Short abstract
Heart valves reside in a dynamic mechanical environment experiencing a multitude of forces. These forces have been shown to play a key role in valvular pathophysiology. However, knowledge of proteins involved in valvular mechanobiology is limited to only a few proteins of interest, namely, α-smooth muscle actin, transforming growth factor β, etc. Valvular endothelium mediates valvular homeostasis and controls valvular interstitial cell phenotype transformation. But, how endothelium mediates valvular response to dynamic forces is also unknown. In this study, proteomic analysis of mitral valve anterior leaflets under 10% cyclic radial strain was performed. Endothelium from these samples was removed to test how endothelium mediates mitral valve response to stretch. Results show that stretch downregulated cytoskeletal proteins and proteins involved in energy metabolism such as glycolysis and oxireductase activity. Endothelium removal resulted in downregulation of extracellular matrix and cell-matrix adhesion proteins. Removal of endothelium also resulted in upregulation of translation-related and chaperone proteins. Overall, this high throughput study provides insights into new protein groups that may be involved in mitral valve response to mechanical stretch and loss of endothelium.
Impact statement
This work is important to the field of heart valve pathophysiology as it provides new insights into molecular markers of mechanically induced valvular degeneration as well as the protective role of the valvular endothelium. These discoveries reported here advance our current knowledge of the valvular endothelium and how its removal essentially takes valve leaflets into an environmental shock. In addition, it shows that static conditions represent a mild pathological state for valve leaflets, while 10% cyclic stretch provides valvular cell quiescence. These findings impact the field by informing disease stages and by providing potential new drug targets to reverse or slow down valvular change before it affects cardiac function.
Keywords: Mitral valve, cyclic stretch, endothelial cells, mechanotransduction, proteome, interactome
Introduction
Heart valves consist of leaflets, which are made of two types of cells: valvular endothelial cell (VEC) and valvular interstitial cell (VIC). Both cell types play active roles in maintaining valvular homeostasis.1,2 VICs reside within the 3D leaflet matrix. During valvulogenesis, VICs maintain an activated or myofibroblastic phenotype while proliferating and secreting extracellular matrix (ECM) to form the leaflets.3 In adult valves, VICs maintain a quiescent or fibroblastic phenotype, neither proliferating nor synthesizing ECM.3 However, with the onset of injury or abnormal mechanical forces, these quiescent phenotypes can become activated myofibroblasts and take part in valve repair and remodeling.1,4 Activated VICs are contractile and proliferative, and synthesize and remodel the ECM.1,5 VICs remaining in activated state culminate in degenerative disorders such as myxomatous degeneration and calcification.1,3 Another VIC phenotype, named osteoblastic VIC, shows osteoblastic properties and take part in dystrophic and osteogenic valve calcification.4,6 The osteoblastic phenotype has been shown to arise from quiescent VICs in osteogenic medium in vitro.1
Heart valves reside in a dynamic hemodynamic environment experiencing different types of forces. Mechanical forces have been shown to be a major cause of degenerative diseases of the valves.7–10 In physiological conditions, porcine mitral valve anterior leaflets experience peak tensile strains of 2.5–3.3% in the circumferential direction and 16–22% in the radial direction.11,12 During pathological conditions such as hypertension or ventricular remodeling, mitral valves experience up to 50% increase in peak strains.9,13 VICs have been extensively studied under mechanical stretch in vitro.14–20 Aortic10,21–23 and mitral7–9,24 valve leaflets have also been mechanically stretched in vitro. Mitral valve tissue stretch in vitro resulted in elevated expression of activated phenotype markers α-smooth muscle actin, matrix catabolic enzymes, ECM components, neurotransmitter serotonin, and vasoconstrictors.7–9,24
The valvular endothelium plays a key role in valvular pathophysiology. The endothelium maintains valvular homeostasis via cellular transformation,25 mechanotransduction,2 and VEC–VIC interaction.26–29 VECs line the outer surface of valve leaflets and experience shear, tensile, and compressive forces.2,30 VEC–VIC interaction plays a protective role in valvular pathophysiology. VECs suppress VIC transformation to activated26–28 and osteoblastic29 phenotypes via paracrine signaling. VEC-derived nitric oxide is involved in paracrine signaling possibly working via cyclic guanosine 3′,5′-monophosphate pathway.26,29 VECs, in osteogenic media in vitro, have been shown to transform into osteoblastic VICs via endothelial to mesenchymal transition.31 VIC–VEC interaction also suppresses osteoblastic transition of VECs.31 Endothelial dysfunction, due to abnormal mechanical forces, denudation and inflammatory responses, also results in valvular pathogenesis.2,32
Studies of valvular mechanical stretch and the protective endothelial role during stretch are limited to a few proteins of interest.14–17,26–29 A thorough study of valvular protein expression changes due to mechanical stretch with or without endothelium could open the door to new findings and insights into new valvular pathological pathways. To test how mechanical stretch and VEC–VIC interactions affect protein expression in mitral valves, anterior leaflet sections underwent 10% radial cyclic strain for 48 h with or without endothelium. Label-free nanoflow liquid chromatography tandem-mass spectrometry (LC-MS/MS) was used to analyze protein expression. Significantly differentially expressed proteins were clustered into three groups:
Stretch effect. This group of proteins is affected by cyclic stretch and is potential key members and regulators of mitral valve response to stretch.
Endothelium removal (ER) effect. This group of proteins is affected by the removal of endothelium from mitral valve tissue. These proteins may be linked to VIC–VEC interactions, endothelium maintenance and endothelium mediated valvular homeostasis.
Interaction effect. This is the effect of interaction between stretch and ER. This group represents proteins that are affected by stretch, only when the endothelium is not present. They represent proteins involved in mitral valve response to stretch without endothelium protection.
Proteomic analyses showed that stretch downregulated cytoskeletal proteins and proteins involved in energy metabolism such as glycolysis and oxireductase activity. Stretch also upregulated calcification proteins. Endothelium removal resulted in downregulation of extracellular matrix and cell-matrix adhesion proteins. Removal of endothelium also resulted in upregulation of translation and chaperone proteins.
Methods
Tissue preparation
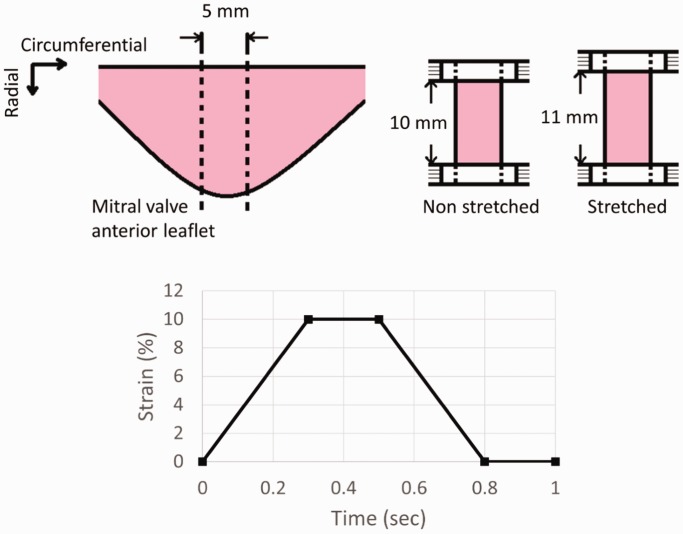
Porcine hearts were collected aseptically within 4 h of death. Mitral valve anterior leaflets were excised and washed in sterile phosphate buffer saline (137 mM sodium chloride, 2.7 mM potassium chloride, 4.3 mM sodium phosphate dibasic and 1.46 mM potassium phosphate monobasic, all Fisher Scientific, Waltham, MA); 5 mm-wide radial strips from the central region of the anterior leaflets were excised (Figure 1). In case of ER samples, strips were incubated in 600 U/mL collagenase (Sigma-Aldrich, St. Louis, MO) in cell culture medium for 10 min at 37°C. Dulbecco's modified Eagle medium (Mediatech, Corning, Manassas, VA) was supplemented with 10% fetal calf serum (Sigma-Aldrich) and 1% antibiotics/antimycotics (10,000 U/mL penicillin G, 10 mg/mL streptomycin sulfate and 25 μg/mL amphotericin B (Quality Biologicals, Gaithersburg, MD)). After incubation with collagenase, the strips were briefly vortexed. Endothelium from the tissue strips was scrubbed off using a cell scrubber on both sides of the strip.
Figure 1.
Schematics of tissue stretch and stretch regime; 5 mm wide radial strips from porcine mitral valve anterior leaflets were excised. Samples were stretched radially at 10% strain and 1 Hz for 48 h according to the stretch regime shown in the bottom figure.
Stretch
Tissue sections with intact or removed endothelium were placed between two clamps of CellScale MCT6 uniaxial stretcher (CellScale, Ontario, Canada) (Figure 1). Tautness of the strips between the clamps was ensured. The gap between clamps was set at 10 mm. Mitral valve anterior leaflet has been shown to experience radial strain ranging from 0% to 16–22% under physiological conditions.11 In this experiment, we provided a radial strain of 10% to tissue strips at a frequency of 1 Hz for 48 h. During one cycle, tissue strips were elongated to 10% strain in 0.3 s and remained under 10% strain for 0.2 s. After that they were returned back to 0% strain in 0.3 s and remained under 0% strain for 0.2 s (Figure 1). This stretch regime was selected to mimic a cardiac cycle lasting 1 s at 1 Hz frequency with systolic and diastolic phases each lasting 0.5 s. The non-stretched tissue sections both with intact or removed endothelium were cultured in 6-well plate culture plates for 48 h. Both stretched and non-stretched cultures were cultured with enough culture medium to submerge tissue samples and maintain overall conditions similar to the ones in the CellScale device. Samples were classified into four treatments: NS (non-stretched with intact endothelium), ST (stretched with intact endothelium), NSER (non-stretched with endothelium removed) and STER (stretched with endothelium removed).
Protein extraction
Treated tissues were minced and incubated with 0.5 mL extraction buffer per 0.1 g of tissue at 4°C for 1 h with rocking. Extraction buffer contained 150 mM sodium chloride, 50 mM HEPES salt, 2 mM dithiothreitol, 25 µg/mL digitonin (all Fisher Scientific) and 1% IGEPAL CA630 (Sigma-Aldrich). Samples were centrifuged and pellets were removed; 1% protease inhibitor cocktail (Active Motif, Carlsbad, CA) was added to extracted soluble protein samples prior to storage at −20°C. Bicinchoninic acid assay (Fisher Scientific) was used to determine protein concentration and manufacturer protocol was followed.
LC-MS/MS sample preparation
For each treatment, 50 μg of protein was fractionated by polyacrylamide gel electrophoresis. Three replicate wells were used for each treatment. Coomassie blue stain was performed using 1 g/L of Coomassie R250 dye in 45% ethanol and 10% glacial acetic acid (all Fisher Scientific) for 1 h. The stain was washed in excess 10% ethanol and 10% glacial acetic acid. Coomassie-stained gel bands were cut in 4 sections according to molecular weight. Bands were washed with water for 5 min and with acetonitrile (Fisher Scientific) for 5 min. Then, bands were treated with 10 mM dithiothreitol and 40 mM ammonium bicarbonate (Fisher Scientific) at 56°C 1 h. Samples were further alkylated using 55 mM iodoacetamide (Sigma-Aldrich) and 4 mM ammonium bicarbonate for 1 h in the dark. A final wash with acetonitrile for 1 min was performed. Bands were digested using sequencing grade modified trypsin (33 µg/mL) (Promega Corporation, Madison, WI) at 37°C overnight to fragment proteins into peptides. Peptides were extracted from gel bands twice using 50% acetonitrile and 0.1% formic acid (Fisher Scientific) for 15 min and 100% acetonitrile for 1 min.
LC-MS/MS
Methods in this section were adapted from Kottapalli et al.33 Nano flow LC-MS/MS was performed at the Center for Biotechnology and Genomics, Texas Tech University. Dionex nano high-performance liquid chromatography was used for chromatographic separation with a trapping column (C18, 3 μm, 100 Å, 75 μm × 2 cm), equilibrated with solvent A (2% acetonitrile and 0.1% formic acid) and a reverse phase column (C18, 2 μm, 100 Å, 75 μm × 15 cm). Equal sample sizes of all treatment groups were individually injected first into the trapping column and washed for 10 min with solvent A at a flow rate of 300 nL/min. Reverse phase chromatography was used to elute peptides using a linear gradient with decreasing solvent A (2% acetonitrile and 0.1% formic acid) and increasing solvent B (98% acetonitrile and 0.1% formic acid). The gradient was 120 min, kept constant for the first 10 min at 5% solvent B, followed by a linear increase to 20% solvent B in 55 min. Solvent B was further increased to 30% in 25 min, followed by another increase to 50% over 20 min. Solvent B was then immediately increased to 80% in 1 min and kept at 80% for another 4 min. Peptides were fed to linear ion trap mass spectrometer (Fisher Scientific) at 2 kV capillary voltage. Scanning of resulting spectra was performed over mass range of 400–2000 atomic mass units. Scan settings included choosing six most intense ions with dynamic exclusion list size of 200, exclusion duration of 90 s, mass width of ±1.5 m/z, repeat count of 2, and repeat duration of 30 s. Collision-induced dissociation at a normalized collision energy of 35% was used while generating mass spectra.
Bioinformatic analyses of mass spectrometry data
‘.raw’ files from LC−MS/MS were uploaded to MaxQuant software (Max Planck Institute of Biochemistry, Germany) and searched against a protein database of Sus scrofa obtained from UniProt. Default search parameters in MaxQuant for label-free quantification were used except minimum ratio count was set to 1 and in advanced identification setting, match between runs was turned on. Setting minimum ratio count to 1 means only one peptide ratio between samples is necessary for intensities of a protein to be compared across samples.34 Matching between runs allowed peptides not identified in all samples to be used for protein quantification by matching their mass and aligned retention times.34 The search result detected total 3782 proteins. This protein list was then uploaded to Perseus (Max Planck Institute of Biochemistry) for subsequent analyses. MaxQuant identified proteins that are potential contaminants, identified by modified and reverse sequences (false positives). After filtering out these three types, 3437 proteins remained. Only the proteins which had at least two replicates of intensity values across all treatment groups were selected. This resulted in a list of 1333 proteins. The missing intensity values of the third replicate were replaced with random numbers drawn from a normal distribution. The default parameters for this operation from Perseus were used.
The list of 1333 proteins was uploaded to Multi-Experiment Viewer35 (MeV) (TM4 Microarray Software Suite) to perform statistical analysis. Assuming normal distribution of data, two-factor analysis of variance was performed with stretch and ER being the two factors. A cutoff P-value of 0.01 and a minimum fold change in expression level of 1.5 was chosen. This analysis resulted in three groups of significantly differentially expressed proteins. We identify them throughout this study as stretch effect, ER effect, and interaction effect groups. From a list of 1333 proteins, numbers of significantly differentially expressed proteins were 202 due to stretch, 155 due to ER, and 111 due to interaction between stretch and ER. Among these proteins, there are 21, 25, and 12 uncharacterized proteins in stretch, ER, and interaction effect groups, respectively. Differentially expressed proteins from each group were visualized by generating heat maps (Supplementary Figure 1, Supplementary Figure 2, and Supplementary Figure 3). Proteins from each group (except uncharacterized ones) were uploaded onto STRING v1036 (Search Tool for the Retrieval of Interacting Genes/Proteins, STRING Consortium 2017) for the generation of protein interaction maps. STRING identifies and predicts functional interactions between proteins based on previous database knowledge as well as genomic context predictions, high-throughput lab experiments, co-expression, automated textmining, etc.37 STRING provides a confidence score between 0 and 1 for each interaction that estimates the validity of the interaction.37 In this experiment, a minimum confidence score of 0.9 was set. Markov Cluster Algorithm using proximity of predicted functional partners was also performed to produce clusters of proteins in the interaction maps. PANTHER (Protein ANalysis THrough Evolutionary Relationships, http://www.pantherdb.org/) classification system was used with Gene Ontology (GO, http://www.geneontology.org) to identify the molecular functions of proteins38,39 (Figure 2, Supplementary Table 1). For proteins with multiple functions, a function resulting in lowest P-value in PANTHER classification system was selected.
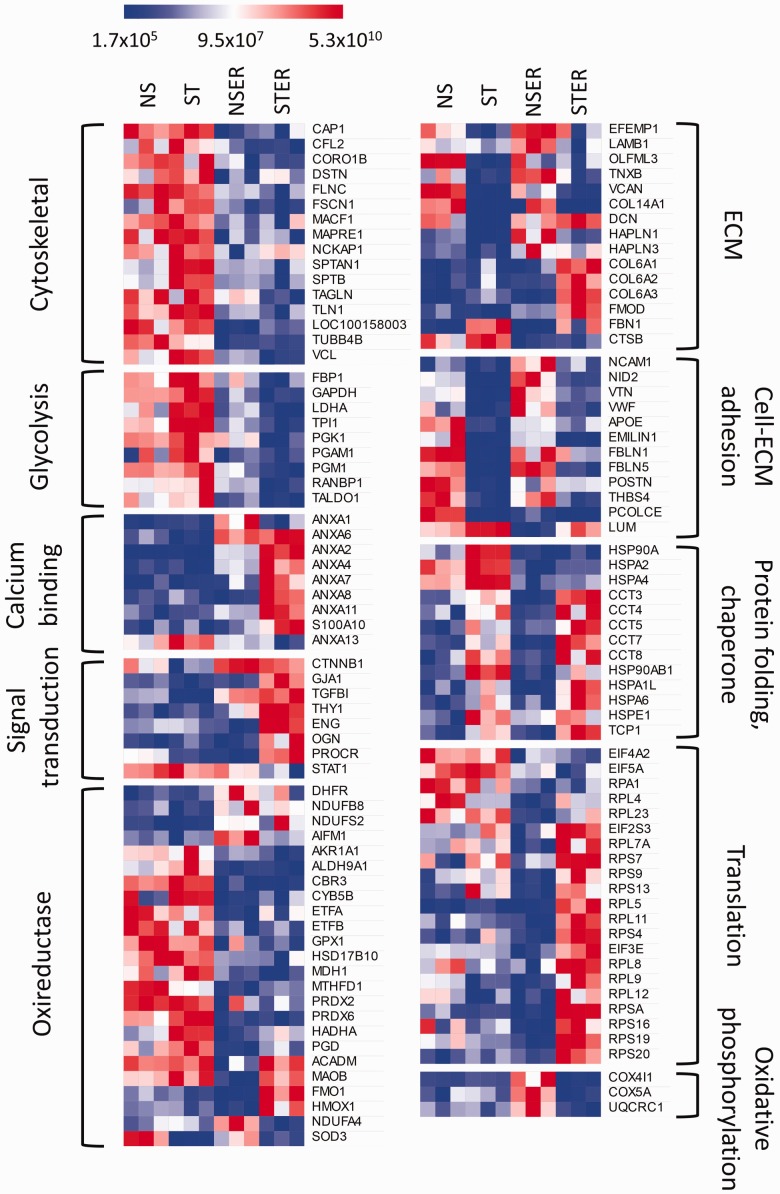
Figure 2.
Heat map showing expression of functionally classified significantly (P < 0.01) differentially expressed proteins across all four treatment groups. Treatment groups are NS (non-stretched with intact endothelium), ST (stretched with intact endothelium), NSER (non-stretched with endothelium removed), and STER (stretched with endothelium removed). Each treatment group has three replicates. Each row represents a protein. Red, white, and blue corresponds to high, medium, and low intensity values. Proteins related to same function are clustered together. Protein functions are mentioned on both sides.
Results
Proteins identified
After selecting proteins with at least two replicates of intensity values for each treatment, 1333 proteins were identified and quantified in this study. After two factor analysis of variance, three groups of proteins were generated according to the effects of stretch, ER, and their interaction. Heat maps show intensities of proteins differentially expressed (P < 0.01) in these groups. The number of significantly differentially expressed proteins with a minimum 1.5-fold intensity change is 202 for stretch effect (Supplementary Figure 1), 155 for ER effect (Supplementary Figure 2) and 111 for interaction effect (Supplementary Figure 3). Among these proteins, there are 21, 25, and 12 uncharacterized proteins in stretch, ER, and interaction effect groups, respectively.
Protein functions
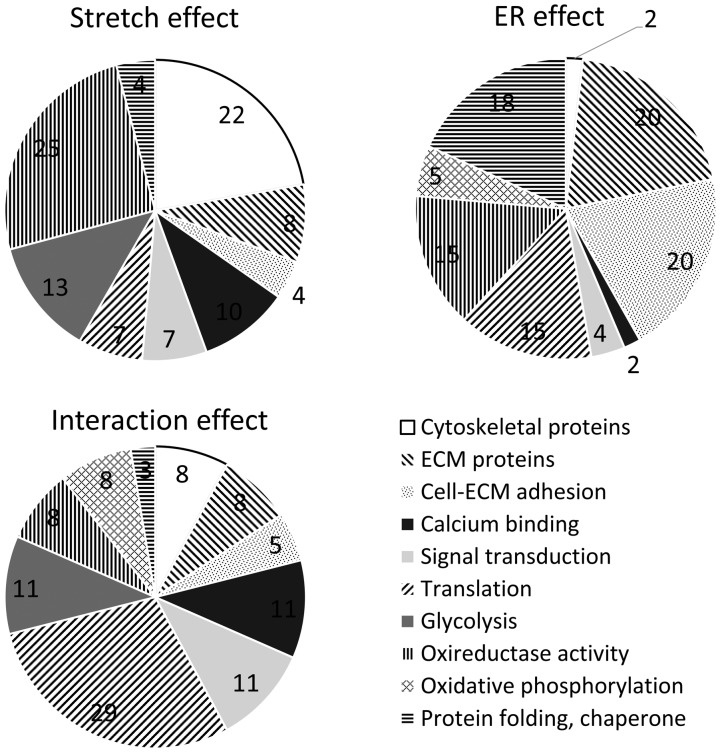
Proteins were further classified according to their primary functions in Figure 2 and Supplementary Table 1. Stretched tissue had lower expression of cytoskeletal, translational, metabolic, and chaperone proteins than static tissue, perhaps suggesting a switch to a more physiological condition. Stretched tissue had higher expression of ECM, calcium binding, and signal transduction proteins than static tissue. Removal of endothelium downregulated adhesion and oxidative phosphorylation proteins, while upregulating translation and chaperone proteins, indicating a need for repair. Except collagen VI and fibrillin 1, ECM proteins were downregulated due to endothelium removal. Interaction effect between stretch and ER groups upregulated ECM, calcium binding, signal transduction, and translation proteins. Interaction effect generally downregulated glycolysis and oxidative phosphorylation proteins. Pie charts reporting relative distribution of proteins of these functional groups are shown in Figure 3. In terms of number of proteins differentially expressed, stretch affected cytoskeletal, calcification, glycolysis, and redox proteins to a greater extent. Endothelium removal affected ECM, adhesion, translation, redox, and chaperone proteins. The interaction effect largely affected calcium binding, signal transduction, translation, and glycolysis.
Figure 3.
Pie charts showing percentages for number of proteins of different functions affected by stretch, endothelium removal (ER), and interaction between stretch and ER. Stretch affects cytoskeletal, calcification, glycolysis, and oxireductase proteins in greater percentages. ER mostly affects ECM, adhesion, translation, oxireductase, and chaperone proteins. Interaction effects have higher percentages of calcium binding, signal transduction, translation, and glycolysis proteins.
Protein interaction maps
Pie charts in Figure 3 provide functional information about proteins affected by stretch, ER, and stretch–ER interaction. However, they do not provide any insight into how proteins with similar function interact with each other. Protein interaction maps (Figures 4 to 6) produced using STRING show how differentially expressed proteins in this study interact in the same and across functional groups. Although all significantly differentially expressed proteins (except uncharacterized proteins) from each group were used for analysis in STRING, not all of them appear in the interaction maps. This is due to lack of evidence in literature for their confident functional association with other proteins in the group.
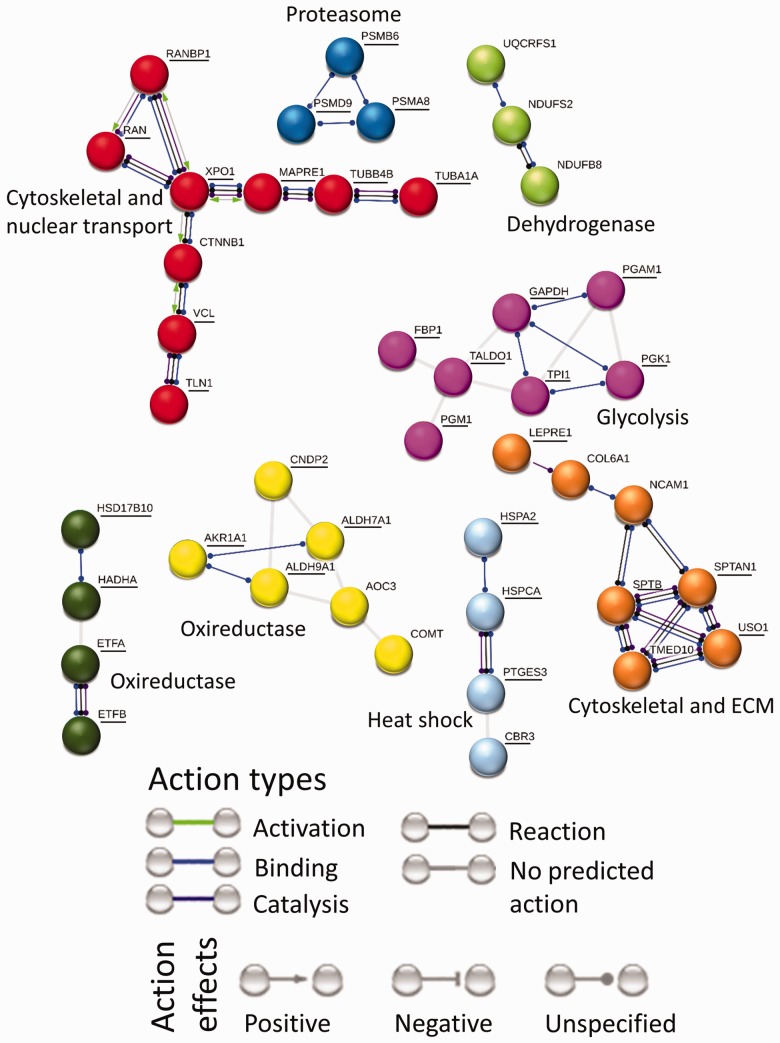
Figure 4.
Interaction network for proteins affected by stretch. Downregulated proteins are underlined, whereas upregulated proteins are not. Protein interaction map for stretch effect contains eight clusters. There are cytoskeletal-nuclear transport and cytoskeletal-ECM clusters where cytoskeletal proteins are downregulated. There is an energy metabolism cluster in the form of downregulated glycolysis proteins. There are two oxireductase, one heat shock and one proteasome cluster where proteins are downregulated. A cluster with upregulated proteins contains dehydrogenases.
The protein interaction map for stretch effect (Figure 4) contains eight clusters. There is a mixed function cluster where cytoskeletal proteins microtubule-associated protein (MAPRE1) and vinculin (VCL) are connected to nuclear transport protein Exportin-1 (XPO1) via signal transduction protein β-catenin (CTNNB1). Another mixed function cluster connects ECM protein collagen (COL6A1) to neural cell adhesion molecule 1 (NCAM1) to cytoskeletal protein spectrin (SPTB and SPTAN1) to vesicular transport proteins (TMED10 and USO1). An oxireductase cluster shows interaction between hydroxyacyl and hydroxysteroid dehydrogenases (HADHA and HAD17B10) with electron transfer flavoproteins (ETF-A, B). Another oxireductase cluster connects alcohol and aldehyde dehydrogenases (AKR1A1 and ALDH-7A1, 9A1) to cytosolic dipeptidase (CNDP2) and metabolic enzymes (amine oxidase (AOC3) and catechol O-methyltransferase (COMT)). There is an energy metabolism cluster in the form of downregulated glycolysis proteins. A protein folding related cluster contains molecular chaperone heat shock proteins (HSP-A2, CA) and chaperone prostaglandin E synthase (PTGES3) connected to oxireductase protein carbonyl reductase 3 (CBR3). A cluster with upregulated proteins contains dehydrogenases.
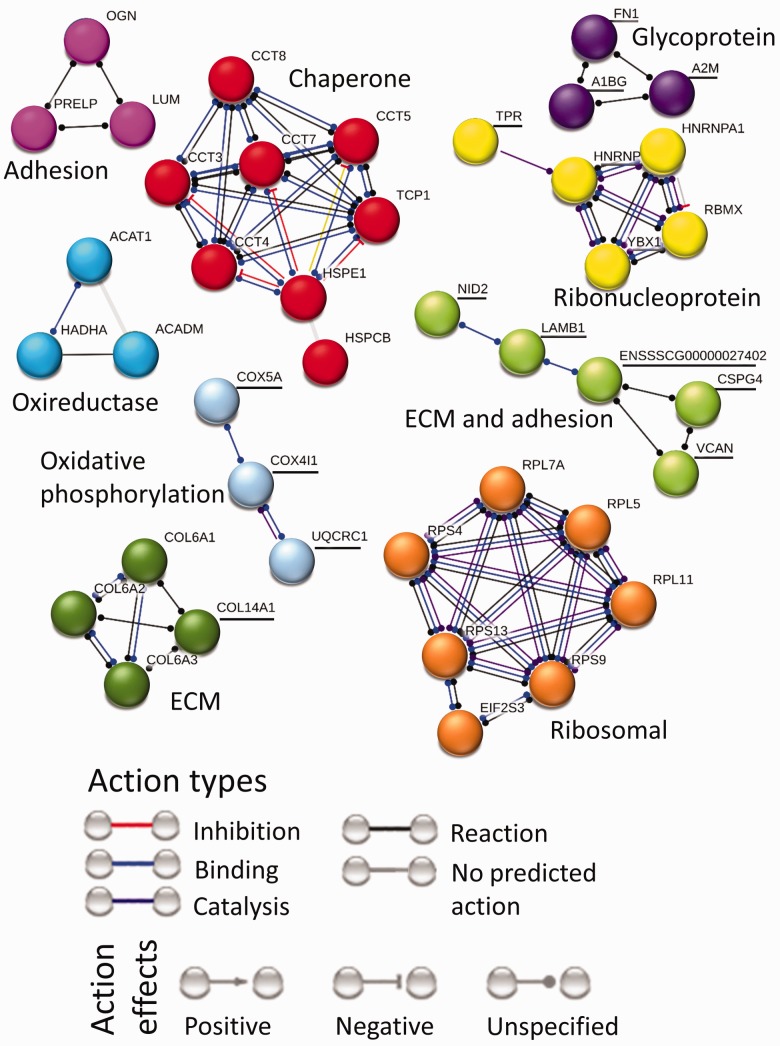
The protein interaction map for ER effect (Figure 5) contains nine clusters. The two big clusters contain upregulated molecular chaperones and upregulated translation-related ribosomal proteins. The chaperone cluster connects molecular chaperone T-complex proteins (CCT-3, 4, 5, 7, 8 and TCP1) to molecular chaperone heat shock proteins (HSP-E1, CB). The translation-related cluster shows interaction between ribosomal proteins (RPL-5, 7A, 11 and RPS-3, 9, 13) and eukaryotic translation initiation factor (EIF2S3). There are four ECM and adhesion protein clusters. The ECM-adhesion cluster shows interaction between adhesion protein nidogen 2 (NID2) and ECM proteins laminin (LAMB2), versican (VCAN) and chondroitin sulfate proteoglycan 4 (CSPG4). An oxireductase cluster contains proteins involved in acetyl-CoA transferase and acyl-CoA dehydrogenase activities (ACAT1, ACADM and HADHA).
Figure 5.
Interaction network for proteins affected by ER. Downregulated proteins are underlined, whereas upregulated proteins are not. Protein interaction map for ER effect contains nine clusters. The two big clusters contain upregulated molecular chaperone and translation-related ribosomal protein. There are four ECM and adhesion protein clusters. The dark green collagen and pink adhesion protein clusters are upregulated, whereas light green ECM-adhesion and purple glycoprotein clusters are downregulated. Other clusters in this map include upregulated oxireductase proteins, downregulated oxidative phosphorylation proteins and mixed ribonucleoproteins.
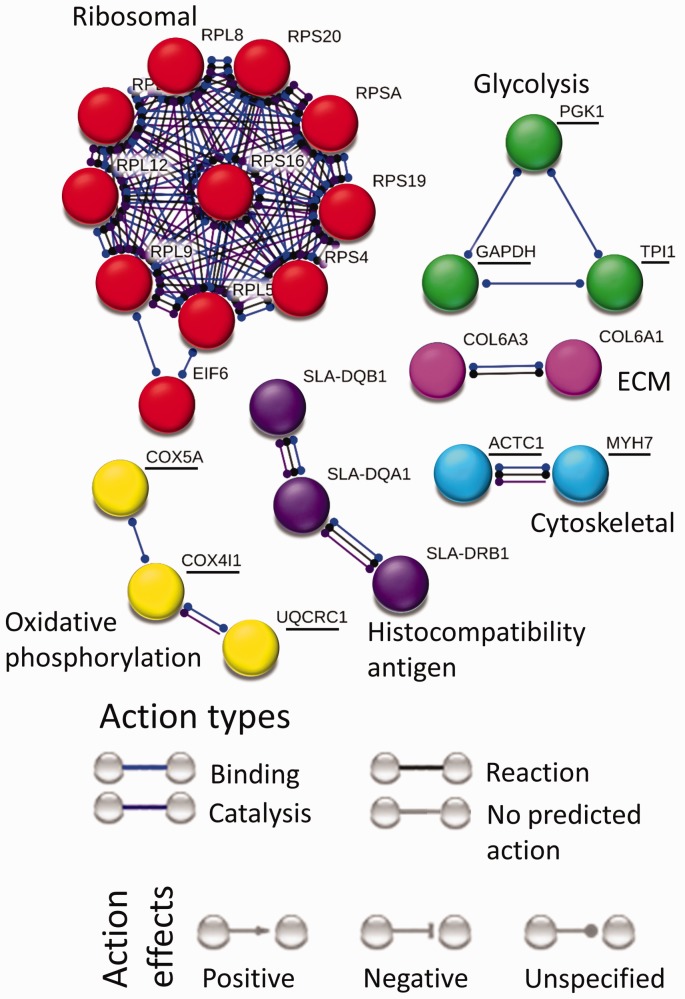
The protein interaction map for interaction effect (Figure 6) contains six clusters largely disconnected from one another. The largest cluster contains upregulated translation-related proteins connecting ribosomal proteins (RPL-5,8,9,11,12, RPS-A,4,16,19,20) to eukaryotic translation initiation factor 6 (EIF6).
Figure 6.
Interaction network for proteins affected by interaction between stretch and ER. Downregulated proteins are underlined, whereas upregulated proteins are not. Protein interaction map for interaction effect contains six clusters. The biggest cluster contains upregulated translation related ribosomal proteins. Among downregulated clusters are cytoskeletal, glycolysis and oxidative phosphorylation protein clusters. Upregulated clusters include ECM and histocompatibility antigen proteins.
Discussion
Stretch effect
Myxomatous degeneration is a form of mitral valve disease, associated with aging, which results in thickened leaflets and consequent valve failure. The mechanical environment plays a crucial role in mediating mitral valve degeneration.7,40 Pathological cyclic tensile strain induces myxomatous effector proteins and degenerative pathogenesis in mitral valves.7,8 However, the molecular mechanisms and signaling pathways involved in mitral valve mechanobiology and mechanotransduction are not well understood. Here, we compared proteomes of physiologically stretched mitral valves with those in pathological static environments to provide insights into novel molecular mechanisms involved in mitral valve mechanotransduction.
Physiological radial strain applied to mitral valve anterior leaflets downregulated the expression of cytoskeletal proteins in this study (Figure 2). Cytoskeletal protein expression suggests VIC quiescence under stretch and VIC activation under static conditions.5 Previous studies of valve leaflets and VICs under physiological stretch also report VIC quiescence in vitro,14,20,22 whereas proteomic studies of myxomatous mitral valves reveal abnormal upregulation of cytoskeletal proteins.41 One cytoskeletal protein with higher static expression compared to stretch is filamin C. Mutations in filamin genes promote the development of myxomatous valves in human and mice.42–44 Mutations in filamin A leads to VIC’s inability to efficiently organize ECM in fetal stages resulting in adult myxomatous valves.44 Filamin C is part of a filamin family of proteins, which crosslink actin filaments into a dynamic 3D structure.42,45 Filamins also facilitate transmembrane and cytoplasmic signaling molecule adhesion to cytoskeleton in mechanical stress-related cytoskeletal remodeling.44,46 Here, upregulation of filamin C in static conditions suggests filamin induces cytoskeletal reorganization in a mechanical stress environment. Compared to static controls, physiological stretch also reduced the expression of ribosomal proteins and stress related heat shock proteins (Figures 2 and 4). Ribosomal proteins are involved in protein translation. Heat shock proteins, involved in protein folding after translation, are upregulated when cells are under stressful conditions.47 Together, their downregulation suggests a quiescent state of VICs represented by reduced protein synthesis rates.
We further observed that energy metabolism proteins have lower expression under physiological stretch and higher expression under static conditions (Figures 2 and 4). Among those are glycolytic proteins. Much is lacking in the knowledge connecting valvular pathophysiology to energy metabolism. However, a previous proteomic study has identified metabolic proteins to be upregulated in myxomatous mitral valve from dogs.41 A glycolysis protein, L-lactate dehydrogenase A, downregulated by stretch in this experiment was found to be upregulated in myxomatous mitral valves.41 Proteomic analysis of atrial tissue of patients suffering from combined atrial fibrillation and valvular disease have shown downregulation in metabolic proteins compared to healthy tissue.48,49 Dysregulated metabolic activity in metabolic syndrome has also been implicated in progression of aortic valve calcification50,51 and degeneration of bioprosthetic valve.52 This suggests that metabolic proteins may be key regulators of valvular pathophysiology. This may also suggest that the tissue under physiological stretch is in a quiescent state and there is less energy demand. A differentially expressed protein in the glycolysis cluster (Figure 4) is glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH has been widely used as loading control in immunoblot experiments. However, a transcriptomics analysis of valvular cells53 and a gene array study54 have also found GAPDH to be differentially expressed across treatments. Cytoskeletal protein tubulin is also often used as loading control. However, tubulin was downregulated by stretch in this experiment, despite our overall protein load controls. Tubulin, a cytoskeletal protein, has been shown to be affected by mechanical stretch previously.55 Lung adenocarcinoma cells show a 50% reduction in polymerized tubulin when 10% strain is applied.55 Tubulin has been shown to be a key member in cellular realignment process due to cyclic strain.56,57 These results suggest that cytoskeletal and metabolic proteins should be reconsidered as loading controls in experiments involving valvular mechanobiology.
Upregulation of collagen (COL6A1), hyaluronan and proteoglycan link proteins, decorin and collagen fibrillogenesis protein, fibromodulin58,59 (Figure 2) upon stretch suggests that there is active ECM synthesis. Previous experiments of cyclic stretch on mitral VICs also show upregulated ECM synthesis.17,60 In addition to upregulating ECM proteins, downregulation of catabolic enzyme cathepsin B (Figure 2), which degrade and remodel ECM,61 was also observed. Downregulation of a catabolic enzyme also suggests greater ECM reorganization. Upregulation of adhesion proteins neural cell adhesion molecule 1 and nidogen 2 (Figure 2) suggests elevated cellular interactions with surrounding ECM due to mechanical stretch.
Calcium binding proteins or calcium-mediated phospholipid and membrane binding proteins were upregulated under stretch compared to static conditions (Figure 2). They are annexin 1, 2, 4, 6, 7, and S100A10.62,63 Annexin activity is regulated by calcium ion (Ca2+) concentration.64–66 Annexins themselves also function as ion channel regulators, regulating intracellular Ca2+ concentration.66,67 Transformation of aortic qVIC to obVIC, in vitro, is known to be mediated by mechanical stretch18,19,23 and by stretch-mediated intracellular calcium ion concentration.19 Recently, studies have identified the role of VIC-derived matrix vesicles in aortic valve calcification.68–70 Annexin 1, 2, 3, 4, 5, 6, 7, and 11 overexpression was identified in matrix vesicles from calcifying VICs in high Ca2+ medium.68 Annexin 6 was also found to be colocalized with matrix vesicles in human calcific aortic valves.68 Annexin and matrix vesicle-mediated calcification follows an osteogenic rather than dystrophic mechanism as suggested by high expression of osteogenic markers in these studies68 and abundant annexin expression in osteogenic cell types.69 However, these studies are of aortic valve biology. Here, we propose that physiological stretch-mediated upregulation of annexins is explained by mitral VICs having an annexin-mediated pro-calcific mechanism similar to their aortic VIC counterparts. A previous proteomic study on mitral valves identified annexin A-1, 5, 6 to be downregulated in myxomatous valves.41 This matches our results as annexins were upregulated only in physiologically stretched mitral valve suggesting an important role for annexin in physiology. However, it is important to note that these conclusions need further investigation.
We also observed higher expression of signal transduction protein, transforming growth factor β (TGFβ), after stretch compared to static conditions (Figure 2). TGFβ is a well-known inducer and marker of VIC myofibroblastic activation.71,72 Its upregulation with stretch suggests a basal level of expression required even in valvular physiology. To support this argument, there are many published works, which question TGFβ as marker of myofibroblastic activation.73–75 Upregulation of TGFβ and other cell-ECM adhesion proteins together due to stretch suggest a secondary role of TGFβ in stretched mitral valve leaflets, perhaps due to differences in the surrounding valvular environment.
Figures 2 and 4 also show a downregulation in oxireductase activity under stretch compared to static conditions. Previous studies have shown that oxidative stress potentiates calcification in aortic valves.29,76,77 Oxidative stress in aortic valves has been shown to increase due to downregulation of antioxidant enzymes.76 Oxireductase enzymes being downregulated by stretch, specially two antioxidants peroxiredoxins (PRDX-2, 6) (Figure 2), suggest that physiological stretch reduces the need for antioxidant pathways.
Endothelium removal effect
Among differentially expressed proteins due to endothelium removal, the major impact is seen in ECM proteins and cell-ECM adhesion proteins (Figure 2). Downregulation of ECM proteins with loss of endothelium suggests an endothelial role in maintaining an intact basal layer and ECM.78,79 Downregulation of cell–ECM interaction proteins seen with endothelium removal further supports an endothelial role in maintaining ECM homeostasis. However, ECM proteins collagen VI and fibrillin 1 were upregulated with endothelium removal. The synthesis of these proteins could be the first response to loss of basement membrane by VICs. A transcriptomics study of valvular cells experiencing variable stiffness also identified similar results where some ECM proteins, including fibrillin 1, are upregulated and some are downregulated with VIC activation.53 Fibrillin 1 assembles into microfibrils that play important role in elastic fiber formation and maintenance.80,81 Fibrillin 1 also plays crucial role in regulating cytokine signaling.82 Fibrillin 1 mutations leading to disruption in its structural and cytokine signal-regulating functions result in valvular diseases such as Marfan syndrome.83,84 Endothelium removal related fibrillin 1 upregulation in this study points to VIC’s response to upregulate ECM elastic structure reorganization and cytokine signaling. Similar to static conditions, endothelium removal upregulated translation via ribosomal proteins and protein folding activity via chaperone proteins compared to physiologically stretched intact leaflets (Figure 2). Together, these suggest upregulated protein synthesis, and hence an activated state in VICs due to the removal of endothelium.
Effect of interaction between stretch and endothelium removal
Proteins in this group are differentially expressed due to the combined action of stretch and endothelium removal. Similarly to stretch effect, the interaction effect downregulated glycolysis proteins and upregulated calcium binding and signal transduction proteins. And similarly to ER effect, the interaction effect downregulated adhesion and oxidative phosphorylation proteins and upregulated translation related ribosomal and collagen VI proteins (Figure 2). These suggest that interaction between stretch and ER augments the effect that stretch or endothelium removal alone have on proteins of these particular functions. We also observed that if an interaction effect protein is also in stretch effect or ER effect group, the fold change in interaction effect is greater (Figure 2 and Supplementary Table 1). For example, interaction effect has a greater fold change in glycolysis proteins L-lactate dehydrogenase A, phosphoglycerate kinase 1, and triosephosphate isomerase compared to stretch effect. And interaction effect has a greater fold change in translation related ribosomal proteins RPL5, RPL11 and RPS4 compared to ER effect. An exception to this observation is signal transduction protein TGFβ. This suggests that valvular endothelium may play an important role in signal transduction via this protein.
The downregulation of cytoskeletal proteins suggests a quiescent VIC phenotype due to interaction effect (Figure 2). We saw that calcium binding proteins are upregulated by both stretch and ER effect (Figure 2). This means that proteins with this function are upregulated by stretch with or without the presence of endothelium. This suggests endothelium has minimal effect on the pro-calcific state observed due to stretch in this experiment. Translation-related ribosomal proteins were upregulated in both ER and interaction group. A few of these ribosomal proteins (RPL-5,11, RPS4) show greater fold change in interaction effect than ER effect. We conclude that stretch and ER combined augment the upregulation in protein synthesis activity seen due to ER alone.
Trends in expression changes of several protein functions in this study suggest particular VIC phenotypes. However, widely used marker proteins for VIC phenotypes such as α-smooth muscle actin and several matrix metalloproteinases were not identified in this study. Excised mitral valve leaflet strips under static culture for two days were used as controls for this study. But this duration away from the in vivo native valvular environment may have significantly impacted their physiology. Despite these limitations, results from this study provide insights into new protein groups that may be involved in valvular mechanotransduction and endothelial regulation of mechanotransduction.
Conclusions
Thorough studies of mitral valve response to dynamic mechanical stimuli are lacking. Knowledge of how the endothelium affects mitral valve responses to mechanical stimuli is also missing. In this study, proteomic analysis of mitral valve anterior leaflets under 10% physiological radial cyclic strain was performed. The endothelium from these samples was removed to test how endothelium mediates mitral valve mechanoresponse. Differentially expressed proteins due to these treatments were classified into three groups: stretch effect, endothelium removal effect, and effect of interaction between stretch and endothelium removal. Results showed downregulation in cytoskeletal and energy metabolism proteins due to stretch suggesting quiescent state. There is upregulation in calcium binding proteins due to stretch suggesting pro-calcific state. Endothelium removal downregulated ECM and cell-ECM adhesion proteins suggesting protective role of endothelium in ECM homeostasis. Endothelium removal also upregulated protein synthesis activities. Interaction between stretch and endothelium resulted in quiescent and pro-calcific state as well as upregulated protein synthesis activity. Overall, high throughput omics studies can provide insights into molecular mechanisms regulating mechanotransduction.
Supplemental Material
Supplemental Material for The effect of physiological stretch and the valvular endothelium on mitral valve proteomes by Mir S Ali, Xinmei Wang and Carla MR Lacerda in Experimental Biology and Medicine
ACKNOWLEDGEMENTS
We thank Dr. Masoud Zabet and Adnan Ahmed from Center for Biotechnology and Genomics, Texas Tech University, Lubbock, TX for their help in LC-MS/MS methods and data analysis. We also thank Jackson Brothers Meat Locker, Post, TX for providing us with porcine hearts.
Authors’ contributions
Mir S. Ali: Data collection, Data analysis and interpretation, Manuscript writing. Xinmei Wang: Data collection, Data analysis and interpretation. Carla M. R. Lacerda: Conception and design, Financial support, Data analysis and interpretation, Manuscript writing, Final approval of the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This study was supported by startup funds provided to C.M.R.L. by Texas Tech University.
References
- 1.Liu AC, Joag VR, Gotlieb AI. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am J Pathol 2007; 171:1407–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher JT, Nerem RM. Valvular endothelial cells and the mechanoregulation of valvular pathology. Philos Trans R Soc Lond B Biol Sci 2007; 362:1445–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David Merryman W. Mechano-potential etiologies of aortic valve disease. J Biomech 2010; 43:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Leinwand LA, Anseth KS. Cardiac valve cells and their microenvironment – insights from in vitro studies. Nat Rev Cardiol 2014; 11:715–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabkin-Aikawa E, Farber M, Aikawa M, Schoen FJ. Dynamic and reversible changes of interstitial cell phenotype during remodeling of cardiac valves. J Heart Valve Dis 2004; 13:841–7 [PubMed] [Google Scholar]
- 6.Ruiz JL, Hutcheson JD, Aikawa E. Cardiovascular calcification: current controversies and novel concepts. Cardiovasc Pathol 2015; 24:207–12 [DOI] [PubMed] [Google Scholar]
- 7.R. Lacerda C M, Orton EC. Evidence of a role for tensile loading in the pathogenesis of mitral valve degeneration. J Clin Exp Cardiolog 2012; 1:1–5 [Google Scholar]
- 8.Lacerda CMR, MacLea HB, Kisiday JD, Orton EC. Static and cyclic tensile strain induce myxomatous effector proteins and serotonin in canine mitral valves. J Vet Cardiol 2012; 14:223–30 [DOI] [PubMed] [Google Scholar]
- 9.Lacerda CMR, Kisiday J, Johnson B, Orton EC. Local serotonin mediates cyclic strain-induced phenotype transformation, matrix degradation, and glycosaminoglycan synthesis in cultured sheep mitral valves. AJP Hear Circ Physiol 2012; 302:H1983–90 [DOI] [PubMed] [Google Scholar]
- 10.Thayer P, Balachandran K, Rathan S, Yap CH, Arjunon S, Jo H, Yoganathan AP. The effects of combined cyclic stretch and pressure on the aortic valve interstitial cell phenotype. Ann Biomed Eng 2011; 39:1654–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacks MS, Enomoto Y, Graybill JR, Merryman WD, Zeeshan A, Yoganathan AP, Levy RJ, Gorman RC, Gorman JH. In-vivo dynamic deformation of the mitral valve anterior leaflet. Ann Thorac Surg 2006; 82:1369–77 [DOI] [PubMed] [Google Scholar]
- 12.Sacks MS, Yoganathan AP. Heart valve function: a biomechanical perspective. Philos Trans R Soc Lond B Biol Sci 2007; 362:1369–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunzelman KS, Einstein DR, Cochran RP. Fluid-structure interaction models of the mitral valve: function in normal and pathological states. Philos Trans R Soc 2007; 362:1393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould RA, Chin K, Santisakultarm TP, Dropkin A, Richards JM, Schaffer CB, Butcher JT. Cyclic strain anisotropy regulates valvular interstitial cell phenotype and tissue remodeling in three-dimensional culture. Acta Biomater 2012; 8:1710–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KE, Metzler SA, Warnock JN. Cyclic strain inhibits acute pro-inflammatory gene expression in aortic valve interstitial cells. Biomech Model Mechanobiol 2010; 9:117–25 [DOI] [PubMed] [Google Scholar]
- 16.Merryman WD, Lukoff HD, Long RA, Engelmayr GC, Hopkins RA, Sacks MS, Sacks MS. Synergistic effects of cyclic tension and transforming growth factor-beta1 on the aortic valve myofibroblast. Cardiovasc Pathol 2007; 16:268–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V, Tseng H, Lawrence BD, Jane Grande-Allen K. Effect of cyclic mechanical strain on glycosaminoglycan and proteoglycan synthesis by heart valve cells. Acta Biomater 2009; 5:531–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher CI, Chen J, Merryman WD. Calcific nodule morphogenesis by heart valve interstitial cells is strain dependent. Biomech Model Mechanobiol 2013; 12:5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutcheson JD, Venkataraman R, Baudenbacher FJ, David Merryman W. Intracellular Ca 2+ accumulation is strain-dependent and correlates with apoptosis in aortic valve fibroblasts. J Biomech 2012; 45:888–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali MS, Wang X, Lacerda CMR. A survey of membrane receptor regulation in valvular interstitial cells cultured under mechanical stresses. Exp Cell Res 2017; 351:150–6 [DOI] [PubMed] [Google Scholar]
- 21.Balachandran K, Konduri S, Sucosky P, Jo H, Yoganathan AP. An ex vivo study of the biological properties of porcine aortic valves in response to circumferential cyclic stretch. Ann Biomed Eng 2006; 34:1655–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balachandran K, Sucosky P, Jo H, Yoganathan AP. Elevated cyclic stretch alters matrix remodeling in aortic valve cusps: implications for degenerative aortic valve disease. Am J Physiol Heart Circ Physiol 2009; 296:H756–64 [DOI] [PubMed] [Google Scholar]
- 23.Balachandran K, Sucosky P, Jo H, Yoganathan AP. Elevated cyclic stretch induces aortic valve calcification in a bone morphogenic protein-dependent manner. Am J Pathol 2010; 177:49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen LG, Zhao J, Yang J, Thomsen PD, Gregersen H, Hasenkam JM, Smerup M, Pedersen HD, Olsen LH. Increased expression of endothelin B receptor in static stretch exposed porcine mitral valve leaflets. Res Vet Sci 2007; 82:232–8 [DOI] [PubMed] [Google Scholar]
- 25.Mahler GJ, Farrar EJ, Butcher JT. Inflammatory cytokines promote mesenchymal transformation in embryonic and adult valve endothelial cells. Arterioscler Thromb Vasc Biol 2013; 33:121–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gould ST, Matherly EE, Smith JN, Heistad DD, Anseth KS. The role of valvular endothelial cell paracrine signaling and matrix elasticity on valvular interstitial cell activation. Biomaterials 2014; 35:3596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butcher JT, Nerem RM. Valvular endothelial cells regulate the phenotype of interstitial cells in co-culture: effects of steady shear stress. Tissue Eng 2006; 12:905–15 [DOI] [PubMed] [Google Scholar]
- 28.Shapero K, Wylie-Sears J, Levine R. A, Mayer JE, Bischoff J. Reciprocal interactions between mitral valve endothelial and interstitial cells reduce endothelial-to-mesenchymal transition and myofibroblastic activation. J Mol Cell Cardiol 2015; 80:175–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards J, El-Hamamsy I, Chen S, Sarang Z, Sarathchandra P, Yacoub MH, Chester AH, Butcher JT. Side-specific endothelial-dependent regulation of aortic valve calcification: interplay of hemodynamics and nitric oxide signaling. Am J Pathol 2013; 182:1922–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balachandran K, Alford PW, Wylie-Sears J, Goss J a., Grosberg a., Bischoff J, Aikawa E, Levine R a., Parker KK. Cyclic strain induces dual-mode endothelial-mesenchymal transformation of the cardiac valve. Proc Natl Acad Sci U S A 2011; 108:19943–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hjortnaes J, Shapero K, Goettsch C, Hutcheson JD, Keegan J, Kluin J, Mayer JE, Bischoff J, Aikawa E. Valvular interstitial cells suppress calcification of valvular endothelial cells. Atherosclerosis 2015; 242:251–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leask RL, Jain N, Butany J. Endothelium and valvular diseases of the heart. Microsc Res Tech 2003; 60:129–37 [DOI] [PubMed] [Google Scholar]
- 33.Kottapalli KR, Zabet-Moghaddam M, Rowland D, Faircloth W, Mirzaei M, Haynes PA, Payton P. Shotgun label-free quantitative proteomics of water-deficit-stressed midmature peanut (Arachis hypogaea L.) seed. J Proteome Res 2013; 12:5048–57 [DOI] [PubMed] [Google Scholar]
- 34.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteom 2014; 13:2513–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Meth Enzymol 2006; 411:134–93 [DOI] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43:D447–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, Von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45:D362–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8:1551–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 2017; 45:D183–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards JM, Farrar EJ, Kornreich BG, Mose NS, Butcher JT. The mechanobiology of mitral valve function, degeneration, and repair. J Vet Cardiol 2012; 14:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacerda CMR, Disatian S, Orton EC. Differential protein expression between normal, early-stage, and late-stage myxomatous mitral valves from dogs. Proteomics 2009; 3:1422–9 [DOI] [PubMed] [Google Scholar]
- 42.Norris RA, Moreno-Rodriguez R, Wessels A, Merot J, Bruneval P, Chester AH, Yacoub MH, Hagège A, Slaugenhaupt SA, Aikawa E, Schott JJ, Lardeux A, Harris BS, Williams LK, Richards A, Levine RA, Markwald RR. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev Dyn 2010; 239:2118–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyndt F, Gueffet J-P, Probst V, Jaafar P, Legendre A, Le Bouffant F, Toquet C, Roy E, McGregor L, Lynch SA, Newbury-Ecob R, Tran V, Young I, Trochu J-N, Le Marec H, Schott J-J. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation 2006; 115:40–9 [DOI] [PubMed] [Google Scholar]
- 44.Sauls K, De Vlaming A, Harris BS. Williams K, Wessels A, Levine RA, Slaugenhaupt SA, Goodwin RL, Pavone LM, Merot J, Schott JJ, Le Tourneau T, Dix T, Jesinkey S, Feng Y, Walsh C, Zhou B, Baldwin S, Markwald RR, Norris RA. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc Res 2012; 96:109–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 2004; 6:1034–8 [DOI] [PubMed] [Google Scholar]
- 46.Duval D, Lardeux A, Le Tourneau T, Norris RA, Markwald RR, Sauzeau V, Probst V, Le Marec H, Levine R, Schott JJ, Merot J. Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation. Biochim Biophys Acta 2014; 1843:324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature 2011; 475:324–32 [DOI] [PubMed] [Google Scholar]
- 48.Lamirault G, Gaborit N, Le Meur N, Chevalier C, Lande G, Demolombe S, Escande D, Nattel S, Léger JJ, Steenman M. Gene expression profile associated with chronic atrial fibrillation and underlying valvular heart disease in man. J Mol Cell Cardiol 2006; 40:173–84 [DOI] [PubMed] [Google Scholar]
- 49.Tu T, Zhou S, Liu Z, Li X, Liu Q. Quantitative proteomics of changes in energy metabolism-related proteins in atrial tissue from valvular disease patients with permanent atrial fibrillation. Circ J 2014; 78:993–1001 [DOI] [PubMed] [Google Scholar]
- 50.Katz R, Wong ND, Kronmal R, Takasu J, Shavelle DM, Probstfield JL, Bertoni AG, Budoff MJ, O’Brien KD. Features of the metabolic syndrome and diabetes mellitus as predictors of aortic valve calcification in the multi-ethnic study of atherosclerosis. Circulation 2006; 113:2113–9 [DOI] [PubMed] [Google Scholar]
- 51.Briand M, Lemieux I, Dumesnil JG, Mathieu P, Cartier A, Després JP, Arsenault M, Couet J, Pibarot P. Metabolic syndrome negatively influences disease progression and prognosis in aortic stenosis. J Am Coll Cardiol 2006; 47:2229–36 [DOI] [PubMed] [Google Scholar]
- 52.Briand M, Pibarot P, Després J-PP, Voisine P, Dumesnil JG, Dagenais F, Mathieu P. Metabolic syndrome is associated with faster degeneration of bioprosthetic valves. Circulation 2006; 114:512–7 [DOI] [PubMed] [Google Scholar]
- 53.Mabry KM, Payne SZ, Anseth KS. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials 2016; 74:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Jonge HJM, Fehrmann RSN, de Bont ESJM, Hofstra RMW, Gerbens F, Kamps WA, de Vries EGE, van der Zee AGJ, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS One. 2007; 2:e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geiger RC, Taylor W, Glucksberg MR, Dean DA. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther 2006; 13:725–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubitschke H, Schnauss J, Nnetu KD, Warmt E, Stange R, Kaes J. Actin and microtubule networks contribute differently to cell response for small and large strains. New J Phys 2017; 19:093003 [Google Scholar]
- 57.Morioka M, Parameswaran H, Naruse K, Kondo M, Sokabe M, Hasegawa Y, Suki B, Ito S. Microtubule dynamics regulate cyclic stretch-induced cell alignment in human airway smooth muscle cells. PLoS One 2011; 6:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiberg C, Heinegård D, Wenglén C, Timpl R, Mörgelin M. Biglycan organizes collagen VI into hexagonal-like networks resembling tissue structures. J Biol Chem 2002; 277:49120–6 [DOI] [PubMed] [Google Scholar]
- 59.Kalamajski S, Oldberg A. The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol 2010; 29:248–53 [DOI] [PubMed] [Google Scholar]
- 60.Gupta V, Werdenberg JA, Lawrence BD, Mendez JS, Stephens EH, Grande-Allen KJ. Reversible secretion of glycosaminoglycans and proteoglycans by cyclically stretched valvular cells in 3D culture. Ann Biomed Eng 2008; 36:1092–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation 2001; 104:2525–32 [DOI] [PubMed] [Google Scholar]
- 62.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 2005; 6:449–61 [DOI] [PubMed] [Google Scholar]
- 63.Yáñez M, Gil-Longo J, Campos-Toimil M. Calcium binding proteins. In: Islam MS, ed. Advances in experimental medicine and biology Vol. 740. ■: Springer, 2012, pp.461–482 [DOI] [PubMed]
- 64.Rescher U, Gerke V. Annexins – unique membrane binding proteins with diverse functions. J Cell Sci 2004; 117:2631–9 [DOI] [PubMed] [Google Scholar]
- 65.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev 2002; 82:331–71 [DOI] [PubMed] [Google Scholar]
- 66.Hawkins TE, Merrifield CJ, Moss SE. Calcium signaling and annexins. Cell Biochem Biophys 2000; 33:275–96 [DOI] [PubMed] [Google Scholar]
- 67.Lizarbe MA, Barrasa JI, Olmo N, Gavilanes F, Turnay J. Annexin-phospholipid interactions. Functional implications. Int J Mol Sci 2013; 14:2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui L, Rashdan NA, Zhu D, Milne EM, Ajuh P, Milne G, Helfrich MH, Lim K, Prasad S, Lerman DA, Vesey AT, Dweck MR, Jenkins WS, Newby DE, Farquharson C, Macrae VE. End stage renal disease-induced hypercalcemia may promote aortic valve calcification via Annexin VI enrichment of valve interstitial cell derived-matrix vesicles. J Cell Physiol 2017; 232:2985–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakhshian Nik A, Hutcheson JD, Aikawa E. Extracellular vesicles as mediators of cardiovascular calcification. Front Cardiovasc Med 2017; 4:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krohn JB, Hutcheson JD, Martínez-Martínez E, Aikawa E. Extracellular vesicles in cardiovascular calcification: expanding current paradigms. J Physiol 2016; 594:2895–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walker GA, Masters KS, Shah DN, Anseth KS, Leinwand L. A. Valvular myofibroblast activation by transforming growth factor-β: implications for pathological extracellular matrix remodeling in heart valve disease. Circ Res 2004; 95:253–60 [DOI] [PubMed] [Google Scholar]
- 72.Liu AC, Gotlieb AI. Transforming growth factor-beta regulates in vitro heart valve repair by activated valve interstitial cells. J Pathol 2008; 173:1275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AMT, Billiar KL. Investigating the role of substrate stiffness in the persistence of valvular interstitial cell activation. J Biomed Mater Res A 2012; 100 A:2474–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C, Gotlieb AI. Transforming growth factor-β regulates the growth of valve interstitial cells in vitro. Am J Pathol 2011; 179:1746–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ali MS, Deb N, Wang X, Rahman M, Christopher GF, Lacerda CMR. Correlation between valvular interstitial cell morphology and phenotypes: a novel way to detect activation. Tissue Cell 2018; 54:38–46 [DOI] [PubMed] [Google Scholar]
- 76.Miller JD, Chu Y, Brooks RM, Richenbacher WE, Peña-Silva R, Heistad DD. Dysregulation of antioxidant mechanisms contributes to increased oxidative stress in calcific aortic valvular stenosis in humans. J Am Coll Cardiol 2008; 52:843–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liberman M, Bassi E, Martinatti MK, Lario FC, Wosniak J, Pomerantzeff PMA, Laurindo FRM. Oxidant generation predominates around calcifying foci and enhances progression of aortic valve calcification. Arterioscler Thromb Vasc Biol 2008; 28:463–70 [DOI] [PubMed] [Google Scholar]
- 78.Young EWK, Wheeler AR, Simmons CA, Kallenbach K, Rebe P, Oberbeck A, Herden T, Haverich A, Mertsching H, Kaushal S, Vacanti JP, Schoen FJ, Mayer JE. Matrix-dependent adhesion of vascular and valvular endothelial cells in microfluidic channels. Lab Chip 2007; 7:1759. [DOI] [PubMed] [Google Scholar]
- 79.Davis GE, Senger DR. Endothelial extracellular matrix: biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ Res 2005; 97:1093–107 [DOI] [PubMed] [Google Scholar]
- 80.Muiznieks LD, Keeley FW. Molecular assembly and mechanical properties of the extracellular matrix: a fibrous protein perspective. Biochim Biophys Acta 2013; 1832:866–75 [DOI] [PubMed] [Google Scholar]
- 81.Kielty CM. Elastic fibres in health and disease. Expert Rev Mol Med 2006; 8:1–23 [DOI] [PubMed] [Google Scholar]
- 82.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat Genet 2003; 33:407–11 [DOI] [PubMed] [Google Scholar]
- 83.Judge DP, Dietz HC. Marfan’s syndrome. Lancet 2005; 366:1965–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verstraeten A, Alaerts M, Van Laer L, Loeys B. Marfan syndrome and related disorders: 25 years of gene discovery. Hum Mutat 2016; 37:524–31 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for The effect of physiological stretch and the valvular endothelium on mitral valve proteomes by Mir S Ali, Xinmei Wang and Carla MR Lacerda in Experimental Biology and Medicine