Abstract
The aim of this study was to investigate implementation of multiplex PCR assays (broad screening PCR) on the distribution and characteristics of notified Shiga toxin-producing Escherichia coli (STEC) cases in Norway, 2007–2017. We described STEC cases notified to the Norwegian Surveillance System for Communicable Diseases (MSIS), 2007–2017 and categorised cases as high-virulent, low-virulent or unclassifiable STEC infections based on guidelines for follow-up of STEC cases. We conducted descriptive analysis and time series analysis allowing for trends and seasonality, and calculated adjusted incidence rate ratios (aIRR) using negative binomial regression for laboratories with and without broad screening PCR. A total of 1458 STEC cases were notified to MSIS (2007–2017), median age 21 years, 51% female. Cases were categorised as having 475 (33%) high-virulent, 652 (45%) low-virulent, and 331 (23%) unclassifiable STEC infections. We observed a higher increasing monthly trend in cases (aIRR = 1.020; 95% CI 1.016–1.024) notified from laboratories with broad screening PCR (n = 4) compared to laboratories (n = 17) without (aIRR = 1.011; 95% CI 1.007–1.014). Notification of low-virulent STEC infections increased from laboratories with broad screening PCR. The increase in notified STEC cases was prominent in cases categorised with a low-virulent STEC infection and largely attributable to unselective screening methods. We recommend NIPH to maintain differentiated control measures for STEC cases to avoid follow-up of low-virulent STEC infections. We recommend microbiological laboratories in Norway to consider a more cost-effective broad screening PCR strategy that enables differentiation of high-virulent STEC infections.
Electronic supplementary material
The online version of this article (10.1007/s10096-019-03475-5) contains supplementary material, which is available to authorized users.
Keywords: STEC diagnostic, Multiplex PCR panels, Incidence of STEC, High-virulent STEC, Low-virulent STEC
Introduction
Shiga toxin-producing Escherichia coli (STEC) infection may lead to mild gastroenteritis, haemorrhagic colitis or the life-threatening complication haemolytic-uraemic syndrome (HUS) [1]. An estimated 5–10% of patients with a STEC infection develop HUS, a number that may be higher when related to outbreaks [2]. Factors related to both the host and STEC have been associated with an increased risk for development of HUS. Young age, as well as the presence of Shiga toxin-producing gene stx2, especially the subtypes stx2a and stx2d, and the intimin-encoding gene eae (E. coli attaching and effacing), have been documented as factors associated with increased risk of HUS [3–6]. Classification of STEC has traditionally been based on seropathotypes, classifying serotypes according to association with severity of illness, outcome and outbreaks [7]. Knowledge of the evolution of pathogenic STEC has led to alternative classifications based on virulence factors, especially the stx genes, and their association with the development of HUS [4, 8].
In Norway, STEC infections have been mandatory notifiable since 1995 and reported via the Norwegian Surveillance System for Communicable Diseases (MSIS). Mandatory notification of diarrhoea-associated HUS was added to MSIS in 2006 following a national outbreak of STEC O103:H25 the same year [9, 10]. The National Reference Laboratory for Enteropathogenic Bacteria (NRL) at the Norwegian Institute of Public Health (NIPH) receives presumptive STEC isolates for verification and characterisation from all the Norwegian medical microbiological laboratories. Historically, laboratories have identified STEC by culturing, with focus on the identification of O157 [3, 11]. In the years following the 2006 outbreak, a majority of medical microbiological laboratories in Norway implemented PCR detection of stx. In recent years, multiplex PCR assays (in this paper referred to as “broad screening PCR”) have been introduced into routine primary diagnostics as a screening tool for gastrointestinal pathogens in some laboratories.
The number of notified STEC cases in Norway has increased in recent years, while the number of notified HUS cases has remained stable. This has challenged the existing system for infection control and follow-up of STEC cases. In 2016, the national guidelines on follow-up of STEC infections were revised in accordance with evidence on the association of HUS with STEC virulence factors [12]. Consequently, all STEC cases were assigned into either “high-virulent” or “low-virulent” STEC infection categories. To limit the socioeconomic consequences and the psychological impact of infection control measures for the patients and their families, only cases with high-virulent STEC infections identified in high-risk groups for disease transmission (e.g. food handlers, kindergarten children and staff) are now subject to follow-up.
The aim of this study was to investigate the observed increase of notified STEC cases in Norway from 2007 to 2017 in order to assess the effect of broad screening PCR implementation at the medical microbiological laboratories on the distribution and characteristics of notified STEC cases.
Materials and methods
Data collection
The notification criteria of STEC to MSIS are a clinically compatible case that is epidemiologically linked or is laboratory confirmed by (a) isolation of STEC positive for stx1 or stx2 gene(s), (b) detection of stx1 or stx2 gene(s) without isolation of strain, (c) detection of Stx in faeces without isolation of strain or (d) detection of STEC-specific antibodies in a HUS case. In absence of stx, a HUS patient with eae-positive E. coli and a patient with eae-positive E. coli with a known genotype (MLVA, multiple-locus variable-number of tandem repeat analysis), that has previously been identified in a HUS case, are also notifiable to MSIS. The latter is notified by the NRL as a probable case of STEC, which has lost its stx gene (STEC-LST).
We extracted data on all STEC cases notified to MSIS from 2007 to 2017 including demographics (age, sex, place), clinical presentation (symptoms, hospitalisation) and laboratory findings (date of sampling, diagnosing laboratory, serotype, stx subtype, presence of eae and ehxA, MLVA-type). Incomplete laboratory data in MSIS was supplemented with data from the NRL where available. In addition, we extracted data on all HUS (acute renal failure and at least microangiopathic haemolytic anaemia and/or thrombocytopenia) cases with an epidemiological link notified to MSIS in the same study period.
We gathered information on the implementation of broad screening PCR methodology at the medical microbiological laboratories from a national survey on laboratory practice from 2017, and through personal communication with the laboratories.
We extracted data on concomitant bacterial infections for all reported STEC cases from laboratories with broad screening PCR methodology. We defined a concomitant bacterial infection as notification of a pathogen included in the broad screening PCR panel (Salmonella spp., Campylobacter spp., Shigella spp., Yersinia spp. and/or other enteropathogenic E. coli) from the same laboratory and same sampling date as the STEC case.
Categorisation of STEC cases
We categorised STEC cases into high- or low-virulent infections based on the 2016 revised guidelines [12].
A case was categorised as having a high-virulent STEC infection if
-
i)
positive for stx2 subtypes 2a, 2c, 2d, or
-
ii)
positive for stx1 subtype 1a in a patient ≤ 5 years with bloody diarrhoea, or
-
iii)
notified as a HUS patient, or
-
iv)
negative for stx, but eae-positive E. coli strain (STEC-LST) with a genotype (MLVA-type) previously seen in a HUS case
A case was categorised as having a low-virulent STEC infection if
-
i)
positive for stx1 (not 1a in a patient ≤ 5 years with bloody diarrhoea), or
-
ii)
positive for stx2 subtypes 2b, 2e, 2f, 2g
Cases that did not fulfil any of the above-mentioned criteria due to missing and/or insufficient data were categorised as having an unclassifiable STEC infection.
Statistical analysis
We described cases in terms of demographic, clinical and microbiological characteristics. Incidence rates of notified STEC cases were calculated using population numbers provided by Statistics Norway registries (www.ssb.no).
We used chi-squared test for categorical variables to examine the distribution of demographics (sex, age, seasonality and place of infection), clinical (hospitalisation) and microbiological (serogroups and virulence profile) characteristics between cases with high-virulent and low-virulent STEC infections. We applied the Wilcoxon’s rank sum test to examine the differences between the two groups with respect to continuous variables (age).
We conducted time series analysis allowing for trends and seasonality (1 year periodicity) and calculated adjusted incidence rate ratios (aIRRs) with 95% confidence intervals (CIs) using negative binomial regression on 2007–2017 data for cases reported from laboratories that implemented broad screening PCR and from laboratories that did not implement this screening method.
We considered a p value of ≤ 0.05 as statistically significant. We performed all statistical analysis in Stata version 14 (Stata Corporation, College Station, Texas, USA).
Results
Notified STEC cases and categorisation of the cases
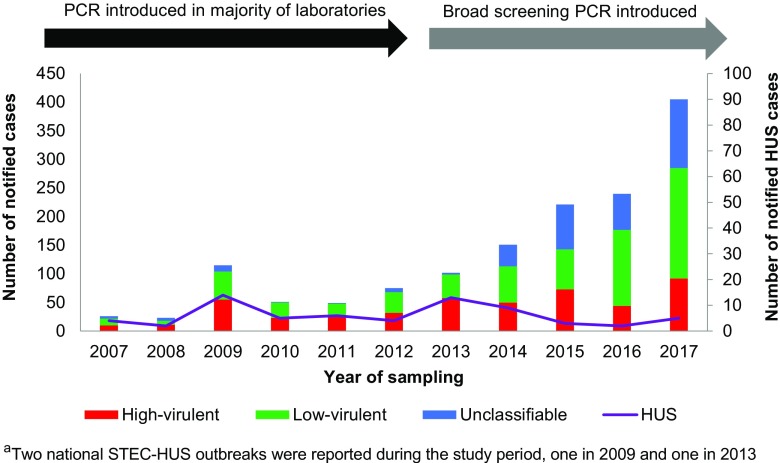
From 2007 to 2017, 1458 STEC cases were notified to MSIS. The median age of the cases was 21 years (range 0–97 years) and 51% of cases were female. The most frequent age group was ≤ 5 years with 37% of the cases. Among cases with known clinical outcome (1278), 5% developed HUS, 25% reported bloody diarrhoea as the worst clinical outcome and 11% were asymptomatic infections. At the time of notification, 26% of the cases were reported as hospitalised. For cases with available data on place of acquisition, 71% (902/1280) reported a domestically acquired infection. One or multiple stx subtype(s) was identified in 64% (936), eae in 55% (796) and ehxA in 48% (705) of all notified cases. The notified cases were categorised as 475 (33%) high-virulent, 652 (45%) low-virulent and 331 (23%) as unclassifiable STEC infections (Fig. 1).
Fig. 1.
Annual distribution of cases categorised with high-virulent, low-virulent or unclassifiable Shiga toxin-producing Escherichia coli (STEC) infections notified to the Norwegian Surveillance System for Communicable Diseases (MSIS), 2007–2017 (N = 1458), and the number of HUSa cases (purple line, N = 67). The time periods when the majority of clinical medical laboratories in Norway introduced PCR detection of stx and implemented broad screening PCR in five of the laboratories are indicated with a black and grey arrow, respectively
The estimated annual STEC notification rate increased from 0.6 cases per 100,000 population in 2007 to 7.6 in 2017. In children (< 16 years of age), the estimated annual notification rate increased from 1.3 cases per 100,000 population in 2007 to 14.0 in 2017. In children ≤ 5 years of age, the estimated annual notification rate increased from 2.9 cases per 100,000 population in 2007 to 28.9 in 2017.
The NRL at NIPH received sample material (isolate or a mixed culture positive for stx) for 1135 (78%) of the notified cases including 99% of the cases with a high-virulent, 87% of the cases with a low-virulent and 30% of the cases with an unclassifiable STEC infection. The proportion of cases with sample material received decreased over the study period, from 96% (324/339) in the years 2007–2012 to 72% (811/1119) in 2013–2017. The lowest yearly proportion was recorded in 2017 (64%, 260/405).
Comparing cases with high-virulent versus low-virulent STEC infection
Demographic and clinical data
We observed a difference in the age distribution between cases with high-virulent and low-virulent STEC infections, with an estimated median age of 5 years (range 0–97 years) compared to 22 years (range 0–93 years) respectively in the two groups (p < 0.001). Furthermore, we identified a difference in seasonality between the two groups with a higher proportion of cases with high-virulent STEC infections during summer (36% vs 29%) and less during winter (14% vs 21%) (p = 0.008). No difference was observed between the two groups in terms of distribution of sex or place of infection (domestically acquired or infected abroad). Cases with high-virulent STEC infection were more frequently reported as hospitalised than cases with a low-virulent infection (42% vs 21%, p < 0.001) (Table 1).
Table 1.
Demographic and clinical characteristics of cases categorised with high-virulent (number of cases; N = 475) versus low-virulent (N = 652) Shiga toxin-producing Escherichia coli (STEC) infections, notified to the Norwegian Surveillance System for Communicable Diseases (MSIS), Norway, 2007–2017 (N = 1127)
| Variable | Category | All casesa (N = 1127) |
High-virulent STEC casesa (N = 475) |
Low-virulent STEC casesa (N = 652) |
Chi-square p valueb |
|||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |||
| Sex | Female | 585 | 52 | 258 | 54 | 327 | 50 | 0.167 |
| Male | 542 | 48 | 217 | 46 | 325 | 50 | ||
| Age group | ≤ 5 | 458 | 41 | 242 | 51 | 216 | 33 | < 0.001 |
| 6–15 | 101 | 9 | 39 | 8 | 62 | 9 | ||
| 16–41 | 295 | 26 | 108 | 23 | 187 | 29 | ||
| 42–64 | 163 | 14 | 48 | 10 | 115 | 18 | ||
| 65+ | 110 | 10 | 38 | 8 | 72 | 11 | ||
| Seasonality | Winter | 204 | 18 | 66 | 14 | 138 | 21 | 0.008 |
| Spring | 168 | 15 | 71 | 15 | 97 | 15 | ||
| Summer | 360 | 32 | 170 | 36 | 190 | 29 | ||
| Autumn | 395 | 35 | 168 | 35 | 227 | 35 | ||
| Reported infected abroad | No | 747 | 74 | 332 | 76 | 415 | 73 | 0.255 |
| Yes | 260 | 26 | 105 | 24 | 155 | 27 | ||
| Hospitalised | No | 750 | 71 | 260 | 59 | 490 | 79 | < 0.001 |
| Yes | 311 | 29 | 182 | 41 | 129 | 21 | ||
aThe numbers and proportions reported per column for each characteristic use the number of cases with available (known) information regarding each characteristic
bA p value of ≤ 0.05 (italicised) was considered statistically significant
Microbiological characteristics
Subtype of stx was available for 85% (403/475) of the cases with high-virulent STEC and for 82% (532/652) of the cases with low-virulent STEC infections. In the former group, the most commonly identified subtypes were stx2a (224/403; 56%) and Stx2c (157/403; 39%), whereas stx1a (278/532; 52%) and stx2b (159/532; 30%) were more frequently seen in the low-virulent group (Online Resource 1). Furthermore, serogroups O157 (43% vs 1%), O145 (15% vs 5%) and O26 (17% vs 9%) were more commonly identified in cases with high-virulent STEC infection than in the low-virulent group, while the opposite was observed for serogroup O103 (4% vs 23%) (p < 0.001). Additionally, virulence genes eae and ehxA were more prevalent in the high-virulent group (87% vs 51%, p < 0.001 and 77% vs 51%, p < 0.001, respectively) (Table 2).
Table 2.
Microbiological characteristics of cases categorised with high-virulent (number of cases; N = 475) versus low-virulent (N = 652) Shiga toxin-producing Escherichia coli (STEC) infections, notified to the Norwegian Surveillance System for Communicable Diseases (MSIS), Norway, 2007–2017 (N = 1127)
| Characteristics | Category | All casesa (N = 1127) |
High-virulent STEC casesa (N = 475) |
Low-virulent STEC casesa (N = 652) |
Chi-square p valueb |
|||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |||
| Serogroup distribution | O157 | 195 | 20 | 192 | 43 | 3 | 1 | < 0.001 |
| O103 | 136 | 14 | 16 | 4 | 120 | 23 | ||
| O26 | 126 | 13 | 78 | 17 | 48 | 9 | ||
| O145 | 96 | 10 | 67 | 15 | 29 | 5 | ||
| Other | 418 | 43 | 96 | 21 | 322 | 62 | ||
| eae c | Positive | 710 | 67 | 406 | 87 | 304 | 51 | < 0.001 |
| Negative | 352 | 33 | 60 | 13 | 292 | 49 | ||
| ehxA d | Identified | 700 | 62 | 365 | 77 | 335 | 51 | < 0.001 |
| Not identified | 427 | 38 | 110 | 23 | 317 | 49 | ||
aThe numbers and proportions reported per column for each characteristic use the number of cases with available (known) information regarding each characteristic
bA p value of ≤ 0.05 (italicised) was considered statistically significant
cIntimin-encoding gene (Escherichia coli attaching and effacing)
dEnterohaemolysin-encoding gene (enterohaemolysin)
Implementation of broad screening PCR
Five medical microbiological laboratories implemented broad screening PCR during the study period. The different laboratories implemented broad screening PCR on the following dates: November 1st 2013, June 1st 2014, March 16th 2015, August 4th 2015 and April 1st 2017. The second laboratory had no record of notified STEC cases prior to 2013 and was therefore excluded from the time series analysis. The remaining 17 medical microbiological laboratories in Norway did not implement broad screening PCR during the study period.
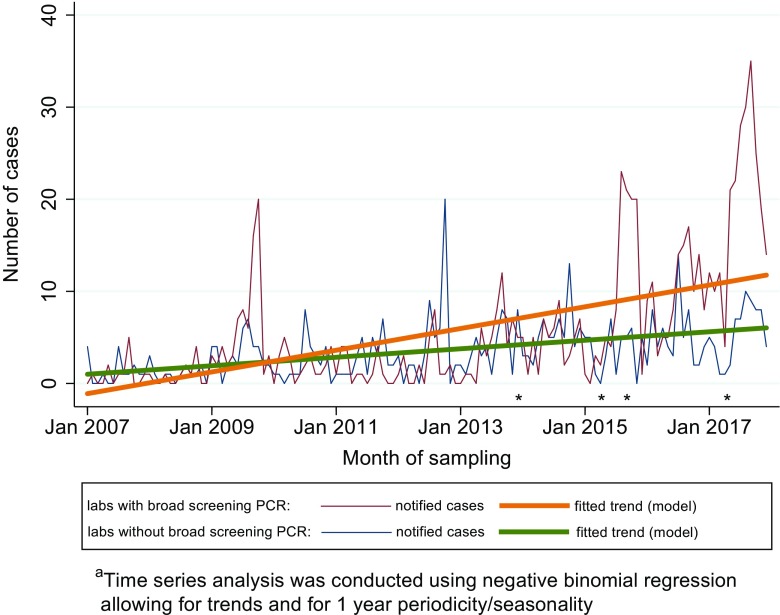
Adjusted for 1-year periodicity (significant in both models; sine-wave p < 0.001, cosine-wave p < 0.001), we observed a higher increasing monthly trend in STEC cases (aIRR = 1.020; 95% CI 1.016–1.024) notified from the four laboratories that had implemented broad screening PCR, compared to laboratories that had not implement this method (aIRR = 1.011; 95% CI 1.007–1.014, non-overlapping confidence intervals) (Fig. 2).
Fig. 2.
Monthly distribution of notified Shiga toxin-producing Escherichia coli (STEC) cases with fitted trend based on time series analysis modela for the four medical microbiological laboratories that implemented broad screening PCR (N = 728 cases) and for the 17 laboratories that did not implement broad screening PCR (N = 461 cases), Norway, 2007–2017. The different time points that the four laboratories started implementing broad screening PCR are marked with an asterisk (*)
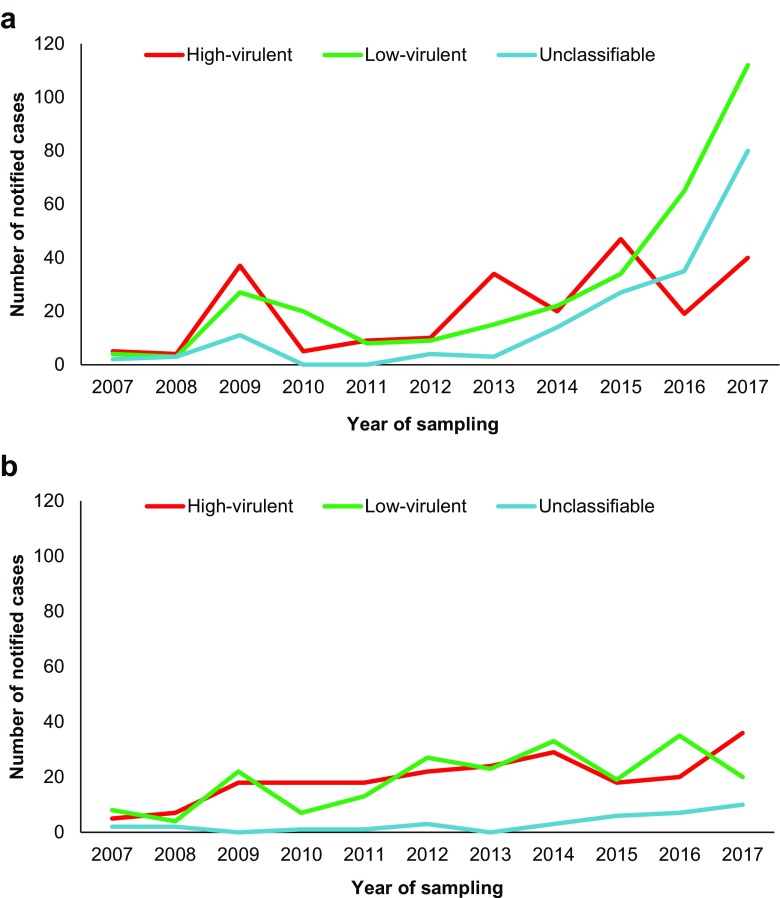
The difference in annual number of cases categorised as high-virulent, low-virulent or unclassifiable STEC infections was assessed in laboratories with and without broad screening PCR (Fig. 3). Throughout the study period, laboratories that implemented broad screening PCR notified 260 (26%) cases with high-virulent, 441 (44%) cases with low-virulent and 296 (30%) cases with unclassifiable STEC infections, while the remaining laboratories notified 215 (47%), 211 (46%) and 35 (8%) cases in the different categories, respectively (Online Resource 2). From 2013, when the first laboratory introduced broad screening PCR, there was an increase in the proportion of cases with low-virulent and unclassifiable STEC infections notified from these laboratories. In the laboratories without broad screening PCR, the distribution of categorised cases was relatively stable throughout the study period. In 2017, the proportion of cases with high-virulent STEC infection was 17% in cases reported from laboratories with broad screening PCR, and 55% in cases reported from laboratories without broad screening PCR. Before 2013, both groups of laboratories had comparable distribution of notified cases categorised as high-virulent, low-virulent or unclassifiable STEC infections.
Fig. 3.
Annual distribution of cases notified and categorised with high-virulent, low-virulent or unclassifiable Shiga toxin-producing Escherichia coli (STEC) infections from a four laboratories that implemented broad screening PCR (N = 728) and b 17 laboratories that did not (N = 461), Norway, 2007–2017
Concomitant bacterial infections
During the study period, 997 STEC cases were notified from the five laboratories that had implemented broad screening PCR. For 115 (12%) of these cases, one or more concomitant enteropathogenic bacteria were identified. An increasing proportion of cases with concomitant bacteria was observed after the introduction of broad screening PCR (15%) compared to before (7%). In 2017, concomitant bacteria were detected in 15% (51/339) of all notified STEC cases, which were 44% of all concomitant bacteria identified during the study period. After the implementation of broad screening PCR, concomitant bacteria were identified in 11 (9%) cases with high-virulent, 26 (8%) cases with low-virulent and 59 (23%) cases with unclassifiable STEC infections. The most common groups of concomitant bacteria were Campylobacter spp. (37%, 43/115 cases), followed by atypical enteropathogenic Escherichia coli (aEPEC) (31%, 36/115), and Salmonella spp. (12%, 14/115).
Discussion
In this study, we have investigated the increase of notification of STEC cases to MSIS in Norway from 2007 to 2017. We observed an overall increase throughout the study period, with annual number of STEC notifications relatively stable until 2014, followed by a sharp increase in the following years. A similar observation was seen in our neighbouring countries, Sweden and Denmark [13, 14]. Furthermore, an overall steady increase of notified STEC cases has been reported in Europe and the US over several years [15, 16], mainly due to identification of non-O157 STEC infections [16, 17]. The 2016 notification rate in Norway (4.6 per 100,000 population) was the fourth highest reported in the European Union summary report, following Ireland, Sweden and Switzerland (15.6, 6.5 and 5.5, respectively) [15].
The recent upsurge in Norway coincided with the implementation of broad screening PCR for enteropathogenic bacteria at five of the larger medical microbiological laboratories in the country. This was prominent in cases categorised with a low-virulent STEC infection. In 2016 and 2017, the number of notified cases with low-virulent STEC was more than double than that of high-virulent STEC infections, largely attributed to cases notified from laboratories that had implemented broad screening PCR during the study period. Meanwhile, the annual distribution of cases categorised as high- or low-virulent STEC infections was quite stable during the study period in the remaining 17 laboratories. In addition, the annual number of notified HUS cases remained stable. HUS surveillance can be used to monitor STEC occurrence [15, 18, 19]. Based on this, a previous study on paediatric HUS and STEC in Norway strongly suggested an underestimation of STEC incidence [20]. From this, one would expect an increased detection rate of low-virulent STEC in Norwegian laboratories when implementing unselected screening, as seen in our study. Other reports have shown similar effect; a study from Denmark noted an 88% increase of STEC in an associated laboratory after implementation of non-selective stool screening [17]. This likely reflects both the effect of a broader diagnostic approach and improved detection for non-O157 STEC over the last decade [16, 17, 21]. A moderate increase of STEC cases notified to MSIS was also observed in laboratories without broad screening PCR diagnostics, probably due to implementation of selective PCR methods for detection of STEC following the 2006 outbreak [10, 22].
Furthermore, we observed a marked increase in notified cases without identification of toxin subtype, which is also reflected by trends reported in the European surveillance data [15]. The majority of these cases were categorised with unclassifiable STEC infections in our study. Mostly, these were stx1/2 positive and culture negative, which is a common finding in culture-independent (PCR-based) STEC detection [23]. Such cases are often associated with high stx CT values, which may suggest the presence of non-viable STEC [8], or represent identification of stx from free temperate bacteriophages [23]. Other bacteria, such as Escherichia albertii and Citrobacter freundii, may also carry stx [24, 25]. Furthermore, the unclassifiable STEC infections were highly represented in cases where concomitant enteropathogenic bacteria had been notified, predominantly in laboratories that had implemented broad screening PCR. Studies have shown that concomitant enteropathogens occur frequently in stx-positive samples compared to samples with other common enteropathogens [8, 26]. These are all potential sources of notified stx findings that may not yield positive cultures, and likely contribute to the increase of cases with unclassified STEC infections when non-selective screening is applied. In addition, the prevalence of STEC or stx in healthy carriers is mostly unknown, but important to consider when evaluating the clinical impact of a stx-positive finding. A recent study reported an incidence rate of STEC infection in asymptomatic adults as high as 84.2 per 100,000 population [27]. Interestingly, many of these STEC belonged to O serogroups that were untypeable or rarely found in symptomatic patients and > 80% were eae negative. The stx-positive but culture-negative STEC cases pose a growing challenge to the STEC surveillance system, as no cultures are available to the national reference laboratories for molecular characterisation and cluster detection. In Norway, most of these cases would require to be followed up as a probable high-virulent STEC infection until three consecutive stool samples are negative or a positive culture can confirm a low-virulent STEC [12]. Consequently, there is an ongoing debate regarding the increasing workload related to cases with unclassifiable STEC infections. As the differentiation of STEC is predominantly based on stx subtype, standardised subtyping directly from DNA obtained from enriched broth from positive samples rather than STEC isolates could improve subtype determination rates regardless of culture yield [8]. More specific methods, such as microfiltration of samples, have also been suggested to avoid interference of free bacteriophages in STEC identification [23]. Regardless, studies assessing the clinical relevance of such cases are needed.
Broad screening PCR provide fast and sensitive identification and allow for rapid exclusion of possible enteropathogens [26, 28]. However, laboratories using broad screening PCR methodology test stool samples against a panel of common enteropathogenic bacteria instead of a selective diagnostic approach based on clinical assessment. Consequentially, they contribute to higher identification rates of both primary enteropathogens and concomitant bacteria [26, 28, 29]. Higher identification rates contribute to an increased socioeconomic burden for public health services, those directly affected, and the society [8, 9, 30]. According to our findings, a large proportion of STEC infections can effectively be categorised as low-virulent, thus largely decreasing the number of cases in need of strict follow-up and control measures. We consider this an important and necessary response to the constant improvement of STEC detection methods. Others have suggested more drastic measures, such as reserving multiplexed panels to specific patient populations to improve test utilisation [28].
There are multiple broad screening PCRs commercially available and medical microbiological laboratories in Norway are autonomous in their choice of diagnostic methodology for STEC infections. This can lead to variability in the capacity to detect different STEC between laboratories as the methodologies differ in sensitivity. In addition, the laboratories are not required to inform the NRL of any changes in diagnostic methodologies, including implementation of broad screening PCR. While the increase of STEC cases following the introduction of broad screening PCR can be observed in the number of notified cases, the exact date of implementation was unknown for one laboratory. Although we contacted the laboratory to confirm the date of introduction, it could only provide the month of implementation. This may have resulted in minimal errors in the grouping of cases pre- or post-introduction of broad screening PCR.
Conclusions
The increase in notified STEC cases in Norway from 2007 to 2017 is largely attributable to implementation of broad screening PCR at five of the larger medical microbiological laboratories in the country. The increase was prominent in cases categorised with a low-virulent STEC infection. We recommend NIPH to maintain differentiated control measures for STEC cases to avoid follow-up of low-virulent STEC infections. We recommend microbiological laboratories in Norway to consider a more cost-effective broad screening PCR strategy that enables differentiation of high-virulent STEC infections.
Electronic supplementary material
(DOCX 29 kb)
Acknowledgments
We would like to acknowledge the clinicians, laboratory workers and secretaries of the medical microbiological laboratories included and the implicated staff at the NRL and MSIS at the NIPH who contribute invaluably to the continuous identification and surveillance of STEC in Norway.
Abbreviations
- aEPEC
Atypical enteropathogenic Escherichia coli
- E. coli
Escherichia coli
- eae
E. coli attaching and effacing
- ehxA
Enterohaemolysin
- HUS
Haemolytic-uraemic syndrome
- MLVA
Multiple-locus variable-number of tandem repeat analysis
- MSIS
The National Surveillance System for Communicable Diseases
- NIPH
Norwegian Institute of Public Health
- NRL
National Reference Laboratory for Enteropathogenic Bacteria
- NSF
Non-sorbitol fermenting
- SF
Sorbitol fermenting
- Stx
Shiga toxin
- STEC
Shiga toxin-producing E. coli
- STEC-LST
STEC who have lost their toxin(s)
- VTEC
Verocytotoxigenic E. coli
Authors’ contributions
LTB and UN had the project idea. LTB led the work on the methodological and structural design. GRJ wrote the protocol, assembled and processed the data, wrote the initial manuscript draft and led the manuscript writing and submission process. LVe and GRJ conducted the statistical analysis. All authors participated in the work on the methodological and structural design, participated in the design and interpretation of the data, reviewed literature, revised the manuscript critically with important conceptual contributions and approved the final version of the manuscript.
Funding
This study was funded by the Norwegian Institute of Public Health.
Ethical approval and informed consent
All STEC cases and strains are routinely notified to and collected at the NIPH for disease surveillance, outbreak detection and further characterisation, respectively. This study is based on data from notifications to MSIS and bacterial isolates from the strain collection at the NRL. We present summarised data on demographic, clinical and surveillance aspects. The Norwegian Communicable Disease Control Act and its companying regulations oblige the NIPH to perform national surveillance of communicable diseases, including STEC infections. In accordance with this, the present study and its potential findings were considered as assessment of the surveillance and guidelines provided by the NIPH. This qualifies as quality control of one of the imposed tasks of the NIPH. Accordingly, ethical approval from a Regional Ethical Committee was not required and informed consent was not required from the patients involved.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2(1):15–38. doi: 10.1128/CMR.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fakhouri F, Zuber J, Fremeaux-Bacchi V, Loirat C. Haemolytic uraemic syndrome. Lancet. 2017;390(10095):681–696. doi: 10.1016/S0140-6736(17)30062-4. [DOI] [PubMed] [Google Scholar]
- 3.Brandal LT, Wester AL, Lange H, Lobersli I, Lindstedt BA, Vold L, Kapperud G. Shiga toxin-producing Escherichia coli infections in Norway, 1992-2012: characterization of isolates and identification of risk factors for haemolytic uremic syndrome. BMC Infect Dis. 2015;15:324. doi: 10.1186/s12879-015-1017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheutz F (2014) Taxonomy meets public health: the case of Shiga toxin-producing Escherichia coli. Microbiol Spectr 2(3). 10.1128/microbiolspec.EHEC-0019-2013 [DOI] [PubMed]
- 5.Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185(1):74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- 6.Naseer U, Lobersli I, Hindrum M, Bruvik T, Brandal LT (2017) Virulence factors of Shiga toxin-producing Escherichia coli and the risk of developing haemolytic uraemic syndrome in Norway, 1992-2013. Eur J Clin Microbiol Infect Dis. 10.1007/s10096-017-2974-z [DOI] [PMC free article] [PubMed]
- 7.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol. 2003;41(11):4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer RF, Ferdous M, Ott A, Scheper HR, Wisselink GJ, Heck ME, Rossen JW, Kooistra-Smid AM. Assessing the public health risk of Shiga toxin-producing Escherichia coli by use of a rapid diagnostic screening algorithm. J Clin Microbiol. 2015;53(5):1588–1598. doi: 10.1128/JCM.03590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norwegian Institute of Public Health (2017) E. coli-enteritis (including EHEC-infection and HUS) [In Norwegian]. http://www.fhi.no/artikler/?id=82709. Accessed 11 Nov 2017
- 10.Schimmer B, Nygard K, Eriksen HM, Lassen J, Lindstedt BA, Brandal LT, Kapperud G, Aavitsland P. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect Dis. 2008;8:41. doi: 10.1186/1471-2334-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne L, Vanstone GL, Perry NT, Launders N, Adak GK, Godbole G, Grant KA, Smith R, Jenkins C. Epidemiology and microbiology of Shiga toxin-producing Escherichia coli other than serogroup O157 in England, 2009-2013. J Med Microbiol. 2014;63(Pt 9):1181–1188. doi: 10.1099/jmm.0.075895-0. [DOI] [PubMed] [Google Scholar]
- 12.Norwegian Institute of Public Health (2016) Oppfølging av tilfeller med Shigatoksin (Stx) produserende Escherichia coli (STEC/EHEC) og hemolytisk-uremisk syndrom (HUS) i Norge [In Norwegian]. Norwegian Institute of Public Health,. https://www.fhi.no/globalassets/dokumenterfiler/usortert/oppfolging_av_ehecpasienter_2016.pdf. Accessed 11.11.2017 2017
- 13.The Public Health Agency of Sweden (2017) Enterohemorragisk E. coli infektion (EHEC) [In Swedish]. The Public Health Agency of Sweden,. https://www.folkhalsomyndigheten.se/folkhalsorapportering-statistik/statistikdatabaser-och-visualisering/sjukdomsstatistik/enterohemorragisk-e-coli-infektion-ehec/. Accessed 22.02.2018 2018
- 14.Statens Serum Institut (2018) VTEC - HUS, Individuelle anmeldelsespligtige sygdomme [In Danish]. Statens Serum Institut,. https://www.ssi.dk/Smitteberedskab/Sygdomsovervaagning/Sygdomsdata.aspx?sygdomskode=VTEC&xaxis=Aar&show=&datatype. Accessed 23.02.2018 2018
- 15.EFSA (European Food Safety Authority), ECDC (European Centre for Disease Prevention and Control) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017;15(12):5077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marder EP, Cieslak PR, Cronquist AB, Dunn J, Lathrop S, Rabatsky-Ehr T, Ryan P, Smith K, Tobin-D'Angelo M, Vugia DJ, Zansky S, Holt KG, Wolpert BJ, Lynch M, Tauxe R, Geissler AL. Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2013-2016. MMWR Morb Mortal Wkly Rep. 2017;66(15):397–403. doi: 10.15585/mmwr.mm6615a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen RM, Nielsen MTK, Moller S, Ethelberg S, Skov MN, Kolmos HJ, Scheutz F, Holt HM, Rosenvinge FS (2017) Shiga toxin-producing Escherichia coli: incidence and clinical features in a setting with complete screening of patients with suspected infective diarrhoea. Clin Microbiol Infect. 10.1016/j.cmi.2017.10.002 [DOI] [PubMed]
- 18.Espie E, Grimont F, Mariani-Kurkdjian P, Bouvet P, Haeghebaert S, Filliol I, Loirat C, Decludt B, Minh NN, Vaillant V, de VH. Surveillance of hemolytic uremic syndrome in children less than 15 years of age, a system to monitor O157 and non-O157 Shiga toxin-producing Escherichia coli infections in France, 1996–2006. Pediatr Infect Dis J. 2008;27(7):595–601. doi: 10.1097/INF.0b013e31816a062f. [DOI] [PubMed] [Google Scholar]
- 19.Kuehne A, Bouwknegt M, Havelaar A, Gilsdorf A, Hoyer P, Stark K, Werber D, HUSasn G. Estimating true incidence of O157 and non-O157 Shiga toxin-producing Escherichia coli illness in Germany based on notification data of haemolytic uraemic syndrome. Epidemiol Infect. 2016;144(15):3305–3315. doi: 10.1017/S0950268816001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenssen GR, Hovland E, Bjerre A, Bangstad HJ, Nygard K, Vold L. Incidence and etiology of hemolytic-uremic syndrome in children in Norway, 1999–2008—a retrospective study of hospital records to assess the sensitivity of surveillance. BMC Infect Dis. 2014;14:265. doi: 10.1186/1471-2334-14-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Cartter M, Tobin-D’Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Wolpert B, Henao OL. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2014. MMWR Morb Mortal Wkly Rep. 2015;64(18):495–499. [PMC free article] [PubMed] [Google Scholar]
- 22.Health CAfDaTi . Rapid response report: summary with critical appraisal. Ottawa (ON): CADTH Rapid Response Reports; 2015. Screening of Shiga-toxigenic Escherichia coli in clinical fecal samples: a review of diagnostic accuracy, clinical utility, cost-effectiveness and guidelines. [PubMed] [Google Scholar]
- 23.Martinez-Castillo A, Muniesa M. Implications of free Shiga toxin-converting bacteriophages occurring outside bacteria for the evolution and the detection of Shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol. 2014;4:46. doi: 10.3389/fcimb.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, Kaneko A, Isobe J, Yamaguchi K, Horikawa K, Gomes TA, Linden A, Bardiau M, Mainil JG, Beutin L, Ogura Y, Hayashi T. Clinical significance of Escherichia albertii. Emerg Infect Dis. 2012;18(3):488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt H, Montag M, Bockemuhl J, Heesemann J, Karch H. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect Immun. 1993;61(2):534–543. doi: 10.1128/iai.61.2.534-543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer RF, Ott A, Kesztyus B, Kooistra-Smid AM. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol. 2010;48(11):4140–4146. doi: 10.1128/JCM.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita-Ishihara T, Iyoda S, Iguchi A, Ohnishi M. Secondary Shiga toxin-producing Escherichia coli infection, Japan, 2010-2012. Emerg Infect Dis. 2016;22(12):2181–2184. doi: 10.3201/eid2212.160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binnicker MJ. Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness. J Clin Microbiol. 2015;53(12):3723–3728. doi: 10.1128/JCM.02103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luna-Gierke RE, Wymore K, Sadlowski J, Clogher P, Gierke RW, Tobin-D'Angelo M, Palmer A, Medus C, Nicholson C, McGuire S, Martin H, Garman K, Griffin PM, Mody RK. Multiple-aetiology enteric infections involving non-O157 Shiga toxin-producing Escherichia coli—FoodNet, 2001-2010. Zoonoses Public Health. 2014;61(7):492–498. doi: 10.1111/zph.12098. [DOI] [PubMed] [Google Scholar]
- 30.Toljander J, Dovarn A, Andersson Y, Ivarsson S, Lindqvist R. Public health burden due to infections by verocytotoxin-producing Escherichia coli (VTEC) and Campylobacter spp. as estimated by cost of illness and different approaches to model disability-adjusted life years. Scand J Public Health. 2012;40(3):294–302. doi: 10.1177/1403494811435495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 29 kb)