/0009-2797(93)90117-H [DOI] [PubMed] [Google Scholar]
- 34.Bolton JL, Valerio LG Jr., and Thompson JA (1992) The enzymatic formation and chemical reactivity of quinone methides correlate with alkylphenol-induced toxicity in rat hepatocytes. Chem. Res. Toxicol. 5, 816–822, DOI: 10.1021/tx00030a014 [DOI] [PubMed] [Google Scholar]
- 35.Toteva MM, Moran M, Amyes TL, and Richard JP (2003) Substituent effects on carbocation stability: the pK(R) for p-quinone methide. J. Am. Chem. Soc. 125, 8814–8819, DOI: 10.1021/ja029588v [DOI] [PubMed] [Google Scholar]
- 36.McCrane MP, Hutchinson MA, Ad O, and Rokita SE (2014) Oxidative quenching of quinone methide adducts reveals transient products of reversible alkylation in duplex DNA. Chem. Res. Toxicol. 27, 1282–1293, DOI: 10.1021/tx500152d [DOI] [PubMed] [Google Scholar]
- 37.McCrane MP, Weinert EE, Lin Y, Mazzola EP, Lam YF, Scholl PF, and Rokita SE (2011) Trapping a labile adduct formed between an ortho-quinone methide and 2’-deoxycytidine. Org. Lett. 13, 1186–1189, DOI: 10.1021/ol200071p [DOI] [PubMed] [Google Scholar]
- 38.Weinert EE, Dondi R, Colloredo-Melz S, Frankenfield KN, Mitchell CH, Freccero M, and Rokita SE (2006) Substituents on quinone methides strongly modulate formation and stability of their nucleophilic adducts. J. Am. Chem. Soc. 128, 11940–11947, DOI: 10.1021/ja062948k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolton JL, Turnipseed SB, and Thompson JA (1997) Influence of quinone methide reactivity on the alkylation of thiol and amino groups in proteins: studies utilizing amino acid and peptide models. Chem.-Biol. Interact. 107, 185–200, DOI: 10.1016/S0009-2797(97)00079-3 [DOI] [PubMed] [Google Scholar]
- 40.Stich TA, Myers WK, and Britt RD (2014) Paramagnetic intermediates generated by radical S-adenosylmethionine (SAM) enzymes. Acc. Chem. Res. 47, 2235–2243, DOI: 10.1021/ar400235n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graf E (1992) Chinese Drugs of Plant Origin Chemistry, Pharmacology, and Use in Traditional and Modern Medicine, Springer-Verlag, Berlin. [Google Scholar]
- 42.Zhang J, Mohamad H, Wong JH, Bilal M, Lloyd AJ, Yusoff AAM, Osman H, Wong KT, Idris Z, and Abdulluh JM (2017) The Effect of 4-hydroxybenzaldehyde on the γ-aminobutyric Acid Type A Receptor. Maylays J. Med. Sci. 24, 94–99, DOI: 10.21315/mjms2017.24.2.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manley SL and Chapman DJ (1979) Metabolism of L-Tyrosine to 4-Hydroxybenzaldehyde and 3-Bromo-4-Hydroxybenzaldehyde by Chloroplast-containing Fractions of Odonthalia floccosa (Esp.) Falk. Plant Physiol. 64, 1032–1038, DOI: 10.1104/pp.64.6.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakas A and Horvat S (2003) Study of degradation pathways of Amadori compounds obtained by glycation of opioid pentapeptide and related smaller fragments: stability, reactions, and spectroscopic properties. Biopolymers 69, 421–431, DOI: 10.1002/bip.10338 [DOI] [PubMed] [Google Scholar]
- 45.Podstolski A, Havkin-Frenkel D, Malinowski J, Blount JW, Kourteva G, and Dixon RA (2002) Unusual 4-hydroxybenzaldehyde synthase activity from tissue cultures of the vanilla orchid Vanilla planifolia. Phytochemistry 61, 611–620, DOI: 10.1016/S0031-9422(02)00285-6 [DOI] [PubMed] [Google Scholar]
- 46.Sinha AK, Sharma UK, and Sharma N (2008) A comprehensive review on vanilla flavor: extraction, isolation and quantification of vanillin and others constituents. Int. J. Food Sci. Nutr. 59, 299–326, DOI: 10.1080/09687630701539350 [DOI] [PubMed] [Google Scholar]
- 47.Pérez Silva A, Gunata Z, Lepoutre J, and Odoux E (2011) New insight on the genesis and fate of odor-active compounds in vanilla beans (Vanilla planifolia G. Jackson) during traditional curing. Food Res. Int. 44, 2930–2937, DOI: 10.1016/j.foodres.2011.06.048 [DOI] [Google Scholar]
- 48.Janes D and Kreft S (2008) Salicylaldehyde is a characteristic aroma component of buckwheat groats. Food Chem. 109, 293–298, DOI: 10.1016/j.foodchem.2007.12.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


