Breast cancer is a recurrent type of cancer among women worldwide. Despite remarkable progress in the prevention, detection, and treatment of breast cancer, it still remains a major chronic problem worldwide and poses significant challenges, like metastasis to distant organs, demanding the need for novel biomarkers and therapeutic targets.
KEYWORDS: breast cancer, FAK, c-Fos, miRNA 551a
ABSTRACT
Breast cancer is a recurrent type of cancer among women worldwide. Despite remarkable progress in the prevention, detection, and treatment of breast cancer, it still remains a major chronic problem worldwide and poses significant challenges, like metastasis to distant organs, demanding the need for novel biomarkers and therapeutic targets. Focal adhesion kinase (FAK), a member of the protein tyrosine kinases, has been shown to be expressed in high levels in breast tumors. Of late, FAK has emerged as an impending curative target in breast carcinoma, with few of the small molecular inhibitors reaching the clinical trial stage. In the current study, we established that microRNA 551a (miR-551a) precisely regulates FAK by binding to the complementary sequences in the 3′ untranslated region (UTR) of mRNAs of FAK and inhibits its expression in breast carcinoma cell lines. Further, results from human breast carcinoma samples illustrated that miR-551a levels were substantially downregulated in tumor samples, with a concurrent rise in the expression of FAK. Functional experimental studies using miR-551a-overexpressing breast cancer cells and nude mouse xenograft models revealed the tumor suppressor role of miR-551a. We also found that miR-551a expression decreased the invasion and migratory ability of breast carcinoma cells by inhibiting MMP-9 activity. Regulation studies performed utilizing promoter luciferase assays, chromatin immunoprecipitation (ChIP), and electrophoretic mobility shift assay (EMSA) revealed that c-Fos binds to the miR-551a promoter and activates it. Further, we observed a considerable increase in the amount of miR-551a levels upon c-Fos overexpression. All of these results showed that miR-551a can be of clinical relevance in understanding the regulation of FAK in breast tumorigenesis.
INTRODUCTION
Breast cancer is a frequent cancer among women worldwide and is at present the number one cancer in females. Presently, research efforts in breast carcinoma are focused on the domains of recognition of new biomarkers, which will impact early diagnosis, on disease behavior, including aggressiveness and treatment response, and on promoting research on unravelling novel molecular targets. Conventional chemotherapeutic modalities have demonstrated efficacy in the treatment of several types of epithelial tumors although the responses to almost all DNA damage-inducing chemotherapeutic agents seem to be varied. Despite similar treatment regimens of breast cancer patients, the results are mixed, with some patients benefiting whereas others have minimal responses. These results from the clinics have remained the cornerstone for providing the impetus to search for molecular markers to categorize patients. Among the nonreceptor protein tyrosine kinases, focal adhesion kinase (FAK) is a member specified to be differentially expressed in tumors (1–3). FAK is said to be engaged in transducing signals from integrins to focal adhesion sites involved in cellular contact with the extracellular matrix (ECM) (4). FAK is now considered to be a favorable target in carcinomas, owing to its differential expression at both levels (transcriptional and translational) and to its contribution in tumorigenesis, particularly in the process of invasion and metastasis (5–8). Although overexpression/activation of FAK is widely known in solid tumors, its mechanism of regulation is still under intense study.
It is known that target mRNA expression in cells is efficiently controlled by endogenously expressed, small noncoding RNAs, termed microRNAs (miRNAs). miRNAs work by binding to the cellular mRNA targets and quenching their expression, and recent studies have shown miRNAs to be probable biomarker candidates for cancers. Recently, Kurozomi et al. reviewed the association between miRNA and breast cancer subtypes and its association in cancer development, progression, and metastases (9). There are increasing reports of miRNAs being secreted as exosomes in blood and of their application as signature molecules (9–13). miRNA expression in cancer cells has been correlated to chemoresistance, altered metabolism, and the most aggressive form of breast cancer, namely, the triple-negative form (14–17). Ascertaining the presence of miRNA in circulation was found to be useful in early breast cancer detection (12, 18, 19). Dysregulation of miRNA has been reported as a biomarker for patient management in breast cancer (20). A recent study in fibroblast cells showed that miRNA 218 (miR-218) controlled myofibroblast differentiation by transforming growth factor β (TGF-β) and was mediated by FAK (21). Another miRNA, miR-7 was shown to regulate invasion of breast carcinoma cells, mediated by FAK (22). Considering these facts, we decided to study the regulation of FAK by miRNA and identified that miR-551a silences FAK, and we further validated the functional outcome of miR-551a overexpression in breast carcinoma cells and nude mouse xenograft models. In addition, we proposed a mechanism of deregulation of miR-551a in breast tumors.
RESULTS
miR-551a targets FAK.
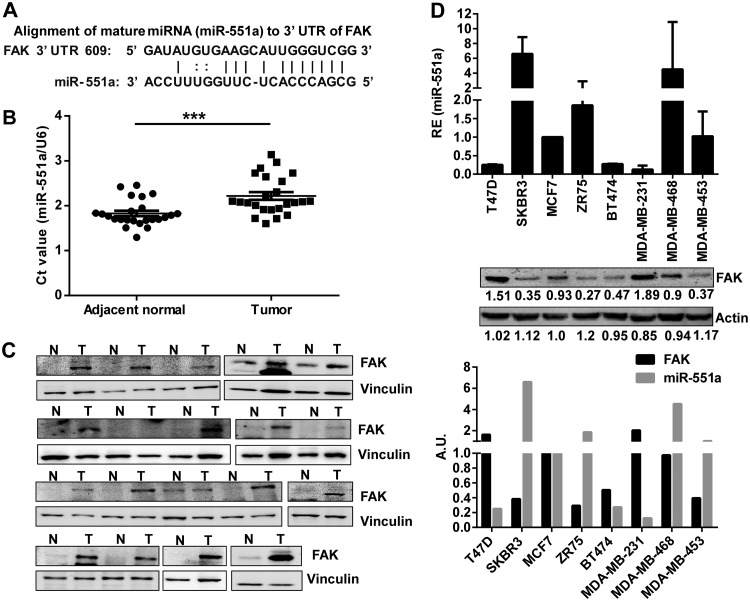
In an effort to identify microRNAs that might target the oncogenic FAK, we screened public databases such as miRANDA, TargetScan, and DIANA-microT for probable complementation of a bare minimum of 6 bp to the seed domain of miRNAs. This effort culminated in recognition of miR-551a as a candidate microRNA that could target FAK (data not shown). We also used FindTar and RNAhybrid to understand the change in free energy (ΔG) and possible hybridization of miR-551a and the 3′ untranslated region (UTR). Analysis of the possible hybrid structure revealed that ΔG is –21.3 kcal and that miR-551a binds the 3′ UTR of FAK by a 7-mer seed sequence and 14 bp complementarity in total (Fig. 1A) (23–28). Further, we showed by using in silico tools that miR-551a precisely regulates FAK by binding to the complementary sequences in the 3′ UTR of mRNAs across different species, which were found to be conserved (data not shown). Based on these bioinformatics data, we analyzed 24 paired clinical samples of human breast carcinomas for expression of miR-551a using quantitative PCR (qPCR) to emphasize the physiological and functional importance of miR-551a. Among the 24 samples of human breast carcinomas analyzed, 22 showed considerable downregulation of miR-551a, and this was found to be statistically significant compared with levels in adjacent normal tissue samples (Fig. 1B). It was noteworthy that we found a concomitant increase in FAK expression in 17 of the 19 tumor samples tested (Fig. 1C). Further, we screened a panel of breast carcinoma cells to ascertain the endogenous levels of miR-551a using qPCR and the target protein FAK by immunoblotting. Screening results showed that FAK and miR-551a levels had an inverse correlation in the majority of the breast cancer cell lines (Fig. 1D).
FIG 1.
Inverse relation between FAK and miR-551a expression in breast cancer. (A) Representation of the complementarity between miR-551a and the 3′ UTR of FAK. (B) Scatter plot showing the expression of miR-551a in terms of the threshold cycle (CT) value normalized to that of U6 in breast tumors and adjacent normal breast tissues (n = 24; ***, P = 0.0006, unpaired t test). (C) Western blot images showing FAK expression in breast tumors. (D) Bar graph and Western blot showing the expression of miR-551a and FAK, respectively, in breast cancer cell lines. The bar graph represents the inverse relation between FAK, normalized to the level of actin, and miR-551a expression (bottom panel). AU, arbitrary units; RE, relative expression; N, normal tissue; T, tumor.
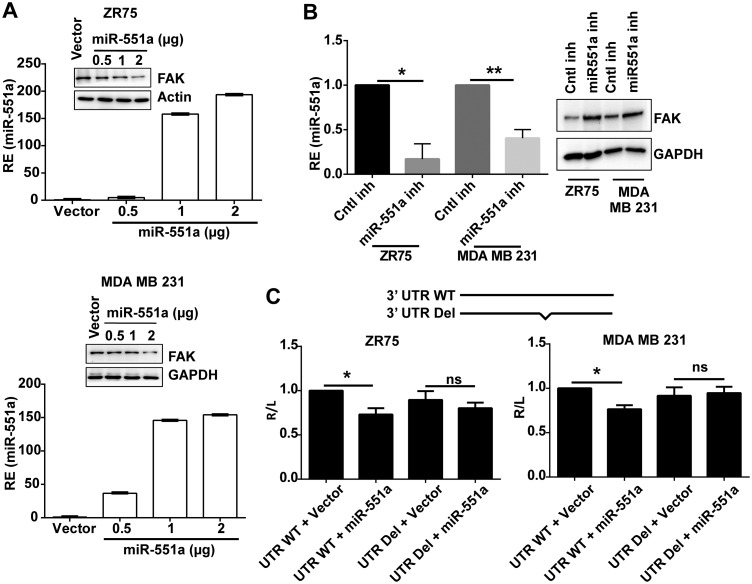
The above results were further backed by the observation that transient overexpression of miR-551a brought about reduction in the target protein FAK in the breast cancer cell lines ZR75 and MDA-MB-231 (Fig. 2A), and this reduction was found to be dependent on the concentration of miR-551a. The same is the case with other breast cancer cell lines (data not shown). In further support of this finding, when cells were transfected with miR-551a inhibitors (antagomirs), expression of FAK was increased. (Fig. 2B). To authenticate the direct interaction of the target miR-551a and the FAK 3′ UTR, luciferase assays were performed. For this, we cloned the 3′ UTR of FAK in the psiChek2 dual-luciferase vector and then cotransfected cells with miR-551a or the empty vector. The data revealed a decline in luciferase activity with miR-551a compared to that with the empty vector, suggesting that miR-551a directly binds to the 3′ UTR of FAK. To confirm this binding, we made deletion constructs and performed luciferase assays, which showed that the decline in luciferase activity was eradicated upon deletion of the miR-551a binding region, confirming that miR-551a binds to the mRNA of FAK at the 3′ UTR (Fig. 2C).
FIG 2.
miR-551a inhibits the expression of FAK and targets its 3′ UTR. (A) Western blots showing the expression of FAK in concentration-dependent transfection of miR-551a and expression of miR-551a by qPCR. (B) Increased expression of FAK with miR-551a inhibitor (inh) transfection (*, P = 0.0139; **, P = 0085; n = 3). (C) Bar graphs representing the relative luciferase activity (normalized to that of firefly luciferase) of the 3’ UTR wild type (WT) or a 3′ UTR deletion mutant (UTR Del) upon cotransfection with empty vector or miR-551a (n = 3; *, P values of 0.0136 and 0.0320 for MDA-MB-231 and ZR75 cell lines, respectively; ns, not significant, by one-way analysis of variance).
Tumor suppressor property of miR-551a in cell line models.
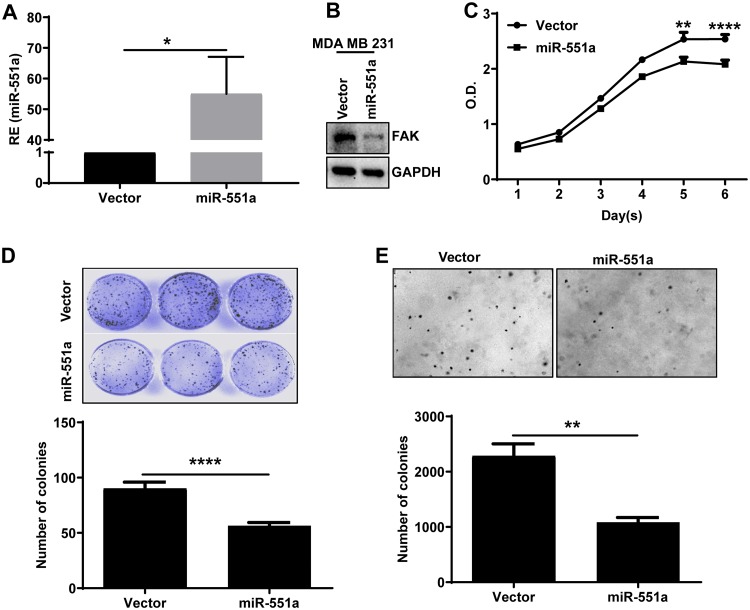
Based on the above results, we decided to examine the operative role of miR-551a in breast cancers. For this, we generated stable lines of MDA-MB-231 and ZR75 cells overexpressing miR-551a using a retroviral system. Results from qPCR and Western blotting showed that upon stable overexpression of miR-551a, there was a subsequent downregulation of the target protein FAK (Fig. 3A and B) (ZR75 cell line data not shown). Further, miR-551a overexpression significantly reduced cell growth and colony-forming ability, as determined by clonogenic and soft-agar assays of MDA-MB-231 cells (Fig. 3C to E).
FIG 3.
miR-551a inhibits proliferation, clonogenic cell survival, and anchorage-independent growth in MDA-MB-231 cells. (A and B) Bar graph and Western blot showing the overexpression of miR-551a and FAK downregulation in MDA-MB-231 cells stably expressing vector or miR-551a (n = 3; *, P =0.0133, unpaired t test). (C) Graph representing the growth of cells expressing either vector or miR-551a by MTT assay (n = 3; **, P < 0.005; ****, P < 0.0001, unpaired t test). (D and E) Representative images and bar graph showing the effect of miR-551a on clonogenic cell survival and anchorage-independent growth, respectively (n = 3; ****, P < 0.0001; **, P = 0.0033, unpaired t test).
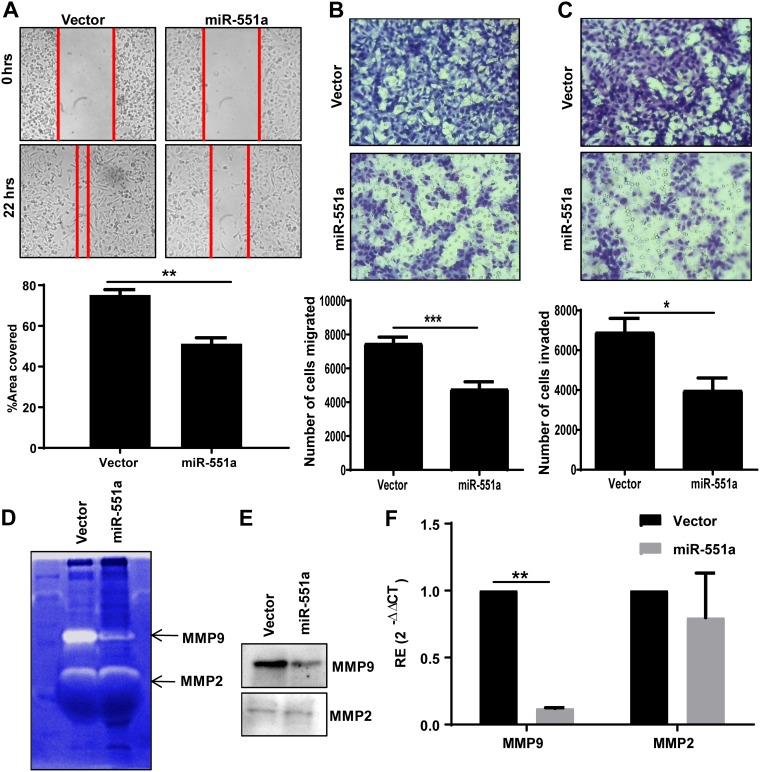
The metastatic property of cancer cells requires cellular separation from the primary site, migration, and finally adherence to a favored distant site. Given that the role of FAK in metastasis is well recognized (6), we examined the capability of miR-551a-overexpressing MDA-MB-231 cells in conventional wound healing and Boyden chamber assays. Data from wound healing assays revealed a significant reduction in wound healing capacity, indicating the reduced migration ability of miR-551a-overexpressing cells (Fig. 4A). This was further confirmed by compromised migration and invasion competencies of MDA-MD 231-cells stably expressing miR-551a as evaluated against a control group (Fig. 4B and C) in a transwell Boyden chamber. These experimental data collectively signify that miR-551a can reduce invasion and migration.
FIG 4.
miR-551a reduces migration and invasion by inhibiting the expression of MMP-9. (A) Representative images and bar graph showing the migration of cells expressing either vector or miR-551a in a wound healing assay. (n = 3; **, P = 0.0036, unpaired t test). (B and C) Representative images and bar graphs showing the migration and invasion of cells through transwell Boyden chambers (***, P = 0.0006 [n = 6]; *, P = 0.0131 [n = 5], unpaired t test). (D) Representative image of gelatin zymogram showing the activity of MMP-9 and MMP-2 in conditioned medium. (E) Western blotting for MMP-2 and MMP-9 expression in conditioned medium. (F) Bar graph showing relative expression of MMP-2 and MMP-9 in MDA-MB-231 cells expressing miR-551a (n = 3; **, P = 0.0044, unpaired t test). CT, threshold cycle.
Matrix-degrading enzymes are known to be secreted by cancer cells to initiate migration and invasion, and FAK is recognized as enhancing invasion of cancers by raising the cellular levels of matrix metalloproteinase 9 (MMP-9) (29). MDA-MB-231 cells stably expressing miR-551a showed a decline in MMP-9 activity, as ascertained by the gelatin zymography (Fig. 4D). This decline in activity could be attributed to a reduction in the protein as well as mRNA levels of MMP-9, as determined by qPCR and immunoblotting. The expression levels were compared with levels in vector control cells (Fig. 4E and F). It was interesting that there was no change in the MMP-2 activity.
To confirm that the effect of miR-551a is, indeed, through FAK, rescue experiments were performed. Data from MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) and wound closure assays showed that overexpression of FAK in miR-551a-expressing cells could rescue the phenotype of cell growth and migration (data not shown). These findings further validate that miR-551a indeed acts by targeting FAK.
Ectopic expression of miR-551a inhibits tumor formation in the nude mouse.
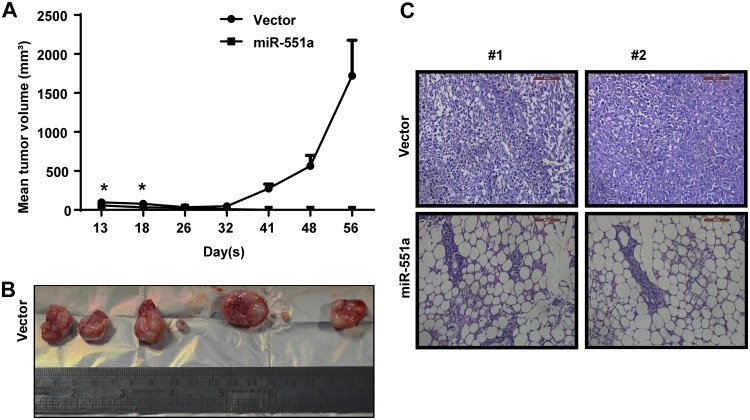
In addition to the in vitro study results, we performed xenograft studies on 6-week-old female athymic nude mice using the MDA-MB-231 cells stably expressing miR-551a and vector control cells. Mice injected with cells expressing miR-551a showed less tumor volume than those receiving empty vector transfected cells. Initially, we observed miniscule tumors in the mice injected with miR-551a-overexpressing cells, and these later regressed. To quantify tumor growth, tumor volumes (in cubic millimeters) were measured at seven time points following the initial xenograft (Fig. 5A and B). Further, study results showed that mice with miR-551a xenografts remained healthy. Neither the control vector- nor miR-551a-expressing mouse group showed any signs of metastasis at the time of autopsy. Further, hematoxylin and eosin (H&E) staining of the excised tumors of the control mice showed that the tumor cells were highly pleomorphic, with darkly stained nuclei and many mitotic figures apparent in the high-power field (Fig. 5C). Thus, the mouse xenograft study convincingly demonstrated that miR-551a inhibited tumor formation.
FIG 5.
miR-551a inhibits the formation of tumor in nude mice. (A) Line plot representing the growth of tumor volume (5 mice per group). (B) Representative images of the excised tumors. (C) Representative images of the H&E staining performed on the excised tumors.
c-Fos regulates miR-551a.
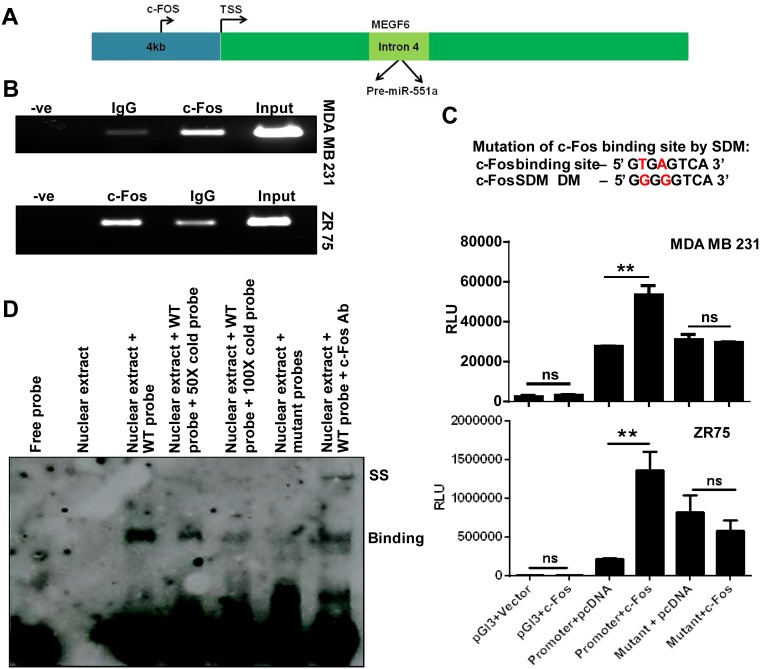
miR-551a was shown to be downregulated in acute myeloid leukemia (AML) with central nervous system (CNS) relapse (30) and in breast cancer (current study). miR-551a is an intragenic miRNA released from intron 4 of the gene MEGF6 located on chromosome 1. Using the tool miRStart, we found that miR-551a shared its transcriptional start site (TSS) with the host gene MEGF6. To further unravel miR-551a transcriptional regulation, we adopted an in silico tool, the ConSite program. Using this program, we investigated the upstream 4-kb putative promoter region of miR-551a for binding motifs for several transcriptional factors and located the binding site for the transcription factor c-Fos within the −2066 and −2073 regions (Fig. 6A). Since c-Fos had the highest score among the transcriptional factors and since c-Fos is a transcriptional coactivator, we concentrated on c-Fos as a prospective upstream modulator of miR-551a expression.
FIG 6.
c-Fos binds to the promoter of miR-551a. (A) Schematic showing the binding of c-Fos to the predicted binding site of the putative promoter region of miR-551a. (B) Representative ChIP images of agarose gel showing the PCR amplification for the putative promoter region of miR-551a upon immunoprecipitation with IgG or c-Fos antibody. (C) Bar graph representing the relative luciferase activity of cloned miR-551a promoter or mutant upon cotransfection with vector (pcDNA) or c-Fos (**, P < 0.05; ns, not significant, by one-way analysis of variance). (D) EMSA showing the binding of biotinylated wild-type (WT) probes (lane 3) spanning the c-Fos binding region in miR-551a promoter, competitive inhibition of band intensity with nonbiotinylated probes (lanes 4 and 5). The binding was abolished when biotinylated mutant probes were used (lane 6). The supershift was observed when the lysate was incubated with anti-c-Fos antibody (lane 7). −ve, negative (empty); RLU, relative light units; Ab, antibody; DM, double mutant.
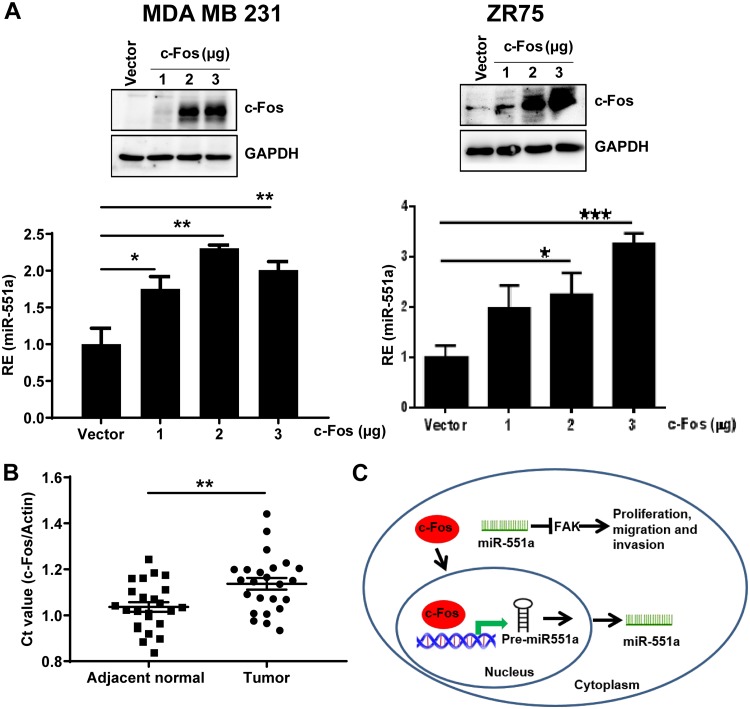
To understand the regulation of miR-551a by c-Fos, an miR-551a promoter was cloned into pGL3 (promoter less) and expressed in breast cancer cells. A chromatin immunoprecipitation (ChIP) experiment confirmed that c-Fos is recruited to the promoter region comprising positions −2066 to −2073 (8 bp) (Fig. 6B). Furthermore, to corroborate the importance of c-Fos binding sequence to the miR-551a promoter, luciferase assays were performed. For this, we cotransfected the full-length promoter of miR-551a and plasmids mutated at the c-Fos binding sequences with pcDNA/c-Fos. Results showed a significant increase in the relative luciferase activity levels when the putative promoter was cotransfected with c-Fos compared to levels with the pcDNA empty vector (Fig. 6C, third and fourth bars). Data analysis of the promoter luciferase levels did not show any significant alterations in the promoter activity when the mutations were incorporated in the putative promoter of miR-551a (Fig. 6C, fifth and sixth bars), confirming the c-Fos binding site. To provide strong evidence for the c-Fos binding sequence in the miR-551a promoter, an electrophoretic mobility shift assay (EMSA) was performed. EMSA results showed a shift matching the c-Fos site whereas a supershift was noted in the presence of a c-Fos antibody (Fig. 6D). To provide conclusive evidence for this regulation, c-Fos overexpression in MDA-MB-231 and ZR-75 breast carcinoma cells produced a considerable increase in miR-551a expression (Fig. 7A). Based on this experimental evidence, we conclude that c-Fos directly interacts with the presumed promoter sequence of miR-551a and regulates its expression. To further supplement the correlation between c-Fos and miR-551a expression, we screened the same set of clinical breast cancer samples (used earlier for FAK and miR-551a) for c-Fos expression. Data analysis (Fig. 7B) showed a measurable decline in c-Fos expression (mean fold change of 0.5) in breast carcinoma samples in contrast to levels in normal breast tissue. In-depth analysis of the expression patterns of miR-551a and c-Fos showed a positive correlation (data not shown).
FIG 7.
c-Fos promotes the miR-551a expression in breast cancer cell lines. (A) Bar graph showing relative increase in miR-551a expression by qPCR upon c-Fos transfection in a dose-dependent manner. c-Fos transfection was confirmed by Western blotting in MDA-MB-231 and ZR75 cell lines. (*, P < 0.05; **, P < 0.005; ***, P < 0.0005, by one-way analysis of variance) (B) Scatter plot showing the expression of c-Fos in terms of the threshold cycle (CT) value normalized to that of actin in breast tumors and adjacent normal breast tissues (n = 24; **, P = 0.0039, by an unpaired t test). (C) Proposed mechanism of action of miR-551a and targeting of FAK.
DISCUSSION
With improvements in the diagnosis and therapy of breast cancer, the major challenges still lie in treating triple-negative breast cancer (TNBC), metastatic cancer, and chemoresistant tumors. Recently, miRNAs have emerged as new therapeutic tools for the management of cancer (31–33). In this paper, we identified and validated that miR-551a targets FAK in its 3′ UTR. Earlier, miR-551a had also been shown to target PRL3 (phosphatase of regenerating liver 3) and CKB (creatine kinase, brain type) in gastric cancer and colorectal cancer, respectively (34, 35). Despite these studies, the role of miR-551a in breast carcinomas was still not clarified. Therefore, we focused on studying the outcome of regulation of FAK by miR-551a in breast cancer. Clinical significance for this study arose from the data showing that breast tumor samples demonstrated elevation in the expression of FAK in the majority of the samples. The increased FAK expression in tumor samples may probably in part be attributed to decreased miR-551a levels. Additional studies on engineered cell lines and in vivo xenograft models revealed miR-551a’s tumor suppressor role in breast carcinomas. From these data, we further understood the regulation of miR-551a in breast cancer cells. The facts that miR-551a levels are low in breast cancer samples and that c-Fos binds to the miR-551a promoter prompted us to investigate c-Fos-mediated miR-551a regulation. In the breast cancer samples analyzed, c-Fos and miR-551a were found to be deregulated, and c-Fos was found to have a positive correlation with miR-551a. Deregulation of c-Fos has been reported in ovarian, gastric, and breast cancer. A reduced c-Fos level was reported to be coupled with poor progression-free survival. Similarly, loss of c-Fos has been correlated with advanced disease stages of gastric cancer, lymphatic invasion, poor survival, and metastasis (36–40).
It is well known that FAK signaling controls motile and invasive cell phenotypes in cancer cells by regulating MMPs (6, 29, 41–43). Our results demonstrated that miR-551a-overexpressing cells, via downregulation of FAK, showed decreased migratory and invasive potential, as evident by reduced MMP-9 activity selectively. This suggests that miR-551a expression can act to prevent metastasis in breast carcinomas by reducing the levels of MMP-9. MMP-9 is well known for its role in promoting invasive characteristics of carcinoma cells and has been reported to have clinical significance in breast cancer. These results were further supported by Boyden chamber assays.
It was earlier reported that miR-551a inhibits in vitro gastric carcinoma cell migration and invasion and also liver metastasis in mouse models (34, 35). The current study shows an important tumor-suppressive role of miR-551a in breast carcinoma cells under both in vitro and in vivo conditions. Recently, it was shown that demethoxycurcumin inhibits ovarian tumor cell proliferation by upregulating miR-551a expression, further indicating its growth-inhibitory role (44). Loo et al. have reported that endogenous miR-551a and miR-483-5p inhibit liver metastasis but have no effect on tumor growth (35), and this was probably attributable to differences in the origins of the cell lines and the cancer types under study. The overall effect of the miRNA is determined by the balance between these tumor suppressors and tumor promoters. Considering the current study and earlier reports, miR-551a can be of clinical relevance in understanding breast tumorigenesis. Based on our findings, we propose that miR-551a has a tumor-suppressive function targeting FAK in its 3′ UTR. We further demonstrate that c-Fos stimulates the expression of miR-551a by binding to its putative promoter (Fig. 7C).
MATERIALS AND METHODS
Tumor samples, cell lines, plasmids and reagents.
Cell lines T47D, ZR75, MDA-MB-453, MCF7, and MDA-MB-468 were purchased from the National Centre for Cell Science (NCCS), Pune, India. SKBR3 and MDA-MB-231 were gifts from Asha Nair from the Rajiv Gandhi Centre for Biotechnology (RGCB), India. BT474 was a gift from the Cancer Institute, Adyar, Chennai, India. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose and 10% fetal bovine serum (FBS). Breast tumor and matched adjacent normal breast tissues were obtained from Sri Ramachandra Medical College and Research Institute (SRMC), Chennai, India, with ethical clearance. All of the tumors were infiltrating carcinoma, nonmetastatic, grade 2 or 3, and patients had a mean age of 50 years. The same sets of tumors and matched adjacent normal tissues were used for studying the expression of miR-551a, FAK, and c-Fos. psiCHECK-2 from Promega was used for cloning of the 3′ UTR of FAK in NotI and XhoI restriction sites. pGL3 basic from Promega (Madison, WI) was used for cloning a 4-kb promoter region of miR-551a. Site-directed mutagenesis (SDM) in the c-Fos binding site of the promoter region was done using a QuikChange II SDM kit (number 200523) from Agilent Technologies (Santa Clara, CA). An expression plasmid for miR-551a was purchased from Origene (SC400530; Rockville, MD). Control and miR-551a power inhibitors were purchased from Exiqon. For retroviral transduction, miR-551a was cloned into pMSCV-Puro-GFP (where MSCV is mouse stem cell virus and GFP is green fluorescent protein) (gift from Erik Flemington, Tulane University). Retroviral packaging plasmids pUMVC and pCMV-VSV-G were gifts from Bob Weinberg (8449 and 8454, respectively; Addgene) (45). pcDNA 3-Flag-c-Fos was a gift from Yosef Shaul, Weizmann Institute of Science, Israel. Transfections were done using Fugene HD from Promega (E2311; Madison, WI) according to the manufacturer’s instructions. Puromycin from MP Biomedicals (194539; Santa Ana, CA) was used at 1 μg/ml for generating stable cells in MDA-MB-231 cells.
Antibodies for FAK (3285S), c-Fos (4384S), MMP-2 (4022), and MMP-9 (3852) were purchased from Cell Signaling Technology (Danvers, MA). Actin (A2228), vinculin (V9131), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; G8795) antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Actin, GAPDH, or vinculin was used as an internal control for Western blot analyses.
RNA isolation was done using TRIzol reagent (15596026; Life Technologies, Invitrogen, Carlsbad, CA). cDNA synthesis was done using a high-capacity reverse transcription kit from Applied Biosystems (4368814; CA). For miR-551a and U6 snRNA, stem-loop primers were used for cDNA synthesis. U6 snRNA and actin were used as reference genes for qPCR. qPCR was performed using SYBR green chemistry using SYBR Premix Ex Taq II (RR820A; Clontech, Fremont, CA).
ChIP and EMSA.
ChIP was performed using a ChIP assay kit (17-295; EMD, Millipore, MA) and c-Fos antibody from Santa Cruz (SC-253; Dallas, TX). EMSA was performed according to the manufacturer’s instructions (20148; Pierce Biotechnology, Rockford, IL).
MTT, wound healing, soft-agar, and clonogenic assays.
For the MTT assay, 3,000 cells were seeded in 96-well plates, and absorbance was measured at 570 nm using MTT reagent (33611; SRL Pvt., Ltd.). For the wound healing assay, a scratch was made in a monolayer of confluent cells, and multiple images were acquired at 0 h and 22 h using a Carl Zeiss microscope. The area was calculated using Zen 2011 software. For the soft-agar assay, 5,000 cells were seeded in 35-mm plates with top and bottom layers containing agarose at 0.5% and 0.8%, respectively. Cells were stained with crystal violet and counted after 3 weeks. For the clonogenic assay, 500 cells were initially seeded in 60-mm plates, and colonies were counted after staining with crystal violet after 3 weeks. For gelatin zymography, conditioned medium was collected and concentrated, and the protocol described by Toth and Fridman was followed (46).
Transwell migration and invasion assays.
A total of 25,000 cells were seeded in the upper chamber of a transwell for migration and invasion assays in serum-free medium. Serum-containing medium was used as a chemoattractant in the bottom chamber. Cells were stained using HemaColor stain after 48 h and counted.
Xenograft study.
For a xenograft study, 7 million cells (approximately) were mixed with Matrigel and injected into the mammary fat pad of nude mice. Five mice were used for each group, i.e., vector controls and miR-551a-expressing cells. Tumor volumes were measured over a period of 8 weeks. Mice were euthanized, and tumors were excised at the end of the study. All the animal experiments were performed as per the CPCSEA guidelines after prior approval.
Statistical analyses.
All the data are expressed as means ± standard errors of the means (SEM) using Prism (GraphPad Software, Inc.).
ACKNOWLEDGMENTS
We profusely thank Moshe Oren, Weizmann Institute of Science, Israel, for his help and support for completing this study. We thank Archana Kanakarajan for helping us procure the primary breast tumors and adjacent normal breast tissues. We thank Venu, S. Thanikachalam, and Y. Murkunde, CEFT, SRMC, for their help in performing xenograft study. We thank Nitish R. Mahapatra for the suggestions and providing the c-Fos antibody, Erik Flemington for the PMSCV-Puro-GFP vector, and Yosef Shaul for the c-Fos expression plasmid. We thank Arathy S Kumar, Swetha Raghavan, Shabeer Ahmed, Rakesh Nankar, Krishna Kumar, and Sansrity Sinha for their help and suggestions.
We thank the Department of Biotechnology (grant BT/PR3180/NNT/28/553/2011), Government of India, for financial support to S.K.R. and Indian Institute of Technology Madras (IITM) for all other facilities. Anuj thanks CSIR for a fellowship.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Owens LVM, Xu L, Craven RJ, Dent GA, Weiner TM, Kornberg LJ, Liu E, Cance WG. 1995. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res 55:2752–2755. [PubMed] [Google Scholar]
- 2.Lark AL, Livasy CA, Dressler L, Moore DT, Millikan RC, Geradts J, Iacocca M, Cowan D, Little D, Craven RJ, Cance W. 2005. High focal adhesion kinase expression in invasive breast carcinomas is associated with an aggressive phenotype. Mod Pathol 18:1289–1294. doi: 10.1038/modpathol.3800424. [DOI] [PubMed] [Google Scholar]
- 3.Lightfoot HM, Lark AL, Livasy CA, Moore DT, Cowan D, Dressler L, Craven RJ, Cance W. 2004. Upregulation of focal adhesion kinase (FAK) expression in ductal carcinoma in situ (DCIS) is an early event in breast tumorigenesis. Breast Cancer Res Treat 88:109–116. doi: 10.1007/s10549-004-1022-8. [DOI] [PubMed] [Google Scholar]
- 4.Mitra SK, Schlaepfer DD. 2006. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol 18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Nimwegen MJv, Verkoeijen S, Buren L, Burg D, Water B. 2005. Requirement for focal adhesion kinase in the early phase of mammary adenocarcinoma lung metastasis formation. Cancer Res 65:4698–4706. doi: 10.1158/0008-5472.CAN-04-4126. [DOI] [PubMed] [Google Scholar]
- 6.Mitra SK, Lim ST, Chi A, Schlaepfer DD. 2006. Intrinsic focal adhesion kinase activity controls orthotopic breast carcinoma metastasis via the regulation of urokinase plasminogen activator expression in a syngeneic tumor model. Oncogene 25:4429–4440. doi: 10.1038/sj.onc.1209482. [DOI] [PubMed] [Google Scholar]
- 7.Provenzano PP, Inman DR, Eliceiri KW, Beggs HE, Keely PJ. 2008. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am J Pathol 173:1551–1565. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurio N, Shimo T, Fukazawa T, Takaoka M, Okui T, Hassan NMM, Honami T, Hatakeyama S, Ikeda M, Naomoto Y, Sasaki A. 2011. Anti-tumor effect in human breast cancer by TAE226, a dual inhibitor for FAK and IGF-IR in vitro and in vivo. Exp Cell Res 317:1134–1146. doi: 10.1016/j.yexcr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J. 2017. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet 62:15–24. doi: 10.1038/jhg.2016.89. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi RU, Miyazaki H, Ochiya T. 2015. The roles of microRNAs in breast cancer. Cancers (Basel) 7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, Alajez NM. 2016. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep 6:25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ. 2016. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire A, Brown JA, Kerin MJ. 2015. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev 34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, Cai Y, Li X, Xing H, Shi M, Chen Y, Bruce CL, Wang T, Jin L, Qi X, Hua D, Jin J, Ma X. 2014. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett 588:2009–2015. doi: 10.1016/j.febslet.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. 2008. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 16.Eichner LJ, Perry M-C, Dufour CR, Bertos N, Park M, St-Pierre J, Giguère V. 2010. miR-378* mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab 12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, Chang G, Li X, Li Q, Wang S, Wang W. 2014. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One 9:e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. 2013. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 19.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT, Kwong A. 2013. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 21.Guo F, Carter DE, Leask A. 2014. miR-218 regulates focal adhesion kinase–dependent TGFβ signaling in fibroblasts. Mol Biol Cell 25:1151–1158. doi: 10.1091/mbc.e13-08-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W, Wu Z, Chen T, Wu W, Lobie PE, Zhu T. 2012. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS One 7:e41523. doi: 10.1371/journal.pone.0041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. 2009. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellwanger DC, Buttner FA, Mewes HW, Stumpflen V. 2011. The sufficient minimal set of miRNA seed types. Bioinformatics 27:1346–1350. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. 2008. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One 3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betel D, Wilson M, Gabow A, Marks DS, Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsia DA, Mitra SK, Hauck CR, Streblow DN, Nelson JA, Ilic D, Huang S, Li E, Nemerow GR, Leng J, Spencer KS, Cheresh DA, Schlaepfer DD. 2003. Differential regulation of cell motility and invasion by FAK. J Cell Biol 160:753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. 2009. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One 4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. 2013. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther 21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naidu S, Magee P, Garofalo M. 2015. MiRNA-based therapeutic intervention of cancer. J Hematol Oncol 8:68. doi: 10.1186/s13045-015-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rupaimoole R, Slack FJ. 2017. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu G, Liu Z, Tu Y, Peng G, Lee DW, Park SS. 2012. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett 323:41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, Saltz L, Paty PB, Tavazoie SF. 2015. Extracellular metabolic energetics can promote cancer progression. Cell 160:393–406. doi: 10.1016/j.cell.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Zhang JS, Yu JC, Cui QC, Zhou WX, Kang WM, Ma ZQ. 2010. Negative association of c-fos expression as a favorable prognostic indicator in gastric cancer. Arch Med Res 41:201–206. doi: 10.1016/j.arcmed.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Kharman-Biz A, Gao H, Ghiasvand R, Zhao C, Zendehdel K, Dahlman-Wright K. 2013. Expression of activator protein-1 (AP-1) family members in breast cancer. BMC Cancer 13:1–10. doi: 10.1186/1471-2407-13-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahner S, Baasch C, Schwarz J, Hein S, Wolber L, Janicke F, Milde-Langosch K. 2008. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer 99:1269–1275. doi: 10.1038/sj.bjc.6604650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira-Ferrer L, Rößler K, Haustein V, Schröder C, Wicklein D, Maltseva D, Khaustova N, Samatov T, Tonevitsky A, Mahner S, Jänicke F, Schumacher U, Milde-Langosch K. 2014. c-FOS suppresses ovarian cancer progression by changing adhesion. Br J Cancer 110:753–763. doi: 10.1038/bjc.2013.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Zhang L, Yang H, Huang X, Otu H, Libermann TA, DeWolf WC, Khosravi-Far R, Olumi AF. 2007. c-Fos as a proapoptotic agent in TRAIL-induced apoptosis in prostate cancer cells. Cancer Res 67:9425–9434. doi: 10.1158/0008-5472.CAN-07-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen T, Dutta A, Chatterjee A. 2010. Epigallocatechin-3-gallate (EGCG) downregulates gelatinase-B (MMP-9) by involvement of FAK/ERK/NFκB and AP-1 in the human breast cancer cell line MDA-MB-231. Anti-Cancer Drugs 21:632–644. doi: 10.1097/CAD.0b013e32833a4385. [DOI] [PubMed] [Google Scholar]
- 42.Ganguly KK, Sen T, Pal S, Biswas J, Chatterjee A. 2012. Studies on focal adhesion kinase in human breast cancer cell MDA-MB-231. Abc 02:29–42. doi: 10.4236/abc.2012.21004. [DOI] [Google Scholar]
- 43.Shibata K, Kikkawa F, Nawa A, Thant AA, Naruse K, Mizutani S, Hamaguchi M. 1998. Both focal adhesion kinase and c-Ras are required for the enhanced matrix metalloproteinase 9 secretion by fibronectin in ovarian cancer cells. Cancer Res 58:900–903. [PubMed] [Google Scholar]
- 44.Du Z, Sha X. 2017. Demethoxycurcumin inhibited human epithelia ovarian cancer cells growth via up-regulating miR-551a. Tumour Biol 39 doi: 10.1177/1010428317694302. [DOI] [PubMed] [Google Scholar]
- 45.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, Weinberg RA, Novina CD. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth M, Fridman R. 2001. Assessment of gelatinases (MMP-2 and MMP-9) by gelatin zymography. Methods Mol Med 57:163–174. doi: 10.1385/1-59259-136-1:163. [DOI] [PMC free article] [PubMed] [Google Scholar]