Abstract Abstract
Several new Arthrinium specimens were collected from various locations in Mediterranean and temperate Europe. A collection of the type species, A.caricicola, was obtained from dead leaves of Carexericetorum in Berlin. Sequences of four genetic markers, ITS, 28S rDNA, tef1 and tub2 were produced from almost all collections and analyzed with those available in public databases. Results are employed to support six new species: A.balearicum, A.descalsii, A.esporlense, A.ibericum, A.italicum and A.piptatheri. The type species, A.caricicola, is related to other species occurring on Carex sp.; these might represent an independent lineage from Apiospora and the remaining species of Arthrinium. Finally, the sexual morph of A.marii is described and illustrated for the first time.
Keywords: Apiosporaceae , Ascomycota , Sordariomycetes , Xylariales , ITS, 28S rDNA, tef1, tub2
Introduction
The genus Arthrinium Kunze (Apiosporaceae, Sordariomycetes) differs from other anamorphic genera because of the presence of basauxic conidiophores, which arise from structures called conidiophore mother cells (Schmidt and Kunze 1817; Hughes 1953; Minter 1985). This infrequent type of conidiogenesis can be found also in Cordella Speg., Dictyoarthrinium S. Hughes, Pteroconium Sacc. ex Grove, and Spegazzinia Sacc. (Ellis 1971), but Pteroconium and Cordella are now considered synonyms of Athrinium (Seifert et al. 2011; Crous and Groenewald 2013). Apiospora Sacc., the sexual state of Arthrinium, is also considered a synonym based on the one fungus-one name policy (Hawksworth et al. 2011; Crous and Groenewald 2013), and Nigrospora Zimm. is thought to be the closest relative (Wang et al. 2017).
There are about 80 valid species names of Arthrinium. The most significant contributions to species diversity of Arthrinium before the DNA-era were those of Schmidt and Kunze (1817), Kunze and Schmidt (1823), Fuckel (1870, 1874), Ellis (1963, 1965, 1971, 1976), and Larrondo and Calvo (1990, 1992). Genetic evidence allowed to confirm some of these taxa and propose multiple new species, e.g. Crous and Groenewald (2013), Singh et al. (2013), Dai et al. (2016, 2017), Jiang et al. (2018), and Wang et al. (2018). Smith et al. (2003) produced the first genetic data (18S and 28S rDNA) of A.phaeospermum (Corda) M.B. Ellis, supporting that this genus, as well as Apiospora, represent a separate family within Xylariales. This was later confirmed by Spatafora et al. (2006) and Zhang et al. (2006) who added new information from gene-coding DNA markers (18S and 28S rDNA, tef1, rpb2). Singh et al. (2013) published a ITS rDNA phylogeny including several type sequences obtained by Ogawa et al. (unpublished), such as those of A.marii Larrondo & Calvo, A.hispanicum Larrondo & Calvo, A.mediterranei Larrondo & Calvo, A.serenense Larrondo & Calvo, and A.phaeospermum, and introduced the new species A.rasikravindrae Shiv M. Singh, L.S. Yadav, P.N. Singh, Rah. Sharma & S.K. Singh (as rasikravindrii). Soon afterwards, Crous and Groenewald (2013) published a comprehensive re-evaluation of Arthrinium based on multigenic data, introducing eight new species and providing genetic data from several type strains of other taxa. They formally proposed the synonymy between Arthrinium and Apiospora, giving priority to Arthrinium, but provided no data of the type species, A.caricicola Kunze & J.C. Schmidt. Sharma et al. (2014) published the new species A.jatrophae R. Sharma, G. Kulk. & Shouche and built a phylogenetic tree based on rDNA that showed three main clades: one formed by A.urticae M.B. Ellis, a second including A.puccinioides Kunze & J.C. Schmidt and A.japonicum Pollack & C.R. Benj., and a third including the remaining known species of Arthrinium and Apiospora. Multigenic data of the first two clades was first obtained by Ogawa et al. (unpublished), and also Crous and Groenewald (2013), although they did not include these data in their phylogenetic analyses. Some new species of Arthrinium were described in the next years (Crous et al. 2015; Senanayake et al. 2015; Hyde et al. 2016; Dai et al. 2016, 2017; Wang et al. 2018; Jiang et al. 2018), and the multilocus phylogenetic analysis revealed that the sister clade of Arthrinium was Nigrospora in Apiosporaceae (Wang et al. 2017).
Morphological features traditionally employed to discriminate between species of Arthrinium include conidial shape, conidiophores, presence or absence of sterile cells and the presence of setae. Two great groups of species can be discriminated: 1) those with irregularly shaped conidia (including the type species A.cariciola and several others mainly associated with Carex spp. (Cyperaceae, Poales), such as A.austriacum Petr., A.fuckelii Gjaerum, A.globosum Koskela, A.japonicum, A.kamtschaticum Tranzschel & Woron., A.morthieri Fuckel, A.muelleri M.B. Ellis, A.naviculare Rostr., A.puccinioides and A.sporophleum Kunze), and 2) the remaining species with globose to ellipsoid conidia, mainly associated with other plants in the Poales (Cyperaceae, Poaceae, Restionaceae), e.g. A.pterospermum (Cooke & Massee) Arx, A.phragmitis Crous, A.sacchari (Speg.) M.B. Ellis, A.saccharicola F. Stevens, A.kogelbergense Crous, and A.hysterinum (Sacc.) P.M. Kirk, or even a wider diversity of potential hosts, such as A.arundinis (Corda) Dyko & B. Sutton, A.phaeospermum, A.rasikravindrae and A.malaysianum Crous.
Spatafora et al. (2006) and Zhang et al. (2006) were the first to obtain genetic data from the type species of Apiospora, Ap.montagnei Sacc. (CBS 212.30, AFTOL-ID 951) and suggested that it belongs in a distinct family within Xylariales. Sequences of a few other species of Apiospora are also available, including Ap.sinensis K.D. Hyde, J. Fröhl. & Joanne E. Taylor (HKUCC 3143 in Smith et al. 2003), Ap.setosa Samuels, McKenzie & D.E. Buchanan (ICMP 6888 /ATCC 58184 ex type PDD 41017 in Huhndorf et al. 2004), and Ap.tintinnabula Samuels, McKenzie & D.E. Buchanan (ICMP 6889-96 ex type PDD 41022 in Jaklitsch and Voglmayr 2012). Jaklitsch and Voglmayr (2012) produced a 28S rDNA phylogeny where the type species Ap.montagnei seemed not significantly different from Ap.sinensis but distinct from the other species sequenced. In addition, some Apiospora sexual morphs have been biologically linked with putatively prioritary Arthrinium taxa: A.hysterinum = Ap.bambusae (Turconi) Sivan. (Sivanesan 1983; Kirk 1986; Réblová et al. 2016), A.arundinis = Ap.montagnei (Hyde et al. 1998), and A.sinense = Ap.sinense (Réblová et al. 2016). However, none of these putative synonymies has been confirmed with genetic data, as some type collections are missing or too old for standard DNA analysis.
The aim of the present study was to study new Arthrinium samples found in temperate and southern Europe, including one specimen of A.caricicola and several putatively new species, and compare them morphologically and genetically with existing taxa. In some cases, e.g. Ap.tintinnabula, type collections were loaned and additional sequences obtained to delimit the genetic boundaries of some species.
Materials and methods
Pure culture isolation
During the surveys conducted in 2017 and 2018, 34 fresh specimens were collected from various plant hosts in Germany, Italy, Portugal and Spain. To isolate the sexual morph, ascomata were removed from the stromata using a sterile razor blade, transferred to a water droplet mounted on a microscope slide, torn apart with forceps to release the ascospores from asci, and pipetted on a 2% malt extract agar (MEA) plate supplemented with 200 mg/L penicillin G and streptomycin sulphate. Germinated ascospores were then transferred to MEA 2% plates, which were sealed with plastic film and incubated at room temperature. To isolate the asexual morph, plate cultures were superficially scrapped with a needle to dislodge conidia that were transferred to a drop of water. The suspension was then picked up with a syringe, and small droplets sown on a MEA 2% plate supplemented with 200 mg/L penicillin G and streptomycin sulphate. The germinated conidia were then transferred to 2% MEA plates, which were sealed with laboratory film and incubated at room temperature. Cultures were deposited at CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands (CBS).
Morphological observations
Hand sections of stromata or conidiomata were made using a razor blade and mounted in water on a microscope slide. Observations were made with a Zeiss Axioscop microscope using differential interference contrast (DIC), images were taken with a FLIR camera with A. Coloma open source software. Measurements were taken with FIJI ImajeJ software, reported with maximum and minimum values in parentheses, and the range representing the mean plus and minus the standard deviation, followed by the number of measurements in parentheses. For certain images of conidiophores, the image stacking software Zerene Stacker v. 1.04 (Zerene Systems LLC, Richland, WA, USA) was used. Morphological descriptions were based on cultures sporulating on 2% MEA medium at room temperature. The original specimens were deposited at the fungarium of the Real Jardin Botanico de Madrid (MA-Fungi).
DNA isolation, amplification and phylogenetic analyses
Total DNA was extracted from dry specimens employing a modified protocol based on Murray and Thompson (1980). PCR amplification was performed with the primers ITS1F and ITS4 (White et al. 1990; Gardes and Bruns 1993) for ITS region, while LR0R and LR5 (Vilgalys and Hester 1990; Cubeta et al. 1991) were used to amplify the 28S rDNA region, T1, Bt2a, and Bt2b (Glass and Donaldson 1995; O’Donnell and Cigelnik 1997) for the β-tubulin gene (tub2), and EF1-728F, EF1-983F and EF1-1567R (Rehner and Buckley 2005) for the translation elongation factor 1a (tef1) gene. PCR reactions were performed under a program consisting of a hot start at 95 °C for 5 min, followed by 35 cycles at 94 °C, 54 °C and 72 °C (45, 30 and 45 s respectively) and a final 72 °C step 10 min. PCR products were checked in 1% agarose gels, and positive reactions were sequenced with one or both PCR primers. Chromatograms were checked searching for putative reading errors, and these were corrected.
BLAST (Altschul et al. 1990) was used to select the most closely related sequences from INSDC public databases. Sequences came mainly from Crous and Groenewald (2013), Singh et al. (2013), Sharma et al. (2014), Crous et al. (2015), Senanayake et al. (2015), Dai et al. (2016, 2017), Hyde et al. (2016), Réblová et al. (2016), Jiang et al. (2018), and Wang et al. (2018), as well as Ogawa et al. (unpublished). Two distinct alignments were built in MEGA 5.0 (Tamura et al. 2011) and aligned with Clustal W with manual corrections: 1) a multigenic alignment including ITS, 28S rDNA, tub2 and tef1 data (without introns) from all Apiosporaceae and related families, and 2) a second alignment built with the same DNA markers (with introns) including only species related with A.sacchari (/saccharii clade). Introns were removed from tef1 and tub2, and GBlocks (Castresana 2000) was employed to remove 201 ambiguously aligned sites from ITS rDNA in the Apiosporaceae alignment, but not in the alignment of the /sacchari clade, in order to resolve this complex with all the phylogenetic signal available. The final alignment of the Apiosporaceae included five partitions with 217/461 (ITS rDNA), 229/846 (28S rDNA), 78/252 (tub2), 43/147 (tef1 EF1-728F to EF1-983F), and 76/413 (tef1 EF1-983F to EF1-1567R) variable sites, while the final alignment of the /sacchari clade had 35/535 (ITS rDNA), 18/837 (28S rDNA), 99/719 (tub2), 68/429 (tef1 EF1-728F to EF1-983F), and 4/407 (tef1 EF1-983F to EF1-1567R) variable sites. The aligned loci were loaded in PAUP* 4.0b10 (Swofford 2001) and subjected to MrModeltest 2.3 (Nylander 2004). Model GTR+G+I was selected and implemented in all partitions in MrBayes 3.2.6 (Ronquist and Huelsenbeck 2003), where a Bayesian analysis was performed (data partitioned, two simultaneous runs, six chains, temperature set to 0.2, sampling every 100th generation) until convergence parameters were met after about 3.43M generations (Apiosporaceae) or 0.9M (/sacharii clade), standard deviation having fell below 0.01. Finally, a full search for the best-scoring maximum likelihood tree was performed in RAxML (Stamatakis 2006) using the standard search algorithm (data partitioned, GTRMIX model, 2000 bootstrap replications). Significance threshold was set above 0.95 for posterior probability (PP) and 70% bootstrap proportions (BP).
Results
Phylogeny
The analysis of ITS, 28S rDNA, tef1 and tub2 data from the entire family Apiosporaceae (Fig. 1) produced a phylogeny with two main significantly supported clades: 1) composed of A.puccinioides, A.japonicum and newly sequenced specimens matching the species A.cariciola, A.curvatumvar.minus, and A.sporophleum, and 2) a second clade containing all other sequences of Arthrinium and Apiospora. Among the other specimens analyzed, some matched the genetic concept of A.hysterinum, A.phragmitis, A.arundinis, A.rasikravindrae, or A.marii. Five new lineages were also found, which are formally proposed as new taxa below.
Figure 1.
50% majority rule consensus phylogram obtained in MrBayes from 25725 trees after the analysis of ITS rDNA, 28S rDNA, tef1 and tub2 sequences (introns excluded) of the family Apiosporaceae. Nodes were annotated if supported by > 70% ML BP or > 0.95 bayesian PP, but non-significant support values are exceptionally represented inside parentheses. Bold names represent samples sequenced in the present study.
The analysis of ITS, 28S rDNA, tef1 and tub2 of the species around A.sacchari (/sacchari clade) (Fig. 2) showed that the clade of A.marii contains the types of A.hispanicum and A.mediterranei, but receives low overall support, maybe because of the incomplete data from these two species. Samples CBS 113535 and CBS 114803 were identified as A.marii too, but seem to represent an independent lineage.
Figure 2.
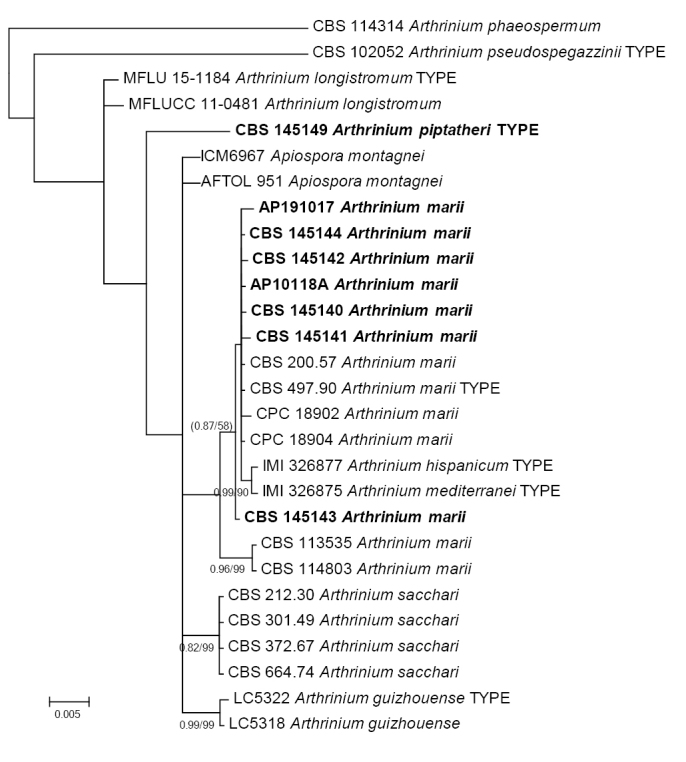
50% majority rule consensus phylogram obtained in MrBayes from 6750 trees after the analysis of ITS rDNA, 28S rDNA, tef1 and tub2 sequences (introns included) of the /sacchari clade. Nodes were annotated if supported by > 70% ML BP or > 0.95 bayesian PP, but non-significant support values are exceptionally represented inside parentheses. Bold names represent samples sequenced in the present study.
Table 1.
Details of strains included in this study. Types are in bold.
| Species | Isolate | CBS culture | Herbarium code | Host | ITS rDNA | 28S rDNA | tef1 | tub2 |
| A. arundinis | AP11118A | CBS 145128 | MA-Fungi 91722 | Bambusa sp. | MK014835 | MK014868 | MK017945 | MK017974 |
| A.balearicum, holotype | AP24118 | CBS 145129 | MA-Fungi 91723 | Undetermined poaceae | MK014836 | MK014869 | MK017946 | MK017975 |
| A. caricicola | AP23518 | CBS 145127 | MA-Fungi 91725 | Carex ericetorum | MK014838 | MK014871 | MK017948 | MK017977 |
| A.curvatumvar.minus. | AP25418 | CBS 145131 | MA-Fungi 91726 | hojas de Carex sp. | MK014839 | MK014872 | MK017949 | MK017978 |
| A.descalsii, holotype | AP31118A | CBS 145130 | MA-Fungi 91724 | Ampelodesmos mauritanicus | MK014837 | MK014870 | MK017947 | MK017976 |
| A.esporlense, holotype | AP16717 | CBS 145136 | MA-Fungi 91727 | Phyllostachys aurea | MK014845 | MK014878 | MK017954 | MK017983 |
| A. hysterinum | AP15318 | CBS 145132 | MA-Fungi 91728 | Phyllostachys aurea | MK014840 | MK014873 | MK017950 | MK017979 |
| ICMP6889 | Bambusa | MK014841 | MK014874 | MK017951 | MK017980 | |||
| AP29717 | CBS 145133 | MA-Fungi 91729 | Phyllostachys aurea | MK014842 | MK014875 | MK017952 | MK017981 | |
| AP2410173 | CBS 145134 | MA-Fungi 91730 | Phyllostachys aurea | MK014843 | MK014876 | |||
| AP12118 | CBS 145135 | MA-Fungi 91731 | Phyllostachys aurea | MK014844 | MK014877 | MK017953 | MK017982 | |
| A.ibericum, holotype | AP10118 | CBS 145137 | MA-Fungi 91732 | Arundo donax | MK014846 | MK014879 | MK017955 | MK017984 |
| A.italicum, holotype | AP221017 | CBS 145138 | MA-Fungi 91733 | Arundo donax | MK014847 | MK014880 | MK017956 | MK017985 |
| AP29118 | CBS 145139 | MA-Fungi 91734 | Phragmites australis | MK014848 | MK014881 | MK017957 | MK017986 | |
| A. marii | AP13717 | CBS 145140 | MA-Fungi 91735 | Arundo donax | MK014849 | MK014882 | MK017958 | MK017987 |
| AP10118A | Phragmites australis | MK014850 | MK014883 | MK017959 | MK017988 | |||
| AP11717A | CBS 145141 | MA-Fungi 91737 | Ampelodesmos mauritanicus | MK014851 | MK014884 | MK017960 | MK017989 | |
| AP191017 | MA-Fungi 91738 | Phragmites australis | MK014852 | MK014885 | MK017961 | MK017990 | ||
| AP261017 | CBS 145142 | MA-Fungi 91739 | Piptatheri miliaceum | MK014853 | MK014886 | MK017962 | MK017991 | |
| Vog2 | CBS 145143 | MA-Fungi 91740 | Phragmites australis | MK014854 | MK014887 | MK017963 | MK017992 | |
| AP31118 | CBS 145144 | MA-Fungi 91736 | Ampelodesmos mauritanicus | MK014855 | MK014888 | MK017964 | MK017993 | |
| A. phragmitis | AP281217A1 | CBS 145145 | MA-Fungi 91741 | Phragmites australis | MK014856 | MK014889 | MK017965 | MK017994 |
| AP2410172A | CBS 145146 | MA-Fungi 91742 | Phragmites australis | MK014857 | MK014890 | MK017966 | MK017995 | |
| AP3218 | CBS 145147 | MA-Fungi 91743 | Phragmites australis | MK014858 | MK014891 | MK017967 | MK017996 | |
| AP29717A | CBS 145148 | MA-Fungi 91744 | Arundo donax | MK014859 | MK014892 | MK017968 | MK017997 | |
| A.piptatheri, holotype | AP4817A | CBS 145149 | MA-Fungi 91745 | Piptatherum miliaceum | MK014860 | MK014893 | MK017969 | |
| A. puccinioides | AP26418 | CBS 145150 | MA-Fungi 91746 | Carex arenaria | MK014861 | MK014894 | MK017970 | MK017998 |
| A. rasikravindrii | AP8817 | CBS 145151 | MA-Fungi 91747 | Phyllostachys aurea | MK014862 | MK014895 | ||
| AP10418 | CBS 145152 | MA-Fungi 91748 | Phyllostachys aurea | MK014863 | MK014896 | MK017971 | MK017999 | |
| AP2410171 | CBS 145153 | Phyllostachys aurea | MK014864 | MK014897 | MK017972 | MK018000 | ||
| A. sporophleum | AP21118 | CBS 145154 | MA-Fungi 91749 | Juncus sp. | MK014865 | MK014898 | MK017973 | MK018001 |
Table 2.
Details of all strains included in the phylogenetic analyses. Sequences generated in this study are shown in bold.
| Species | voucher/culture | ITS rDNA | 28S rDNA | tub2 | tef1 |
| Apiospora setosa | ICMP 4207 | DQ368631 | DQ368620 | ||
| Apiospora tintinnabula | ICMP6889 | MK014841 | MK014874 | MK017951 | MK01980 |
| Arthrinium ‘vietnamense’ | IMI 99670 | KX986096 | KX986111 | KY019466 | |
| Arthrinium arundinis | CBS 106 12 | KF144883 | KF144927 | KF144973 | KF145015 |
| Arthrinium arundinis | CBS 145128 | MK014835 | MK014868 | MK017945 | MK017974 |
| Arthrinium arundinis | CBS 449 92 | KF144887 | KF144931 | KF144977 | KF145019 |
| Arthrinium arundinis | CBS 450 92 | AB220259 | KF144932 | KF144978 | KF145020 |
| Arthrinium arundinis | CBS 124788 | KF144885 | KF144929 | KF144975 | KF145017 |
| Arthrinium arundinis | CBS 133509 | KF144886 | KF144930 | KF144976 | KF145018 |
| Arthrinium arundinis | CBS 114316 | KF144884 | KF144928 | KF144974 | KF145016 |
| Arthrinium arundinis | CBS 464 83 | KF144888 | KF144933 | KF144979 | KF145021 |
| Arthrinium arundinis | CBS 732 71 | KF144889 | KF144934 | KF144980 | KF145022 |
| Arthrinium arureum | CBS 24483 | AB220251 | KF144935 | KF144981 | KF145023 |
| Arthrinium balearicum | CBS 145129 | MK014836 | MK014869 | MK017946 | MK017975 |
| Arthrinium camelliae-sinensis | LC8181 | KY494761 | KY494837 | KY705229 | KY705157 |
| Arthrinium camelliae-sinensis | LC5007 | KY494704 | KY494780 | KY705173 | KY705103 |
| Arthrinium caricicola | CBS 145127 | MK014838 | MK014871 | MK017948 | MK017977 |
| Arthrinium curvatum var. minus | CBS 145131 | MK014839 | MK014872 | MK017949 | MK017978 |
| Arthrinium descalsii | CBS 145130 | MK014837 | MK014870 | MK017947 | MK017976 |
| Arthrinium dichotomanthi | LC8175 | KY494755 | KY494831 | kY705223 | KY705151 |
| Arthrinium dichotomanthi | LC4950 | KY494697 | KY494773 | KY705167 | KY705096 |
| Arthrinium esporlense | CBS 145136 | MK014845 | MK014878 | MK017954 | MK017983 |
| Arthrinium euphorbiae | IMI 285638b | AB220241 | AB220335 | AB220288 | |
| Arthrinium garethjonesii | JHB004 | KY356096 | KY356091 | ||
| Arthrinium garethjonesii | HKAS 96289 | NR_154736 | NG_057131 | ||
| Arthrinium guizhouense | LC5322 | KY494709 | KY494785 | KY705178 | KY705108 |
| Arthrinium guizhouense | LC5318 | KY494708 | KY494784 | KY705177 | KY705107 |
| Arthrinium hydei | CBS 114990 | KF144890 | KF144936 | KF144982 | KF145024 |
| Arthrinium hydei | LC7103 | KY494715 | KY4947911 | KY705183 | KY705114 |
| Arthrinium hyphopodii | MFLUCC 15-003 | NR_154699 | |||
| Arthrinium hyphopodii | JHB003 Art | KY356098 | KY356093 | ||
| Arthrinium hysterinum | CBS 145133 | MK014842 | MK014875 | MK017952 | MK01981 |
| Arthrinium hysterinum | CBS 145135 | MK014844 | MK014877 | MK017953 | MK01982 |
| Arthrinium hysterinum | CBS 145132 | MK014840 | MK014873 | MK017950 | MK01879 |
| Arthrinium hysterinum | CBS 145134 | MK015843 | MK014876 | ||
| Arthrinium ibericum | CBS 145137 | MK014846 | MK014879 | MK017955 | MK017984 |
| Arthrinium italicum | CBS 145138 | MK014847 | MK014880 | MK017956 | MK017985 |
| Arthrinium italicum | CBS 145139 | MK014848 | MK014881 | MK017957 | MK017986 |
| Arthrinium japonicum | IFO30500 | AB220262 | AB220309 | AB220309 | |
| Arthrinium japonicum | IFO31098 | AB220264 | AB220311 | AB220311 | |
| Arthrinium jatrophae | CBS 134262 | NR_154675 | |||
| Arthrinium jatrophae | MMI00051 | AB743995 | |||
| Arthrinium jiangxiense | LC4577 | KY494693 | KY494769 | KY705163 | KY705092 |
| Arthrinium jiangxiense | LC4494 | KY494691 | KY494766 | KY705160 | KY705089 |
| Arthrinium kogelbergense | CBS 113332 | KF144891 | KF144937 | KF144983 | KF145025 |
| Arthrinium kogelbergense | CBS 113333 | KF144892 | KF144938 | KF144984 | KF145026 |
| Arthrinium kogelbergense | CBS 113335 | KF144893 | KF144939 | KF144985 | KF145027 |
| Arthrinium kogelbergense | CBS 117206 | KF144895 | KF144941 | KF144987 | KF145029 |
| Arthrinium longistromum | MFLU 15-1184 | NR_154716 | |||
| Arthrinium longistromum | MFLUCC 11-0481 | KU940141 | KU863129 | ||
| Arthrinium malaysianum | CBS 102053 | KF144896 | KF144942 | KF144988 | KF145030 |
| Arthrinium malaysianum | CBS 251.29 | KF144897 | KF144943 | KF144989 | KF145031 |
| Arthrinium marii | CPC 18902 | KF144901 | KF144948 | ||
| Arthrinium marii | CBS 145140 | MK014849 | MK014882 | MK017958 | MK017987 |
| Arthrinium marii | CBS 114803 | KF144899 | KF144945 | KF144991 | KF145033 |
| Arthrinium marii | CBS 113535 | KF144898 | KF144944 | KF144990 | KF145032 |
| Arthrinium marii | CBS 145141 | MK014851 | MK014884 | MK017960 | MK017989 |
| Arthrinium marii | CBS 145142 | MK014853 | MK014886 | MK017962 | MK017991 |
| Arthrinium marii | CBS 145143 | MK014854 | MK014887 | MK017963 | MK017992 |
| Arthrinium marii | CBS 145144 | MK014855 | MK014888 | MK017964 | MK017993 |
| Arthrinium mediterranei | IMI 326875 | AB220243 | AB220337 | AB220290 | |
| Arthrinium neosubglobosa | HKAS 96354 | NR_154737 | NG_057131 | ||
| Arthrinium neosubglobosa | JHB006 | KY356089 | KY356094 | ||
| Arthrinium obovatum | LC8177 | KY494757 | KY494833 | KY705225 | KY705153 |
| Arthrinium obovatum | LC4940 | KY494696 | KY494772 | KY705166 | KY705095 |
| Arthrinium ovatum | CBS 115042 | KF144903 | KF144950 | KF144995 | KF145037 |
| Arthrinium phaeospermum | CBS 114317 | KF144906 | KF144953 | KF144998 | KF145040 |
| Arthrinium phaeospermum | CBS 114318 | KF144907 | KF144954 | KF144999 | KF145041 |
| Arthrinium phaeospermum | CBS 114315 | KF144905 | KF144952 | KF144997 | KF145039 |
| Arthrinium phaeospermum | CBS 114314 | KF144904 | KF144951 | KF144996 | KF145038 |
| Arthrinium phragmitis | CBS 145145 | MK014856 | MK014889 | MK017965 | MK017994 |
| Arthrinium phragmitis | CBS 145146 | MK014857 | MK014890 | MK017966 | MK017995 |
| Arthrinium phragmitis | CBS 135458 | KF144909 | KF144956 | KF145001 | KF145043 |
| Arthrinium phragmitis | CBS 145147 | MK014858 | MK014891 | MK017967 | MK017996 |
| Arthrinium phragmitis | CBS 145148 | MK014859 | MK014892 | MK017968 | MK017997 |
| Arthrinium piptatheri | CBS 145149 | MK014860 | MK014893 | MK017969 | |
| Arthrinium pseudosinense | CBS 135459 | KF144910 | KF144957 | KF145044 | |
| Arthrinium pseudospegazzinii | CBS 102052 | KF144911 | KF144958 | KF145002 | KF145045 |
| Arthrinium pterospermum | CBS 123185 | KF144912 | KF144959 | KF145003 | |
| Arthrinium pterospermum | CBS 134000 | KF144913 | KF144960 | KF145004 | KF145046 |
| Arthrinium puccinioides | CBS 145150 | MK014861 | MK014894 | MK017970 | MK017998 |
| Arthrinium rasikravindrae | CBS 33761 | KF144914 | KF144961 | ||
| Arthrinium rasikravindrae | CBS 145151 | MK014862 | MK014895 | ||
| Arthrinium rasikravindrae | CPC 21602 | KF144915 | |||
| Arthrinium rasikravindrae | CBS 145152 | MK014863 | MK014896 | MK017971 | MK017999 |
| Arthrinium rasikravindrae | LC7115 | KY494721 | KY494797 | KY708159 | KY705118 |
| Arthrinium rasikravindrae | CBS 145153 | MK014864 | MK014897 | MK017972 | MK018000 |
| Arthrinium sacchari | CBS 30149 | KF144917 | KF144963 | KF145006 | KF145048 |
| Arthrinium sacchari | CBS 21230 | KF144916 | KF144962 | KF145005 | KF145047 |
| Arthrinium sacchari | CBS 66474 | KF144919 | KF144965 | KF145008 | KF145050 |
| Arthrinium sacchari | CBS 37267 | KF144918 | KF144964 | KF145007 | KF145049 |
| Arthriniumsaccharicola (1) | CBS 19173 | KF144920 | KF144966 | KF145009 | KF145051 |
| Arthriniumsaccharicola (1) | CPC 18977 | KF144923 | |||
| Arthriniumsaccharicola (2) | CBS 33486 | AB220257 | KF144967 | KF145010 | KF145052 |
| Arthriniumsaccharicola (2) | CBS 83171 | KF144922 | KF144969 | KF145012 | KF145054 |
| Arthriniumsaccharicola (2) | CBS 46383 | KF144921 | KF144968 | KF145011 | KF145053 |
| Arthrinium serenense | ATCC 76309 | AB220240 | AB220334 | AB220287 | |
| Arthrinium serenense | IMI 326869 | AB220250 | AB220344 | AB220297 | |
| Arthrinium sporophleum | CBS 145154 | MK014865 | MK014898 | MK017973 | MK018001 |
| Arthrinium subglobosa | MFLUCC 11-0397 | KR069112 | NG_057070 | ||
| Arthriniumsubglobosa (‘hyphopodii’) | MFLUCC 15-003 | KR069111 | |||
| Arthrinium subroseum | LC7292 | KY494752 | KY494828 | KY705220 | KY705148 |
| Arthrinium subroseum | LC7215 | KY494740 | KY494816 | KY705208 | KY705236 |
| Arthrinium thailandicum | LC5630 | KY494714 | KY494790 | KY806200 | KY705113 |
| Arthrinium thailandicum | MFLUCC 15-0202 | KU940145 | KU863133 |
Taxonomy
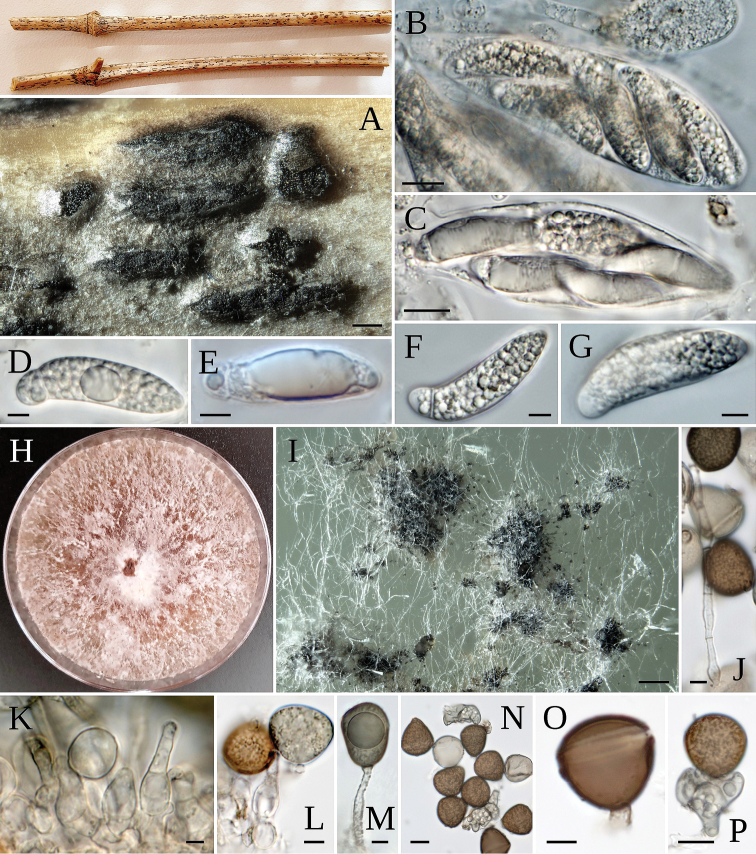
Arthrinium balearicum
Pintos & P. Alvarado sp. nov.
828866
Figure 3.
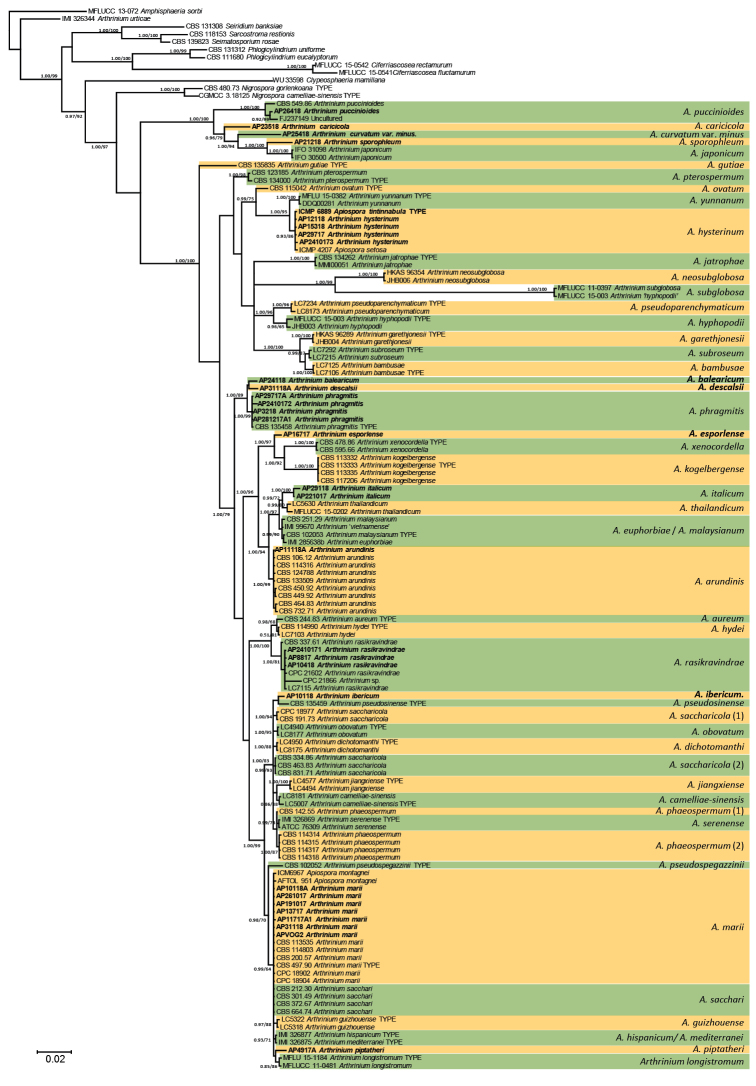
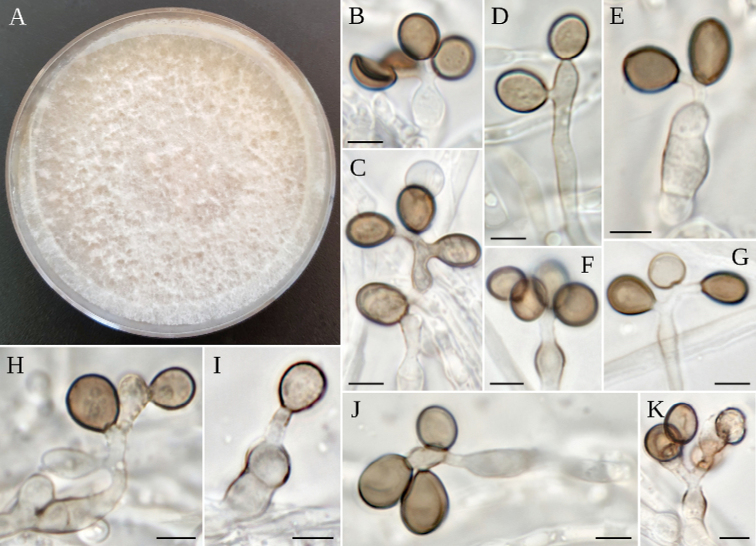
A.balearicumA stromata on host; B asci C–F ascospores G colony on MEA. Scale bars: 200 µm (A); 20 µm (B); 5 µm (C–F).
Etymology.
Refers to the Balearic Islands (Spain), where the holotype was found.
Diagnosis.
Sexual morph: Stromata forming black, linear, confluent raised areas on host surface, with the longer axis broken at the apex, (500–)600–1500(–2000) µm × (200–)320–450(–500) µm (n = 20). Ascomata globose to subglobose, with flattened base, blackish brown, (120–) 140–180 (–200) µm in diameter (n = 30). Peridium 8–15 µm thick, consisting of 4–5 layers of cells arranged in textura angularis, externally dark brown, hyaline in the inner part. Ostiole single, central, 30–60 µm in diameter, with a periphysate channel 20–30 µm long. Peryphises broad, colourless. Hamathecium composed of dense hypha-like, broad septate paraphyses, deliquescing early, 4–6 µm thick. Asci 8-spored, unitunicate, clavate, broadly cylindrical, with an inconspicuous pedicel, rounded apex, thin-walled, without an apical apparatus, measuring (77–)80–98(–105) × (14–)15–19(–21) µm (n = 22). Ascospores 1–3-seriate, hyaline, apiospore smooth-walled, fusiform, elliptical, reniform, straight or curved, bicellular, wider at the center of the longest cell, measuring (23–)26–30(–32) × (7–)9–10(–12) µm (n = 35), basal cell 3–6 µm long, sometimes containing a droplet. Asexual morph: not observed. Culture characteristics: colonies flat spreading on MEA 2%, with moderate aerial mycelium, reverse withish.
Type.
Spain: Balearic Islands: Mallorca, Llucmajor, on undetermined Poaceae, 24 Jan. 2018, A. Pintos (MA-Fungi 91723 holotype, AP24118 isotype, CBS 145129 ex-type culture).
Notes.
Arthriniumbalearicum is related with A.descalsii, but has some genetic differences with this species having only 93% (482/518 bp) of its ITS rDNA, 99% (821/823 bp) of 28S rDNA, 97% (688/707 bp) of tef1, and 98% (406/413 bp) of tub2 similar. It is also phylogenetically close to A.phragmitis, a species with a similar ascospore size, (23–)26–30(–32) × (7–)9–10(–12) µm in A.balearicum and (22–)23–28(–30) µm × (6–)7–9(–10) µm in A.phragmitis. Unfortunately, the asexual morph of A.balearicum could not be studied to compare it with that of A.phragmitis.
Arthrinium caricicola
Kunze & J.C. Schmidt, Mykologische Hefte (Leipzig) 1: 9 (1817)
Figure 4.
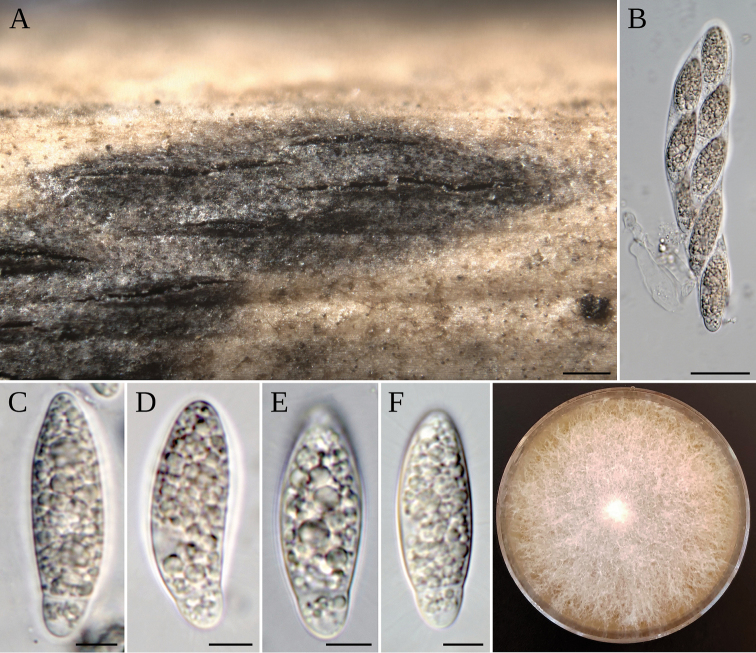
A.caricicolaA colony on host B colony on MEAC conidiophore mother cell D, E conidiophore mother cell, conidiophore bearing conidia, conidia F–H conidia I conidia with scar J lobate sterile cells. Scale bars: 200 µm (A); 5 µm (C–I); 10 µm (J). KA.caricicola syntype, colonies on host; L, M conidia.
Description.
Asexual morph: colonies on the host punctiform, pulvinate, 140–400 µm in diameter, blackish brown. Mycelium formed by hyaline smooth, branched hyphae, 2–5 µm in diameter. Conidiophore mother cells arising from a superficial or erumpent mycelial mat, subspherical to lageniform in shape, hyaline with brown pigments at the base, measuring (4–)5–7(–8) × (8–)9–11(–12) µm (n = 45). Conidiophores erect or ascending, simple, straight or flexuous, cylindrical, smooth-walled, colourless excepting for the thick, brown to dark brown, transversal septa, 15–100 × 3–5 µm (n = 50). Conidia fusiform or broadly spindle-shaped, smooth-walled, broader at the middle, tapering towards the narrowly rounded ends, dark brown with a hyaline rim, (37–)44–51(–55) µm in frontal view, (8–)9–11(–12) µm in side view (n = 50). Sterile cells smaller, 15–19 × 10–13 µm, and paler than conidia, bicuspidate or irregularly lobed. Culture characteristics: flat colonies spreading on MEA 2%, with moderately abundant, white cottony aerial mycelium, reverse whitish too, circular in shape with irregular edge.
Notes.
The conidia of A.caricicola and A.japonicum have a similar fusiform shape and length, but differ in width ((8–)9–11(–12) µm vs 12–16(–20) µm). Conidia of A.mytilimorphum have also a similar shape, but turns out shorter and thinner (20–30 × 6–8.5 µm). The morphological characters of the syntype of A.caricicola deposited by Fries in the Herbarium of Uppsala University as Fung. Scleromyc. Suecici, fully match the specimen collected in this study. The closely related species A.sporophleum has very different lemon-shaped conidia, while those of A.curvatumvar.minus are curved, and those of A.puccinioides are polygonal.
Specimens examined.
Germany: Brandenburg: south of Liberose, on dead leaves of Carexericetorum, 14 May 2018, R. Jarling (MA-Fungi 91725).
Arthrinium curvatum var. minus
M.B. Ellis, Trans. Brit. Mycol. Soc. 34: 501 (1951)
Figure 5.
A.curvatumvar.minusA colony on host B conidiophore mother cell C, D conidiophore mother cell, conidiophore bearing conidia E, F curved conidia G colony on MEA. Scale bars: 200 µm (A); 5 µm (B–F).
Physalospora scirpi Arx, Gen. Fungi Sporul. Cult. (Lehr): 116 (1970).
Pseudoguignardia scirpi Gutner, Mater. Mikol. Fitopat. Ross. 6(1): 311 (1927).
Description.
Asexual morph: Colonies are compact, round, dark to black, 80–320 in diameter. Mycelium is composed of hyaline to pale brown smooth hyphae 2–7 µm in diameter. Conidiophore mother cells spherical to lageniform, hyaline with brown pigments at the base, measuring (4–)5–7(–8) × (4–)5–6(–7) µm (n = 30). Conidiophores cylindrical unbranched, straight or flexuous, hyaline and smooth walled, with a single brown transversal septa, measuring 30–100 × 2–4 µm. (n = 30). Conidiogenous cells cylindrical 1–1.5 × 1–1.5 µm (n = 20). Conidia borne along the sides of conidiophores, curved, rounded at the ends, brown, with a hyaline germ slit and a clearly visible scar, (8–)9–10(–11) µm long in frontal view, (5–)6–7(–8) µm in side view (n = 30). Sterile cells rounded, paler than conidia. Culture characteristics flat colonies spreading on MEA 2% with moderate aerial mycelium, reverse withish.
Notes.
Arthriniumcurvatumvar.minus can be confused with A.curvatumvar.curvatum , but conidia of var. minus measure (8–)9–10(–11) × (5–)6–7(–8) µm, while those of A.curvatumvar.curvatum measure 11–15 × 6–8 µm. Gutner (1927) described Pseudoguignardiascirpi, a sexual morph of A.curvatum, later combined as Physalosporascirpi (Arx 1970). Arthriniumcurvatumvar.minus is closely related with A.sporophleum (with lemon-shaped conidia) and A.japonicum (with larger fusiform conidia) and to a lesser extent also with A.caricicola (with larger fusiform conidia) and A.puccinioides (with polygonal conidia). Ellis et al. (1951) described A.curvatumvar.minus, a taxon with similarly shaped but smaller conidia than A.curvatum. The specimen studied in the present work matches the shape and size of conidia reported by Ellis et al. (1951) for A.curvatumvar.minus, rather than those of A.curvatumvar.curvatum.
Specimens examined.
Germany: Brandenburg: south of Liberose, on dead leaves of Carex sp., 28 Mar. 2018, R. Jarling (MA-Fungi 91726).
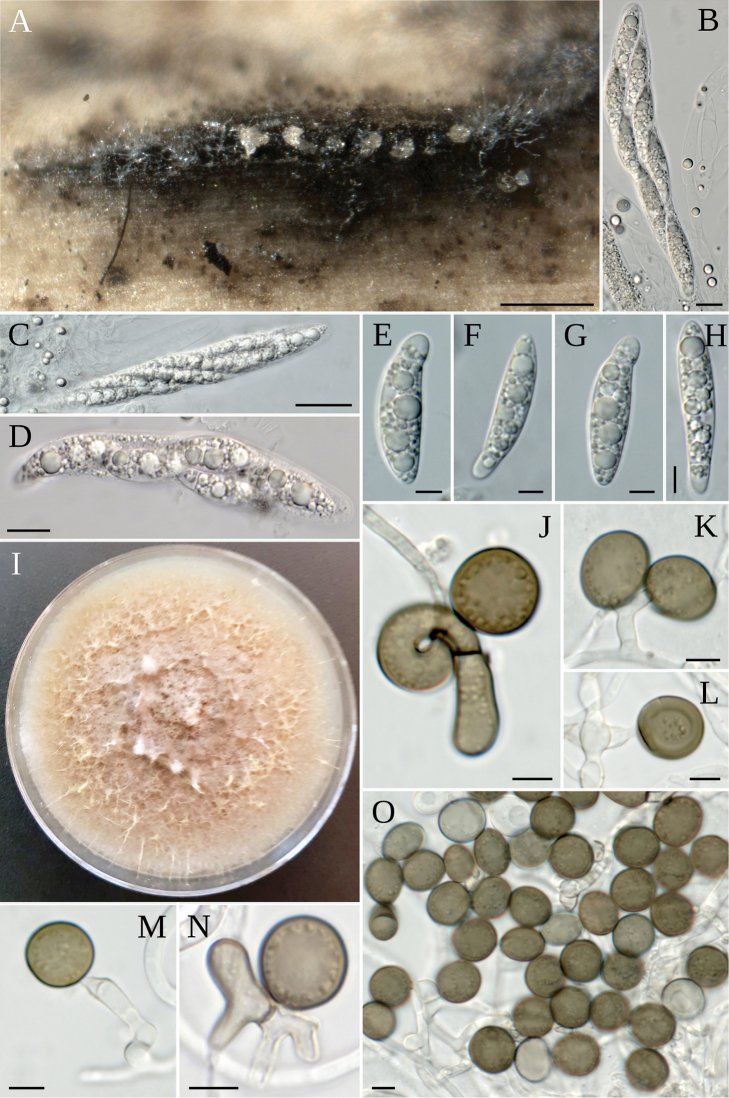
Arthrinium descalsii
Pintos & P. Alvarado sp. nov.
828867
Figure 6.
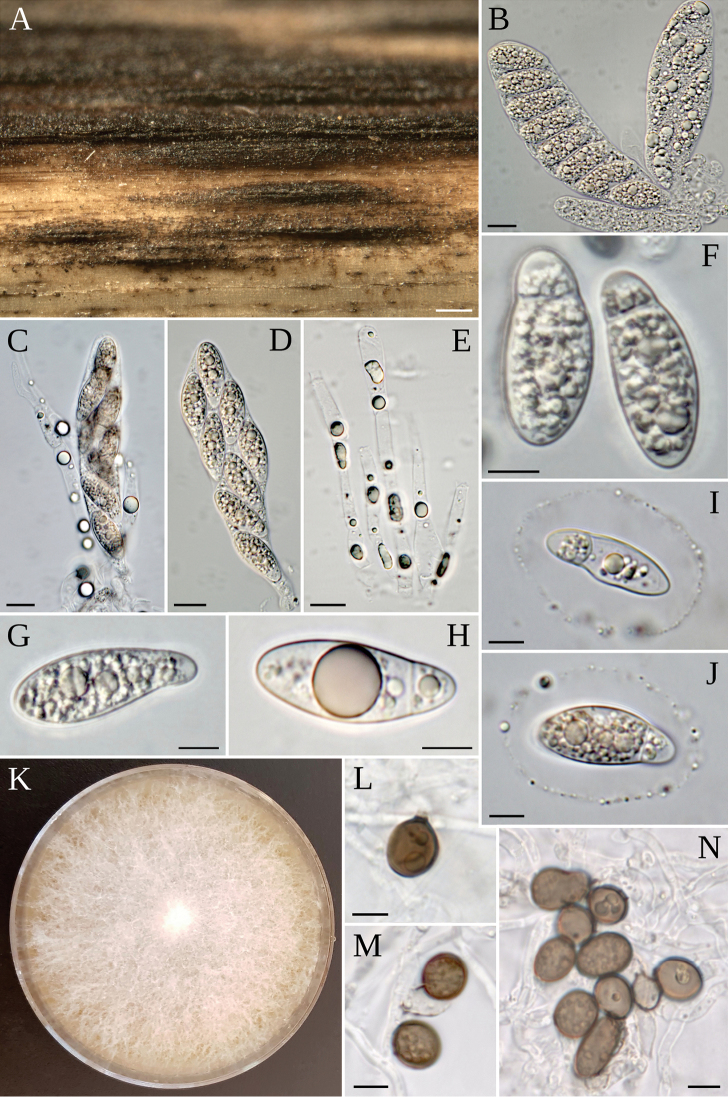
A.descalsiiA stromata on host B–D asci with ascospores E paraphyses F, G ascospores I, J ascospores with sheath K colony on MEA 2%; coniogenous cell giving rise to conidia; conidiogenous cells giving rise to conidia and conidia cluster G conidia. Scale bars: 200 µm (A); 10 µm (B–E); 5 µm (F–J); 5 µm (L–N).
Etymology.
Named to honor the eminent mycologist Enric Descals Callisen.
Diagnosis.
Sexual morph: Stromata forming black fusiform spots that merge with each other with age, forming an erumpent black mass visible at the naked eye, 2–10 × 0.2–0.5 mm in size, with the long axis broken at the top revealing the ostioles of pseudothecia. Ascomata pseudothecia, subglobose with a flattened base, arranged in rows, brown to dark brown, 150–220 μm high × 150–250 μm wide (n = 20). Peridium with several layers of cells arranged in textura angularis, with a conspicuous ostiole 50–80 μm in diameter, periphysate. Hamathecium paraphyses hyphae-like, septate, hyaline. Asci cylindrical, clavate, with a short or indistinct pedicel, with rounded apices, measuring (73–)82–95(–111) × (16–)17–20(–23) μm (n = 30). Ascospores uniseriate to biseriate, hyaline, smooth-walled, apiosporic, composed of a large curved upper cell and smaller lower cell, fusiform to slightly curved in shape with narrowly rounded ends, guttulated, sometimes with a thick gelatinous sheath, (17–)18–22(–24) × (6–)7–9(–10) μm, and a basal cell 3–5 μm (n = 45). Asexual morph: Mycelium hyaline, septate, branched, hyphae 1.5–4.5 μm in diameter Conidiophores reduced to the conidiogenous cells. Conidiogenous cells solitary on hyphae, ampuliform, hyaline to brown, 5 × 4 μm. Conidia brown, smooth, guttulate, globose to ellipsoid (5–)7(–8) µm long (n = 20) in face view, lenticular with a paler equatorial slit and 6-7 μm long in side view (n = 10). Sterile cells elongated, sometimes mixed among conidia. Culture characteristics: ascospores germinating on MEA 2% within 24–48 h. Colonies flat, spreading, with sparse aerial mycelium, pale siena.
Notes.
Arthriniumdescalsii is closely related with A.phragmitis and A.balearicum. It was found in the Mediterranean grass Ampelodesmosmauritanicus, although additional samples are needed before concluding if it could be exclusively associated with this endemic host. Ascospore size is often smaller than that of A.balearicum, (23–)26–30(–32) × (7–)9–10(–12) µm, but it matches that reported in the protologue of A.phragmitis, (20–)22–24(–25) × (7–)8–9(–10) µm. However, the conidiophores of A.descalsii are reduced to conidiogenous cells, while those of A.phragmitis measure about 10–45 × 1.5–2 µm, and conidia are slightly smaller in face view, measuring (5–)7(–8) µm long in A.descalsii and up to 8–10(–11) µm in A.phragmitis.
Type.
Spain: Balearic Islands: Mallorca, es Capdella, on dead stems of Ampelodesmosmauritanicus, 31 Jan. 2018, A. Pintos (MA-Fungi 91724 holotype, AP31118A isotype, CBS 145130 ex-type culture).
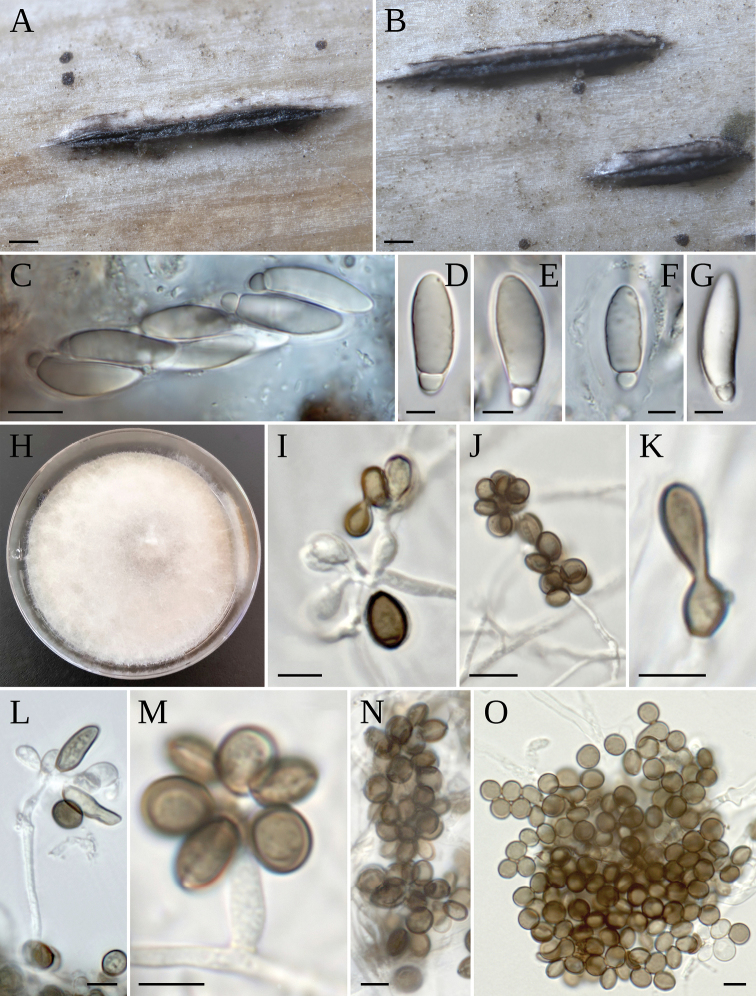
Arthirnium esporlense
Pintos & P. Alvarado sp. nov.
828868
Figure 7.
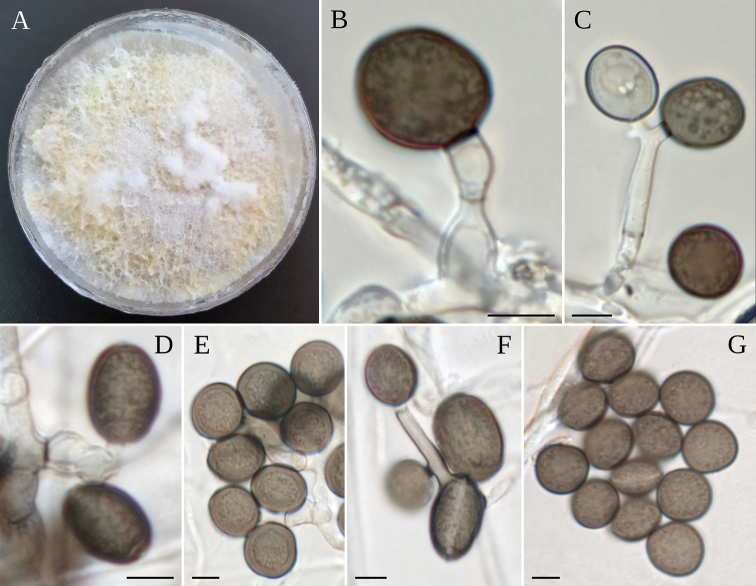
A.esporlenseA colony on MEAB–F coniogenous cell giving rise to conidia G conidia. Scale bars: 5 µm (B–G).
Etymology.
In reference to Esporles, the village of Mallorca (Spain) where it was found.
Diagnosis.
Asexual morph: Mycelium consisting of smooth, hyaline, branched septate hyphae about 1.5–4 µm in diameter. Conidiophores reduced to conidiogeous cells. Conidiogenous cells polyblastic, aggregated in clusters on hyphae, smooth, hyaline to pale brown, ampuliform, cylindrical or lageniform, measuring 4–22 × 4–8 μm. Conidia brown, smooth, globose with a pale equatorial slit and (8–)9–12(–13) µm long in frontal view, lenticular and 6–8 μm long in side view (n = 30). Sterile cells elongated, sometimes mixed among conidia, paler than them. Culture characteristics: colonies flat, spreading, with moderate aerial mycelium, on MEA 2% surface white with yellowish patches, reverse concolour with age.
Type.
Spain: Balearic Islands: Mallorca, Esporles, on dead culms of Phyllostachysaurea, 16 July 2017, A. Pintos (MA-Fungi 91727 holotype, AP16717 isotype, CBS 145136 ex-type culture).
Notes.
Arthriniumesporlense is closely related with A.xenocordella and A.kogelbergense. However, A.esporlense does not produce brown setae as A.xenocordella, a species until now known only from soil samples (Crous and Groenewald 2013). Arthriniumesporlense morphologically differs from A.kogelbergense by producing slightly bigger conidiogenous cells (4–22 × 4–8 μm vs 5–12 × 4–5 μm). These three species are genetically related (1.00 PP, 96 BP) to the group formed by A.arundinis, A.thailandicum D.Q. Dai & K.D. Hyde, A.malaysianum and the new species A.italicum proposed below.
Arthrinium hysterinum
(Sacc.) P.M. Kirk, Trans. Brit. Mycol. Soc. 86: 409 (1986)
Figure 8.
A.hysterinum lenticular-shaped colonies on host A stromata and conidiomata B, C asci D–G ascospores H colony on MEAI black masses of conidia in culture K, L conidiophore mother cell M rugose conidiogenous cell N–P conidia with lobate sterile cells O conidia. Scale bars: 200 µm (A); 10 µm (B, C); 5 µm (D–G); 200 µm (I); 5 µm (K, M, O); 10 µm (P).
Melanconium hysterinum Sacc., Bolm Soc. broteriana, Coimbra, sér. 1 11: 21 (1893) [Basionym].
Scyphospora hysterina (Sacc.) Sivan., Trans. Brit. Mycol. Soc. 81: 331 (1983).
Melanconium bambusae Turconi, Atti Ist. bot. R. Univ. Pavia, sér. 2 16: 251 (1916).
Scirrhia bambusae Turconi, Atti Ist. bot. R. Univ. Pavia, sér. 2 16: 531 (1916).
Scirrhodothis bambusae (Turconi) Trotter, in Saccardo, Syll. Fung. 24: 611 (1926).
Placostroma bambusae (Turconi) R. Sprague, Diseases Cereals Grasses N. Amer.: 121 (1950).
Apiospora bambusae (Turconi) Sivan., Trans. Brit. Mycol. Soc. 81: 331 (1983).
Scyphospora phyllostachydis L.A. Kantsch., Bolêz. Rast. 17: 88 (1928).
Cordella johnstonii M.B. Ellis, Mycol. Pap. 103: 31 (1965).
Apiospora setosa Samuels et al., New Zealand J. Bot. 19: 142 (1981).
Apiospora tintinnabula Samuels et al., New Zealand J. Bot. 19: 142 (1981).
Description.
Sexual morph: Stromata black, fusiform, forming rows of densely arranged perithecial ascomata parallel to the main axis of the host, measuring (400–) 600–2500(–3000) × (250–)320–450(–550) µm (n = 30). Ascomata globose to subglobose, with a flattened base, blackish brown, (130–)250–290(–320) µm in diameter (n = 30). Peridium consisting of 3 or 4 layers of cells arranged in textura angularis, dark brown in the external side, hyaline in the inside, ostiole single, central, 10–30 µm in diameter, with a periphysate channel 20–35 µm long. Peryphises broad, colourless. Hamathecium composed of dense hypha-like, broad septate paraphyses, early deliquescing. Asci 8-spored, unitunicate, clavate, broadly cylindrical, pedicel indistinct, apical rounded, thin-walled, without an apical apparatus, measuring (76–) 85–98(–115) × (20–)22–26(–28) µm (n = 22). Ascospores uni- to tri-seriate, hyaline, apiosporic, smooth-walled, fusiform, elliptical, reniform, straight or curved, smooth-walled, sometimes with an internal droplet, bicellular, the widest part located in the central part of the longest cell, some ascospores have a mucose sheath covering them, (28–)32–34(–38) × (8–)9–11(–13) (n = 35) µm, basal cell 5–7 µm. Asexual morph: Mycelium branched, septate. Conidiomata on host surrounding the stromata of the sexual phase, parallel to the longitudinal axis of the stem, subepidermal, opening by longitudinal splitting of the epidermis and revealing a black conidial mass, (450–) 630–950(–1000) × (275–)345–550 (–600) µm (n = 35). Conidiophore mother cell arising from the stroma, ampuliform, lageniform, cupulate or cylindrical, sometimes with granular pigments at the apex, (5)6–10(–16) × (3–)5–7(–8) µm (n = 24). Conidiophores basauxic, polyblastic, cylindrical, hyaline to light brown, smooth or with granular pigments in all their length, straight or flexuous, septate or not, sometimes exceeding 90 μm in length × 2–4 μm wide (n = 43). Conidia globose to obovoid, dark brown, with a central scar at the base, (15–)16–20(–21) in frontal view, (14–)15–18(–19) in side view (n = 40). Sterile cells gray, irregularly angled and lobed, (15–)17–41(–42) × (10–)14–23(–25) µm (n = 30). Culture characteristics: colonies in MEA 2% flat, spreading, first white and cottony, later became dark pink, mycelium branched, septate, hyaline, reverse dark.
Notes.
After the works of Samuels (1981), Sivanesan (1983), Kirk (1986) and Réblová et al. (2016), Ap.bambusae, Ap.setosa and Ap.tintinnabula, as well as Scyphosporaphyllostachydis, are all considered synonyms of A.hysterinum. Arthriniumhysterinum is phyllogenetically close to A.yunnanum D.Q. Dai & K.D. Hyde, but morphologically differs from the latter because of its thinner asci (76–115 × 20–28 vs 85–100 × 30–35 μm). In addition, A.hysterinum has longer conidiophores up to 90 μm long, and lobed sterile cells while in A.yunnanum conidiophores do not exceed 50 μm, and sterile cells are lacking.
Specimens examined.
New Zealand: Waikato: Paeroa, on dead culm of Bambusa sp., 28 Feb. 1980, E.H.C. McKenzie & P.R. Johnston (ICMP 6889 ex-type culture).
Spain: Galicia: Santiago de Compostela, on dead culms of Phyllostachysaurea, 12 Jan. 2018, A. Pintos (MA-Fungi 91731, AP12118). Balearic Islands: Mallorca, Esporlas, on dead culms of Phyllostachysaurea, 29 July 2017, A. Pintos (MA-Fungi 91729, AP29717). Mallorca, Jardin Botanico de Soller, on dead culms of Phyllostachysaurea, 24 Oct. 2017, A. Pintos (MA-Fungi 91730, AP2410173). Mallorca, Soller, on dead culms of Phyllostachysaurea, 15 Mar. 2018, A. Pintos (MA-Fungi 91728, AP15318).
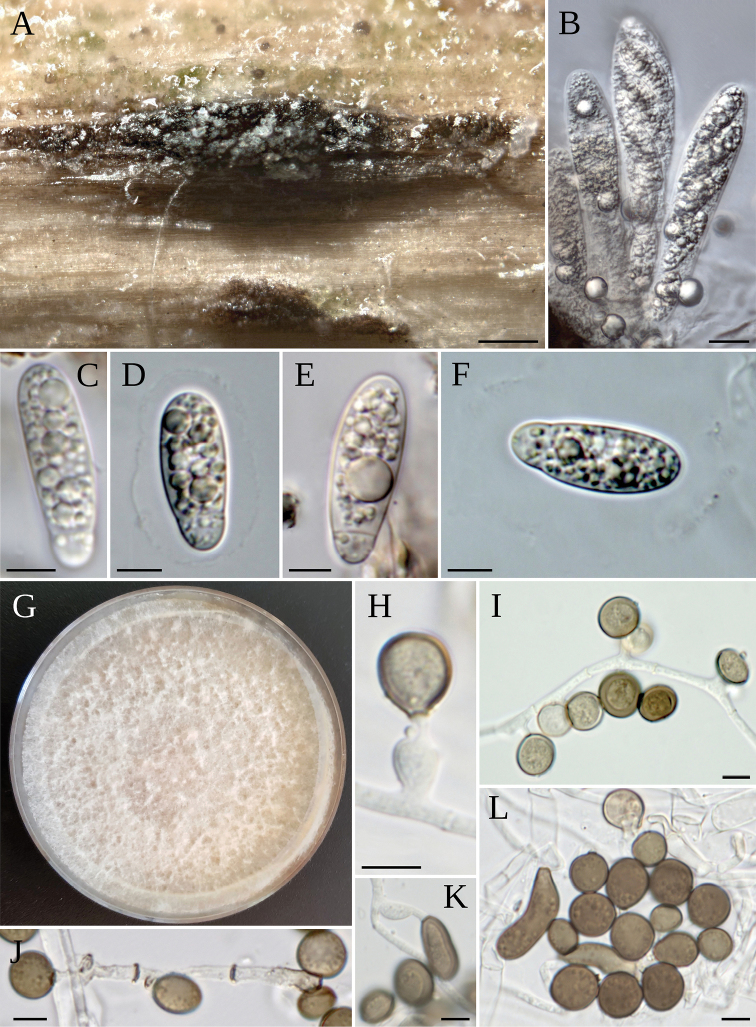
Arthrinium ibericum
Pintos & P. Alvarado sp. nov.
828869
Figure 9.
A.ibericumA ascomata with oozing ascospores B–D asci E–H ascospores I colony on MEAJ–M conidiogenous cells giving rise to conidia N sterile cell with conidia O conidia. Scale bars: 200 µm (A); 10 µm (B–D); 20 µm (C); 5 µm (E–H); 5 µm (J–O).
Etymology.
In reference to the Iberian Peninsula, where the holotype was collected.
Diagnosis.
Sexual morph: Stromata solitary to gregarious, immersed or semi-immersed, fusiform to ellipsoid in shape, black, with the long axis broken at the top, 2–5 × 0.5–1 mm. Ascomata perithecial, subglobose with a flattened base, arranged in rows, brown to dark brown, exudating a white cirrhus of ascospores, 170–300 µm in diameter and 200–300 µm high. Peridium consisting in 3 or 4 layers of cells arranged in textura angularis. Ostiole single, central, 12–30 µm in diameter, with a periphysate channel. Hamathecium composed of dense, septate, branched paraphyses. Asci 8-spored, clavate or cylindrical, lacking an apical apparatus, shortly pedicelate, measuring (82–)90–125(–128) × (14–)15–19(–21) μm (n = 30). Ascospores uniseriate to biseriate, hyaline, smooth-walled, apiosporic, composed of a large curved upper cell and small lower cell, fusiform or slightly curved in shape with narrowly rounded ends, uniguttulated, lacking a gelatinose sheath, measuring (28–)29–34(–37) × (5–)6–8(–9) μm, and a basal cell 5–7 μm (n = 45). Asexual morph: Mycelium hyaline, septate, branched, hyphae 2–4 μm in diameter. Conidiophores reduced to the conidiogenous cells. Conidiogenous cells aggregated in clusters on hypha or solitary, ampuliform or cylindrical, 6–12 × 3 μm. Conidia brown, smooth, globose to ellipsoid (9–)10(–12) µm long (n = 30) in face view, lenticular, with a paler equatorial slit, and (6–)7(–8) μm long (n = 40) in side view. Sterile cells elongated, rolled up, sometimes mixed among conidia. Culture characteristics: ascospores germinating on MEA 2% within 24–48 h. Colonies flat, spreading, with sparse aerial mycelium, pale siena with white patches.
Type.
Portugal. Viana do Castelo: Valença do Minho, on dead culms of Arundodonax. 10 Jan. 2018, A. Pintos (MA-Fungi 91732 holotype, AP10118 isotype, CBS 145137 ex-type culture).
Notes.
Arthriniumibericum belongs to the large clade around A.sacchari, where it shows a relation with the subclade of A.phaeospermum, A.saccharicola, and the modern species A.serenense, A.camelliae-sinensis, A.jiangxiense, A.dichotomanthi, A.obovatum and A.pseudosinense. The size of conidia is more or less similar to that of A.camelliae-sinensis, where these measure about 9.0–13.5 μm in frontal view, but conidiogenous cells are a bit smaller in this species, measuring about 4.0–9.5 × 3.0–6.0 μm. Arthriniumpseudosinense has slightly smaller asci measuring 85–100 × 15–20 µm, and ellipsoid conidia covered with a mucilaginous sheath. Arthriniumsaccharicola has hyphae slightly wider, about 3–5 µm. The genetic identity of A.phaeospermum is still dubious because of the lack of a proper type, but the lineages of this species in the work of Crous and Groenewald (2013) have slightly smaller conidiogenous cells measuring 5–10 × 3–5 μm, and a different iron-grey colour of colonies in MEA.
Arthrinium italicum
Pintos & P. Alvarado sp. nov.
828870
Figure 10.
A.italicumA, B stromata on host C asci D, E, G ascospores F ascospores with sheath H colony on MEAI–M conidiogenous cell giving rise to conidia N, O conidia. Scale bars: 200 µm (A, B); 5 µm (D–G); 5 µm (H–L, N, O); 10 µm (M).
Etymology.
In reference to Italy, the country where the holotype was found.
Diagnosis.
Sexual morph: Stromata solitary to gregarious, inmersed to erumpent, fusiform, with long axis broken at the top by one or two cracks, 0.5–4 × 0.2–0.5 mm (n = 20). Ascomata uniseriate or irregularly arranged beneath stromata, pseudothecial, black, globose to subglobose with a flattened base, 150–200 μm high × 230–300 μm wide. Peridium composed of 5 or 6 layers of brown cells arranged in textura angularis, with a conspicuous peryphisate ostiole. Hamathecium paraphyses hyphae-like. Asci broadly cylindrical, clavate or subglobose, pedicel indistinct, apically rounded (70–)72–93(–96) × (14–)15–18(–20) μm (n = 30). Ascospores apiosporic, clavate to fusiform with narrowly rounded ends, composed of a large upper cell and small lower cell, hyaline, smooth-walled, surrounded by a gelatinose sheath, measuring (20–)21–25(–26) × (5–)6– 9(–10) μm, basal cell 3–5 μm (n = 45). Asexual morph: Mycelium consisting of smooth, hyaline, branched, septate hyphae 1.5–4 µm in diameter. Conidiophores straight or flexuous, cylindrical, colourless except for the thick brown transversal septa, smooth-walled, 10–50 × 1–3 μm. Conidiogenous cells ampuliform, cylindrical or doliform, hyaline to brown, (3–)4–7(–9) × (1.5–)2–3(–5) μm (n = 30). Conidia brown, smooth, globose in face view, lenticular in side view, 4–6 × 3–4 μm (n = 65), with a pale equatorial slit. Culture characteristics: on MEA 2%, sparse aerial mycelia, surface dirty white, reverse pale yellowish.
Type.
Italy: Sicily: On dead culms of Arundodonax, 19 June 2016, H. Voglmayr (MA-Fungi 91733 holotype, AP221017 isotype, CBS 145138 ex-type culture).
Notes.
Arthriniumitalicum is phylogenetically close to A.thailandicum, and to a lesser extent to A.malaysianum. Stromata of A.thailandicum are smaller than those of A.italicum, measuring 0.45–0.99 × 0.3–0.55 mm, ascomata are perithecical, its conidiogenous cells are longer (11.5–39 × 2–3.5 μm) and branched, and conidia measure 5–9 × 5–8 μm. The conidia of A.malaysianum are similar in size, but this species does not produce conidiophores.
Other specimens examined.
Spain: Balearic Islands: Mallorca, Puerto de Andratx, on dead culms of Phragmitesaustralis, 29 Jan. 2018, A. Pintos (MA-Fungi 91734, AP29118).
Arthrinium marii
Larrondo & Calvo, Mycologia 82: 397 (1990)
Figure 11.
A.mariiA stromata on host B asci C–F ascospores G colony on MEAH–I, K conidiogenous cells giving rise to conidia J conidiophore bearing conidia L conidia and sterile cells. Scale bars: 200 µm (A); 10 µm (B); 5 µm (C–F); 5 µm (H–L).
Description.
Sexual morph: Stromata forming black fusiform spots, visible at the naked eye, with a long axis broken at the top revealing the ostioles of pseudothecia, 2–6 × 0.2–0.5 mm in size. Ascomata subglobose, sometimes with a flattened base, brownish to reddish brown, 150–190 μm high × 160–250 μm wide (n = 20). Peridium with several layers of cells arranged in textura angularis, with a conspicuous ostiole 50–7–80 μm diameter, periphysate. Hamathecium paraphyses not prominent, hyphae-like, septate, hyaline. Asci 8-spored, unitunicate, broadly cylindrical to clavate, with rounded apex and a short pedicel, (60–)70–100(–115) × (16–)18–20(–22) μm (n = 30). Ascospores fusiform to elliptical, with narrowly rounded ends, hyaline, with multiple guttules, surrounded by a mucilaginous sheath, (16)19–23(–24) × (6–)7–8(–10) μm, basal cell 2–5 (n = 30). Asexual morph: Mycelium consisting of smooth, hyaline, branched, septate hyphae measuring 1.5–5 µm in diameter. Conidiophores straight or flexuous, cylindrical, colourless except for the thick brown transverse septa, measuring 10–40 × 2–3 μm. Conidiogenous cells ampuliform to cylindrical, hyaline to brown, (3–)4–7(–11) × (1.4–)2–4(–5) μm (n = 30). Conidia, brown, smooth, granular, globose in face view, lenticular in side view, measuring (6–)7–8(–9) × 4–5(–6) µm, with a pale equatorial slit. Sterile cells elongated, brown. Culture characteristics: ascospores germinating on MEA 2% within 24–48 h. Colonies flat, spreading, with sparse aerial mycelium, reverse concolour with agA.
Notes.
Arthriniummarii was proposed by Larrondo and Calvo (1990) who described its asexual morph. This apparently frequent species has been isolated from the atmosphere, pharmaceutical excipients, home dust, and beach sand, as well as from various plant hosts (Crous 2013). In the present work the sexual morph is described for the first time. Genetically, samples identified as A.marii seem to represent two distinct clades (Fig. 2), with differences in tub2 and tef1 genes, but it should be further investigated with additional data before concluding if these clades should be interpreted as intraspecific variability, partially isolated lineages, or fully isolated species. Similarly, the incomplete data from the type specimens of A.hispanicum and A.mediterranei do not allow one to conclude if these apparently related species represent a single taxon or even belong to A.marii.
Specimens examined.
Austria: Oberösterreich: St. Willibald, on dead culms of Phragmitesaustralis, 10 July 2016, H. Voglmayr, (MA-Fungi 91738, AP191017).
Italy: Sicily: casa de la Monache, on dead culms of Phragmitesaustralis, 16 July 2016, H. Voglmayr (MA-Fung 91740, APVog2).
Portugal: Viana do Castelo: Valença do Minho, on dead culms of Phragmitesaustralis, 10 Jan. 2018, A. Pintos (AP10118A).
Spain: Balearic Islands: Mallorca, Esporlas, on dead culms of Arundodonax, 13 July 2017, A. Pintos (MA-Fungi 91735, AP13717). Ibidem., 29 July 2017, A. Pintos (AP29717). Palma de Mallorca, on Ampelodesmosmauritanicus, 11 July 2017, A. Pintos (MA-Fungi 91737, AP11717A). Palma de Mallorca, on dead culms of Phragmitesaustralis, 26 July 2017, A. Pintos (MA-Fungi 91739, AP261017).
Arthrinium piptatheri
Pintos & P. Alvarado. sp. nov.
828871
Figure 12.
A.piptatheriA colony on MEAB–K conidiogenous cells giving rise to conidia. Scale bars: 5 µm (B–K).
Etymology.
Named after Piptatherum, the host plant from which it was first isolated.
Diagnosis.
Asexual morph: Mycelium consisting of smooth, hyaline, branched, septate hyphae measuring 1–4 µm in diameter. Conidiophore mother cells hyaline to brown, aggregated in clusters or solitary on hyphae, ampuliform, cylindrical or doliform, 4–11 × 2–5 µm, growing above one or several hyaline cylindrical cells. Conidiophore reduced to a conidiogenous cell. Conidiogenous cells basauxic, polyblastic, sympodial, cylindrical, discrete, sometimes branched, smooth-walled, measuring 6–27 × 2–5 μm (n = 25). Conidia globose to ellipsoidal, pale brown to brown, with a thin hyaline germ-slit, 6–8 × 3–5 μm (n = 30). Sterile cells eloganted, brown, sometimes mixed among conidia, 13–16 × 4–5 μm (n = 30). Culture characteristics: on MEA 2%, colonies flat, spreading, with sparse aerial mycelium, reverse concolour with agar.
Type.
Spain: Balearic Islands: Mallorca: Llucmajor, on dead stems of Piptatherummiliaceum, 4 Aug. 2017, A. Pintos (MA-Fungi 91745 holotype, AP4817A isotype, CBS 145149 ex-type culture).
Notes.
Arthriniumpiptatheri is genetically close, but genetically distinct from A.marii, A.sacchari, A.guizhouense, A.hispanicum, A.mediterranei, A.longistromum D.Q. Dai & K.D. Hyde, and to a lesser extent A.pseudospegazzinii (Fig. 2) and the clade around A.phaeospermum (Fig. 1). The incomplete genetic data available is probably the cause behind the lack of significant support for some of these taxa. Morphologically, A.piptatheri differs from A.marii because of its sympodial, branched conidiogenous cells. Arthriniumguizhouense has shorter conidiogenous cells (3.5–8.0 μm). Finally, some sequences of Ap.montagnei are related also with this group (Fig. 2), but this species is considered the sexual morph of A.arundinis, with a very different genetic profile in Crous and Groenewald (2013), so its actual identity should be further investigated.
Arthrinium puccinioides
Kunze & J.C. Schmidt, Mykologische (Leizpig) 2: 103 (1823)
Figure 13.
A.puccinioidesA colony on host B colony on MEAC conidiophore mother cell D–F conidiophore bearing conidia G–H conidia in side view. Scale bars: 100 µm (A); 5 µm (C–H).
Conoplea puccinioides DE Candolle, 1905, Flore Francaise, Ed. 3, Tome 2, p.73, ex Mérat, Novuvelle Flore des environs de Paris, 1821, p. 16.
Goniosporium puccinioides (Kunze & J. C.Schmidt) Link, in Willdenow, Sp.pl., Edn 4 6(1): 44 (1824).
Gonatosporium puccinioides (Kunze & J. C.Schmidt) Corda, Icon. Fung. (Prague) 3:8 (1839).
Description.
Asexual morph: Mycelium consisting on smooth hyaline, branched, septate hyphae measuring 1.5–5 µm in diameter. Colonies are small, rounded or ovoid, dark brown, 50–400 µm in diameter. Conidiophore mother cells subspherical, lageniform or barrel-shaped, 4–5 × 3–5 µm (n = 30). Conidiophores cylindrical, straight or flexuous, septate, hyaline excepting for the thick brown or dark brown transversal septa, 20–140 × 3–4 µm (n = 30). Conidiogenous cells cylindrical, occurring between the conidiophore septa, 0.9–1.8 µm. Conidia dark brown, smooth, polygonal with rounded angles to hemispherical, measuring (8–)9–11(–12) × 8–9 µm, with one or two concentric pale rings. Sterile cells spherical, triangular or polygonal, with refractive bodies inside, paler than conidia, 6–9 µm in diameter. Culture characteristics colonies flat spreading on MEA 2%, with moderate aerial mycelium, reverse whitish, no esporulate on culture.
Notes.
Arthriniumpuccinioides is the only species of Arthrinium with polygonal conida. It shows a genetic relationship with other species found in Carex sp. hosts, such as A.caricicola, A.curvatumvar.minus, A.japonicum or A.sporophleum. The present sample fits the original description of A.puccinioides by Kunze and Schmidt (1823) as well as those by Ellis et al. (1951), Ellis (1965), and Scheuer (1996).
Specimens examined.
Germany: Berlin: Köpenick, Stellingdamm, on dead leaves of Carexarenaria, 26 April 2017, R. Jarling (MA-Fungi 91746, AP26418).
Arthrinium sporophleum
Kunze, 1823, in Kunze & Schmidt's Mykologische Hefte, 2, p. 104; Fries, 1832, Systema Mycol., 3, p. 377
Figure 14.
A.sporophleumA colony on host B conidiophore mother cells C–E conidiophore mother cells with conidiophore bearing conidia, F with sterile cell F–H conidia I colony on MEA. Scale bars: 100 µm (A); 5 µm (B–H).
Sporophleum gramineum Nees, 1824, apud Link in Linne, Species Plantarum, ed. 4 (Willdenow's), 6, 1, p. 45.
Torula eriophori Berkeley, 1836, Fungi in J. E. Smith's English Flora, 5 (2), p. 359.
Arthrinium sporophleoides Fuckel, Jb. nassau. Ver. Naturk. 27–28: 78 (1874) [1873–74]
Description.
Asexual morph: Mycelium consisting on smooth hyaline branched hyphae, 2–5 µm in diameter. Colonies oval to irregular, dark blakish brown, 300–1200 × 150–650 µm. Conidiophore mother cells sub-cylindrical, hyaline to pale brown, measuring 5–7 × 5–7 µm (n = 20). Conidiophores straight to flexuous, cylindrical, hyaline except for the thick brown to dark brown transversal septa, 30–130 × 2–4 µm (n = 20). Conidia brown, smooth, lemon-shaped in face view, measuring (10–)11–14(–15) × (5–)6–8(–9) µm (n = 45), triangular with the outer edge curved and rounded angles in side view, measuring 5–8 µm thick. Sterile cells paler than conidia, subspherical or triangular, 5–8 µm wide. Culture characteristics: on MEA 2% colonies cottony, white with grey patches, reverse pale grey.
Notes.
Arthriniumsporophleum is the only species of Arthrinium with lemon-shaped conidia. Kunze (1823) considered that Sporophleumgramineum represents a synonym of this species, and Cooke (1954) considered A.sporophleoides Fuckel a synonym of this species too. The only sample analyzed in the present work fits the descriptions of this species by Kunze (1823), Ellis et al. (1951), Ellis (1965) and Scheuer (1996). This sample was found in Juncus sp., but this remarkable species has been often reported from Carex sp. hosts (Ellis 1965). Interestingly, other species occurring in Carex sp. present also conidia with unusual shapes, e.g. A.puccinioides (polygonal), A.curvatumvar.minus (curved), and A.caricicola or A.japonicum (fusiform).
Specimens examined.
Spain: Balearic Islands: Mallorca, Escorca, on dead leaves of Juncus sp., 21 Feb. 2018, A. Pintos 21218 (MA-Fungi 91749).
Other specimens studied.
Arthriniumarundinis: Spain: Galicia: Santiago de Compostela, city garden, culms of Bambusa sp., 11 Jan. 2018, A. Pintos 11118A (MA-Fungi 91722). Arthriniumphragmitis: Spain: Balearic Islands: Mallorca, Esporles, on dead culms of Arundodonax, 29 July 2017, A. Pintos (MA-Fungi 91744, AP29717A). Ibidem., on dead stem of Phragmitesaustralis, 3 Feb. 2018, A. Pintos (MA-Fungi 91743, AP3218). Jardin Botanico de Soller, on dead culms of Arundodonax, 24 Oct. 2017, A. Pintos (MA-Fungi 91742, AP2410172A). Puigpunyent, on dead culms of Phragmitesaustralis, 28 Dec. 2017, A. Pintos (MA-Fungi 91741, AP281217A1). Arthriniumrasikravindrii: Spain: Balearic Islands: Mallorca, Esporlas, on dead culms of Phyllostachysaurea, 8 Aug. 2017, A. Pintos (MA-Fungi 91747, AP8817). Jardin Botanico de Soller, on dead culms of Bambusa sp., 24 Oct. 2017, A. Pintos (AP2420171). Soller, on dead culms of Phyllostachysaurea, 10 Apr. 2018, A. Pintos (MA-Fungi 91748, AP10418).
Discussion
Arthrinium is thought to represent the asexual morph of Apiospora because genetic data of Ap.montagnei (type species of Apiospora, Müller and Arx 1962) grouped together with other species of Arthrinium (Crous and Groenewald 2013; Senanayake et al. 2015; Réblová et al. 2016). Unfortunately, no data from the type species of Arthrinium, A.caricicola, was available to confirm this synonymy. In the present work, a phylogenetic relationship was found between a specimen identified as A.caricicola and other species of Arthrinium mainly occurring in Carex sp., such as A.curvatumvar.minus, A.japonicum, A.puccinioides and A.sporophleum. Moreover, this clade was not significantly related with all other species of Arthrinium and Apiospora found in other hosts or substrates, suggesting that both clades could be interpreted as independent genera sister to Nigrospora. In this case, the synonymy between Arthrinium and Apiospora could be rejected, requiring new combinations. However, this hypothesis should be further confirmed after the analysis of the remaining known species occurring in Cyperaceae hosts, such as A.austriacum, A.fuckelii, A.globosum, A.kamtschaticum, A.morthieri, A.muelleri, or A.naviculare.
Arthrinium species have been found in several different plant hosts (Ramos et al. 2010; Sharma 2014), where they sometimes cause plant diseases (Martínez-Cano et al. 1992; Mavragani et al. 2007; Chen et al. 2014; Li et al. 2016). They are also isolated from lichens (He and Zhang 2012), marine algae (Suryanarayanan 2012), soil (Singh et al. 2013) and can even cause infections in humans (Rai 1989; Zhao et al. 1990; Hoog et al. 2000). In the present study six new species of Arthrinium are proposed: A.balearicum, A.descalsii, A.esporlense, A.ibericum, A.italicum, and A.piptatheri, all of them found in the Mediterranean biogeographical region, excepting for A.ibericum, which was found in the Atlantic areas of Spain. All these new taxa were found growing on plant hosts of the Poaceae family, such as Arundodonax or Piptatherummiliaceum. However, A.marii was the species most frequently found in the surveys, occurring on the Poaceae grasses Ampelodesmosmauritanicus and Phragmitesaustralis, in agreement with the data reported by Crous and Groenewald (2013). Arthriniumphragmitis was found also on Phragmitesaustralis and less commonly in Arundodonax, while A.hysterinum and A.rasikravindrae were associated with the Poaceae bamboos Phyllostachysaurea and Bambusa sp. Several colonies of A.rasikravindrae were found growing on Phyllostachysaurea as well, where they developed acervular conidiomata, a feature not observed in the protologue of this species, and therefore not considered diagnostic, in the same way as conidial shape, presence of setae, or lobate sterile cells.
Apiosporatintinnabula (Samuels et al. 1981) is considered a synonym of A.hysterinum (Sivanesan 1983; Kirk 1986). Multigenic data from the ex-type culture ICMP 6889 of Ap.tintinnabula was obtained so as to compare it with the newly found specimens of A.hysterinum, and no significant difference could be found. Interestingly, the collections of A.hysterinum studied in the present work presented sterile lobed cells, a feature not mentioned in the protologue of Ap.tintinnabula. The genetic data available from Ap.setosa and Ap.bambusae (28S and tub2) are not significantly different from those of A.hysterinum and Ap.tintinnabula, although additional markers would be needed to confirm a putative synonymy.
Supplementary Material
Acknowledgements
We thank Dr Hermann Voglmayr for his valuable advice, Chris Yeates and Martin Bemmann for providing literature, and Äsa Kruys for providing details about type collection of Arthriniumcaricicola.
Citation
Pintos A, Alvarado P, Planas J, Jarling R (2019) Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys 49: 15–48. https://doi.org/10.3897/mycokeys.49.32115
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arx JA von. (1970) The Genera of Fungi Sporulating in Pure Culture (3rd edn). Cramer Vaduz, 424 pp.
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chen K, Wu XQ, Huang MX, Han YY. (2014) First report of brown culm streak of Phyllostachyspraecox caused by Arthriniumarundinis in Nanjing, China. Plant Disease 98: 1274. 10.1094/PDIS-02-14-0165-PDN [DOI] [PubMed]
- Cooke WB. (1954) The gennus Arthrinium Mycologia 46 (6) 815–822. 10.1080/00275514.1954.12024418 [DOI]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4: 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeta MA, Echandi E, Abernethy T, Vilgalys R. (1991) Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81: 1395–1400. 10.1094/Phyto-81-1395 [DOI] [Google Scholar]
- Dai DQ, Jiang HB, Tang LZ, Bhat DJ. (2016) Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7: 1332–1345. 10.5943/mycosphere/7/9/7 [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82: 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- de Hoog GS, Guarro J, Gené J, Figueras MJ. (2000) Atlas of Clinical Fungi (2nd edn). CBS, Utrecht, The Netherlands, and Universitat Rovira i Virgili, Reus, 1126 pp. [Google Scholar]
- Ellis MB, Ellis EA, Ellis JP. (1951) British marsh and fen fungi. II. Transactions of the British Mycological Society 34: 497–514. 10.1016/S0007-1536(51)80034-2 [DOI] [Google Scholar]
- Ellis MB. (1963) Dematiaceous Hyphomycetes. IV. Mycological Papers 87: 1–42. [Google Scholar]
- Ellis MB. (1965) Dematiaceous Hyphomycetes. VI. Mycological Papers 103: 1–46. [Google Scholar]
- Ellis MB. (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, 608 pp. [Google Scholar]
- Ellis MB. (1976) More Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, 507 pp. [Google Scholar]
- Fuckel L. (1870) Symbolae mycologicae. Beiträge zur Kenntniss der Rheinischen Pilze. Jahrbücher des Nassauischen Vereins für Naturkunde 23–24: 1–459.
- Fuckel L. (1874) Symbolae mycologicae. Beiträge zur Kenntniss der rheinischen Pilze Jahrbücher des Nassauischen Vereins für Naturkunde 27–28: 1–99.
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, et al. (2011) The Amsterdam declaration on fungal nomenclature. IMA Fungus 2: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang Z. (2012) Diversity of organism in the Usnealongissima lichen. African Journal of Microbiology Research 6: 4797–4804. [Google Scholar]
- Hughes SJ. (1953) Conidiophores, conidia, and classification. Canadian Journal of Botany 31: 577–659. 10.1139/b53-046 [DOI] [Google Scholar]
- Huhndorf SM, Miller AN, Fernandez FA. (2004) Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. 10.1080/15572536.2005.11832982 [DOI] [PubMed] [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, Baghela A. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Fröhlich J, Taylor JE. (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50: 21–80. [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2012) Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Diversity 52: 75–98. 10.1007/s13225-011-0104-2 [DOI] [Google Scholar]
- Jiang N, Li J, Tian CM. (2018) Arthrinium species associated with bamboo and reed plants in China. Fungal Systematics and Evolution 2: 1–9. 10.3114/fuse.2018.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM. (1986) New or interesting microfungi. XV. Miscellaneous hyphomycetes from the British Isles. Transactions of the British Mycological Society 86: 409–428. 10.1016/S0007-1536(86)80185-1 [DOI] [Google Scholar]
- Kunze G, Schmidt JC. (1823) Mykologische Hefte. 2. Vossische Buchhandlung, Leipzig, 176 pp. [Google Scholar]
- Larrondo JV, Calvo MA. (1990) Two new species of Arthrinium from Spain. Mycologia 82: 396–398. 10.1080/00275514.1990.12025899 [DOI] [Google Scholar]
- Larrondo JV, Calvo MA. (1992) New contributions to the study of the genus Arthrinium. Mycologia 84: 475–478. 10.1080/00275514.1992.12026164 [DOI] [Google Scholar]
- Li BJ, Liu PQ, Jiang Y, Weng QY, Chen QH. (2016) First report of culm rot caused by Arthriniumphaeospermum on Phyllostachysviridis in China. Plant Disease 100: 1013–1013. 10.1094/PDIS-08-15-0901-PDN [DOI] [Google Scholar]
- Martínez-Cano C, Grey WE, Sands DC. (1992) First report of Arthriniumarundinis causing kernel blight on barley. Plant Disease 76: 1077. 10.1094/PD-76-1077B [DOI]
- Mavragani DC, Abdellatif L, McConkey B, Hamel C, Vujanovic V. (2007) First report of damping-off of durum wheat caused by Arthriniumsacchari in the semi-arid Saskatchewan fields. Plant Disease 91: 469. 10.1094/PDIS-91-4-0469A [DOI] [PubMed]
- Minter DW. (1985) A re-appraisal of the relationships between Arthrinium and other hyphomycetes. Proceedings of Indian Academy of Sciences (Plant Science) 94: 281–308. [Google Scholar]
- Müller E, Arx JA von. (1962) Die Gattungen der didymosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz 11: 1–922. [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Uppsala, Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Rai MK. (1989) Mycosis in man due to Arthriniumphaeospermumvar.indicum. First case report. Mycoses 32: 472–475. 10.1111/j.1439-0507.1989.tb02285.x [DOI] [PubMed] [Google Scholar]
- Ramos HP, Braun GH, Pupo MT, Said S. (2010) Antimicrobial activity from endophytic fungi Arthrinium state of Apiosporamontagnei Sacc. and Papulasporaimmersa. Brazilian Archives of Biology and Technology 53: 629–632. 10.1590/S1516-89132010000300017 [DOI] [Google Scholar]
- Réblová M, Miller AN, Rossman AY, et al. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7: 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Samuels GJ, McKenzie EHC, Buchanan DE. (1981) Ascomycetes of New Zealand. 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. New Zealand Journal of Botany 19: 137–149. 10.1080/0028825X.1981.10425113 [DOI] [Google Scholar]
- Schmidt JC, Kunze G. (1817) Mykologische Hefte. 1. Vossische Buchhandlung, Leipzig, 109 pp. [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. (2011) The Genera of Hyphomycetes. [CBS Biodiversity Series 9]. CBSKNAW Fungal Biodiversity Centre, Utrecht, 997 pp. [Google Scholar]
- Senanayake IC, Maharachchikumbura SS, Hyde KD, Bhat JD, Jones EG, McKenzie EH, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S. (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity 73: 73–144. 10.1007/s13225-015-0340-y [DOI] [Google Scholar]
- Sharma R, Kulkarni G, Sonawane MS, Shouche YS. (2014) A new endophytic species of Arthrinium ( Apiosporaceae) from Jatrophapodagrica. Mycoscience 55: 118–123. 10.1016/j.myc.2013.06.004 [DOI] [Google Scholar]
- Singh SM, Yadav LS, Singh PN, Hepat R, Sharma R, Singh SK. (2013) Arthriniumrasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon 122: 449–460. 10.5248/122.449 [DOI] [Google Scholar]
- Sivanesan A. (1983) Studies on Ascomycetes. Transactions of the British Mycological Society 81: 313–332. 10.1016/S0007-1536(83)80084-9 [DOI] [Google Scholar]
- Smith GJD, Liew ECY, Hyde KD. (2003) The Xylariales: a monophyletic order containing 7 families. Fungal Diversity 13: 185–218. [Google Scholar]
- Spatafora JW, Sung G-H, Johnson D, Hesse C, O’Rourke B, et al. (2006) A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. 10.1080/15572536.2006.11832630 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Suryanarayanan TS. (2012) Fungal endosymbionts of seaweeds. In: Raghukumar C. (Ed.) Biology of Marine Fungi.Springer, Dordrecht, 53–70. 10.1007/978-3-642-23342-5_3 [DOI] [PubMed]
- Swofford DL. (2001) PAUP*4.0b10: phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically ampliWed ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu F, Crous PW, Cai L. (2017) Phylogenetic reassessment of Nigrospora: ubiquitous endophytes, plant and human pathogens. Persoonia 39: 118–142. 10.3767/persoonia.2017.39.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tan X-M, Liu F, Cai L. (2018) Eight new Arthrinium species from China. MycoKeys 34: 1–24. 10.3897/mycokeys.34.24221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky J, White TJ (Eds) PCR protocols: A Guide to Methods and Applications. Academic Press, San Diego, 482 pp. [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH. (2006) An overview of the systematics of the Sordariomycetes based on four-gene phylogeny. Mycologia 98: 1076–1087. 10.3852/mycologia.98.6.1076 [DOI] [PubMed] [Google Scholar]
- Zhao YM, Deng CR, Chen X. (1990) Arthriniumphaeospermum causing dermatomycosis, a new record of China. Acta Mycologica Sinica 9: 232–235. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.