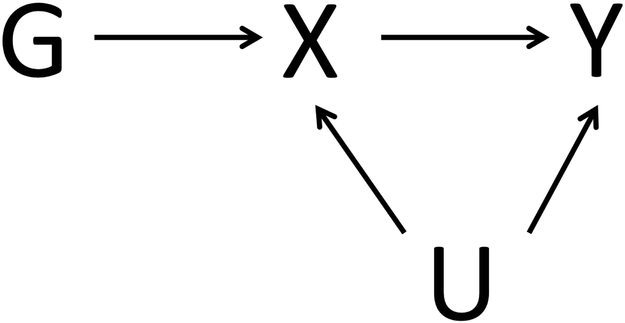Abstract
Mendelian randomization (MR) is defined as the utilization of genetic variants as instrumental variables to assess the causal relationship between an exposure and an outcome (Davey Smith & Ebrahim, 2003). By leveraging genetic polymorphisms as proxy for an exposure, the causal effect of an exposure on an outcome can be assessed while addressing susceptibility to biases prone to conventional observational studies, including confounding and reverse causation, where the outcome causes the exposure (Davey Smith & Ebrahim, 2007). Analogous to a randomized controlled trial where patients are randomly assigned to subgroups based on different treatments, in an MR analysis, the random allocation of alleles during meiosis from parent to offspring assigns individuals to different subgroups based on genetic variants (Davey Smith & Ebrahim, 2007). Recent methods use summary statistics from genome-wide association studies to perform MR, bypassing the need for individual-level data (Burgess et al., 2015). Here, we provide a straightforward protocol for using summary-level data to perform MR and provide guidance for utilizing available software.
Keywords: Mendelian randomization, genetic variation, instrumental variable analysis, TwoSampleMR, summarized genetic data
INTRODUCTION
The aim of many, if not all, observational studies is to associate an exposure and a disease or phenotype to eventually collect evidence to discern a causal relationship. However, observational associations are influenced by biases such as measured and unmeasured confounding, which can occur when an outside variable is associated with the exposure and the disease, and reverse causality and therefore can lack ability to establish a directional effect(Greenland, Robins, & Pearl, 1999). The principle underlying Mendelian randomization (MR) methodology is that such biases can be circumvented by leveraging genetic variants associated with an exposure as an “instrumental variable” (IV) to estimate the effect of genetic variation within an exposure on an outcome(Davey Smith & Ebrahim, 2007). An IV is defined as an external variable G that is associated with the exposureXand independent of outcome Yas well as any factors associated with outcome Y, other than via X(Greenland, 2018). Genetic variants can be utilized as “IVs”, thereby serving the role of randomizing “exposure”.
To utilize a genetic variant as an IV, three assumptions must be satisfied(Davey Smith & Hemani, 2014) (see Figure 1): (i) the genetic variant must be associated with the exposure, (ii) the genetic variant must be independent of any confounder of the exposure-outcome, and (iii) the genetic variant must be independent of the outcome, except via a possible association with the exposure.
Figure 1.
Directed acyclic graph depicting the IV assumptions for conducting Mendelian randomization. G, the genetic variant, must be (i) associated with exposure X, (ii) independent of any confounder U, and (iii) independent of outcome Y.
In the simplest MR technique (for one genetic variant), the presence of an association between a genetic variant and an exposure and the genetic variant and an outcome may imply causal effect of the exposure on the outcome(D. A. Lawlor, Harbord, Sterne, Timpson, & Davey Smith, 2008). MR can be performed with individual-level participant data, obtained from the genetic data for each participant, or with summary-level data, which usually contains per-allele regression coefficients and standard errors analyzed over all individuals within a study(Haycock et al., 2016; D. A. Lawlor, 2016). In summary data MR, summary-level data can either be obtained from publicly available summary level data or by consortia of genome-wide association studies (GWAS), or can be calculated from individual-level participant information(Burgess et al., 2015).
MR can be performed in a “one-sample” or a “two-sample” setting. One-sample MR is performed when the data on the exposure and the outcome are derived from a single dataset(Burgess, Davies, & Thompson, 2016). Two-sample MR is performed when the data on the exposure and the outcome are derived from two non-overlapping and independent datasets, allowing one dataset to be used for performing the summary-level instrument-exposure analysis and the other dataset for performing the instrument-outcome association analysis(Burgess et al., 2016; Hartwig, Davies, Hemani, & Davey Smith, 2016).
Here, we present a protocol to perform MR using summary-level data, which can be performed in the one-sample or two-sample setting, and we provide an RStudio markdown file to demonstrate how to use the TwoSampleMR package in R. The code and implementation of MR in the protocols below are inspired by and utilize resources provided by the MRC Integrative Epidemiology Unit and the MR-Base Collaboration(Hemani, Haycock, Zheng, Gaunt, & Elsworth, n.d.; Hemani et al., 2018).
BASIC PROTOCOL 1: Performing a Mendelian randomization analysis in R using summarized genetic data
In this protocol, we show how to perform MR using summary statistics using different methods of analysis. In the simplest method, the causal effect of the exposure on the outcome can be calculated by a “2-stage least-squares” (2SLS) regression, where the exposure is regressed on the genetic instrument, and the outcome is regressed over the exposure values (where linear or logistic regression is used for continuous or binary outcome variables, respectively)(Haycock et al., 2016).
In the inverse variance weighted (IVW) method, the causal effect of the exposure on the outcome for a single genetic variant can be estimated as a ratio of the association estimate for the outcome and the exposure (Bowden, Davey Smith, Haycock, & Burgess, 2016; Burgess & Thompson, 2017). For multiple independent genetic variants, the ratio estimates from each genetic variant can be meta-analyzed to form the overall causal estimate (Bowden, Davey Smith, et al., 2016; Burgess & Thompson, 2017).
MR-Egger can be used when the IV assumptions do not hold or weakly hold, and entails a modification to the IVW estimate calculation where the intercept term is calculated as part of the MR-Egger estimate, instead of setting the intercept term of the regression to zero (Bowden, Davey Smith, & Burgess, 2015). In MR-Egger, the intercept serves as a test for directional pleiotropy (meaning the genetic variants exert pleiotropic effects on the outcome)(Burgess & Thompson, 2017). In the protocol below, we describe how to conduct an MR analysis using these methods and provide guidance for utilizing MR software in R in order to perform, interpret, and visualize results of MR analyses.
Necessary Resources
Hardware
Computer running Linux, Mac OS, or Windows
Software
R package version >= 3.1.0 (Team, 2014)
Files
GWAS summary statistics (including SNP, major allele, minor allele, allele frequency, effect size, standard error, p-value, and sample size) for the exposure and outcome of interest.
Note that GWAS summary statistics may be available in different kinds of formats-- in this case, look at the header of the GWAS summary statistics file and identify if the following data are included, at a minimum: SNP, major allele, minor allele, allele frequency, effect size, standard error, p-value, and sample size. Remember that some information that may be missing from your summary statistics file, may be present in the paper referencing the GWAS.
The protocol and code below was inspired by the short course offered in the Mendelian Randomization Conference on July 10, 2017 by the MRC Integrative Epidemiology Unit.
Protocol steps —Step annotations
-
1.
Obtain GWAS summary statistics for your exposure (Figure 1, X) and outcome (Figure 1, Y) of interest. Resources such as the NHGRI-EBI Catalog(Burdett et al., n.d.) can be leveraged to search for and download publicly-available GWAS summary statistics.
-
2.
In this approach, genetic variants are utilized as instrumental variables (IVs), or “instruments” for the exposure. Determine usability of GWAS summary statistics from Step 1 by ensuring that the instrument-exposure data and the instrument-outcome data have listed the effect allele, allele frequency, beta, standard error, p-value, and sample size (as shown in Figure 2).
-
3.
Determine if the IV assumptions hold for conducting an MR analysis. The first assumption can be evaluated by linear regression of the exposure on the instrument and calculating the F-statistic for your instrument (Palmer et al., 2011; Teumer, 2018). This can be calculated as,, for N sample size, Knumber of genetic variants, and R2the proportion of the variance of the exposure explained by the IV (Burgess, Thompson, & CRP CHD Genetics Collaboration, 2011). An F statistic less than 10 denotes a weak instrument (Teumer, 2018).
The second and third assumptions are more challenging to formally validate due to the possibility of unknown effects(Palmer et al., 2011; Teumer, 2018). In assessing the second assumption, consider any potential confounding variables (Figure 1, U) that may play a role in the association between your exposure and outcome, and in assessing the third assumption, consider potential issues such as pleiotropy or population substructure that may serve as a violation (Palmer et al., 2011; Teumer, 2018).
-
4.Run R package. Input exposure and outcome GWAS summary statistic data, using the read.table function.
exposure_data<-read.table(“exposure_filename.txt”, head=T, sep=“\t”) outcome_data<-read.table(“outcome_filename.txt”, head=T, sep=“\t”)
Figure 2.
Shown are the first few rows of the body mass index GWAS summary statistics published from the UK Biobank and The Genetic Investigation of ANthropometric Traits (GIANT) Consortium meta-analysis(Yengo et al., 2018).
-
5.
Identify instruments. Find independent SNPs that are GWAS significant (P < 5.0 × 10−8) for the exposure and identify the effects for these “instrument” SNPs from the outcome GWAS. Independent SNPs that are GWAS significant for the exposure are “instruments” – or proxies for exposure -- in this analysis.
-
6.
Harmonize the exposure and outcome datasets. Ensure that the effect alleles from both files are the same. If not, then “flip” the log odds ratio of the effect allele of one of the datasets (multiply by −1). Ensure that the effect in the exposure file reflects the trait-increasing allele.
Note that the steps listed below for the ratio of coefficients (Steps 7-8), the inverse-variance weighted method (Step 9), and MR-Egger (Step 10) are independent and do not have to be performed consecutively (the results from one analysis do not affect the results of another analysis).
Ratio of coefficients (or Wald) method
-
7.Calculate the ratio of coefficients, or the Wald ratio. This is the simplest method for estimating the causal effect of the exposure on the outcome, and is the coefficient of the genetic variant in the regression of the outcome (represented here as outcome_data$beta) divided by the coefficient of the genetic variant in the regression of the exposure (represented here as exposure_data$beta)(Burgess, Small, & Thompson, 2017).
wald_ratio <- outcome_data$beta/exposure_data$beta wald_ratio_standard_error <- outcome_data$SE/exposure_data$beta z_statistic <- wald_ratio/wald_ratio_standard_error p_value <- 2*pnorm(abs(z_statistic) ,lower.tail=F)
Note: The Wald ratio corresponds to the log odds ratio for the outcome per unit change of the exposure.
-
8.Perform a fixed-effects meta-analysis using the Wald ratio.
effect <- sum(wald_ratio*wald_ratio_standard_error^−2)/ (sum(wald_ratio_standard_error^−2)) standard_error <- sqrt(1/sum(wald_ratio_standard_error^−2)) Z_statistic <- effect/standard_error p_value <- 2*pnorm(abs(Z_statistic) ,lower.tail=F)
Inverse-variance weighted (IVW) method
-
9.Perform an inverse-variance weighted (IVW) linear regression to estimate the effect of the exposure on the outcome.
IVW_weights <- outcome_data$SE^−2 inverse_weighted_LR <- lm(outcome_data$beta ~ exposure_data$beta - 1 ,weights=IVW_weights)
The command summary(inverse_weighted_LR) displays the effect, standard error, and p-value of the exposure on the outcome.
Note that the intercept term here is zero in order to calculate the IVW estimate (Burgess & Thompson, 2017). In the case that a single genetic variant satisfies the IV assumptions, the effect of the exposure on the outcome can be estimated as a ratio of the estimated coefficient for the outcome to the estimated coefficient for the exposure for the genetic variant (Burgess & Thompson, 2017).
MR-Egger Regression
-
10.Perform an MR-Egger regression to estimate the effect of the exposure on the outcome.
MR_egger_regression <- lm(outcome_data$beta ~ exposure_data$beta, weights=1/IVW_weights)
The command summary(MR_egger_regression) displays the effect, standard error, and p-value of the exposure on the outcome. Note that the intercept term here is calculated in the MR-Egger analysis (Bowden et al., 2015; Burgess & Thompson, 2017).
ALTERNATE PROTOCOL 1: Performing Mendelian randomization using the TwoSampleMR package in R.
The TwoSampleMR package in R facilitates conducting two-sample MR analyses by offering access to the large MR-Base repository of GWAS summary statistics and providing easy-to-use software for proper harmonization of datasets, estimating the causal effect using a range of MR methods, conducting sensitivity analyses, and visualizing results (Hemani et al., n.d., 2018).
This protocol and code below was inspired by the TwoSampleMR documentation provided by the MRC Integrative Epidemiology Unit and the MR-Base Collaboration, which can be found on https://mrcieu.github.io/TwoSampleMR/(Hemani et al., n.d., 2018).
Necessary Resources
Hardware
Computer running Linux, Mac OS, or Windows
Software
R package version >= 3.1.0 (Team, 2014) with the following libraries installed: devtools(Wickham, Hester, & Chang, 2018), TwoSampleMR(Hemani et al., n.d., 2018), MRInstruments(Hemani’, n.d.),and tidyverse(Wickham, 2017).
Files
GWAS summary statistics (including SNP, major allele, minor allele, allele frequency, effect size, standard error, p-value, and sample size) for the exposure and outcome of interest OR these files can be obtained by browsing through existing catalogues from the MR Base databases accessible through the MRInstruments package(Hemani’, n.d.). Note that some information that may be missing from your summary statistics file, may be present in the paper referencing the GWAS or may be calculated using the information in the file. Further note that your data can be formatted in the correct manner for use in the TwoSampleMR package by using the function format_data (as described in step #2 of the protocol below)(Hemani et al., n.d., 2018).
The .Rmd file “TwoSampleMR_protocol.Rmd” included in this manuscript will serve as a guide through the protocol below.
Protocol steps—Step annotations
-
1.Load the TwoSampleMR package in R (Hemani et al., n.d., 2018). You can install the
devtools package from CRAN-like repositories with the install.packages(“devtools”) command in order to utilize the install_github function(Wickham et al., 2018). install.packages(“devtools”) library(devtools) install_github(“MRCIEU/TwoSampleMR”) library(TwoSampleMR)
-
2.
Identify and obtain GWAS summary statistics. You can either obtain your own summary statistics or browse through the MR Base GWAS database(Hemani et al., 2018) (available_outcomes() can show the list of available GWASs).
External summary statistics can be read in and converted to the correct format using format_data. For example, the body mass index (BMI) GWAS summary statistics as shown in Figure 2 can be converted as follows:
exposure_converted_dataframe <- format_data(exposure_dataset, type = “exposure”, snp_col = “SNP”, beta_col = “BETA”, se_col = “SE”, effect_allele_col = “Tested_Allele”, other_allele_col = “Other_Allele”, eaf_col = “Freq_Tested_Allele_in_HRS”, pval_col = “P”, samplesize_col = “N”)
The R package MRInstruments contains data sources to search for genetic instruments that can be used for your MR analysis(Hemani’, n.d.). In this demonstration, we use data from the gwas_catalog to search for the instruments from the 2010 GWAS on BMI published in Nature Genetics by Speliotes et al (Speliotes et al., 2010). This data can be searched for and installed as follows:
devtools::install_github(“MRCIEU/MRInstruments”) library(MRInstruments) data(gwas_catalog) exposure_data <- subset(gwas_catalog, PubmedID == “20935630”)
-
3.Ensure that your data is presented in the correct input format as required by the package by running the format_data function and perform linkage disequilibrium (LD) clumping to remove any non-independent SNPs.
exposure_data <- format_data(exposure_data) exposure_data <- clump_data(exposure_data)
-
4.Extract the instrumental SNPs for your outcome of interest. In this example, we are using the 2014 GWAS summary statistics for type 2 diabetes susceptibility as published in Nature Genetics by the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium (DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al., 2014).
outcome_data <- extract_outcome_data( snps = exposure_data$SNP, outcomes = 23 ) -
5.Harmonize exposure and outcome datasets to ensure the reference alleles from both datasets match. Prune your harmonized dataset. Here, the exposure and outcome datasets are harmonized (shown in Figure 3) and renamed as dat.
dat <- harmonise_data( exposure_dat = exposure_data, outcome_dat = outcome_data ) dat <- power.prune(dat) -
6.Perform an MR analysis (results shown in Figure 4) and specify the types of method in method_list() of the mr()function.
results <- mr(dat)
It is conventional to report results from multiple methods. The full list of available MR methods can be identified from mr_method_list().
-
7.Conduct sensitivity analyses. Check for heterogeneity and test for directional horizontal pleiotropy.
mr_heterogeneity(dat) mr_pleiotropy_test(dat)
-
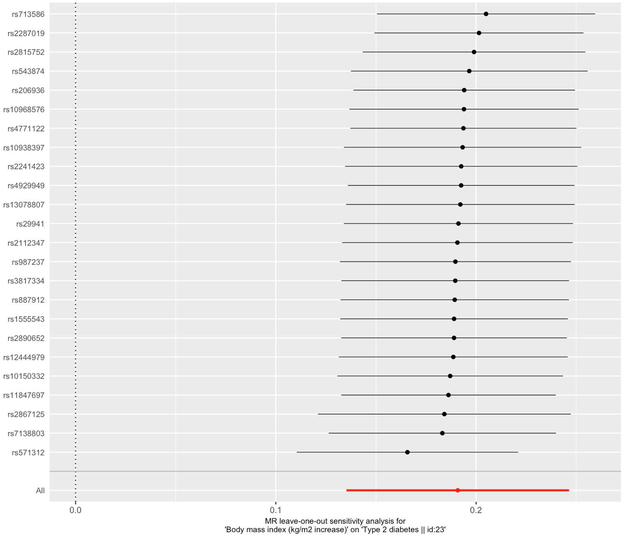
8.Perform a leave-one-out sensitivity analysis (by sequentially removing each SNP from the MR analysis and running MR) and visualize results from this sensitivity analysis (shown in Figure 5).
results_leaveoneout <- mr_leaveoneout(dat). mr_leaveoneout_plot(results_leaveoneout) plot_leaveonout[[1]]
-
9.Visualize MR results.
scatter_plot <- mr_scatter_plot(results, dat) scatter_plot[[1]]
Figure 3.
Shown are the first few rows of the harmonized dataset.
Figure 4.
The causal effects, standard errors, and p-values obtained from the MR analysis using the default methods of MR Egger, weighted median, inverse variance weighted, simple mode, and weighted mode, are shown.
Figure 5.
The results from the leave-one-out sensitivity analyses are shown on the scatterplot. The estimated causal effect is shown for each excluded SNP and the overall estimate using all the SNPs is shown in red. The error bars represent the 95% confidence intervals.
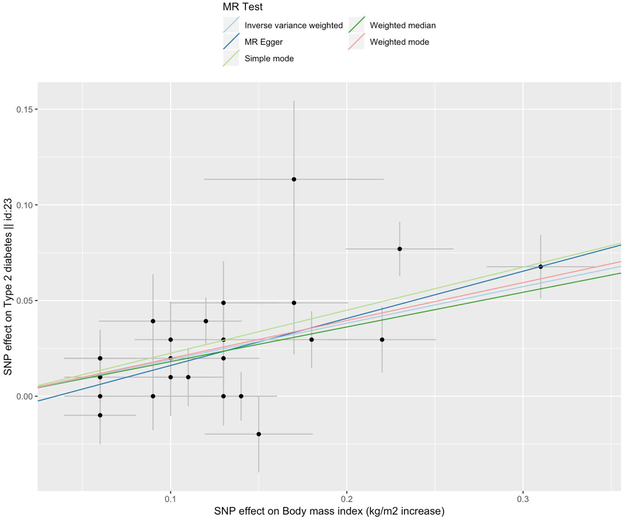
The command mr_scatter_plot(results, dat) creates a scatterplot for each exposure-outcome association (shown in Figure 6). A specification of the method in method_list() visualizes the estimated causal effect according to the specified MR method.
Figure 6.
The scatterplot suggests a positive causal relationship of the SNP effects on BMI against the SNP effects on type 2 diabetes. Each point displayed on the graph represents a single genetic variant. The horizontal and vertical lines extending from each point represent the 95% confidence interval for the genetic associations. The horizontal axis of the graph displays the estimated genetic associations with the exposure (BMI), and the vertical axis displays the estimated genetic associations with the outcome (type 2 diabetes). The color of the lines indicate the type of MR test used (light blue for inverse variance weighted, dark blue for MR Egger, light green for simple mode, dark green for weighted median, and red for weighted mode).
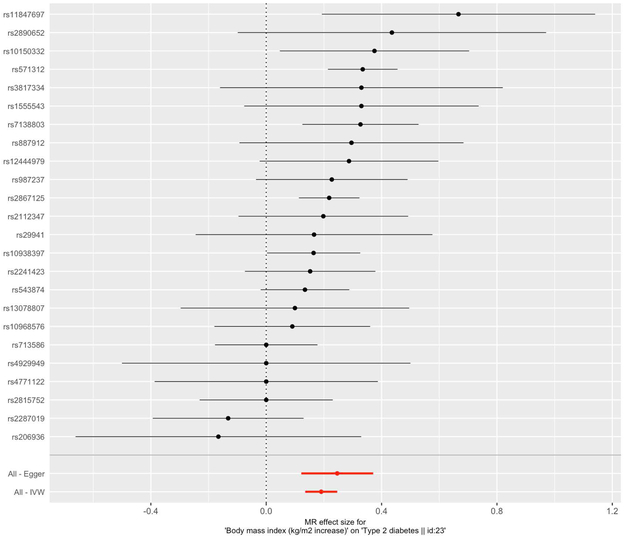
Additionally, a forest plot can be made to compare the MR estimates derived from the different MR methods (shown in Figure 7).
Figure 7.
The forest plot shows the causal estimate using each SNP alone as well as the overall causal estimate using all the SNPs with MR-Egger and IVW. The error bars represent the 95% confidence intervals.
single_snp_analysis <- mr_singlesnp(dat) forest_plot <- mr_forest_plot(single_snp_analysis) forest_plot[1]
GUIDELINES FOR UNDERSTANDING RESULTS
By leveraging a genetic approach as demonstrated in our example above, we were able to provide evidence in support of a positive causal effect of BMI on type 2 diabetes, which was consistent across all MR methods. We obtained effect sizes of 0.25, 0.18, and 0.19 for MR Egger, weighted median, and inverse variance weighted, respectively, which correspond to the estimated causal effect on type 2 diabetes per unit increase in BMI (kg/m2). In a “leave-one-out” sensitivity analysis, where we sequentially excluded a SNP and performed MR, we observe that the causal estimate remains robust. The forest plot compares the estimated causal effects for all the SNPs as determined by MR-Egger and IVW to the estimated causal effect as determined per each SNP. While the MR-Egger and IVW estimates agree in our demonstrated example, the IVW estimate can substantially differ from the MR-Egger estimate, suggesting the possibility of directional pleiotropy (Burgess & Thompson, 2017). Directional pleiotropy is the phenomena when genetic variants affect multiple traits on different causal pathways, potentially resulting in a violation of the instrumental variable assumptions necessary for conducting an MR analysis (Burgess & Thompson, 2017). In summary, we highlight the utility of MR in assessing causal relationships, while accounting for limitations prone to many conventional observational epidemiological studies.
COMMENTARY
Background Information
The concept of utilizing IVs to examine causal effects was first introduced in econometrics 90 years ago, and applied to disease outcomes in 1986 by Martijn Katan (Thomas & Conti, 2004). In assessing the causal role of low serum cholesterol levels and cancer, Katan explained that the relationship was likely not affected by diet or other confounding factor, but that the relationship can be elucidated by observation of the number of cancer patients who carry the E-2 isoform of the apolipoprotein (ApoE) gene, which is associated with lower serum density lipoprotein than major isoforms E-3 and E-4 (Katan, 1986). Since then, there have been many studies that have attempted to assess causal relationships using MR for a range of exposures and outcomes, including biomarkers (i.e. C reactive protein in association with coronary heart disease (C Reactive Protein Coronary Heart Disease Genetics Collaboration (CCGC) et al., 2011)), clinical traits (i.e. BMI in association with cardiometabolic traits (Holmes, Lange, et al., 2014)), disease phenotypes (i.e. a range of biomarkers in association with coronary heart disease (Bennett & Holmes, 2017)), socioeconomics (i.e. educational attainment in association with coronary heart disease (Tillmann et al., 2017)), behavioral characteristics (i.e., alcohol consumption in association with cardiovascular disease (Holmes, Dale, et al., 2014)), and intrauterine effects on offspring outcomes (D. Lawlor et al., 2017) (i.e., maternal homocysteine levels in association with offspring birthweight (Lee et al., 2013)).Results from these studies attempt to assess causality for a broad range of exposures and have shown feasibility of use of MR to explore promising areas for therapeutic intervention.
For example, an MR study demonstrated that genetic variants in the gene encoding the target of statin therapy, HMG-CoA reductase or HMGCR, is associated with increased risk for type 2 diabetes and related traits such as higher body weight and waist circumference, highlighting a potential pharmacological application of MR(Swerdlow et al., 2015). In another example, MR was used to determine that tobacco smoking may cause a reduced BMI and a higher resting heart rate, but did not find a strong causal association between smoking and adverse blood pressure, serum lipids, and glucose levels(Åsvold et al., 2014). MR promises to be a valuable method for identifying disease risk factors and areas for intervention and can be leveraged to inform public health policy.
Critical Parameters
There are a number of statistical and methodological challenges and limitations to MR that have been discussed at length in other articles (Burgess, 2012; Haycock et al., 2016; VanderWeele, Tchetgen Tchetgen, Cornelis, & Kraft, 2014). Possible limitations include linkage disequilibrium (i.e., when different loci within a population have correlated allelic states(D. A. Lawlor et al., 2008)), population stratification (i.e., when a population can be broken into subpopulations that exhibit different frequencies of genetic variants or disease(D. A. Lawlor et al., 2008)), or pleiotropy (i.e., when a genetic variant is associated with more than one phenotype(D. A. Lawlor et al., 2008)). Challenges may arise from utilizing a weak instrument (F statistic less than 10), or from situations where the core assumptions are violated or weakly satisfied, and even from cases where the core assumptions are satisfied, but an external factor is at play (i.e., canalization) (Zheng et al., 2017). In fact, the development of novel MR approaches and extensions to the conventional methodology to account for these limitations is a rapidly growing field (Bowden, Del Greco M, et al., 2016; Bowden et al., 2017, 2018; van Kippersluis & Rietveld, 2017; Verbanck, Chen, Neale, & Do, 2018).
For a description of potential limitations that may affect interpretation of MR findings and recommended practices in those situations, we recommend referring to Table 2 from a review article by Zheng (Zheng et al., 2017) and Table II from Lawlor (D. A. Lawlor et al., 2008). We also recommend referring to Table 2 from the review article by Burgess for descriptions of various sensitivity analyses and situations where they would be of relevance (Burgess, Bowden, Fall, Ingelsson, & Thompson, 2017).
Significance Statement.
Conventional observational epidemiological studies aimed at assessing the effect of a modifiable exposure on a disease phenotype can be subject to confounding such as reverse causation, where the disease precedes the exposure(Smith & Ebrahim, 2002). A technique termed ‘Mendelian randomization’ (MR) can overcome this limitation by leveraging genetic variants such as single-nucleotide polymorphisms (SNPs) as instrumental variables to estimate exposure-outcome associations (Smith & Ebrahim, 2004). Summary statistics from genome-wide association studies (GWAS) facilitate conducting an MR analysis without the need for costly direct genotyping or obtaining individual-level data (Burgess et al., 2015). We describe here a protocol for assessing exposure-outcome associations in an MR framework using published GWAS summary statistics.
ACKNOWLEDGEMENT
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE1745303. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. C.J.P is supported by a National Institutes of Health NIAID R01AI127250, National Institute of Environmental Health Sciences (NIEHS) R00 ES023504, R21 ES025052 and National Science Foundation Big Data Spoke (1636870).
We thank Dr. George Davey Smith and his team at the MRC Integrative Epidemiology Unit at the University of Bristol for offering the short course at the 2017 Mendelian Randomization Conference and for providing resources for conducting an MR study. We also thank the MR-Base Collaboration for providing the extended documentation for the TwoSampleMR package (accessible at: https://mrcieu.github.io/TwoSampleMR/).
LITERATURE CITED
- Åsvold BO, Bjørngaard JH, Carslake D, Gabrielsen ME, Skorpen F, Smith GD, & Romundstad PR (2014). Causal associations of tobacco smoking with cardiovascular risk factors: a Mendelian randomization analysis of the HUNT Study in Norway. International Journal of Epidemiology, 43(5), 1458–1470. [DOI] [PubMed] [Google Scholar]
- Bennett DA, & Holmes MV (2017). Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart , 103(18), 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, & Burgess S (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International Journal of Epidemiology, 44(2), 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Haycock PC, & Burgess S (2016). Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genetic Epidemiology, 40(4), 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan NA, & Thompson JR (2016). Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. International Journal of Epidemiology, 45(6), 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, & Thompson J (2017). A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Statistics in Medicine, 36(11), 1783–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Spiller W, Del Greco M F, Sheehan N, Thompson J, Minelli C, & Davey Smith G (2018). Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. International Journal of Epidemiology. 10.1093/ije/dyy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett T, Hastings E, Welter D, SPOT, EMBL-EBI, & NHGRI. (n.d.). GWAS Catalog. Retrieved December 13, 2017, from https://www.ebi.ac.uk/gwas/downloads/summary-statistics [Google Scholar]
- Burgess S (2012). Statistical issues in Mendelian randomization: use of genetic instrumental variables for assessing causal associations. University of Cambridge. Retrieved from https://www.repository.cam.ac.uk/handle/1810/242184 [Google Scholar]
- Burgess S, Bowden J, Fall T, Ingelsson E, & Thompson SG (2017). Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology , 28(1), 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Davies NM, & Thompson SG (2016). Bias due to participant overlap in two-sample Mendelian randomization. Genetic Epidemiology, 40(7), 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, & EPIC- InterAct Consortium. (2015). Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. European Journal of Epidemiology, 30(7), 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Small DS, & Thompson SG (2017). A review of instrumental variable estimators for Mendelian randomization. Statistical Methods in Medical Research, 26(5), 2333–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, & Thompson SG (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. European Journal of Epidemiology, 32(5), 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG, & CRP CHD Genetics Collaboration. (2011). Avoiding bias from weak instruments in Mendelian randomization studies. International Journal of Epidemiology, 40(3), 755–764. [DOI] [PubMed] [Google Scholar]
- C Reactive Protein Coronary Heart Disease Genetics Collaboration (CCGC), Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, … Danesh J (2011). Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ , 342, d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, & Ebrahim S (2003). “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology, 32(1), 1–22. [DOI] [PubMed] [Google Scholar]
- Davey Smith G, & Ebrahim S (2007). Mendelian randomization: genetic variants as instruments for strengthening causal inference in observational studies. Bio-Social Surveys: Current Insight and Future Promise. The National Academies Press, National Research Council, Washington, DC. [Google Scholar]
- Davey Smith G, & Hemani G (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics, 23(R1), R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, … Morris AP (2014). Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature Genetics, 46(3), 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland S (2018). An introduction to instrumental variables for epidemiologists. International Journal of Epidemiology, 47(1), 358. [DOI] [PubMed] [Google Scholar]
- Greenland S, Robins JM, & Pearl J (1999). Confounding and Collapsibility in Causal Inference. Statistical Science: A Review Journal of the Institute of Mathematical Statistics, 14(1), 29–46. [Google Scholar]
- Hartwig FP, Davies NM, Hemani G, & Davey Smith G (2016). Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. International Journal of Epidemiology, 45(6), 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, & Davey Smith G (2016). Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. The American Journal of Clinical Nutrition, 103(4), 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Haycock P, Zheng J, Gaunt T, & Elsworth B (n.d.). TwoSampleMR: Two Sample MR functions and interface to MR Base database. [Google Scholar]
- Hemani gibran. (n.d.). MRInstruments: Data sources for genetic instruments to be used in MR. Retrieved from https://github.com/MRCIEU/MRInstruments [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, … Haycock PC (2018). The MR-Base platform supports systematic causal inference across the human phenome. eLife, 7 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MV, Dale CE, Zuccolo L, Silverwood RJ, Guo Y, Ye Z, … InterAct Consortium. (2014). Association between alcohol and cardiovascular disease: Mendelian randomisation analysis based on individual participant data. BMJ , 349, g4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MV, Lange LA, Palmer T, Lanktree MB, North KE, Almoguera B, … Keating BJ (2014). Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. American Journal of Human Genetics, 94(2), 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan MB (1986). Apolipoprotein E isoforms, serum cholesterol, and cancer. The Lancet, 1(8479), 507–508. [DOI] [PubMed] [Google Scholar]
- Lawlor DA (2016). Commentary: Two-sample Mendelian randomization: opportunities and challenges. International Journal of Epidemiology, 45(3), 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JAC, Timpson N, & Davey Smith G (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in Medicine, 27(8), 1133–1163. [DOI] [PubMed] [Google Scholar]
- Lawlor D, Richmond R, Warrington N, McMahon G, Davey Smith G, Bowden J, & Evans DM (2017). Using Mendelian randomization to determine causal effects of maternal pregnancy (intrauterine) exposures on offspring outcomes: Sources of bias and methods for assessing them. Wellcome Open Research, 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HA, Park EA, Cho SJ, Kim HS, Kim YJ, Lee H, … Park H (2013). Mendelian randomization analysis of the effect of maternal homocysteine during pregnancy, as represented by maternal MTHFR C677T genotype, on birth weight. Journal of Epidemiology / Japan Epidemiological Association, 23(5), 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TM, Sterne JAC, Harbord RM, Lawlor DA, Sheehan NA, Meng S, … Didelez V (2011). Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. American Journal of Epidemiology, 173(12), 1392–1403. [DOI] [PubMed] [Google Scholar]
- Smith GD, & Ebrahim S (2002). Data dredging, bias, or confounding. BMJ , 325(7378), 1437–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GD, & Ebrahim S (2004). Mendelian randomization: prospects, potentials, and limitations. International Journal of Epidemiology, 33(1), 30–42. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, … Loos RJF (2010). Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics, 42(11), 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JEL, Shah T, … Sattar N (2015). HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. The Lancet, 385(9965), 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC (2014). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Teumer A (2018). Common Methods for Performing Mendelian Randomization. Frontiers in Cardiovascular Medicine, 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, & Conti DV (2004). Commentary: the concept of “Mendelian Randomization.” International Journal of Epidemiology, 33(1), 21–25. [DOI] [PubMed] [Google Scholar]
- Tillmann T, Vaucher J, Okbay A, Pikhart H, Peasey A, Kubinova R, … Holmes MV (2017). Education and coronary heart disease: mendelian randomisation study. BMJ , 358, j3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, & Kraft P (2014). Methodological challenges in mendelian randomization. Epidemiology , 25(3), 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kippersluis H, & Rietveld CA (2017). Pleiotropy-robust Mendelian randomization. International Journal of Epidemiology. 10.1093/ije/dyx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbanck M, Chen C-Y, Neale B, & Do R (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nature Genetics, 50(5), 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2017). tidyverse: Easily Install and Load the “Tidyverse.” Retrieved from https://CRAN.R-project.org/package=tidyverse [Google Scholar]
- Wickham H, Hester J, & Chang W (2018). devtools: Tools to Make Developing R Packages Easier. Retrieved from https://CRAN.R-project.org/package=devtools [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, … Others. (2018). Meta-analysis of genome-wide association studies for height and body mass index in\ 700,000 individuals of European ancestry. bioRxiv, 274654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Baird D, Borges M-C, Bowden J, Hemani G, Haycock P, … Smith GD (2017). Recent Developments in Mendelian Randomization Studies. Current Epidemiology Reports, 4(4), 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]