Abstract
The aging process results in significant epigenetic changes at all levels of chromatin and DNA organization. These include reduced global heterochromatin, nucleosome remodeling and loss, changes in histone marks, global DNA hypomethylation with CpG island hypermethylation, and the relocalization of chromatin modifying factors. Exactly how and why these changes occur is not fully understood, but evidence that these epigenetic changes affect longevity and may cause aging, is growing. Excitingly, new studies show that age-related epigenetic changes can be reversed with interventions such as cyclic expression of the Yamanaka reprogramming factors. This review presents a summary of epigenetic changes that occur in aging, highlights studies indicating that epigenetic changes may contribute to the aging process and outlines the current state of research into interventions to reprogram age-related epigenetic changes.
Keywords: Aging, epigenetics, clock, chromatin, histones, reprogramming, sirtuins, DNA methylation
Introduction
The term “epigenetics” is thrown around a lot. Originally, it was coined to describe heritable changes that were non-mendelian, but use of the term has evolved. These days, “epigenetics” more generally refers to all non-genomic information storage in cells including gene networks, chromatin structure and post-translational modifications to histones. With aging, there are distinct changes across the epigenome from DNA modifications to alterations in global chromatin organization. But key questions remain unanswered: How and why do these changes occur? Do these changes drive disease and aging? Are they reversible?
Genomic organization is determined by the complex structure of chromatin (Figure 1). The basic unit of chromatin is the nucleosome, which is made up of 147 DNA base pairs wrapped around an octamer of histone proteins. This octamer usually comprises two copies each of H2A, H2B, H3 and H4 (Luger et al. 1997; Hansen 2002). Within nucleosomes, both histones and the DNA itself are subject to a range of chemical modifications that affect the chromatin structure and ultimately the expression of genes. Chromatin falls into one of two major subtypes: euchromatin, in which the chromatin is open and transcriptionally active and heterochromatin, in which the chromatin is tightly closed and transcriptionally silent (Wallrath 1998; Grewal and Moazed 2003). Regulating the epigenetic network are factors that modify chromatin including DNA- and histone-modifying enzymes, transcription factors, and the more recently identified noncoding RNAs (ncRNAs).
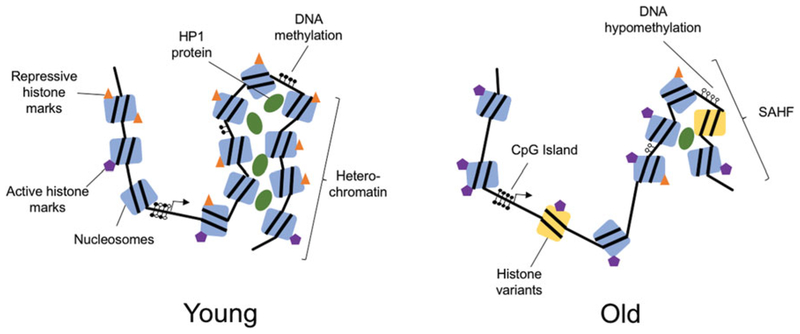
Figure 1.
Age-related changes to chromatin. With increasing age, there are both global and loci-specific changes to chromatin structure. Young chromatin is characterized by predominantly tightly packaged heterochromatin, with repressive histone marks and HP1 protein binding. Nucleosomes are made up of canonical histone proteins and there is an abundance of DNA cytosine methylation. Chromatin of cells from old individuals is characterized by globally reduced heterochromatin with specific areas of heterochromatin known as senescence-associated heterochromatin foci (SAHF). There is a decrease in repressive histone marks and an increase in active histone marks. Non-canonical histone variants are included in nucleosomes and there is general nucleo-some loss. Additionally, there is global DNA hypomethylation except in CpG islands where there is hypermethylation (see the color version of this figure at www.tandfonline.com/ibmg).
With aging there are distinct epigenetic changes including reduced global heterochromatin, formation of distinct heterochromatin foci, remodeling and loss of nucleosomes, changes in the abundance of histone variants, altered histone marks, global hypomethylation of DNA with distinct areas of hypermethylation, changes in ncRNA abundance, and relocalization of chromatin-modifying factors (Figure 1 and Table 1). Consequences of these epigenetic changes include increased genomic instability (Sinclair et al. 1998a; Hu et al. 2014) and distinct changes in gene expression with aging characterized by a loss of silencing, increased translation and increased expression of retrotransposons (Zahn et al. 2007; Budovskaya et al. 2008; Baumgart et al. 2014). Epigenetic changes may contribute to many of the hallmarks of aging including cellular senescence and mitochondrial dysfunction (Lopez-Otin et al. 2013; Booth and Brunet 2016). One of the most exciting recent discoveries in this field is the discovery that age-related epigenetic changes in cells and organisms can be reset with reprogramming and that this results in the reversal of many age-related phenotypes (Ocampo, Reddy, and Belmonte 2016).
Table 1.
Studies of age-related epigenetic changes in pre-clinical and clinical models.
| Epigenetic change with aging | Markers | Model(s) | References |
|---|---|---|---|
| Reduced global heterochromatin | HP1 and H3K9me3 reduced, lamin A changes | C. elegans | (Haithcock et al. 2005; Ni et al. 2012) |
| Drosophila | (Brandt et al. 2008; Wood et al. 2010; Larson et al. 2012) | ||
| Human fibroblasts | (Scaffidi and Misteli 2006) | ||
| Progeria patient cells | (Scaffidi and Misteli 2006; Shumaker et al. 2006; Zhang et al. 2015) | ||
| Senescence-associated heterochromatin foci (SAHF) | HP1 and H3K9me3 increased, macroH2A and HMGA increased | Mice | (Kreiling et al. 2012) |
| Baboons | (Herbig et al. 2006; Kreiling et al. 2012) | ||
| Human fibroblasts | (Narita et al. 2003, 2006; Zhang et al. 2005) | ||
| Nucleosome remodeling and loss | Loss of core histone proteins | Yeast | (Dang et al. 2009; Feser et al. 2010; Hu et al. 2014) |
| Worms | (Ni et al. 2012) | ||
| Human fibroblasts | (O’Sullivan et al. 2010b; Ivanov et al. 2013) | ||
| Histone variants increased | H3.3, H3.3cs1 | Mouse brain | (Maze et al. 2015) |
| Human fibroblasts | (Duarte et al. 2015) | ||
| Changed histone marks | Globally active marks increased (H3K4me3, H4K16ac) and repressive marks decreased (H3K9me3, H3K27me3) | Yeast | (Dang et al. 2009; Feser et al. 2010) |
| Drosophila | (Wood et al. 2010; Larson et al. 2012) | ||
| C. elegans | (Ni et al. 2012) | ||
| Killifish | (Liu, Wang, et al. 2013; Baumgart et al. 2014) | ||
| Rats | (Sarg et al. 2002) | ||
| Mouse brain | (Peleg et al. 2010; Ryu et al. 2011) | ||
| Mouse stem cells | (Sun et al. 2014; Schwoerer et al. 2016) | ||
| Mouse fibroblasts | (Lyu et al. 2018) | ||
| Progeria mouse models | (Wang et al. 2010) | ||
| Progeria patient cells | (Shumaker et al. 2006; Maures et al. 2011; Zhang et al. 2015) | ||
| Human fibroblasts | (O’Sullivan et al. 2010a; Shah et al. 2013) | ||
| Human brain tissue | (Nativio et al. 2018) | ||
| DNA methylation changes | Global DNA hypomethylation, CpG island hypermethylation | Salmon | (Berdyshev et al. 1967) |
| Mice | (Wilson et al. 1987; Cole et al. 2017; Petkovich et al. 2017; Stubbs et al. 2017; Wang et al. 2017) | ||
| Rats | (Vanyushin et al. 1973) | ||
| Dogs | (Thompson et al. 2017) | ||
| Rhesus monkeys | (Maegawa et al. 2017) | ||
| Human fibroblasts | (Koch et al. 2012b) | ||
| Human stem cells | (Fernández and Bayón 2014) | ||
| Humans | (Christensen et al. 2009; Zampieri et al. 2015; Li et al. 2017; Wagner 2017; Horvath and Raj 2018) | ||
| Relocalization of chromatin-modifying factors (RCM) | SIRT-1, PARP-1, REST, HDAC-1 | Yeast | (Kennedy et al. 1997; Sinclair and Guarente 1997; Oberdoerffer et al. 2008) |
| Mice | (Oberdoerffer et al. 2008) | ||
| ncRNA changes | H19, Dicer, lin-4, lin-14, mir-34 | Yeast | (Saka et al. 2013) |
| C. elegans | (Boehm and Slack 2005; Ibáñez-ventoso et al. 2006; Mori et al. 2012) | ||
| Mice | (Fu et al. 2008; Li, Khanna, et al. 2011; Mori et al. 2012) | ||
| AD mouse models | (Wang et al. 2009) | ||
| Humans | (Hooten et al. 2010; Wang, Huang, et al. 2011; Zhang et al. 2014) |
This article will review the literature on the epigenetic hallmarks of aging including evidence from preclinical and clinical studies. We will also discuss the potential to delay or reverse these epigenetic changes with dietary interventions, pharmaceutical approaches, or cellular reprogramming.
Epigenetic changes with aging
Reduced global heterochromatin
Transcriptionally inactive heterochromatin is characterized by the presence of heterochromatin protein (HP1), linker histones (eg H1), decreased histone acetylation and increased histone mark H3K9me3 (Oberdoerffer and Sinclair 2007; Tsurumi and Li 2012). With age there is a global reduction in heterochromatin along with a reduction in HP1 and H3K9me3 levels. This global decrease is observed in Caenorhabditis elegans (Haithcock et al. 2005), Drosophila (Brandt et al. 2008; Wood et al. 2010; Larson et al. 2012), senescent human fibroblasts (Scaffidi and Misteli 2006) and fibroblasts from Hutchinson–Gilford Progeria Syndrome (HGPS) and Werner syndrome progeria patients (Scaffidi and Misteli 2006; Shumaker et al. 2006; Zhang et al. 2015). In particular, regions that are normally heterochromatic, such as telomeres and pericentromeres, become more euchromatic as organisms age, from yeast to mammals (Sinclair et al. 1998b; Sedivy et al. 2008).
The first clues that aging is caused by epigenetic changes came from the discovery that a mutation in a yeast gene called silent information regulator, SIR2, extended lifespan by relocalizing the NAD+-dependent Sir2 histone deacetylase to regions of DNA instability, such as DNA breaks and recombination occurring at the ribosomal DNA (rDNA). In doing so, it relieved silencing at the mating type loci that controls cell identity, resulting in sterility, a hallmark of yeast aging (Kennedy et al. 1997; Sinclair and Guarente 1997; Imai et al. 2000). These findings gave rise to the idea that aging might be caused by a loss of heterochromatin (Sinclair et al. 1997; Villeponteau 1997; Imai and Kitano 1998). Consistent with this, upregulation of Sir2 proteins in yeast and other organisms were found to extend lifespan (Kaeberlein et al. 1999; Lamming 2005). The Relocalization of Chromatin Modifiers Hypothesis of Aging (RCM) is discussed further in Section “Re-localization of chromatin modifiers (RCM)”.
Studies of accelerated aging syndromes or progerias indicate that the nuclear envelope may also have a role in contributing to heterochromatin loss with age. Hutchinson-Gilford progeria syndrome (HGPS) is caused by a truncated form of lamin A and results in a reduction in heterochromatin accompanied by reduced HP1 and lower levels of H3K9me3 (Scaffidi and Misteli 2006; Shumaker et al. 2006). In normal aging, there are also changes to lamin A that may contribute to the reduction in heterochromatin (Scaffidi and Misteli 2006). Whether it is lamin defects, heterochromatin loss, or the two processes combined, that drive human aging remains unknown, though there are tantalizing clues.
Senescence-associated heterochromatin foci (SAHFs)
With increasing age, as cells head toward or enter senescence, domains of heterochromatin form, known as senescent-associated heterochromatin foci (SAHFs). SAHFs are characterized by increased HP1, H3K9me3 marks, macroH2A and high-mobility group A (HMGA) protein binding (Narita et al. 2006).
SAHFs were originally thought to be a distinguishing feature of senescent cells (Narita et al. 2003) but they are now believed to occur in old non-senescent cells as well. SAHFs occur in a wide variety of cell types in culture (Narita et al. 2003, 2006; Zhang et al. 2005) and can be detected in the tissues of old mice (Kreiling et al. 2012) and baboons (Herbig et al. 2006; Kreiling et al. 2012). The formation of SAHFs is a nonstochastic process known as heterochromatin redistribution (Sedivy et al. 2008). This redistribution involves heterochromatin decondensation followed by the formation of heterochromatin foci at previously euchromatin regions, a process driven by H3K9 trimethylation, HP1 binding and the subsequent inclusion of macroH2A. This process involves a variety of heterochromatin proteins including histone chaperones HIRA and Asf1 and H3K9 methyltransferases Suv39 h1 and h2 (Zhang et al. 2005, 2007; Narita et al. 2006; Sedivy et al. 2008; Tsurumi and Li 2012). SAHFs specifically form at loci of proliferative genes such as E2F target genes where they act to silence their expression (Narita et al. 2003; Sedivy et al. 2008).
Nucleosome remodeling and loss
With increasing age, there are changes to nucleosome positioning and occupancy, reduced bulk levels of core histones that make up nucleosomes, and replacement of the canonical histones with histone variants such as H3.3. Our understanding of the mechanisms of nucleo-some loss in aging has primarily come from yeast. During yeast aging, nucleosome occupancy decreases by up to 50% across the genome (Hu et al. 2014) and up to half of the core histone proteins are lost with age (Dang et al. 2009; Feser et al. 2010). Nucleosome occupancy decreases as a result of both a reduction in the amount of core histones produced and a change in the activity of histone chaperones (Feser et al. 2010; Liu, Cheung, et al. 2013). There is a decline in protein synthesis of histones with age despite increased transcription across the genome (Lesur and Campbell 2004; Dang et al. 2009; Feser et al. 2010). Loss of histones may be due to a loss of translation (Feser et al. 2010; O’Sullivan et al. 2010b) or perhaps changes in gene expression as a result of histone mark changes at his-tone-coding genes (see Section “Changing post-translational modifications of histones”). Reduced histone protein synthesis results in a loss of histones particularly at subtelomeric regions (Dang et al. 2009) and ultimately reduced nucleosome occupancy and changed nucleosome positioning on particular DNA sequences (Hu et al. 2014). Reduced core histone proteins with aging are also seen in worms (Ni et al. 2012), aging human primary fibroblasts (O’Sullivan et al. 2010b) and senescent human cells (Ivanov et al. 2013).
During aging in yeast and mammals, the abundance of histone protein variants also increases. These are non-canonical histone proteins that are structurally distinct from the canonical proteins and perform specialized functions (Kamakaka and Biggins 2005; Skene and Henikoff 2013). An example is histone H3.3 which is expressed constitutively in cells unlike the canonical H3.1 which is mostly expressed in S phase. It follows then that in senescent cells, which are no longer dividing, and in the aging mouse brain, that H3.3 is a more common form than H3.1 (Maze et al. 2015). Another noncanonical N-terminal cleaved form of H3.3, H3.3cs1, is also more abundant in senescent cells. H3.3cs1 has a role in the silencing of Rb/E2F target genes which regulate the cell cycle (Duarte et al. 2015), indicating that H3.3cs1 may act as a modulator of senescence. Furthermore, overexpression of H3.3 or H3.3cs1 in cells is sufficient to induce senescence, indicating that these histone changes might be a driver of aging (Duarte et al. 2015; Maze et al. 2015).
Another age-related histone variant is macroH2A. Levels of macroH2A increase with aging in mice, primates and in senescent human fibroblasts (Kreiling et al. 2012). The macroH2A protein promotes transcriptional silencing (Pasque et al. 2011; Gaspar-maia et al. 2013) and is abundant in SAHF (Zhang et al. 2005, 2007). In senescent cells, macroH2A relocalizes from senescence-associated secretory phenotype (SASP)-related genes to SAHF heterochromatic foci, where it promotes transcriptional silencing (Pasque et al. 2011; Gaspar-maia et al. 2013; Chen et al. 2015; Kozlowski and Ladurner 2015). Knockout of macroH2A in senescent cells reduces SASP indicating that macroH2A may promote the SASP response (Chen et al. 2015; Kozlowski and Ladurner 2015). We have suggested that, like the yeast relocalization of sirtuin proteins to DNA damage and the activation of mating type genes that promote DNA repair, this mammalian response may also be part of an ancient stress response, one that results in chronic inflammation (Oberdoerffer and Sinclair 2007).
In yeast, the age-related changes in nucleosome abundance cause a global upregulation of gene expression and increased genomic instability including increased DNA breaks, damaged foci formation, translocations, increases in retrotransposons and the insertion of mitochondrial DNA into nuclear DNA (Hu et al. 2014). Interestingly if the age-related loss of histones H3 and H4 is suppressed by overexpressing their two genes, or the genes that degrade these proteins are deleted, then yeast replicative lifespan increases (Feser et al. 2010; Ivanov et al. 2013; Hu et al. 2014; McCormick et al. 2015) and the genetic instability is prevented (Sinclair and Guarente 1997; Hu et al. 2014).
Nucleosome remodeling complexes are ATP-dependent complexes that work in combination with other proteins such as histone-modifying enzymes to remodel nucleosomes at specific locations, allowing them to become open and dynamic (Clapier and Cairns 2009). Several studies have identified a role for these complexes in aging and longevity. For example, in the nematode C. elegans, knockdown of a specific subunit of the nucleosome remodeling and histone deacetylase (NURD) chromatin remodeling complex (LET-218/Mi2 subunit) resulted in increased lifespan (Vaux et al. 2013). Knockout of other subunits of the complex, however, did not increase longevity, for reasons that are unclear (Vaux et al. 2013). Interestingly, in humans, components of this same NURD chromatin remodeling complex (particularly RBBP4 and RBBP7) are reduced with increasing age, as is seen in cells from progeroid HGPS patients and in aging fibroblasts (Pegoraro et al. 2009). In yeast, knockdown of the nucleosome remodeling complex Isw2 increases lifespan (Dang et al. 2015) and in worms nucleosome remodeling complex SWI/SNF was identifies as a crucial cofactor for the longevity extending effects of DAF-16 (Riedel et al. 2013).
Changing post-translational modifications of histones
During aging, histones are post-translationally modified by a variety of histone-modifying enzymes. By promoting or inhibiting the recruitment of transcriptional complexes, post-translational modifications are essential for the control of gene expression (Lauberth et al. 2013; Pal and Tyler 2016). They also facilitate DNA repair, DNA replication and chromatin condensation (Pal and Tyler 2016). There are thousands of possible histone modifications and although we know the basics, the complete set of functions for each modification is not yet known (Kouzarides 2007; Bannister and Kouzarides 2011; Tan et al. 2011). Histone marks are commonly referred to by the specific modification that is seen at the amino acid target (e.g. lysine, K) on a specific histone protein (e.g. H3). For example, H3K4me3 refers to a trimethylation at lysine 4 on H3. Marks that are associated with transcriptional silencing and the formation of heterochromatin include H3K9me3, H3K27me3, H4K20me2 and H3K56ac. Conversely, some marks are associated with active transcription such as H3K4me3 and H4K16ac.
As organisms age, there are clear changes to both the global and specific histone mark patterns. Globally, there is an overall increase in activating marks and a decrease in repressive marks (Benayoun et al. 2015), although the specific pattern of histone modifications differs between organisms, between tissues of the same individual, and even between cells in the same tissue. A recent single cell analysis of human peripheral blood mononuclear cells detected cell type-specific changes to histone modifications during aging, with an overall increased heterogeneity between both cells and individuals (Cheung et al. 2018). These changing patterns with age may also contribute to the aging process and longevity. Alterations in the activity or abundance of histone modifying proteins, including methyltransferases, demethylases, acetylases and deactylases, have significant effects on lifespan in aging yeast, worm and fly models. For example, a screen for genes that extend the lifespan of C. elegans identified the histone methyltransferases ASH-2, SET-2, SET-9, SET-26 and demethylase UTX-1 (Hamilton et al. 2005; Ni et al. 2012).
H3K27me3 is a repressive histone mark, with a variety of roles. In embryonic stem cells, H3K27me3 is a distinctive mark of poised enhancers which are activated during differentiation and modulate the transcription of specific genes that establish and maintain cell identity (Zentner et al. 2011; Calo and Wysocka 2013). In Drosophila, H3K27me3 also has a role in determining topologically associated domains (TADs) of the genome, which can be broadly classified as either areas of low H3K27me3 marks which contain housekeeping genes, and areas of high H3K27me3 marks which contain regulated genes (El-Sharnouby et al. 2017). H3K27me3 is altered in a variety of cell types and species during aging. For example, in the killifish brain, mouse muscle stem cells (Sun et al. 2014; Schwoerer et al. 2016) and brain tissue from the senescent accelerated mouse SAMP8 (Wang et al. 2010), there are increased levels of H3K27me3 with increasing age (Liu, Cheung, et al. 2013; Baumgart et al. 2014). In C. elegans and fibroblasts from HGPS and Werner syndrome progeria patients, however, there are decreased levels of H3K27me3 (Shumaker et al. 2006; Maures et al. 2011; Ni et al. 2012; Zhang et al. 2015). These differences in global levels across tissues are likely due to locus-specific changes in histone marks. For example, while aged mouse hematopoietic stem cells and senescent human lung fibroblasts have an overall increased prevalence of H3K27me3, there are also areas where this mark decreases (Shah et al. 2014; Sun et al. 2014). Interestingly, long-lived naked mole rats have much higher levels of the repressive mark H3K27me3 than even old mice (Tan et al. 2017), and perhaps increased levels of this repressive mark contribute to the longevity of these rodents.
There is additional evidence that changes in H3K27me3 determine lifespan. The methylation status of H3K27 is controlled by the demethylases UTX-1, KDM6B/JMJD-3, PHF8/JMJD-1.2 and the methyltransferases Polycomb and SET-26. Though it is lethal in mice (Welstead et al. 2012), in C. elegans knockdown of UTX-1 extends lifespan by up to 29% (Maures et al. 2011), indicating that certain levels of methylation may be beneficial. Most other work, however, indicates that longevity benefits come from increased demethylation (Jin et al. 2011; Maures et al. 2011; Booth and Brunet 2016). For example, overexpression of the demethylases KDM6B/JMJD-3 or PHF8/JMJD-1.2 increases lifespan in C. elegans and in mice (Labbadia et al. 2015; Merkwirth et al. 2016). Knockout of methyltransferases SET-26 and polycomb increase lifespan in C. elegans and Drosophila respectively (Siebold et al. 2010; Ni et al. 2012).
H3K4me3 is an active histone mark that also changes in a variety of cell types and organisms during aging. As Drosophila age, there is an overall global decrease in this histone mark (Wood et al. 2010). Conversely, in mouse hematopoietic stem cells, there is a global increase in H3K4me3 marks, most notably at genes that regulate cell identity and self-renewal. As human fibro-blasts senesce, H3K4me3 marks both increase and decrease, depending on the locus (Shah et al. 2013; Benayoun et al. 2015; Chen et al. 2015). Manipulation of enzymes involved in H3K4 methylation alters longevity in simple metazoans, demonstrating a causal role for these marks in lifespan. For example, in C. elegans over-expression of the demethylase RBR-2 (Greer et al. 2010) or knockout of the H3K4 methyltransferase SETD1A/SET-2 increases lifespan (Greer et al. 2010) and in both C. elegans and Drosophila loss of the demethylase KDM5B/RBR-1 shortens lifespan (Greer et al. 2010; Li et al. 2010). Together, these studies imply that reduced H3K4me3 marks contribute to increased longevity, though it likely depends on which loci are demethylated.
H3K36me3 is a histone mark associated with transcriptional elongation and splicing in diverse organisms. Aged Drosophila have lower levels of H3K36me3 (Wang et al. 2010) as does brain tissue of the accelerated senescence mouse SAMP8 (Wood et al. 2010). Low levels of this mark are associated with greater gene expression change, implying that loss of these marks with age may be a contributor to aberrant gene expression that may contribute to aging (Pu et al. 2015). Consistent with this, loss of H3K36me3 marks in budding yeast is correlated with a shorter lifespan (Sen et al. 2015) and the deletion of a H3K36me3 demethylase extends both yeast and C. elegans lifespan (Ni et al. 2012; Sen et al. 2015).
H3K56Ac is a histone mark involved in chromatin assembly, transcriptional regulation, genomic stability and DNA replication (Williams et al. 2008; Liu et al. 2012). It is a classic marker of aging in yeast (Dang et al. 2009; Feser et al. 2010) and aging human fibroblasts (O’Sullivan et al. 2010b). The influence of H3K56Ac on longevity is difficult to assess, in part because altering its levels can seemingly result in toxicity. For example, either permanent acetylation or an acetylation-resistant mutant K56 reduces yeast lifespan (Dang et al. 2009; Feser et al. 2010). In yeast, deletion of the deacetylase genes Hst3 and Hst4 or overexpression of Hst3 shortens lifespan (Tsuchiya et al. 2006; Dang et al. 2009). Additionally deletion of the acetyltransferase Rtt109 or the histone chaperone Asf1 also reduced lifespan in yeast (Feser et al. 2010). Importantly, the addition of one extra copy of Hst3 or Hst4 extends lifespan in yeast by means that are not clear (Dang et al. 2009; Feser et al. 2010). Thus, it could be that precise levels of this mark are required for increased lifespan or the affected loci are specific.
H4K16Ac is a ubiquitous histone mark that displays an opposite pattern to H3K56Ac in aging (Dang et al. 2009; O’Sullivan et al. 2010b). In the lateral temporal lobe of human brain samples, H4K16ac is redistributed across the genome during normal aging, with both gains and losses of this mark but an overall global increase (Nativio et al. 2018). Interestingly, brain samples from patients with Alzheimer’s disease have more losses of this mark than gains suggesting that H4K16ac may be differentially affected by normal aging compared to age-related diseases (Nativio et al. 2018). This same study explored the specific patterns of H4K16ac changes with age and found that in normal aging marks were specifically changed for gene pathways related to insulin, inflammation, phosphorylation and the defense response, whereas for Alzheimer’s disease, there were mark changes for pathways of myeloid differentiation, cell death and Wnt and Ras signaling (Nativio et al. 2018).
In yeast, Sir2 is an H4K16 deacetylase that is required for silencing and chromatin compaction at mating type loci, rDNA, and telomeres (Dang et al. 2009). The reduction in Sir2 with aging may contribute to the increased H4K16Ac during aging, and the increased genomic instability at the rDNA locus (Dang et al. 2009). By preventing the formation of toxic rDNA circles that cause aging (Sinclair and Guarente 1997), overexpression of the SIR2 gene extends lifespan in yeast (Kaeberlein et al. 1999) as does deletion of the histone acetyl transferase SAS2 (Dang et al. 2009). This implies that reduced H4K16Ac, most likely at the rDNA locus, is beneficial for longevity. The effect of H4K16ac on lifespan has also been proposed to be due to its effects on chromatin structure at the telomeres, although yeast lifespan is not limited by telomere loss (Austriaco and Guarente 1997; Dang et al. 2009).
H3K9me3 is a hallmark of heterochromatin. It is globally reduced under a variety of conditions including fly aging (Larson et al. 2012) and in fibroblasts from HGPS patients (Shumaker et al. 2006). In a progeria mouse model of Werner syndrome with defective DNA repair, levels of both H3K9me3 marks and the H3K9 methyltransferase SUV39H1 are decreased and it is hypothesized that the decreased H3K9me3 contributes to an inability of the heterochromatin to remodel to allow for DNA repair (Zhang et al. 2015). The deletion of the methyltransferase SUV39H1 in cells results in reduced H3K9me3 levels, defective DNA repair, and cellular senescence (Zhang et al. 2015). Conversely, in some aging situations, H3K9me3 levels are increased, as in the case of aged muscle stem cells (Schwoerer et al. 2016).
H4K20me3 is also a mark of heterochromatin and transcriptional repression, but it tends to increase in models of aging. For example, the mark is increased in fibroblasts from HGPS patients (Shumaker et al. 2006), in old rats (Sarg et al. 2002) and in senescent human cells, especially at SAHF (Nelson et al. 2016). Again, however, there are likely tissue and context-specific changes, as a recent study saw a decrease in H4K20me3 in mouse embryonic fibroblasts and aging cardiomyocytes (Lyu et al. 2018). In senescent cells, H4K20me3 inversely correlates with gene expression, indicating that increased levels of H4K20me3 may act to reduce the age-related increase in gene expression (Nelson et al. 2016).
H1K26 is a Sirt1 deacetylation target, resulting in the formation of facultative heterochromatin (Vaquero et al. 2004). The role of H1K26Ac, and how it changes in aging, is not well understood. Redistribution of SIRT1 with oxidative stress leads to global changes in H1K26Ac marks (Oberdoerffer et al. 2008). It is possible similar changes occur in age, although one study in mouse hepatocytes saw no change in H1K26Ac marks in old compared to young cells (Ghiraldini et al. 2013). Further research is needed to elucidate the role of H1K26Ac in aging.
There is some evidence for global hypoacetylation of histones with aging. In aging mouse brain, there is reduced histone acetylation especially at repeated DNA elements (Ryu et al. 2011) likely caused by decreased levels or the redistribution of specific histone deactylases such as SIRT1 and histone deacetylase 1 (HDAC1) with increasing age (Oberdoerffer et al. 2008; Madabhushi et al. 2014; Zupkovitz et al. 2018).
Histone ubiquitination, which has a role in the regulation of transcription and the DNA damage response, maybe also be important in aging. A screen for long-lived yeast strains identified a relationship between downregulation of components of the histone deubiquinase module (DUBm) of the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex and increased lifespan (Mccormick et al. 2014). Interestingly, in human cells, the nutrient responsive deacetylase SIRT1 is recruited to SAGA where it deacetylates the ubiquitin-specific protease 22 (USP22), a component of the DUBm, connecting nutrient availability to epigenetic alterations during aging (Armour et al. 2013).
In summary, there are distinct changes in histone marks with aging, including a global increase in active histone marks such as H3K4me3 and H4K16ac, and a decrease in repressive marks such as H3K9me3 and H3K27me3. There is also increasing evidence that manipulating the levels of these marks by either increasing or decreasing the levels of the responsible enzymes can extend lifespan by increasing genome stability and preventing the loss of youthful gene expression caused by ‘epigenetic noise’.
DNA methylation changes
DNA methylation of the cytosine in a CpG nucleotide is critical for the regulation of gene expression and the recruitment of histone-modifying enzymes at specific DNA sites. It is particularly important during development, by silencing genes in tissues where their expression is not needed (Pal and Tyler 2016). Within gene promotors, DNA methylation represses transcription and triggers the formation of more compact repressive chromatin structures, whereas highly expressed genes have little or no DNA methylation in their promoter regions, so called “CpG islands”.
Most CpGs in young cells have methylated cytosines, but during aging, there is a decrease in overall methylation especially at repetitive regions (Vanyushin et al. 1973; Romanov and Vanyushin 1981; Wilson et al. 1987; Bjornsson et al. 2008; Bollati et al. 2009; Christensen et al. 2009; Jintaridth and Mutirangura 2010; Horvath 2013; Benayoun et al. 2015; Zampieri et al. 2015; Bormann et al. 2016). Importantly, with increasing age, in all metazoans studied, there are also specific regions of hypermethylation, especially at CpG islands near gene-rich regions (Rakyan et al. 2010; Benayoun et al. 2015; Zampieri et al. 2015). Areas of DNA hypermethylation are usually at genes that are tissue-specific, control differentiation and development, encode transcription factors or are transcription factor binding sites (Benayoun et al. 2015; Zampieri et al. 2015). As described below, these sites serve as the basis of the DNA methylation clock that can predict biological and chronological age in mammals (Horvath 2013; Wagner 2017; Horvath and Raj 2018).
A recent genome-wide DNA methylation study in two large population cohorts found 5168 CpG sites that were altered in their methylation pattern during aging, with the majority (61%) being demethylated (Li et al. 2017). These same general age-related changes are also seen during replicative senescence in vitro (Koch et al. 2012a), in colon samples from old mice (Maegawa et al. 2010), and in blood samples from aged rhesus monkeys (Maegawa et al. 2017). Levels of DNA methylation increase with age in mouse hematopoietic stem cells (Beerman et al. 2013) but in adults stem cells and somatic tissues, there is global hypomethylation with hyper-methylation at CpG islands (Teschendorff et al. 2010; Cole et al. 2017; Hahn et al. 2017; Maegawa et al. 2017; Petkovich et al. 2017; Stubbs et al. 2017). In aging monozygous twins, there are different patterns of DNA methylation that appear to result from changes in the environment or stochastic errors (Fraga et al. 2005).
If and how DNA methylation changes contribute to aging is a topic of intense investigation, but very little is known (Pal and Tyler 2016). Loss of methylation at the integrin alpha L (ITGAL) and interleukin (IL)-17RC promoters leads to an autoimmune response and contribute to macular degeneration (Zhang et al. 2002; Wei et al. 2012), implying that there are pathological effects of reduced DNA methylation, at least at specific sites. In aged beta cells, there is a correlation between DNA methylation and gene expression changes in enhancer but not promoter regions (Avrahami et al. 2016), although other studies have seen no association between DNA methylation patterns and gene expression (Beerman et al. 2013; Yuan et al. 2015). More research is needed in mammals to understand whether age-related DNA methylation changes are a consequence of the aging process, or whether they contribute to this process. It is also likely that age-related changes in transcription factors and histone-modifying enzymes affect DNA methylation and vice versa (Booth and Brunet 2016). During aging, DNA methylation at specific polycomb group protein (PcG) target genes is altered, perhaps explaining why H3K27 trimethylation changes with age (Maegawa et al. 2010; Teschendorff et al. 2010; Bocker et al. 2011; Horvath 2013). In humans, aging results in increased DNA methylation more commonly in regions with H3K4me3 and H3K27me3, whereas DNA hypomethylation is more common in regions populated by H3K9Ac, H3K27Ac, H3K4me1, H3K4me2, and H3K4me3 (Raddatz et al. 2013; Mcclay et al. 2014).
In simple metazoans, we know more about the cause and effect of DNA methylation changes during aging. Overexpression of the DNA methyltransferase dDnmt2, for example, extends longevity in Drosophila (Lin et al. 2005). Although this work indicates that increased DNA methylation may extend lifespan, it is not known if this effect is actually due to DNA methylation changes (Schaefer et al. 2010). Studies by the Berger lab of genetically identical organisms, such as worker bees and queen bees, have identified very different DNA methylation profiles, that appear to contribute to lifespan (Yan et al. 2015; Sen et al. 2016). Interestingly, the DNA methylation profiles of bees can be made much more similar by silencing the DNA methyltransferase Dnmt3 (Kucharski et al. 2008), but the effect of this on lifespan is not yet known.
In humans and other mammals, age-related DNA methylation changes can be used as an epigenetic clock for chronological or biological age prediction (Horvath 2013a; Levine et al. 2018). There have been several recent comprehensive reviews of the so called “Horvath” DNA methylation clock and its applications (Wagner 2017; Horvath and Raj 2018). The methylation clock concept has also been recently applied to mice (Cole et al. 2017; Petkovich et al. 2017; Stubbs et al. 2017; Wang et al. 2017) and even dogs and wolves (Thompson et al. 2017). In addition to normal aging, the Horvath DNA methylation clock which is based on changes at 353 CpG sites, has been applied to numerous human cohorts, including patients with diseases such as HIV, cancer, and progeria patients (Horvath et al. 2015; Horvath and Levine 2015; Maierhofer et al. 2017; Horvath, Oshima, et al. 2018; Horvath, Stein, et al. 2018). The epigenetic clock can also predict accelerated or delayed aging caused by lifestyle changes such as diet and exercise (Quach et al. 2017).
Little is known about other forms of DNA methylation such as 5-hydroxymetylcytosine and cytosine methylation at non-CpG dinucleotides. Further research in this area would be valuable to the field.
Re-localization of chromatin modifiers (RCM)
Although the essential roadmap of epigenetic changes have been mapped, key questions remain. What drives epigenetic change during aging? Is this epigenetic noise random or is it programmed? Can it be prevented or reversed? The relocalization of chromatin modifying factors (RCM) concept of epigenetic aging was first proposed in 2007 (Oberdoerffer and Sinclair 2007). The RCM hypothesis proposes that in response to DNA damage, chromatin-modifying factors relocalize from their transcription-controlling canonical loci to areas of DNA damage to assist in repair. In young cells, these factors quickly return to their original sites, but these factors do not always completely return and over time gene expression patterns are altered, causing a loss of cellular identity, cellular dysfunction, aging (Figure 2).
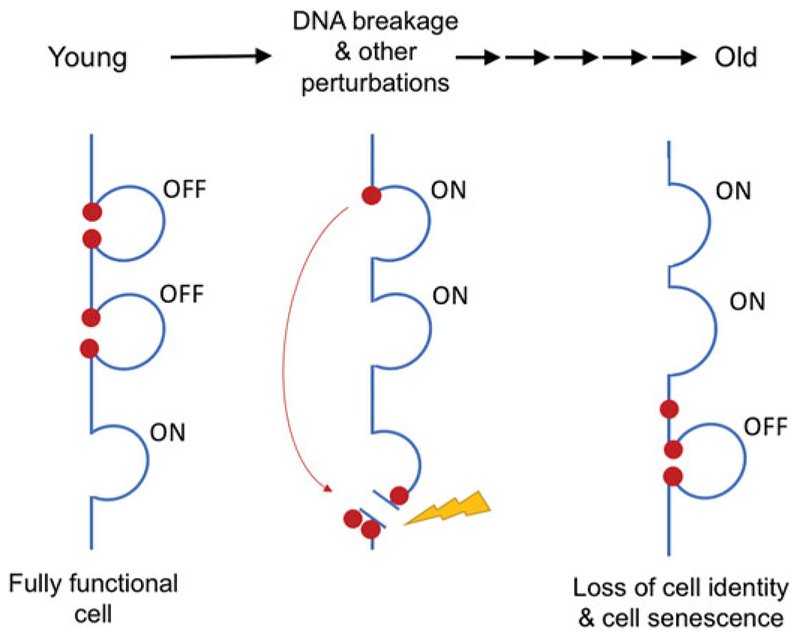
Figure 2.
Relocalization of chromatin modifying factors. In young cells, chromatin modifying factors such as SIRT1 (indicated by red circles) are usually located at canonical loci where they act to modulate transcription. When DNA is damaged these factors are recruited to the site of DNA damage to assist in repair. Relocalization results in altered gene expression or activation of retrotransposons. Usually these factors return to where they came from once the DNA is repaired. After repeated responses to DNA damage, however, not all factors return to their original sites, resulting in the relocalization of chromatin modifying factors (RCM), changes to gene expression, and a loss of cell identity during aging (see the color version of this figure at www.tandfon-line.com/ibmg).
The first evidence for RCM’s role in aging came from studies in yeast. The histone deacetylase Sir2 was identified as an important lifespan determinant in screens for increased replicative aging in yeast (Kaeberlein et al. 1999). Sir2 was then identified as a DNA double-strand break repair protein that relocalizes from silent loci to sites of DNA repair in response to a DNA damage signal from the yeast ATM protein, MEC1 (Kennedy et al. 1997; Sinclair and Guarente 1997; Lee et al. 1999; Martin et al. 1999; Mcainsh et al. 1999; Mills et al. 1999). The absence of Sir2 at silent mating-type loci, however, caused nuclear changes that were similar to those seen in aging yeast (Oberdoerffer and Sinclair 2007). The addition of an extra copy of Sir2 prevented the age-related relocalization (Kaeberlein et al. 1999).
Excitingly, the sirtuins play a similar role in an RCM response in mammals as they do in yeast: in response to a DNA break, they relocalize from promoters and repetitive DNA to sites of damage to facilitate repair (Oberdoerffer et al. 2008; Dobbin et al. 2013). SIRT1 and SIRT6, two nuclear-localized relatives of Sir2, control gene expression in mammals and also play a role in DNA break repair (Oberdoerffer et al. 2008). DNA damage signaling causes SIRT1 and SIRT6 to move away from genes to DNA breaks where they recruit DNA repair factors and modify chromatin (Oberdoerffer et al. 2008; Mao et al. 2011; Dobbin et al. 2013). This relocalization results in transcription of hundreds of genes including changes that are characteristic of aging such as expression of LINE1 elements and developmental genes such as Hoxa9 and Wnt (Logan and Nusse 2004; Clevers 2006; Brack et al. 2007; Oberdoerffer et al. 2008; Singh et al. 2013). These age-related gene expression changes can be prevented in cultured cells and in aging mice by overexpressing SIRT1 or SIRT6 (Mostoslavsky et al. 2006; Oberdoerffer et al. 2008).
The basics of how RCM works in mammals are known. After a DNA break is detected by ATM, it recruits SIRT1 and HDAC1 within seconds which help recruit other DNA repair proteins such as NBS1 and MRE11 (Oberdoerffer et al. 2008; Dobbin et al. 2013). There are likely many chromatin modifying proteins that participate in mammalian RCM, including PARP-1, HDAC1 and REST (Sinclair and Oberdoerffer 2009; Mao et al. 2011; Wang et al. 2013; Lu et al. 2014). Because it is so ancient, RCM likely evolved as an active defense response to co-regulate DNA repair with the expression of genes required for survival (Oberdoerffer and Sinclair 2007; Haigis and Sinclair 2010). In early life, organisms that lacked this mechanism likely went extinct because mating and cell division are deadly events if a broken chromosome is not repaired (Lee et al. 1999; Martin et al. 1999; Mcainsh et al. 1999; Mills et al. 1999). RCM is an established cause of aging in yeast but whether or not RCM is a cause of aging in mammals, and whether it is reversible, are currently unknown. The extent to which the survival circuit is conserved between yeast and humans wasn’t known until 2017, when Bober and colleagues reported that sirtuins stabilize human rDNA by recruiting DNMT1, a DNA methyltransferase that removes methyl marks from the rDNA (Ianni et al. 2017). Then in 2018, Chua and her team found that sirtuins stabilize human rDNA and, in doing so, prevent cellular senescence—essentially the same function as in yeast (Paredes et al. 2018).
ncRNAs
Numerous numbers and types of noncoding RNAs (ncRNAs) exists in the genome (Wilusz et al. 2009) and many of them play essential roles in gene silencing (Costa 2010; Cech and Steitz 2014). For example, the long ncRNAs X inactive-specific transcript (XIST) and HOTAIR can promote gene silencing through their interaction with chromatin-modifying enzymes (Gupta et al. 2010; Shevchenko et al. 2018). A key role of XIST is as the primary driver of X chromosome inactivation, the mechanism in the female sex by which one X chromosome is transcriptionally silenced to ensure appropriate gene dosage (Lee 2009; Froberg et al. 2013; Sado and Brockdorff 2013).
ncRNAs are emerging as having an important role in aging (Esteller 2011; Grammatikakis et al. 2014; Szafranski et al. 2015). In C. elegans, most micro-RNAs (miRNAs) are down regulated with age and reversed by the lifespan-extending intervention calorie restriction (Boehm and Slack 2005; Ibáñez-ventoso et al. 2006; Kato et al. 2011; Mori et al. 2012). In mice and humans, aging is associated with reproducible changes in miRNA levels but not in all organs (Hooten et al. 2010; Li, Khanna, et al. 2011; Wang, Huang, et al. 2011; Mori et al. 2012; Zhang et al. 2014). Levels of the miRNA processing enzyme Dicer decrease in worms and mice, which likely contributes to reduced ncRNAs with increasing age (Maes et al. 2008; Mori et al. 2012; Ungvari et al. 2013) and may contribute to senescence (Zhao et al. 2015).
An example of a C. elegans miRNA that changes with aging is lin-4, which targets the transcription factor lin-14 (Boehm and Slack 2005). Knockout of lin-4 shortens lifespan and overexpression extends it (Boehm and Slack 2005). Lin-4/Lin-14 appear to act through the same pathway as DAF-2 and DAF-16 to modulate the IGF-1 signaling pathway (Boehm and Slack 2005).
In mammals, a classic example of a ncRNA that changes with age is the long-ncRNA H19. H19 interacts with protein MBD1, a methyl-CpG-binding domain protein to repress longevity genes such as IGF-1 (Monnier et al. 2013). Loss of H19-dependent IGF silencing may also contribute to cell senescence (Fu et al. 2008; Venkatraman et al. 2013).
Another example is mir-34, a miRNA that is increased in the brains of AD mouse models (Wang et al. 2009) and in brain samples from AD patients (Sarkar et al. 2016). mir-34 regulates SIRT1 and their expression inversely correlates, indicating mir-34 could be a longevity gene. Whether or not mir-34 is a mediator of caloric restriction and the response to adversity like SIRT1 is not yet known (Lee and Kemper 2010; Jung and Suh 2012).
Transcription factor changes
Though they are not epigenetic factors in the true sense of the word, the transcription factor network is critical for maintaining both short-term and long-term gene expression patterns in youth and as we age. The localization and abundance of dozens of transcription factors change with age, but whether these changes affect chromatin structure or are a result of age-related chromatin structural changes is unknown and, because they are part of the same regulatory network, are very hard to tease apart (Booth and Brunet 2016). A recent meta-analysis of transcription factors from seven in vitro aging gene expression datasets and 18 aged tissue samples, saw distinct patterns of transcription factor changes (Alfego et al. 2018). Two of the most studied transcription factors in aging are FOXO and NF-E2-Related Factor 2 (NRF2) and these appear to both contribute to and respond to age-related chromatin changes (Booth and Brunet 2016).
FOXO proteins are members of a transcription factor family that are associated with and necessary for increased longevity in a variety of species, from worms to mice and possibly humans (Salih and Brunet 2008). FOXO activity is regulated by AMP-activated protein kinase (AMPK), the insulin/insulin-like growth factor 1 (IGF-1) pathways, and oxidative stress to control stress responses, metabolism, proteostasis, and neuronal function (Webb et al. 2016). FOXO typically binds to open chromatin at enhancer regions (Eijkelenboom and Burgering 2013; Webb et al. 2016) and can act as a pioneer factor to open up compact chromatin (Zaret and Carroll 2011). DAF-16, which is the equivalent transcription factor in C. elegans, recruits chromatin remodeling factors to activate the expression of age- and stress-related genes, such as heat-shock proteins (Riedel et al. 2013). In intervertebral discs of mice and humans, expression of FOXO1 and 3 decrease with aging (Alvarez-Garcia et al. 2017), but more research is needed to fully understand its role in epigenetic aging.
Nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) is a mammalian stress-responsive transcription factor that modulates the expression of cellular protection genes (Blackwell et al. 2016; Tebey et al. 2016). Nrf2 orthologs in worms and flies (SKN-1 in C. elegans and CncC in Drosophila) are associated with increased longevity (Sykiotis and Bohmann 2008; Tullet et al. 2017). Nrf2 is activated by reactive oxygen species, nutrient deprivation and metabolic changes by the activity of mTOR and IGF-1, and acts to recruit chromatin remodeling factors such as nucleosome remodeling complex BRG2 to activate the expression of stress response genes (Zhang et al. 2006). Whether or not NRF genes in mammals play a role in longevity is an area of intense research (Kwon et al. 2012; Lewis et al. 2015).
Many other transcription factors also appear to have roles in the aging process including CTCF (Nikolaev et al. 2009; Lake et al. 2016), REST (Lu et al. 2014) and Heat shock factor 1 (HSF-1) (Hsu et al. 2003; Anckar and Sistonen 2011; Brunquell et al. 2016) and p53 (Keizer et al. 2010; Gritsenko et al. 2017).
Transposons
Another aspect that may be either a consequence of epigenetic changes with aging, or contribute to these changes, is increased transcription of transposons. Approximately, 40% of the human genome consists of ancient retrotransposons such as Alu-repeats and Long interspersed element 1 (LINE-1) (Deininger 2011; van Meter et al. 2014; Lander 2001). Transposons are reverse-transcribed to make a cDNA copy that is integrated into the genome elsewhere. Heterochromatin and DNA methylation normally maintain transposons in an inactive state (Slotkin and Martienssen 2007). During aging, however, a loss of heterochromatin results in their increased expression in yeast through to mice (Oberdoerffer et al. 2008; Maxwell et al. 2011; Dennis et al. 2012; Cecco et al. 2013; Li et al. 2013; Wood and Helfand 2013; Hu et al. 2014; Chen et al. 2016), although, until recently, there was little evidence for this being a major mechanism of aging. More recently, however, LINE-1 RNA has been readily detected in the cytoplasm of aged mouse cells and appears to play a major role in hyper-activating the inflammatory response in older animals and even dictating lifespan (Gorbunova et al. 2014, J. Sedivy. personal communication; van Meter et al. 2014).
In yeast, the abundance of Ty RNA, a retrotransposable element, increases by an order of magnitude during replicative aging (Hu et al. 2014). Ty retrotransposition can be partially prevented by overexpression of histones H3/H4 (Hu et al. 2014). In mammals, LINE-1, Alu and satellite repeat RNAs are rare in young individuals but found in abundance during aging (Gorbunova et al. 2014). The SIRT1 deacetylase represses these elements but during aging it is believed to relocalize to sites of DNA instability, leading to de-repression (Oberdoerffer et al. 2008). SIRT6 also acts as a repressor of LINE1 retrotransposons, and depletion of SIRT6 during ageing or in response to DNA damage, results in increased activation of these factors (van Meter et al. 2014). Alu, LINE1 and SVA elements are transcribed in human senescent cells and this may contribute to the inflammatory SASP (Cecco et al. 2013; van Meter et al. 2014). Interestingly, increased transcription of Alu elements promotes the formation of DNA damage foci (Wang, Geesman, et al. 2011), perhaps acting as a positive feedback to enhance the RCM response during aging (Oberdoerffer et al. 2008).
Slowing and reversing epigenetic change
Aging was once considered immutable. In the 2000s interventions began to emerge, first for lower organisms (Howitz et al. 2003) and then in mice (Baur et al. 2006). Calorie restriction, small molecules, and the over-expression of epigenetic regulators such as SIRT1 have been effective at slowing, and even reversing, epigenetic changes and physical decline during aging (Oberdoerffer et al. 2008; Field and Adams 2017). A new approach, called in vivo genetic reprogramming, which seems to reset the epigenome, is a new and promising avenue to pursue (Figure 3), but not without major negative side-effects that must first be overcome.
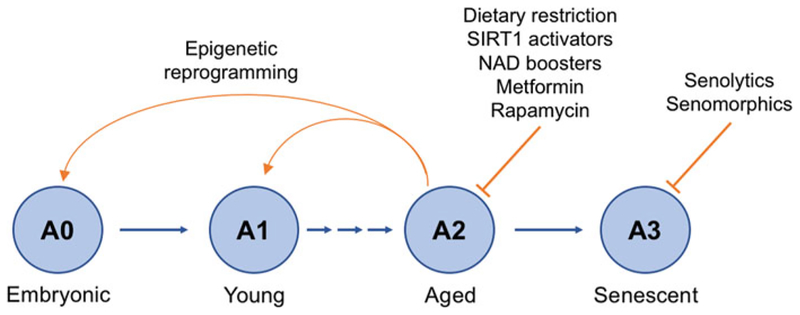
Figure 3.
Interventions to prevent the stages of aging. Aging has four main stages: A0, when an organism/cell is young and healthy; A1, when the organism/cell begins to age; A2, when the organism/cell is aged; and A3, when the organism/cell is senescent. Interventions that delay or prevent aging can act at each of these stages such as calorie restriction, which prevents the aging process at A2. Rapamycin can increase lifespan even when started in later life and acts at A2 and senolytics/senomorphics that act to selectively remove or inhibit the deleterious effects of senescent cells act at stage A3. Reprogramming, the resetting of the epigenetic landscape using specific transcription factors, such as Yamanaka factors, can return cells from the A2 stage to the young A0 stage. Whether or not it is possible to safely reset cells to A1 without risking tumorigenesis is not yet known (see the color version of this figure at www.tandfonline.com/ibmg).
Dietary and pharmacological interventions
Calorie restriction is the most robust intervention that increases lifespan across a wide range of species. Established mechanisms of calorie restriction include modulation of several age-related and nutrient-sensing pathways including AMPK, IGF-1/Insulin, mechanistic target of rapamycin (mTOR) and the sirtuins. Calorie restriction also appears to affect the epigenetic landscape of cells and organisms and prevent many of the epigenetic aging changes discussed above. In aging models, calorie restriction prevents increased retrotransposon expression (Cecco et al. 2013), changes in DNA methylation (Li, Daniel, et al. 2011; Kim et al. 2016), histone post-translational modifications (Li, Daniel, et al. 2011) and the age-related loss of heterochromatin (Jiang et al. 2013).
Sirtuins can slow many aspects of aging at the epigenetic level, including age-related changes in gene expression (Wood et al. 2015), the acetylation of histones and the RCM response (Pal and Tyler 2016). Calorie restriction-induced histone deacetylase activity may also modulate the expression of important age-related genes including upregulation of telomerase reverse transcript-ase (hTERT), and suppression of secreted frizzled related protein 1 (SFRP1) and E-Cadherin (Pruitt et al. 2006; Li, Daniel, et al. 2011). SIRT1 also increases H3K9me3 in calorie restriction by upregulating the methyltransferase SUV39H1 (Vaquero et al. 2007; Li, Daniel, et al. 2011).
Compounds that mimic the beneficial anti-aging and lifespan effects of calorie restriction such as sirtuin-activating compounds (STACs) and rapamycin can also slow epi-genetic changes. SIRT1 activation with STACs suppresses genomic instability and reduces the effect of RCM on aging including reduced H4K16, H3K9 and H3K56 acetylation and increased heterochromatin formation (Oberdoerffer et al. 2008; Chen, Zhao et al. 2012; Sen et al. 2016). By inhibiting TORC1, rapamycin treatment can also prevent age-related epigenetic changes, in part by increasing the occupancy at targets of Rsc9, a subunit of the RSC chromatin remodeling complex (Damelin et al. 2002). Rapamycin also increases expression of the Rpd3-Sin3 HDAC complex that deacetylates H4K5 and H4K12 and promotes condensed chromatin and gene silencing during aging (Rohde and Cardenas 2003). Additionally, rapamycin affects other histone marks which may have important roles in aging including H3K56ac, H3K27me3, H3R2me2, H3K79me3 and H4K20me2 (Chen, Fan et al. 2012; Gong et al. 2015).
Other compounds may also prevent age-related epi-genetic change via their effects on chromatin modifying enzymes (Helin and Dhanak 2013). Remodelin, a compound that inhibits the acetyl-transferase protein NAT10, increases chromatin compaction in fibroblasts from HGPS patients (Larrieu et al. 2014).
Histone acetylation is also able to slow epigenetic changes. For example, spermidine, a polyamine and his-tone deacetylase (HDAC) inhibitor that extends lifespan in a variety of species, may prevent age-related epigenetic changes, in part, by maintaining histone H3 in a hypoacetylated state, and affecting histone and DNA methylation (Eisenberg et al. 2009; Madeo et al. 2018). Similarly, the histone deactylase inhibitor sodium butyrate increased lifespan and reverses hypoacetylation of H4K16 in the progeria HGPS mouse model (Krishnan et al. 2011). Sodium butyrate, a pan-HDAC inhibitor, improves memory potentially by reversing age-related decreases in H3K9 and H3K14 acetylation in aging mouse brains (Singh and Thakur 2018). Another histone deactylase inhibitor called suberoylanilide hydroxamic acid (SAHA) improves cognition in aging mice by preventing deregulated H4K12 acetylation in the brain (Peleg et al. 2010).
Together, these data show that epigenetic changes are preventable by increasing the activity of caloric restriction mimetics and by directly altering the activity of chromatin modifiers. Future work should help establish exactly how these approaches work and whether they are safe enough to be used as broad treatments against age-related decline.
Reprogramming
The reprogramming of cultured cells was first demonstrated by Yamanaka and colleagues who showed the expression of the transcription factors Oct3/4, Sox2, Klf4 and c-Myc (OSKM) in mouse fibroblasts induces them to become pluripotent stem cells that can be differentiated into almost any other cell type (Takahashi and Yamanaka 2006).
Reprogramming of cells with OSKM transcription factors causes epigenetic remodeling over two phases. First is a stochastic phase in which there is differential cell cycle gene expression. This is followed by the deterministic phase which involves activation of pluripotency genes such as Nanog and Oct4. Each of these phases is associated with specific epigenetic changes including modulation of H3K4me3 and H3K27me3 in phase 1 and changes in miRNA expression and DNA methylation at Nanog and Oct4 in the second stage (Hansson et al. 2012; Polo et al. 2012; Buganim et al. 2013; Nashun et al. 2015; Ocampo, Reddy, and Belmonte 2016). Some epigenetic features such as increased H3K27me3 and HP-1gamma and DNA methylation can reduce the reprogramming efficiency, possibly by maintaining heterochromatin (Mikkelsen et al. 2008; Mansour et al. 2012; Sridharan et al. 2013).
In cells, molecular markers of aging can apparently be slowed and even reversed by reprogramming (Table 2) (Mahmoudi and Brunet 2012; Rando and Chang 2012). iPSCs generated from old mice have elongated telomeres and epigenetic profiles more similar to embryonic stem cells, including decreased H3K9me3 and H4K20me3 marks (Marion et al. 2009). Aged mouse hematopoietic stem and progenitor cells are also reprogrammed by OSKM, a treatment that also resets the cell telomere length and gene expression profiles (Wahlestedt et al. 2013). Human senescent and centenarian fibroblasts can also be transformed into iPSCs with transfection of the OSKM factors, which restores gene expression profiles to those of young cells (Lapasset et al. 2011; Yagi et al. 2012). Additionally, the Horvath methylation clock age is reset in iPSCs to that of embryonic stem cells (Horvath 2013). Additionally, fibroblasts isolated from patients with HGPS can be reprogrammed to pluripotency, along with epigenetic changes that include the prevention of progerin accumulation and a restoration of histone marks including H3K9me3 (Liu et al. 2011). These in vitro studies provide promising suggestions that reprogramming may be able to reverse age-related phenotypes in vivo.
Table 2.
Studies of reprogramming age-related epigenetic changes.
| Reprogramming method | Model | Main findings | References |
|---|---|---|---|
| OSKM retrovirus transduction | Mouse fibroblasts | Induction of pluripotent stem cells possible with these four transcription factors. Epigenetics not explored. | (Takahashi and Yamanaka 2006) |
| OSK/OSKM retrovirus transduction | Mouse fibroblasts | Induction of pluripotent stem cells possible - telomeres elongated and have same histone marks as young embryonic stem cells | (Marion et al. 2009) |
| OSKM retrovirus transduction | Aged mouse hematopoietic stem cells | Induction of pluripotent stem cells possible - reset telomere length and gene expression profiles. | (Wahlestedt et al. 2013) |
| OSKMNL/OSKM retrovirus transduction | Human senescent and centenarian fibroblasts | Induction of pluripotent stem cells possible - reset telomere length and gene expression profiles. Epigenetics not explored. | (Lapasset et al. 2011; Yagi et al. 2012) |
| OSKM retrovirus transduction | HGPS patient fibroblasts | Induction of pluripotent stem cells possible - prevented progerin accumulation and reset histone marks eg. H3K9me3 | (Liu et al. 2011) |
| OSKM retrovirus transduction or trans-differentiation | Aging human fibroblasts | Reprogrammed fibroblasts that were differentiated into neurons had no genetic/epigenetic marker of aging. | (Mertens et al. 2018) |
| OSKM retrovirus transduction | Human bone marrow-derived mesenchymal stromal cells and fibroblasts | OSKM induction reset the Horvath DNA methylation epigenetic clock to that of embryonic stem cells. | (Horvath 2013) |
| Inducible OSKM model | Mice | Induction of pluripotent stem cells as indicated by Nanog marker found in many tissues. High mortality from teratomas. | (Abad et al. 2013; Ohnishi et al. 2014) |
| Injection of OSKM plasmids | Mice | Reprogramming markers detected in 24–48 hours, no teratoma evidence. | (Yilmazer et al. 2013) |
| Skeletal muscle injection of OSKM plasmids | Mice | Upregulated pluripotency markers, and improved regeneration potential | (de Lázaro et al. 2017) |
| Inducible OSKM model | Mouse HGPS model (LAKI 4F mice) | Cyclic in vivo induction of factors, restored normal levels of H3K9me3, and H4K20me3, and increased muscle injury recovery and other aging phenotypes | (Ocampo et al. 2016) |
| OSKM | Naked mole rats | Resistant to reprogramming with OSKM as more stable epigenome than mice. | (Tan et al. 2017) |
Recent work has shown that Yamanaka factors can seemingly induce in vivo reprogramming in mice, but apparently not without side effects (Table 2). Whole-body induction of OSKM in an inducible transgenic mouse is associated with Nanog expression in many tissues but these mice have a high rate of teratomas (Abad et al. 2013; Ohnishi et al. 2014). The direct injection of OSKM plasmids into adult mice induces pluripotent stem cells markers in liver within a couple of days (Yilmazer et al. 2013). Local injection of OSKM plasmids into skeletal muscle of adult mice induces pluripotency markers and improves skeletal muscle regeneration with less fibrosis and more myofibers (de Lázaro et al. 2019), but the long-term safety of these treatments is not known beyond 120 days.
Belmonte and colleagues explored the effect of cyclic induction of the OSKM factors in a lamin A-deficient progeria mouse model (Ocampo et al. 2016). Chronic activation of OSKM was not possible because it resulted in mouse lethality within a few days, possibly due to gut toxicity. They hypothesized that cyclic exposure to OSKM would result in partial reprogramming and avoid teratoma formation. Short-term cyclic exposure to OSKM reversed age-related phenotypes in the progeroid mice and increased maximal lifespan by approximately 15% (Ocampo et al. 2016). The treatment restored levels of H3K9me3 and H4K20me3, improved recovery from muscle injury, and reduced gamma-H2AX foci, and there was no indication of teratomas (Ocampo et al. 2016).
Interestingly, the long-lived naked mole rat is resistant to iPSC reprogramming (Tan et al. 2017). Fibroblasts from older naked mole rats have a much lower reprogramming efficiency than mouse fibroblasts, likely due to a more stable epigenome. Naked mole rat cells have higher levels of the repressive mark H3K27me3 and lower levels of the active mark H3K27ac than mice, indicating more heterochromatin (Tan et al. 2017).
Conclusion
There are clear patterns of change during aging that occur at the epigenetic level changes that are conserved from yeast to mammals. It is becoming apparent, however, that there are tissue, cell and even loci-specific differences in epigenetic aging within organisms. New technologies such as single-cell analysis (Cheung et al. 2018) and improved visualization systems (Ren et al. 2017) will increase our understanding of specific, rather than global, epigenetic changes with age.
Further exciting progress in the field will come as we move from the characterization of these epigenetic changes, to focus on understanding the degree to which these epigenetic changes may actually cause aging. There is already evidence that heterochromatin loss, replacement of canonical histones with histone variants, up or down regulation of nucleosome remodeling complexes, and the relocalization of chromatin modifying factors affect lifespan and contribute to the aging process. But what drives these processes? DNA damage? Environmental influences, or simply information loss in the form of epigenetic noise stemming from the chaos of life at the molecular scale?
If epigenetic changes do contribute to the aging process, then there is the exciting possibility that we can prevent or even reverse those changes and intervene in a very upstream cause. Evidence of the prevention of epigenetic changes with aging can be seen with calorie restriction, sirtuin activation and small molecules. Amazingly, reprogramming appears to reverse age-related epigenetic changes and the effects of aging, both in vitro and in vivo. Clearly, in vivo reprogramming to address aging and age-related diseases is an early field but it is growing rapidly and holds considerable promise. There remain a number of uncertainties, however. What is the optimal dose of and combination of reprogramming factors? Will reprogramming ever be safe enough to use in humans? Can in vivo reprogramming be achieved with small molecules instead of gene therapy? And what diseases could reprogramming potentially treat? No doubt, the importance of epigenetics to aging and the potential of epi-genetic reprogramming will only grow in interest in coming years.
Funding
This work was supported by the Glenn Foundation for Medical Research and grants from the NIH (R37 AG028730, R01 AG019719 and R01 DK100263), and Epigenetics Seed Grant (601139_2018) from Department of Genetics, Harvard Medical School. A.E.K is supported by an NHMRC CJ Martin biomedical fellowship (GNT1122542).
Footnotes
Disclosure statement
D.A.S. is a founder, equity owner, advisor to, director of, consultant to, investor in and/or inventor on patents licensed to Vium, Jupiter Orphan Therapeutics, Cohbar, Galilei Biosciences, GlaxoSmithKline, OvaScience, EMD Millipore, Wellomics, Inside Tracker, Caudalie, Bayer Crop Science, Longwood Fund, Zymo Research, EdenRoc Sciences (and affiliates Arc-Bio, Dovetail Genomics, Claret Bioscience, Revere Biosensors, UpRNA and MetroBiotech (an NAD booster company), Liberty Biosecurity). Life Biosciences (and affiliates Selphagy, Senolytic Therapeutics, Spotlight Biosciences, Animal Biosciences, Iduna, Immetas, Continuum Biosciences, Jumpstart Fertility (an NAD booster company), and Lua Communications). Iduna is a cellular reprogramming company, partially owned by Life Biosciences. DS sits on the board of directors of both companies. D.A.S. is an inventor on a patent application filed by Mayo Clinic and Harvard Medical School that has been licensed to Elysium Health; his personal share is directed to the Sinclair lab. For more information see https://genetics.med.harvard.edu/sinclair-test/people/sinclair-other.php. A.E.K. has no conflicts to declare.
References
- Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, et al. 2013. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 502:340–345. [DOI] [PubMed] [Google Scholar]
- Alfego D, Rodeck U, Kriete A. 2018. Global mapping of transcription factor motifs in human aging. PLoS One. 13: e0190457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia O, Matsuzaki T, Olmer M, Masuda K, Lotz MK. 2017. Age-related reduction in the expression of FOXO transcription factors and correlations with intervertebral disc degeneration. J Orthop Res. 35:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 80:1089–1115. [DOI] [PubMed] [Google Scholar]
- Armour SM, Bennett EJ, Braun CR, Zhang X-Y, McMahon SB, Gygi SP, Harper JW, Sinclair DA. 2013. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 33:1487–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austriaco NR, Guarente LP. 1997. Changes of telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 94: 9768–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, Stadler MB, Burger L, Schübeler D, Glaser B, Kaestner KH. 2016. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β-cell function. Cell Metab. 22:619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Groth M, Priebe S, Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, Tozzini T, et al. 2014. RNA-seq of the aging brain in the short-lived fish N. furzeri – conserved pathways and novel genes associated with neurogenesis. Aging Cell. 13:965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. 2006. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 444:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, Rossi DJ. 2013. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 12:413–425. [DOI] [PubMed] [Google Scholar]
- Benayoun B, Pollina EA, Brunet A. 2015. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 16:593–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyshev G, Korotaev G, Boiarskikh G, Vaniushin B. 1967. Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning. Biokhimiia. 32:988–993. [PubMed] [Google Scholar]
- Bjornsson H, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, et al. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 299:2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. 2016. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 88:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. 2011. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 117:182–190. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack F. 2005. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 310: 1954–1958. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. 2009. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 130:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Brunet A. 2016. The aging epigenome. Mol Cell. 62: 728–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann F, Rodríguez-Paredes M, Hagemann S, Manchanda H, Kristof B, Gutekunst J, Raddatz G, Haas R, Terstegen L, Wenck H, et al. 2016. Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell. 15:563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. 2007. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 317: 807–811. [DOI] [PubMed] [Google Scholar]
- Brandt A, Krohne G, Grosshans J. 2008. The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging Cell. 7:541–551. [DOI] [PubMed] [Google Scholar]
- Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. 2016. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics. 17: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 134: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. 2013. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 14: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how and why? Mol Cell. 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecco M, De Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. 2013. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 5:867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 157:77–94. [DOI] [PubMed] [Google Scholar]
- Chen H, Fan M, Pfeffer LM, Laribee RN. 2012. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 40:6534–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ruiz PD, McKimpson WM, Novikov L, Kitsis RN, Gamble MJ. 2015. MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol Cell. 59:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zheng X, Xiao D, Zheng Y. 2016. Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence. Aging Cell. 15: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. 2012. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics. 11:1048–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Vallania F, Warsinske HC, Donato M, Schaffert S, Chang SE, Dvorak M, Dekker CL, Davis MM, Utz PJ, et al. 2018. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell. 173: 1385–1397.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CPG island context. PLoS Genet. 5:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem. 78:273–304. [DOI] [PubMed] [Google Scholar]
- Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127:469–480. [DOI] [PubMed] [Google Scholar]
- Cole JJ, Robertson NA, Rather MI, Thomson JP, McBryan T, Sproul D, Wang T, Brock C, Clark W, Ideker T, et al. 2017. Diverse interventions that extend mouse lifespan suppress shared age-associated epigenetic changes at critical gene regulatory regions. Genome Biol. 18:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F 2010. Non-coding RNAs: meet thy masters.Bioessays. 32:599–608. [DOI] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell. 9:563–573. [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 459:802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry M, Wanat JJ, Saviolaki D, Murakami CJ, Robison B, et al. 2015. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 19:952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lázaro I, Cossu G, Kostarelos K. 2017. Transient transcription factor (OSKM) expression is key towards clinical translation of in vivo cell reprogramming. EMBO Mol Med. 9: 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: Know the SINEs. GenomeBiol. 12:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis S, Sheth U, Feldman JL, English KA, Priess JR. 2012. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8:e1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, et al. 2013. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 16:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte LF, Young ARJ, Wang Z, Wu H, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma A. 2015. Histone H3.3 and its proteolytically processed form drive a cellular senescence program. Nat Commun. 5:5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BMT. 2013. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 14:83–97. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. 2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 11:1305–1314. [DOI] [PubMed] [Google Scholar]
- El-Sharnouby S, Fischer B, Magbanua JP, Umans B, Flower R, Choo SW, Russell S, White R. 2017. Regions of very low H3K27me3 partition the Drosophila genome into topological domains. PLoS One. 12:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2011. Non-coding RNAs in human disease. NatRev Genet. 12:861–874. [DOI] [PubMed] [Google Scholar]
- Fernández A, Bayón G. 2014. H3K4me1 marks DNA regions hypomethylated during aging in stem and differentiated cells. Genome Res. 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2010. Elevated histone expression promotes lifespan extension. Mol Cell. 39:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field AE, Adams PD. 2017. Targeting chromatin aging - The epigenetic impact of longevity-associated interventions. Exp Gerontol. 94:29–33. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Cigudosa JC, Urioste M, Benitez J, Boix-chornet M, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. PNAS. 102:10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froberg JE, Yang L, Lee JT. 2013. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 425: 3698–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu V, Dobosy J, Desotelle J, Almassi N, Ewald J, Srinivasan R, Berres M, Svaren J, Weindruch R, Jarrard D. 2008. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 68:6797–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G, Liu S, Costanzi C, Valle-garcia D, Schaniel C, et al. 2013. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun. 4: 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiraldini FG, Crispim ACV, Mello MLS. 2013. Effects of hyper-glycemia and aging on nuclear sirtuins and DNA damage of mouse hepatocytes. Mol Biol Cell. 24:2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Qian H, Ertl R, Astle CM, Wang GG, Harrison DE, Xu X. 2015. Histone modifications change with age, dietary restriction and rapamycin treatment in mouse brain. Oncotarget. 6:15882–15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Boeke JD, Helfand SL, Sedivy JM. 2014. Human genomics. Sleeping dogs of the genome. Science. 346: 1187–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. 2014. Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY). 6:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E, Maures T, Hauswirth A, Green E, Leeman D, Maro GS, Han S, Banko MR, Gozani O, Brunet A. 2010. Members of the histone H3 lysine 4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 466:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS, Moazed D. 2003. Heterochromatin and epigenetic control of gene expression. Science. 301:798–803. [DOI] [PubMed] [Google Scholar]
- Gritsenko DA, Orlova OA, Linkova NS, Khavinson VK. 2017. Transcription factor p53 and skin aging. Adv Gerontol. 7: 114–115. [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O, Grönke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, Andrews S, Zhang Q, Wakelam MJ, Beyer A, et al. 2017. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 18:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 5: 253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. 2005. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 102:16690–16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. 2005. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19:1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 31:361–392. [DOI] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. 2012. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2:1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Dhanak D. 2013. Chromatin proteins and modifications as drug targets. Nature. 502:480–488. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. 2006. Cellular senescence in aging primates. Science. 311:1257. [DOI] [PubMed] [Google Scholar]
- Hooten NN, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. 2010. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 5:e10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, et al. 2015. Accelerated epigenetic aging in Down syndrome. Aging Cell. 14:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Levine AJ. 2015. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 212: 1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Felton S, Matsuyama M, Lowe D, Kabacik S, Wilson JG, et al. 2018. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging. 10:1758–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Raj K. 2018. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 19:371–384. [DOI] [PubMed] [Google Scholar]
- Horvath S, Stein DJ, Nicole P, Heany SJ, Kobor MS, Lin DT, Myer L, Zar HJ, Evine AJ, Hoare J. 2018. Perinatally acquired HIV infection accelerates epigenetic aging in South African adolescents. Clin Sci. 32:1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman K, Cohen H, Lamming D, Lavu S, Wood J, Zipkin R, Chung P, Kisielewski A, Zhang L, et al. 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 425:191–196. [DOI] [PubMed] [Google Scholar]
- Hsu A, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300:5622. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. 2014. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 28:396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianni A, Hoelper S, Krueger M, Braun T, Bober E. 2017. Sirt7 stabilizes rDNA heterochromatin through recruitment of DNMT1 and Sirt1. Biochem Biophys Res Commun. 492: 434–440. [DOI] [PubMed] [Google Scholar]
- Ibáñez-ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. 2006. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 5: 235–246. [DOI] [PubMed] [Google Scholar]
- Imai S-I, Kitano H. 1998. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp Gerontol. 33:555–570. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 403:795–800. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, Van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 202:129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Du G, Tobias E, Wood JG, Whitaker R, Neretti N, Helfand L. 2013. Dietary and genetic effects on age -related loss of gene silencing reveal epigenetic plasticity of chromatin repression during aging. Aging. 5:813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, et al. 2011. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 14:161–172. [DOI] [PubMed] [Google Scholar]
- Jintaridth P, Mutirangura A. 2010. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics. 41:194–200. [DOI] [PubMed] [Google Scholar]
- Jung HJ, Suh Y. 2012. MicroRNA in aging: from discovery to biology. Curr Genomics. 13:548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Biggins S. 2005. Histone variants: deviants? Genes Dev. 19:295–310. [DOI] [PubMed] [Google Scholar]
- Kato M, Chen X, Inukai S, Zhao H, Slack FJ. 2011. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA. 17:1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keizer PLJ, De Laberge RM, Campisi J. 2010. p53: Pro-aging or pro-longevity? Aging (Albany NY). 2:377–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B, Gotta M, Sinclair D, Mills K, McNab D, Murthy M, Pak S, Laroche T, Gasser S, Guarente L. 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 89:381–391. [DOI] [PubMed] [Google Scholar]
- Kim CH, Lee EK, Choi YJ, An HJ, Jeong HO, Park D, Kim BC, Yu BP, Bhak J, Chung HY. 2016. Short-term calorie restriction ameliorates genomewide, age-related alterations in DNA methylation. Aging Cell. 15:1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Joussen S, Schellenberg A, Lin Q, Zenke M, Wagner W. 2012. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell. 11:366–369. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705. [DOI] [PubMed] [Google Scholar]
- Kozlowski M, Ladurner AG. 2015. ATM, MacroH2A.1, and SASP: the checks and balances of cellular senescence. Mol Cell. 59:713–715. [DOI] [PubMed] [Google Scholar]
- Kreiling JA, Tamamori-adachi M, Sexton AN, Jeyapalan JC, Munoz-najar U, Peterson AL, Manivannan J, Elizabeth S, Pchelintsev NA, Adams PD, et al. 2012. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 10:292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Chow MZY, Wang Z, Zhang L, Liu B, Liu X, Zhou Z. 2011. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci USA. 108: 12325–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 319:1827–1831. [DOI] [PubMed] [Google Scholar]
- Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, Choi YB, Lee AH, Lee KH, Park C, et al. 2012. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 13:150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Richard I, Labbadia J, Morimoto RI. 2015. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol Cell. 59:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Boetefuer EL, Won K, Fan H. 2016. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 44: 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW. 2005. HST2 mediates SIR2-independent lifespan extension by calorie restriction. Science. 309: 1861–1864. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature. 409:860–921. [DOI] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Ait-Hamou N, Leschik J, Pellestor F, Ramirez J-M, De Vos J, et al. 2011. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 25:2248–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. 2014. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 344:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K, Yan S, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX. 2012. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 8:e1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris A, Tang Z, Hughes SH, Roeder RG. 2013. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 152:1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lázaro I, Yilmazer A, Nam Y, Qubisi S, Maizatul F, Degens H, Cossu G, Kostarelos K. 2019. Non-viral, Tumor-free Induction of Transient Cell Reprogramming in Mouse Skeletal Muscle to Enhance Tissue Regeneration. Mol Ther. 1:59–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Klopp RG, Weindruch R, Prolla TA. 1999. Gene expression profile of aging and its retardation by caloric restriction. Science. 285:1390–1394. [DOI] [PubMed] [Google Scholar]
- Lee J, Kemper JK. 2010. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging. 2: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2009. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 23:1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesur I, Campbell JL. 2004. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell. 15:1297–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, et al. 2018. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, Buffenstein R. 2015. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc Natl Acad Sci USA. 112:201417566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Choulet F, Heng Y, Hao W, Paux E, Liu Z, Yue W, Jin W, Feuillet C, Zhang X. 2013. Wheat centromeric retrotransposons: the new ones take a major role in centromeric structure. Plant J. 73:952–965. [DOI] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, Secombe J. 2010. Essential functions of the histone demethylase lid. PLoS Genet. 6: e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Christiansen L, Christensen K, Kruse TA, Redmond P, Marioni RE, Deary IJ, Tan Q. 2017. Identification, replication and characterization of epigenetic remodelling in the aging genome: A cross population analysis. Sci Rep. 7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Khanna A, Li N, Wang E. 2011. Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging (Albany NY). 3:985–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Daniel M, Tollefsbol TO. 2011. Epigenetic regulation of caloric restriction in aging. BMC Med. 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Tang L, Reddy MN, Shen CJ. 2005. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J Biol Chem. 280:861–865. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang Z, Zhang L, Ghosh S, Zheng H, Zhou Z. 2013. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a Progeria mouse model. Nat Commun. 4:1812–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Barkho BZ, Ruiz S, Diep D, Qu J, Yang S, Panopoulos AD, Suzuki K, Kurian L, Walsh C, et al. 2011. Recapitulation of premature aging with iPSCs from Hutchinson Gilford Progeria Syndrome. Nature. 472:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung T, Charville G, Marie Ceniza Hurgo B, Leavitt T, Shih J, Brunet A, Rando T. 2013. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4:189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang D-L, Chen S, Zhao L, Sun F-L. 2012. Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56. J Biol Chem. 287:41469–41480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 20: 781–810. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, López-otín C, Blasco MA, Partridge L, Serrano M. 2013. The hallmarks of aging. Cell. 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. 2014. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 507:448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ, Zu C. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 272:301–311. [DOI] [PubMed] [Google Scholar]
- Lyu G, Guan Y, Zhang C, Zong L, Sun L, Huang X, Huang L, Zhang L, Tian X, Zhou Z, Tao W. 2018. TGF-b signaling alters H4K20me3 status via miR- 29 and contributes to cellular senescence and cardiac aging. Nat Commun. 9:2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Pan L, Tsai LH. 2014. DNA damage and its links to neurodegeneration. Neuron. 83:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Eisenberg T, Pietrocola F, Kroemer G. 2018Spermidine in health and disease. Science. 359:6374. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JJ. 2010. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20:332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, Jelinek J, Colman RJ, Issa JPJ. 2017. Caloric restriction delays age-related methylation drift. Nat Commun. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wu H, Wang E. 2008. Changes in microRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biochem. 105:824–834. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Brunet A. 2012. Aging and reprogramming: a two-way street. Curr Opin Cell Biol. 24:744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer A, Flunkert J, Oshima J, Martin GM, Haaf T, Horvath S. 2017. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY). 9:1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. 2012. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 488: 409–413. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 332: 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. 2009. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 4:141–154. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM, Boveresses C, Epalinges C. 1999. Relocalization of telomeric ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 97:621–633. [DOI] [PubMed] [Google Scholar]
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. 2011. H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent, manner. Aging Cell. 10:980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Burhans WC, Curcio MJ. 2011. Retrotransposition is associated with genome instability during chronological aging. PNAS. 108:20376–20381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh K, Bagot RC, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, Hurd YL, Tamminga CA, et al. 2015. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 87:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcainsh AD, Scott-drew S, Murray JAH, Jackson SP. 1999. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 9: 963–967. [DOI] [PubMed] [Google Scholar]
- Mcclay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, Hudson AD, Harada A, Hultman CM, Magnusson PKE, et al. 2014. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 23:1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC-Z, Ahmed U, Carr D, Murakami CJ, et al. 2015. A comprehensive analysis of replicative lifespan in 4698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 22: 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccormick MA, Mason AG, Guyenet SJ, Dang W, Garza RM, Ting MK, Moller RM, Berger SL, Kaeberlein M, Pillus L, et al. 2014. The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep. 8:477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. 2016. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell. 165:1209–1223.27133168 [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, Mcgrath S, Campbell B, Lee H JR, Gonçalves JT, et al. 2018. Directly reprogrammed human neurons retain aging- associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell. 17:705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature. 454:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 97:609–620. [DOI] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-si-ali S, Dandolo L. 2013. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA. 110:20693–20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell KT, Kahn CR, et al. 2012. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 16: 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. 2006. Genomic instability and aging-like pheno-type in the absence of mammalian SIRT6. Cell. 124: 315–329. [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nuñez S, Chicas A, Hearn SA, Myers MP, Lowe SW. 2006. A Novel role for high-mobility group A proteins in cellular senescence and heterochromatin formation. Cell. 126:503–514. [DOI] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716. [DOI] [PubMed] [Google Scholar]
- Nashun B, Hill PW, Hajkova P. 2015. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. Embo J. 34:1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, Toledo JB, Gosai SJ, Gregory BD, Torres C, et al. 2018. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci. 21:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DM, Jaber-hijazi F, Cole JJ, Robertson NA, Pawlikowski JS, Norris KT, Criscione SW, Pchelintsev NA, Piscitello D, Stong N, et al. 2016. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, Lee SS. 2012. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 11:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev LG, Akopov SB, Didych DA, Sverdlov ED. 2009. Vertebrate protein CTCF and its multiple roles in a large-scale regulation of genome activity. CG Genomics. 10: 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 17:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, Mcvay M, Mostoslavsky R, Park S, Hartlerode A, Stegmuller J, Hafner A, Wright SM, Mills KD, et al. 2008. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 135:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. 2007. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol. 8:692–702. [DOI] [PubMed] [Google Scholar]
- Ocampo A, Reddy P, Belmonte JCI. 2016. Anti-aging strategies based on cellular reprogramming. Trends Mol Med. 22:725–738. [DOI] [PubMed] [Google Scholar]
- Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E, et al. 2016. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 167:1719–1733.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. 2014. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 156:663–677. [DOI] [PubMed] [Google Scholar]
- Pal S, Tyler J. 2016. Epigenetics and aging. Sci Adv. 2: e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S, Angulo-ibanez M, Tasselli L, Carlson SM, Zheng W, Li T, Chua KF. 2018. The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J Biol Chem. 293:11242–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Gillich A, Garrett N, Gurdon JB. 2011. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 30:2373–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Göhler H, Hoffmann K, Misteli T. 2009. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 11:1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-javan S, Agis-balboa RC, Cota P, Wittnam JL, Gogol-doering A, Opitz L, et al. 2010. Associated with age-dependent memory impairment in mice. Science. 328:3–8. [DOI] [PubMed] [Google Scholar]
- Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. 2017. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25:954–960.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 151:1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, Mcgarvey KM, Kang SL, Watkins DN, Herman JG, Baylin SB. 2006. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M, Ni Z, Wang M, Wang X, Wood JG, Helfand SL, Yu H, Lee SS. 2015. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 29:718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM, et al. 2017. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 9:419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Hagemann S, Aran D, Söhle J, Kulkarni PP, Kaderali L, Hellman A, Winnefeld M, Lyko F. 2013. Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics Chromatin. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang T, Beyan H, Whittaker P, Mccann OT, Finer S, Valdes AM, et al. 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20:434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA, Chang HY. 2012. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 148: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Deng L, Xue Y, Suzuki K, Zhang W, Yu Y, Wu J, Sun L, Gong X, Luan H, et al. 2017. Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res. 27:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol. 15:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Cardenas ME. 2003. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Society. 23:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov G, Vanyushin B. 1981. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 653:204–218. [DOI] [PubMed] [Google Scholar]
- Ryu SH, Kang K, Yoo T, Joe CO, Chung JH. 2011. Transcriptional repression of repeat-derived transcripts correlates with histone hypoacetylation at repetitive DNA elements in aged mice brain. Exp Gerontol. 46:811–818. [DOI] [PubMed] [Google Scholar]
- Sado T, Brockdorff N. 2013. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc B Biol Sci. 368:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka K, Ide S, Ganley ARD, Kobayashi T. 2013. Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr Biol. 23:1794–1798. [DOI] [PubMed] [Google Scholar]
- Salih D, Brunet A. 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 20:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. 2002. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 277:39195–39201. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Jun S, Rellick S, Quintana D, Cavendish J, Simpkins J. 2016. Expression of microRNA-34a in Alzheimer’s disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 1646:139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. 2006. Lamin A-dependent nuclear defects in human aging. Science. 312:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoerer S, Becker F, Feller C, Baig A, Koeber U, Henze H, Kraus J, Xin B, Lechel A, Lipka D, et al. 2016. Epigenetic stress responses induce muscle stem cell aging by Hoxa9 developmental signals. Nature. 540:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy J, Banumathy G, Adams P. 2008. Aging by epigenetics-a consequence of chromatin damage? Exp Cell Res. 314:1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Dang W, Donahue G, Dai J, Dorsey J, Cao X, Liu W, Cao K, Perry R, Lee JY, et al. 2015. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 29:1362–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Shah PP, Nativio R, Berger SL. 2016. Epigenetic mechanisms of longevity and aging. Cell. 166:822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, McCormack CE, Bradbury NA. 2014. Do you know the sex of your cells? Am J Physiol Cell Physiol. 306: C3–C18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko AI, Grigor’eva EV, Medvedev SP, Zakharova IS, Dementyeva EV, Elisaphenko EA, Malakhova AA, Pavlova SV, Zakian SM. 2018. Impact of Xist RNA on chromatin modifications and transcriptional silencing maintenance at different stages of imprinted X chromosome inactivation in vole Microtus levis. Chromosoma. 127:129–139. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. PNAS. 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. 2010. Polycomb repressive complex 2 and trithorax modulate Drosophila longevity and stress resistance. PNAS. 107: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D, Mills K, Guarente L. 1998. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 52:533–560. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles-a cause of aging in yeast. Cell. 91:1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. 1997. Accelerated aging and nucleolar fragmentation in yeast SGS1 mutants. Science. 277:1313–1316. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. 1998. Molecular mechanisms of yeast aging. Trends Biochem Sci. 23:131–134. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Oberdoerffer P. 2009. The ageing epigenome: damaged beyond repair? Ageing Res Rev. 8:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh GM, Danaei G, Farzadfar F, Stevens GA, Woodward M, Wormser D, Kaptoge S, Whitlock G, Qiao Q, Lewington S, et al. 2013. The age-specific quantitative effects of metabolic risk factors on cardiovascular diseases and diabetes: a pooled analysis. PLoS One. 8:e65174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Thakur MK. 2018. Histone deacetylase 2 inhibition attenuates downregulation of hippocampal plasticity gene expression during aging. Mol Neurobiol. 55:2432–2442. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2013. Histone variants in pluripotency and disease. Development. 140:2513–2524. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8:272–285. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. 2013. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol. 15:872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs TM, Bonder MJ, Stark AK, Krueger F, von Meyenn F, Stegle O, Reik W, Bolland D, Butcher G, Chandra T, et al. 2017. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull K, Chen R, et al. 2014. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 14: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G, Bohmann D. 2008. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 14:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski K, Abraham K, Mekhail K. 2015. Non-coding RNA in neural function, disease, and aging. Front Genet. 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. [DOI] [PubMed] [Google Scholar]
- Tan L, Ke Z, Tombline G, Macoretta N, Hayes K, Tian X, Lv R, Ablaeva J, Gilbert M, Bhanu NV, et al. 2017. Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Rep. 9:1721–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebey L, Robertson H, Durant S, Vitale S, Penning T, Dinkova-Kostova A, Hayes J. 2016. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med. 88: 108–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, von Holdt B, Horvath S, Pellegrini M. 2017. An epigenetic aging clock for dogs and wolves. Aging (Albany NY). 9:1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. 2006. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 5:505–514. [DOI] [PubMed] [Google Scholar]
- Tsurumi A, Li WX. 2012. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 7:680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JMA, Green JW, Au C, Benedetto A, Thompson MA, Clark E, Gilliat AF, Young A, Schmeisser K, Gems D. 2017. The SKN-1/Nrf2 transcription factor can protect against oxidative stress and increase lifespan in C. elegans by distinct mechanisms. Aging Cell. 16:1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungvari Z, Tucsek Z, Sosnowska D, Toth P, Gautam T, Podlutsky A, Csiszar A, Losonczy G, Valcarcel-ares MN, Sonntag WE, Csiszar A. 2013. Aging-induced dysregulation of dicer1-dependent microRNA expression impairs angiogenic capacity of rat cerebromicrovascular endothelial cells. J Gerontol - Ser A Biol Sci Med Sci. 68:877–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meter M, Kashyap M, R S, Geneva A, Morello T, Seluanov A, Gorbunova V. 2014. SIRT6 represses LINE1 retrotranspo-sons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 5:5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B, Nemirovsky L, Klimenko V, Vasiliev V, Belozersky A. 1973. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia. 19: 138–152. [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. 2007. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 450:440–444. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 16:93–105. [DOI] [PubMed] [Google Scholar]
- Vaux D, Pfefferli C, Passannante M, Belhaj K, Essen A, Von Sprecher SG, Mu F, Wicky C. 2013. The Caenorhabditis elegans LET-418/Mi2 plays a conserved role in lifespan regulation. Aging Cell. 12:1012–1020. [DOI] [PubMed] [Google Scholar]
- Venkatraman A, He X, Thorvaldsen J, Sugimura R, Perry J, Tao F, Zhao M, Christenson M, Sanchez R, Yu J, et al. 2013. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 500: 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B. 1997. The heterochromatin loss model of aging. Exp Gerontol. 32:383–394. [DOI] [PubMed] [Google Scholar]
- Wagner W. 2017. Epigenetic aging clocks in mice and men.Genome Biol. 18:17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt M, Norddahl GL, Sten G, Ugale A, Frisk MM, Mattsson R, Deierborg T, Sigvardsson M, Bryder D. 2013. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 121:4257–4265. [DOI] [PubMed] [Google Scholar]
- Wallrath LL. 1998. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 8:147–153. [DOI] [PubMed] [Google Scholar]
- Wang CM, Tsai SN, Yew TW, Kwan YW, Ngai SM. 2010. Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology. 11:87–102. [DOI] [PubMed] [Google Scholar]
- Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al. 2011. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 10:3016–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tsui B, Kreisberg JF, Robertson NA, Gross AM, Yu MK, Carter H, Brown-borg HM, Adams PD, Ideker T. 2017. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang Q, Hu Y, Stromberg AJ, Nelson PT. 2011. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, MacKenzie IRA, Huang EJ, Tsai LH. 2013. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 16:1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C. 2009. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer ‘ s disease, inhibits bcl2 translation. Brain Res Bull. 80:268–273. [DOI] [PubMed] [Google Scholar]
- Webb AE, Kundaje A, Brunet A. 2016. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 15:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Liu B, Tuo J, Shen D, Chen P, Li Z, Ni J, Dagur P, Sen HN, Jawad S, et al. 2012. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci USA. 109:13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. PNAS. 105:9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. 1987. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 262: 9948–9951. [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Helfand SL. 2013. Chromatin structure and transposable elements in organismal aging. Front Endocrinol (Lausanne). 4:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, et al. 2010. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 9:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SH, Dam SV, Craig T, Tacutu R, Toole AO, Merry BJ. 2015. Transcriptome analysis in calorie-restricted rats implicates epigenetic and post- translational mechanisms in neuroprotection and aging. Genome Biol. 16:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Kosakai A, Ito D, Okada Y, Akamatsu W, Nihei Y, Nabetani A, Ishikawa F, Arai Y, Hirose N, et al. 2012. Establishment of induced pluripotent stem cells from centenarians for neurodegenerative disease research. PLoS One. 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bonasio R, Simola DF, Liebig J, Berger SL, Reinberg D. 2015. DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu Rev Entomol. 60:435–452. [DOI] [PubMed] [Google Scholar]
- Yilmazer A, de Lázaro I, Bussy C, Kostarelos K. 2013. In vivo cell reprogramming towards pluripotency by virus-free overexpression of defined factors. PLoS One. 8:e54754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Jiao Y, Jong SD, Ophoff RA, Beck S. 2015. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 11:e10014996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-mamczarz K, et al. 2007. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 3:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Börkle A, Caiafa P. 2015. Reconfiguration of DNA methylation in aging. Mech Ageing Dev. 151:60–70. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol. 27:2343–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 8:19–30. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al. 2015. A Werner syndrome stem cell model\nunveils heterochromatin alterations\nas a driver of human aging. Science. 348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan M, Zhang XUE, Huang F, Wu K, Zhang J, Liu JUN, Huang Z, Luo H, Tao L, Zhang HUI. 2014. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA. 20:1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. 2006. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 24:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Deng C, Lu Q, Richardson B. 2002. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 123:1257–1268. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wu D, Fei C, Guo J, Gu S, Zhu Y, Xu F, Zhang Z, Wu L, Li X, Chang C. 2015. Down-regulation of Dicer1 promotes cellular senescence and decreases the differentiation and stem cell-supporting capacities of mesenchymal stromal cells in patients with myelodysplastic syndrome. Haematologica. 100:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupkovitz G, Lagger S, Martin D, Steiner M, Hagelkruys A, Seiser C, Schöfer C, Pusch O. 2018. Histone deacetylase 1 expression is inversely correlated with age in the short-lived fish Nothobranchius furzeri. Histochem Cell Biol. 150: 225–269. [DOI] [PMC free article] [PubMed] [Google Scholar]