Abstract
Objective
Somatic TP53 mutation (TP53mut) is a characteristic finding in high-grade serous ovarian cancer (HGSOC). The aim of this study was to assess the clinical efficacy and utility of TP53mut circulating tumor DNA (ctDNA) monitoring as a biomarker for managing HGSOC.
Methods
TP53muts were evaluated in patients who received primary treatment for suspected ovarian cancer at Asan Medical Center. In patients diagnosed with HGSOC and with TP53mut, ctDNA, cancer antigen 125 (CA 125), and computed tomography were followed up according to the treatment course.
Results
Direct sequencing analysis of 103 tumor tissues from 61 HGSOC patients confirmed TP53muts in 41 patients (67.2%). All these patient-specific somatic mutations were detected in plasma cell-free DNA. The mean value of preoperative TP53 mutant allele count (TP53MAC) in stage III patients was 12.2 copies/µL and in stage IV patients was 45.3 copies/µL. TP53MAC was significantly reduced by treatment and there was no significant difference in the rate of decrease compared to CA 125 by the generalized linear mixed model. When patients were divided into a low TP53MAC group (<0.2 copies/µL) and a high TP53MAC group (≥0.2 copies/µL) based on the TP53MAC value at 3 months after the end of chemotherapy, there was a significant difference in time to progression between the two groups (p=0.038).
Conclusion
TP53mut ctDNA shows potential as a tumor-specific biomarker for treatment response monitoring in HGSOC. TP53mut ctDNA levels at 3 months post treatment has a significant prognostic utility than that of CA 125.
Keywords: DNA Mutational Analysis, Tumor Suppressor Protein p53, Ovarian Cancer, Tumor Biomarkers
INTRODUCTION
Ovarian cancer is the sixth most common cancer in women and leading cause of death in gynecologic malignancies [1]. Most women with advanced ovarian cancer experience recurrence within 5 years of diagnosis, despite previously achieving complete remission [2]. To evaluate treatment responses, various surveillance methods have been proposed [3]. In previous studies, serial monitoring of cancer antigen 125 (CA 125) levels showed a sensitivity of 79%–95% and positive predictive value close to 100% [4,5,6]. However, CA 125 has some disadvantages, such as non-specificity for cancer and the inability to conduct evaluation in real-time. Nearly 50% of the ovarian cancer patients with normal CA 125 levels following chemotherapy have persistent disease [7]. To overcome these limitations, liquid biopsy has recently attracted attention as a tool for less invasive, real-time monitoring [8]. In the field of liquid biopsy, circulating tumor cells are exceedingly rare in general solid tumors [9]. In contrast, circulating tumor DNA (ctDNA) in plasma contains tumor-specific mutations, microsatellite instability, loss of heterozygosity, and DNA methylation [10,11,12]. The half-life of cell-free DNA (cfDNA) in the circulation is between 16 minutes and 2.5 hours and can provide ‘real-time’ information regarding cancer genomic alterations during treatment and follow-up [13,14]. Previously published literature described the utility of ctDNA for rapid and accurate monitoring of treatment response in other solid tumors, such as colorectal, breast, skin, and lung cancers [15,16,17,18,19]. However, unlike other cancers, ovarian, fallopian tube, and primary peritoneal carcinomas are typically diagnosed in advanced stages and metastatic tumors often exhibit heterogeneity. Therefore, it is necessary to study the efficacy of ctDNA surveillance methods for ovarian cancer. According to integrated genomic analyses of ovarian carcinoma in The Cancer Genome Atlas (TCGA) project, TP53 mutation (TP53mut) was found in 94.6% of high-grade serous ovarian cancer (HGSOC) patients and is a potential target mutation for surveillance [20].
In this study, we investigated the relationship between TP53mut in primary tumor tissue and ctDNA in the plasma of patients with HGSOC and assessed the clinical efficacy and utility of ctDNA compared to conventional CA 125 for treatment response monitoring during the initial treatment of ovarian cancer.
MATERIALS AND METHODS
1. Ethics and consent
This study included patients enrolled in the prospective study ‘p53 mutations in circulating cell-free tumor DNA of patients with ovarian cancer,’ which was approved by the Institutional Review Board (IRB) at the Asan Medical Center (IRB No. 2013-0572). All patients provided written informed consent for participation in the study and for use of their tissues and blood samples.
2. Study design and patients
1) Patient enrollment and sample collection
In total, 102 patients suspected of ovarian cancer were enrolled in this study from July 2013 to July 2017 at a single institution, Asan Medical Center in Korea. Other inclusion criteria were patients over the age of 18 years, patients who were scheduled for surgery for pelvic mass, and patients who agreed to venous blood collection before surgery. Exclusion criteria were patients who underwent surgical or open biopsy for any reason within the past 28 days, patients with clinically significant medical problems, patients without confirmed HGSOC, patients with metastatic ovarian cancer, patients who received more than 450 mL of blood transfusions within 28 days of the screening day, and patients who died before the 6th administration of chemotherapy.
After being enrolled in the study, 15 mL of whole blood was collected before surgery. DNA was immediately extracted from the plasma or stored at −20°C for several days. Tumor tissues were obtained in the sterile tube at the time of surgery and immediately frozen and stored at −80°C until DNA was extracted. In cases in which sufficient fresh-frozen tissues were not obtained, DNA was extracted with paraffin blocks (formalin-fixed paraffin embedded [FFPE]).
2) Treatment
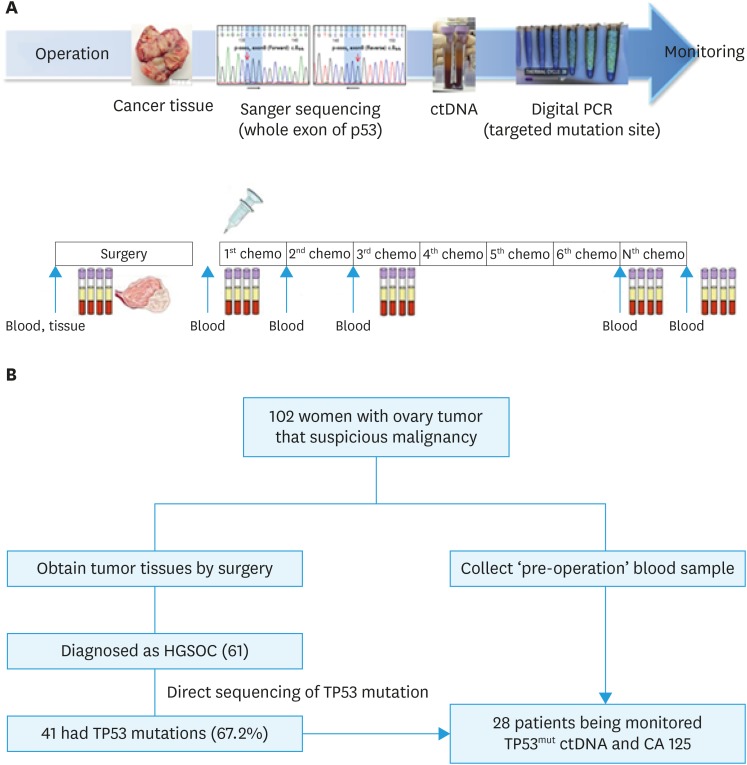
Within 3–6 weeks after surgery, a dose of 175 mg/m2 paclitaxel or 260 mg/m2 Genexol PM and a dose of 5 areas under curve of carboplatin were administered as an intravenous infusion in patients diagnosed with HGSOC. The treatment was repeated every 21 days and delayed in cases of unacceptable toxicities. Chemotherapy was administered for 6 consecutive cycles and then continued until achieving a complete response or disease progression if there was a remnant tumor. Serial blood samples (10 mL) were collected and monitored after the first, second, and sixth cycles of chemotherapy and three months after the end of chemotherapy (Fig. 1A).
Fig. 1. Schema of workflow for ctDNA analysis (A) and diagram showing the flow of patients in this study (B).
CA 125, cancer antigen 125; ctDNA, circulating tumor DNA; HGSOC, high-grade serous ovarian cancer; PCR, polymerase chain reaction; TP53mut, TP53 mutation.
3) Tumor tissue DNA extraction and genomic sequencing
DNA extraction from fresh frozen tissue or FFPE tissue: DNA of fresh frozen tissue obtained during surgery was extracted according to the user manual using a QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany). From FFPE tissue, tumor DNA was extracted using a RecoverAll™ total nucleic acid isolation kit (Ambion, Carlsbad, CA, USA).
DNA quantification and genome sequencing: The concentration of extracted DNA was measured using a Nanodrop ™ 2000 spectrophotometer (Thermo Fisher, Waltham, MA, USA). Genomic sequence analysis was performed by Sanger sequencing. Primers corresponding to exons 2–11 of the TP53 gene were prepared and amplified by polymerase chain reaction (PCR). DNA sequencing was performed using an Applied Biosystems 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Twelve target regions of TP53mut and the primers used in this study are summarized in Supplementary Tables 1 and 2.
4) Circulating cfDNA extraction
Separation of cfDNA from plasma: cfDNA was separated from 1 to 5 mL plasma using a circulating nucleic acid kit (Qiagen) according to the manual. The extracted cfDNA was completely dried in a vacuum concentrator (Eppendorf, Hamburg, Germany) and then dissolved again in a volume of 10 µL.
cfDNA concentration measurement: Extracted cfDNA was assayed using Qubit® dsDNA HS assay kit (Life Technologies, Carlsbad, CA, USA) to detect only double-stranded DNA. The values were measured using a Qubit® 3.0 fluorometer.
5) Measurement of ctDNA and Droplet Digital PCR
A QX200 Droplet Digital PCR system (Bio-Rad, Hercules, CA, USA) was used to quantify mutations in the TP53 gene [21,22]. The level of mutated TP53 ctDNA was quantified by the number of mutated TP53 amplifiable copies, as the TP53 mutant allele count (TP53MAC).
6) CA 125 assay
CA 125 levels were monitored at each cycle. The CA 125 test was performed with an Architect i2000 immunoassay analyzer (Abbott Laboratories, Abbott Park, IL, USA). The reagents were assayed using the ARCHITECT CA125 II assay (Abbott Laboratories).
7) Computed tomography (CT) imaging analysis
Tumor status was assessed by CT scanning after every three cycles of chemotherapy and assessed when signs of disease progression were observed. All CT images were interpreted by experts in radiology and total tumor volume was calculated. Treatment response was assessed according to the response evaluation criteria in solid tumors (RECIST criteria version 1.1) [23].
3. Statistical analysis
Analysis of mutation results in cfDNA was performed using QuantaSoft Software (Bio-Rad). The correlation between tumor volume and TP53MAC and CA 125 was confirmed by linear regression analysis. Changes in TP53MAC and CA 125 values according to the treatment duration were compared using a generalized linear mixed model. In addition, Kaplan-Meier analysis was used to confirm the correlation between time to progression (TTP) and changes in ctDNA and CA 125 at 3 months after chemotherapy. All statistical analyses were performed using SPSS software, version 21.0 (SPSS, Inc., Chicago, IL, USA).
RESULTS
1. Study patients and treatments
Fig. 1B shows the flow of patients in this study. In total, 102 patients with ovarian tumor and suspicious malignancy were assessed. Post-operative diagnosis of 102 suspected ovarian cancer patients are summarized in Supplementary Table 3. In total, 61 patients were eligible for the study based on the criteria and the total follow-up period was 1,489 months (range, 5.6–50.1 months). All patients underwent primary surgery and subsequently received chemotherapy.
2. TP53mut identification and concordance with plasma samples
Direct sequencing analysis of 103 tumor tissues from 61 HGSOC patients confirmed TP53muts in 41 patients (67.2%). The TP53muts identified in 41 patients with HGSOC are summarized in Supplementary Table 4. In total, 52 TP53muts were identified in 41 patients, and 7 patients had TP53muts at 2 different sites and 2 patients had TP53muts at 3 different sites. All these patient-specific somatic mutations were detected in the plasma cfDNA of 38 patients (concordance 100%), except for two patients who were unable to produce a probe and one whose manufactured probe did not function. Among the 41 patients with TP53mut, 13 patients were excluded from tumor response assessment for the following reasons: withdrawal of participation, follow-up loss or early death (n=7), coding silent (n=2), mutation in the site where the probe could not be made (n=2), manufactured probe did not function (n=1), and non-specific mutation (n=1). As a result, 28 patients were monitored for changes according to treatment course for TP53mut ctDNA and CA 125.
3. Baseline characteristics and TP53MAC value
Baseline characteristics of these patients are summarized in Table 1. Stage III and IV were the most common in the initial International Federation of Gynecology and Obstetrics (FIGO) stage, and the mean values of preoperative TP53MAC in stage III and IV patients were 12.2 and 45.3 copies/µL, respectively. Optimal debulking surgery was performed in 60.7% of the patients. The mean value of postoperative TP53MAC and reduction rate compared to preoperative values in patients with optimal debulking surgery and sub-optimal debulking surgery were 1.0 copies/µL and 92.9% and 9.3 copies/µL and 73.9%, respectively. Linear regression analysis was conducted to identify the correlation between tumor volume with preoperative TP53MAC and CA 125. However, both preoperative TP53MAC (adjusted R-square=−0.005) and CA 125 (adjusted R-square=0.0307) showed no correlation with tumor volume. Complete remission was obtained in 75% of the patients after initial chemotherapy, but recurrence occurred in 67.9% of the patients within the study period. The median TTP was 16.9 months and median progression-free survival was 22.7 months in all patients.
Table 1. Baseline characteristics of patients with TP53mut ctDNA and CA 125 monitoring (n=28).
| Variables | Value | |
|---|---|---|
| Age at diagnosis (yr) | 54.6 (49–62) | |
| Follow-up duration (mo) | 31.7 (27.2–37.5) | |
| Pretreatment TP53MAC (copies/µL) | 6.9 (2.2–18.9) | |
| Pretreatment CA 125 (U/mL) | 1,416.5 (904–2,165) | |
| Pretreatment tumor volume (cm3) | 708.9 (353–1,191) | |
| TTP (mo) | 16.9 (11.6–22) | |
| PFS (mo) | 23.0 (17.3–28.6) | |
| Primary site | ||
| Ovarian cancer | 12 (42.9) | |
| Tubal cancer | 12 (42.9) | |
| Primary peritoneal carcinoma | 4 (14.3) | |
| Initial FIGO stage | ||
| II | 2 (7.1) | |
| III | 16 (57.1) | |
| IV | 10 (35.7) | |
| Primary debulking surgery | ||
| R0 (No residual) | 9 (32.1) | |
| Optimal (<1 cm) | 8 (28.6) | |
| Sub-optimal (>1 cm) | 11 (39.3) | |
| Adjuvant chemotherapy cycles | ||
| 6 | 17 (60.7) | |
| 7–9 | 9 (32.1) | |
| 10–12 | 2 (7.1) | |
| Adjuvant chemotherapy response | ||
| CR | 21 (75.0) | |
| PR | 5 (17.9) | |
| SD | 1 (3.6) | |
| PD | 1 (3.6) | |
| Recur | ||
| Yes | 19 (67.9) | |
| No | 9 (32.1) | |
Values are presented as median (interquartile range) or number (%).
CA 125, cancer antigen 125; CR, complete response; ctDNA, circulating tumor DNA; FIGO, International Federation of Gynecology and Obstetrics; PD, progressive disease PFS, progression-free survival; PR, partial response; SD, stable disease; TP53MAC, TP53 mutant allele count; TP53mut, TP53 mutation; TTP, time to progression.
4. ctDNA for treatment response monitoring
Supplementary Fig. 1 is an example of ctDNA and CA 125 monitoring in the P-0005 patient. The TP53-R175H mutation was detected by direct sequencing of ovarian tissue specimens. The same mutation was also detected in plasma sample taken before surgery in the patient. Changes in TP53mut ctDNA and CA 125 levels according to treatment course are shown in this graph, and TP53MAC shows that the value was decreased to 0, unlike that of CA 125.
Supplementary Fig. 2 shows the changes in TP53MAC levels according to treatment progress in each patient. The white part indicates that samples were not collected at an appropriate time or the sample could not be measured correctly because of insufficient sample volume or errors in the experimental procedure. Overall, TP53MAC was significantly reduced by treatment course, indicating that TP53MAC is useful for treatment response monitoring in ovarian cancer. In addition, TP53MAC, which showed a decreasing tendency, was increased in some patients after 3 months from the end of treatment compared to those immediately after chemotherapy. This increase may be related to the resumption of proliferation of remnant cancer cell after the end of chemotherapy.
5. Comparison of ctDNA and CA 125 in treatment response monitoring and its association with TTP
Table 2 shows the percentage changes in the TP53MAC mean value at each treatment course relative to the pre-operative value using the generalized linear mixed model. As the treatment course progressed, both markers significantly decreased based on the treatment response. The p-value was determined by comparing the 2 markers at each time point and there was no significant difference in the rate of decrease between the 2 markers, except after the first chemotherapy.
Table 2. Percentage changes in TP53MAC mean value at each treatment courses relative to pre-operative value.
| Variables | Percentage relative to pre-operative value (%) | |||||
|---|---|---|---|---|---|---|
| Pre-operation | Post-operation | After #1 | After #2 | After #6 | After 3 months | |
| TP53MAC | 100 | 38.3 | 29.4 | 7.2 | 6.1 | 7.4 |
| CA 125 | 100 | 36.9 | 11.7 | 3.5 | 1.4 | 1.8 |
| p-value | 0.815 | 0.011 | 0.602 | 0.432 | 0.360 | |
After #1 means first chemotherapy within 3–6 weeks after surgery; treatment was repeated every 21 days, total 6 consecutive cycles (#1 to #6).
CA 125, cancer antigen 125; TP53MAC, TP53 mutant allele count.
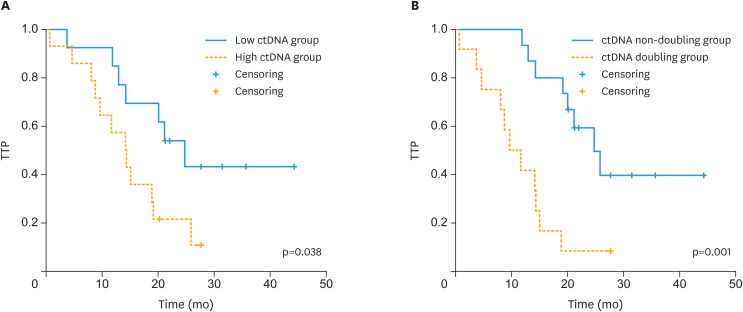
Fig. 2 shows the correlation between TTP and values of ctDNA after 3 months of chemotherapy using Kaplan-Meier analysis. As shown in Supplementary Fig. 1 above, some patients showed increases in TP53MAC at 3 months after the end of treatment compared to those immediately after chemotherapy. Therefore, we performed the analysis by dividing the patients into a low TP53MAC group (<0.2 copies/µL, n=13) and high TP53MAC group (≥0.2 copies/µL, n=14) based on the TP53MAC value at 3 months after the end of chemotherapy, except for one patient whose value was not measured. The analysis showed that there was a significant difference in TTP between the 2 groups (p=0.038). In Univariable analysis, there was no statistically significant correlation between pretreatment clinicopathologic variables and low TP53MAC group and high TP53MAC group (Supplementary Table 5). Among the 27 patients, 12 had more than 2-fold higher TP53MAC levels at 3 months after chemotherapy compared to those immediately after chemotherapy. We performed the analysis by dividing the patients into a TP53MAC doubling group and non-doubling group and there was a significant difference in TTP between the two groups (p=0.001). The probability of recurrence within 12 months after the end of chemotherapy was 6.7% in the TP53MAC non-doubling group and 58.3% in the TP53MAC doubling group. CA 125 levels were also analyzed in the same manner. However, unlike TP53MAC, only 8 patients had CA 125 levels that were more than 2-fold higher at 3 months after chemotherapy compared to those immediately after chemotherapy and there was no significant difference in TTP between the two groups (p=0.674).
Fig. 2. Kaplan-Meier plots for change in ctDNA after 3 months of chemotherapy. (A) Kaplan-Meier curve showing TTP for patients with ctDNA levels <0.2 or ≥0.2 copies/µL at 3 months after the end of chemotherapy. (B) Kaplan-Meier curve showing TTP for patients whose ctDNA levels at 3 months after chemotherapy had not doubled or had doubled compared to those immediately after chemotherapy.
CA 125 = cancer antigen 125; ctDNA, circulating tumor DNA; TTP, time to progression.
DISCUSSION
In this study, direct sequencing analysis of tumor tissues revealed TP53muts in 67.2% of the patients. This is a low value compared to the TP53mut detection rate previously reported in the TCGA data. The actual tumor weight in the tumor specimens used in the analysis of this experiment was unknown. To obtain accurate test results, at least 1 cm of the cancer portion is required for the tumor specimen. The actual tumor volume used in the test may be low, which may have resulted in a low detection rate. Differences in ethnicity may have affected the TP53mut rate. In addition, the TP53mut detection rate varied depending on the number of tumor specimens examined. In the test results, an average of 1.3 TP53mut was detected in a 1 site test, 1.7 TP53mut in a 2-site test, and 2.1 TP53mut in a 3-site test. Therefore, it is recommended that all sites removed by surgery be examined to increase the TP53mut detection rate and reflect heterogeneity between the different sites.
All patient-specific tissue mutations were detected in the plasma cfDNA of patients, showing 100% concordance. This suggests that TP53mut ctDNA monitoring is a useful monitoring technique for the treatment of almost patients with TP53mut in tumors.
We performed linear regression analysis to confirm the correlation between tumor volume and preoperative TP53MAC, but found no significant correlation. Previous studies reported a correlation between the concentration of ctDNA in the plasma and tumor size [24,25]. ctDNA is thought to be released from cancer cells by apoptosis, necrosis, and active release [26,27]. Ovarian cancer has a variety of phenotypes, including main pelvic mass or predominantly lymph node metastasis or peritoneal carcinomatosis, and this difference in the disease pattern and location of the tumor affect the degree of ctDNA release into the blood. And peritoneal seeding less than 1 cm is frequently observed in ovarian cancer patients that cannot be measured sufficiently by CT imaging. These factors may explain why there was no statistically significant correlation between the tumor volume and preoperative level of ctDNA.
In Supplementary Fig. 2, TP53MAC was significantly reduced by treatment, indicating that TP53mut ctDNA is useful for treatment response monitoring. Additionally, compared to CA 125 at each time point during treatment, there was no significant difference in the rate of decrease between the 2 markers.
There was no correlation between preoperative TP53MAC and TTP. It is likely because of differences in resectability (degree of debulking surgery). The mean value of postoperative TP53MAC in patients with optimal debulking surgery was much lower than in patients with sub-optimal debulking surgery. This result emphasizes the importance of a surgeon's efforts to achieve optimal debulking.
We found a significant correlation with TTP when divided into two groups according to the TP53MAC value at 3 months after the end of chemotherapy. In addition, some patients showed more than 2-fold increased TP53MAC levels at 3 months after the end of chemotherapy compared to the levels immediately after chemotherapy, and these patients showed poor prognosis. This means that the TP53MAC value at 3 months after the end of chemotherapy reflects the residual tumor burden and regrowth after the end of treatment. Unlike TP53MAC, CA 125 monitoring did not show this trend and this advantage of TP53mut ctDNA monitoring will help to predict the prognosis and detect relapse earlier after treatment.
Some previous studies have reported the detection of tumor-specific TP53 somatic mutations in ctDNA of patients with epithelial ovarian cancer. Otsuka et al. [28] first studied TP53muts in ovarian cancer and showed TP53mut in cancer tissue in only 44% of the patients and only two cases were detected in ctDNA in preoperative plasma. Swisher et al. [29] found TP53muts in tumors of 50% of the epithelial ovarian cancer patients and identified the corresponding ctDNA in blood and peritoneal fluid in 30% of the patients. In that study, the presence of ctDNA was an independent predictor of survival (p=0.02). Pereira et al. [30] studied gynecologic cancer patients, including 22 ovarian cancer and 4 tubal cancer patients. The detection rate of TP53mut was 66% in all patients with gynecologic cancers. ctDNA was detected in 93.8% of the patients for whom probes were designed and showed that undetectable levels of ctDNA at 6 months after treatment were associated with better prognosis. This result is consistent with our findings that ctDNA levels at 3 months after the end of treatment were associated with TTP. In a recent study of relapsed ovarian cancer, pretreatment ctDNA levels and the extent of ctDNA decrease after chemotherapy were significantly associated with TTP [31]. In the case of recurrent cancer, the proportion of chemo-sensitive cells was greatly different depending on the case. Most initially diagnosed cancer has a large portion of chemo-sensitive cells in the tumor and the degree of debulking has a significant impact on the extent of ctDNA decrease. In addition, the recurrent case showed a similar disseminated pattern, but the various dissemination types can affect the degree of ctDNA release in the initially diagnosed case. Therefore, the change in ctDNA at the initial stage of treatment is highly dependent on the patient in the initially diagnosed case. Therefore, a better approach for initial diagnosis of ovarian cancer is to pay attention to changes in ctDNA levels at the end-point of treatment and after several months, which fully reflect the effects of surgery and chemotherapy.
This is the first study on the efficacy of ctDNA monitoring as a treatment response monitoring tool for initial treatment after initial diagnosis of ovarian cancer, but not in recurrent cases. We demonstrated the efficacy of personalized ctDNA monitoring as a tumor-specific biomarker in the initial treatment of ovarian cancer. In addition, we found that ctDNA monitoring is more useful for prognosis prediction compared to the conventional marker CA 125.
This study had some limitations. Only one patient enrolled in this study died within the study period and the relationship with overall survival was not known. Another limitation is that some data was missing in the ctDNA analysis. Even in specimens obtained from the same site, various TP53muts may be present. Specimen selection bias may lead to inconsistent serial ctDNA changes and disease progression by detecting less important mutation points than pathogenic TP53mut points. Genetic testing should be performed on all removed site tumors to identify stem mutations, which are shared by all tumor regions and have a higher allele fraction in the plasma than do individual mutations. This effort may reduce the effects of intratumoral heterogeneity and clonal diversity [32,33,34,35].
In conclusion, we found that TP53mut ctDNA is useful as a tumor-specific biomarker for treatment response monitoring in HGSOC by significantly reducing the treatment course. Cancer is driven by genomic alterations. Therefore, genomic monitoring is necessary for evaluating early treatment response, monitoring minimal residual disease, and identifying genetic determinants for targeted therapy and evolution of resistance in real-time. Liquid biopsy is a minimally invasive procedure that is useful for serial sample measurements during treatment courses. TP53mut ctDNA is clinically effective and outperforms CA 125 in HGSOC monitoring, as it reflects minimal residual disease and the real-time treatment response. To increase the use of and commercialize ctDNA monitoring, it is necessary to further explore the biology of ctDNA, reduce costs by developing related technologies, establish a standard methodology, conduct quality control of the assays, and validate the method. Further studies are required to confirm the relationship between the change in ctDNA levels and OS through long-term follow-up.
ACKNOWLEDGMENTS
We would like to acknowledge our patients and their caregivers, and the support of the research nurses, members of the Department of Obstetrics and Gynecology, Asan Medical Center, Ulsan University and Asan Institute for Life Sciences.
Footnotes
Funding: This study was supported by a grant (2015-588 & 2016-731) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: K.Y.M., L.S.W.
- Data curation: L.Y.J., L.S.W.
- Formal analysis: L.Y.J.
- Funding acquisition: K.Y.M., C.E.K.
- Investigation: K.Y.M., L.S.W., L.H.Y.
- Methodology: K.Y.M., L.S.W., L.H.Y., L.J.E.
- Project administration: K.Y.M., L.S.W., L.H.Y., L.J.E., C.E.K.
- Resources: K.Y.M., L.H.Y., C.E.K.
- Software: L.J.E.
- Supervision: K.Y.M., C.E.K.
- Validation: K.Y.M., L.S.W., L.H.Y., L.J.E.
- Visualization: L.Y.J., L.S.W., L.H.Y.
- Writing - original draft: L.Y.J.
- Writing - review & editing: K.Y.M., L.Y.J., L.S.W.
SUPPLEMENTARY MATERIALS
Twelve target regions of TP53mut in high-grade serous ovarian carcinoma
Nucleotide sequences of 12 primers used in the study
Post-operative diagnosis of 102 suspected ovarian cancer patients
Summary of TP53muts identified in patients with high-grade serous ovarian carcinoma
Univariable analysis of low- vs. high TP53MAC group for pre-treatment clinicopathological variables
ctDNA and CA 125 monitoring of P-0005 (TP53-R175H mutation). (A) Identification of TP53mut from tissue sample by direct sequencing method. (B) Identification of TP53mut from plasma sample. (C, D) Changes in TP53MAC and CA 125 level according to treatment course.
Changes in TP53 mutated circulating DNA levels according to treatment progress in each patient.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Damak T, Chargui R, Ben Hassouna J, Hechiche M, Rahal K. Results of second-look laparotomy in advanced ovarian cancer: one single center experience. ISRN Obstet Gynecol. 2012;2012:849518. doi: 10.5402/2012/849518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thrall MM, DeLoia JA, Gallion H, Avril N. Clinical use of combined positron emission tomography and computed tomography (FDG-PET/CT) in recurrent ovarian cancer. Gynecol Oncol. 2007;105:17–22. doi: 10.1016/j.ygyno.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 4.Niloff JM, Knapp RC, Lavin PT, Malkasian GD, Berek JS, Mortel R, et al. The CA 125 assay as a predictor of clinical recurrence in epithelial ovarian cancer. Am J Obstet Gynecol. 1986;155:56–60. doi: 10.1016/0002-9378(86)90077-3. [DOI] [PubMed] [Google Scholar]
- 5.Högberg T, Kågedal B. Long-term follow-up of ovarian cancer with monthly determinations of serum CA 125. Gynecol Oncol. 1992;46:191–198. doi: 10.1016/0090-8258(92)90254-g. [DOI] [PubMed] [Google Scholar]
- 6.Rustin GJ, Marples M, Nelstrop AE, Mahmoudi M, Meyer T. Use of CA-125 to define progression of ovarian cancer in patients with persistently elevated levels. J Clin Oncol. 2001;19:4054–4057. doi: 10.1200/JCO.2001.19.20.4054. [DOI] [PubMed] [Google Scholar]
- 7.Bast RC., Jr CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer. 2010;116:2850–2853. doi: 10.1002/cncr.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56:186–197. doi: 10.1515/cclm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 9.Lianidou ES, Strati A, Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. 2014;51:160–171. doi: 10.3109/10408363.2014.896316. [DOI] [PubMed] [Google Scholar]
- 10.Marzese DM, Hirose H, Hoon DS. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn. 2013;13:827–844. doi: 10.1586/14737159.2013.845088. [DOI] [PubMed] [Google Scholar]
- 11.Kuhlmann JD, Schwarzenbach H, Wimberger P, Poetsch M, Kimmig R, Kasimir-Bauer S. LOH at 6q and 10q in fractionated circulating DNA of ovarian cancer patients is predictive for tumor cell spread and overall survival. BMC Cancer. 2012;12:325. doi: 10.1186/1471-2407-12-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci. 2015;2:13. doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene. 2016;590:142–148. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Krawczyk N, Fehm T, Banys-Paluchowski M, Janni W, Schramm A. Liquid biopsy in metastasized breast cancer as basis for treatment decisions. Oncol Res Treat. 2016;39:112–116. doi: 10.1159/000444605. [DOI] [PubMed] [Google Scholar]
- 16.Sanmamed MF, Fernández-Landázuri S, Rodríguez C, Zárate R, Lozano MD, Zubiri L, et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61:297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]
- 17.Olmedillas López S, García-Olmo DC, García-Arranz M, Guadalajara H, Pastor C, García-Olmo D. KRAS G12V mutation detection by Droplet Digital PCR in circulating cell-free dna of colorectal cancer patients. Int J Mol Sci. 2016;17:484. doi: 10.3390/ijms17040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Zhuo M, Ye X, Bai H, Wang Z, Sun Y, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget. 2016;7:20810–20824. doi: 10.18632/oncotarget.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura F, Uchida J, Kukita Y, Kumagai T, Nishino K, Inoue T, et al. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer. 2016;94:68–73. doi: 10.1016/j.lungcan.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamat AA, Bischoff FZ, Dang D, Baldwin MF, Han LY, Lin YG, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther. 2006;5:1369–1374. doi: 10.4161/cbt.5.10.3240. [DOI] [PubMed] [Google Scholar]
- 26.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 27.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 28.Otsuka J, Okuda T, Sekizawa A, Amemiya S, Saito H, Okai T, et al. Detection of p53 mutations in the plasma DNA of patients with ovarian cancer. Int J Gynecol Cancer. 2004;14:459–464. doi: 10.1111/j.1048-891x.2004.014305.x. [DOI] [PubMed] [Google Scholar]
- 29.Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193:662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 30.Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10:e0145754. doi: 10.1371/journal.pone.0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS One. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamal-Hanjani M, Wilson GA, Horswell S, Mitter R, Sakarya O, Constantin T, et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann Oncol. 2016;27:862–867. doi: 10.1093/annonc/mdw037. [DOI] [PubMed] [Google Scholar]
- 33.Popper HH. Commentary on tumor heterogeneity. Transl Lung Cancer Res. 2016;5:433–435. doi: 10.21037/tlcr.2016.08.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol. 2015;9:783–790. doi: 10.1016/j.molonc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Twelve target regions of TP53mut in high-grade serous ovarian carcinoma
Nucleotide sequences of 12 primers used in the study
Post-operative diagnosis of 102 suspected ovarian cancer patients
Summary of TP53muts identified in patients with high-grade serous ovarian carcinoma
Univariable analysis of low- vs. high TP53MAC group for pre-treatment clinicopathological variables
ctDNA and CA 125 monitoring of P-0005 (TP53-R175H mutation). (A) Identification of TP53mut from tissue sample by direct sequencing method. (B) Identification of TP53mut from plasma sample. (C, D) Changes in TP53MAC and CA 125 level according to treatment course.
Changes in TP53 mutated circulating DNA levels according to treatment progress in each patient.